Abstract
Proliferating cell nuclear antigen (PCNA) has been demonstrated to interact with multiple proteins involved in several metabolic pathways such as DNA replication and repair. However, there have been fewer reports about whether these PCNA-binding proteins influence stability of PCNA. Here, we observed a physical interaction between PCNA and MutT homolog2 (MTH2), a new member of the MutT-related proteins that hydrolyzes 8-oxo-7,8-dihydrodeoxyguanosine triphosphate (8-oxo-dGTP). In several unstressed human cancer cell lines and in normal human fibroblast cells, PCNA and MTH2 formed a complex and their mutual binding fragments were confirmed. It was intriguing that PCNA and MTH2 were dissociated dependent on acetylation of PCNA, which in turn induced degradation of PCNA in response to UV irradiation, but not in response to other forms of DNA-damaging stress. To further explore the link between dissociation of PCNA-MTH2 and degradation of PCNA, RNAi against MTH2 was performed to mimic the dissociated status of PCNA to evaluate changes in the half-life of PCNA. Knockdown of MTH2 significantly promoted degradation of PCNA, suggesting that the physiological interaction of PCNA-MTH2 may confer protection from degradation for PCNA, whereas UV irradiation accelerates PCNA degradation by inducing dissociation of PCNA-MTH2. Moreover, secondary to degradation of PCNA, UV-induced inhibition of DNA synthesis or cell cycle progression was enhanced. Collectively, our data demonstrate for the first time that PCNA is protected by this newly identified partner molecule MTH2, which is related to DNA synthesis and cell cycle progression.
Proliferating cell nuclear antigen (PCNA)3 is a member of the DNA sliding clamp family and consists of a ring-shaped trimeric complex (1–3). Three PCNA monomers, each comprising two similar domains, are joined in a head-to-tail arrangement to form a closed ring (4, 5). Because of this unique structure, PCNA encircles the DNA double helix and slides freely along it. PCNA was originally characterized as a DNA polymerase processivity factor and it increases the processivity of DNA synthesis by interacting with polymerase δ (6, 7). Subsequent studies revealed that PCNA plays an important role in DNA replication (8, 9). For example, PCNA not only functions as a protein binding platform to interact with the DNA polymerases, flap endonuclease-1 (Fen1) or DNA ligase I (10–12), but also coordinates complicated processes in DNA replication (2, 13). In addition, PCNA also plays a role in DNA damage repair (14–17) and cell cycle control (18–20).
Because PCNA is essential for DNA synthesis both in DNA replication and repair, a dynamic balance between PCNA synthesis and degradation is critical for maintaining normal DNA synthesis. Up-regulation of PCNA accelerates DNA synthesis and promotes cell proliferation, such that PCNA is regarded as a general proliferation marker in tumor development. On the other hand, degradation of PCNA leads to inhibition of DNA synthesis (9, 21). In this case, in response to inhibition of DNA synthesis by PCNA degradation, both cell proliferation and DNA repair are inhibited, and cells are thus subject to death.
In Escherichia coli, MutT protein encoded by the mutT gene has 8-oxo-dGTPase activity, and hydrolyzes 8-oxo-dGTP to 8-oxo-dGMP, which is nonutilizable for DNA synthesis, thus preventing misincorporation of 8-oxo-dGTP into DNA (22). 8-Oxo-dGTP is a product of dGTP oxidation and can be inserted into opposite dA or dC residues of template DNA at almost equal efficiencies. As a result, G:C to T:A or T:A to G:C transversion mutations occur (22–24). In a mutT-deficient strain, the rate of spontaneous occurrence of A:T to C:G transversion increases by 1000-fold compared with that of cells with wild type mutT (25–27). Therefore, MutT protein is required for preventing mutations and maintaining high fidelity of DNA replication (28). In addition, RibA is a backup enzyme for MutT in E. coli and also plays a role in maintaining high fidelity of DNA replication (29). The MutT homologue MTH1 is the first MutT-related protein found in mammalian cells (30). The spontaneous mutation frequency in MTH1-deficient cells showed an increase of ∼2-fold as compared with that in wild type MTH1 cells (31). Comparing the mutation frequency in mutT-deficient E. coli cells with that in MTH1-deficient mammalian cells suggests that there must be other proteins responsible for preventing occurrence of high numbers of oxidative damage induced mutations in mammalian cells. By searching the GenBankTM EST data base, our research group and others (32) have cloned a new member of MutT-related protein, MTH2. The increased mutation frequency in mutT-deficient cells was significantly reduced by overexpression of MTH2 cDNA (32). Therefore, MTH2 may help to ensure cells achieve accurate DNA synthesis. However, aside from the activity of 8-oxo-dGTPase, the exact mechanism by which MTH2 influences DNA synthesis has not been explored.
The functions of both PCNA and MTH2 partially overlap in DNA synthesis, thus warranting exploration of whether MTH2 works together with PCNA to regulate DNA replication or repair. In this study, we found that MTH2 directly interacts with PCNA, and this interaction enhances PCNA stability. However, when cells were exposed to UV light, the interaction of MTH2 and PCNA was disrupted, and PCNA degradation was accelerated. Consequently, DNA synthesis was reduced, and cell cycling was arrested.
EXPERIMENTAL PROCEDURES
Cell Culture and UV Irradiation
Human lung cancer cell lines A549, H1299, H719 were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and an appropriate amount of penicillin/streptomycin in a 37 °C, 5% CO2, humidified incubator. Human lung diploid fibroblast cells (2BS) or human embryonic kidney 293T cells (HEK293T) were grown in Dulbecco's modified Eagle's medium (10% fetal bovine serum). Cells were plated 24 h prior to UV treatment. Cells were irradiated at the indicated doses with a UV lamp (253.7 nm), and then fresh medium was changed for incubation at 37 °C for 3 to 24 h.
Western Blotting
Protein expression was evaluated by Western blotting as previously described (33). The antibodies used were anti-PCNA, -α-tubulin, -histone-H3 (Santa Cruz Biotechnology), anti-acetyl-lysine, -phosphotyrosine, -phosphoserine, -phosphothreonine (Upstate), anti-Flag (Sigma), anti-ubiquitin (Santa Cruz Biotechnology) and anti-MTH2 (a kind gift from Dr. Mutsuo Sekiguchi, mouse origin).
Co-immunoprecipitation
Co-immunoprecipitation assays were performed as described previously (34). Briefly, total cell extract was incubated with antibodies on ice for 1 h. Agarose A/G was washed three times with Nonidet P-40 buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris, 0.05% SDS, 1 mm PMSF, and a 1% mixture of protease inhibitors), and then added to the samples, which were then rolled at 4 °C for 1 h. After the beads were washed three times with Nonidet P-40 buffer, the pellets were dissolved into 2× SDS loading buffer after centrifugation and boiled 5 min at 100 °C. The protein was analyzed by Western blotting with various antibodies.
Triton Extraction
Cells were rinsed twice in cold phosphate-buffered saline, then incubated with buffer A (100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 10 mm PIPES at pH 6.8, 1 mm EGTA, 0.2 Triton X-100, 25 mm NaF, 2 mm Na3VO4, 5 mm PMSF, 2 μg/ml aprotinin) for 5 min on ice, and the cells were then scraped and centrifuged. The supernatant was the Triton-extractable fraction, and the pellet was then lysed in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris at pH 7.5, 25 mm NaF, 2 mm Na3VO4, 5 mm PMSF, 2 μg/ml aprotinin) and sonicated. After centrifuging, the supernatant was Triton-resistant.
Plasmid Construction
GST-MTH2 or GST-PCNA fusion protein plasmids were generated by inserting MTH2 or PCNA segments into the vector pGEX-4T-2 with BamHI and XhoI (New England Biolabs). pECFP-PCNA or pEYFP- MTH2 was constructed by inserting PCNA or MTH2 segments into the vector pECFP or pEYFP with XhoI and BamHI. Flag-tagged PCNA plasmid was generated by inserting wild-type PCNA into the vector of p3×FLAG- CMV-10. Flag-PCNA mutants was generated using a site-directed mutagenesis kit (Stratagene), following the manufacturer's directions.
Purification of GST Fusion Proteins and GST Pull-down Experiments
GST or GST fusion proteins were expressed in E. coli BL21 (Tiangen Biotechnology, Beijing, China), induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h at 30 °C and purified following the protocol from Amersham Biosciences. Equal amounts of GST or GST fusion proteins were incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) by rocking at 4 °C for 1 h, and the beads were then washed three times with TEN buffer (20 mm Tris-HCl, pH 7.4, 0.1 mm EDTA, and 100 mm NaCl). Total cell extracts from A549 cells were added to the beads and incubated by rocking at 4 °C overnight. The beads were washed three times with TENT buffer (0.5% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 0.1 mm EDTA, and 300 mm NaCl), and then dissolved into 2× SDS loading buffer after centrifugation and boiled 5 min at 100 °C. After centrifuging, the supernatant was extracted and analyzed by Western blotting with antibodies to PCNA and MTH2.
Transcription and Translation in Vitro
Plasmid pHPCNA15 (a kind gift from Dr. Moshe Szyf) was linearized with ClaI. Linear pHPCNA15 was purified and used for in vitro translation employing T7 RNA polymerase (Promega Biotec, Madison, WI) according to the manufacturer's instructions. In vitro translated protein was detected by Western blotting.
Fluorescence Resonance Energy Transfer (FRET)
Both pECFP-PCNA and pEYFP-MTH2 were transfected into HEK293T cells. After 48 h, cells were observed under a TCS SP2 confocal microscope, and the FRET assay was performed with TCS SP2 confocal software (Leica, Germany).
Transient Transfection
Plasmids (pECFP-PCNA, pEYFP-MTH2, p3×FLAG-CMV-PCNA) were transfected into HEK293T cells by use of the Qiagen Effectene Transfection kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RNA Interference (RNAi)
siRNA for MTH2 or PCNA (GeneChem, Shanghai, China) was transfected into A549 cells by use of a Lipofectamine 2000 kit (Invitrogen) for 48 h according to the manufacturer's instructions. To examine the efficiency of siRNA, total protein was extracted for Western blotting using anti-MTH2 or anti-PCNA. MTH2 siRNA: cccagccaucauuuaucuutt; PCNA siRNA: ccuuggcgcuaguauuugatt.
[3H]Thymidine Incorporation Assay
DNA synthesis was assessed by thymidine incorporation assay. Cells were plated onto a 6-well plate 24 h prior to experiments. [3H]Thymidine was added (1 μCi/ml) for 12 h, and DNA was precipitated with cold trichloroacetic acid (50%). The trichloroacetic acid-insoluble DNA was placed on a glass microfiber filter (Whatman, Maidstone, England) and washed twice with cold trichloroacetic acid (5%) and then washed once with ethanol or acetone. After air drying the glass filter, it was put into a vial for counting radioactivity (Beckman LS 5000 Liquid Scintillation Counter).
Cell Cycle Analysis
Cell cycle analysis was performed as described previously (35). UV-irradiated or MTH2-knocked down A549 cells were harvested and stained with propidium iodide. The samples were analyzed with flow cytometry (BD FACS Calibur) using BD CellQuest software.
Immunofluorescence and Confocal Microscopy
A549 cells were plated on coverslips and grown to 50% confluence. After the cells were fixed with 4% paraformaldehyde solution at room temperature for 15 min, they were treated with 0.3% Triton X-100 at 37 °C for 5 min and blocked with 10% goat serum at room temperature for 1 h. The cells were then incubated with 1:20 dilution of anti-8-oxo-dG overnight at 4 °C and then with a 1:100 dilution of Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) for 2 h. Images were captured with TCS SP2 confocal microscopy (Leica, Germany). The fluorescence value was quantified with TCS SP2 confocal software.
RESULTS
PCNA Forms a Complex with MTH2 in Vivo and in Vitro
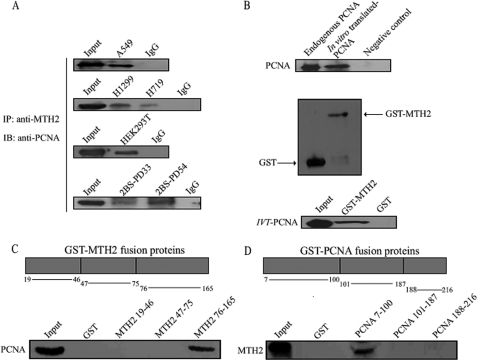
To explore the relationship between PCNA and MTH2, we first tested whether PCNA interacts with MTH2 using the co-immunoprecipitation assay. Endogenous PCNA was co-immunoprecipitated with anti-MTH2 from cell lysates of several human non-small cell lung cancer cell lines (A549, H1299, and H719), human lung diploid fibroblast cells (2BS), and human embryonic kidney 293T cells (HEK293T). It was of interest to observe that PCNA and MTH2 formed a complex in vivo (Fig. 1A). Next, to confirm whether the interaction was a direct interaction, a GST pull-down assay was carried out in vitro using bacterially expressed proteins. Wild-type PCNA translated in vitro (Fig. 1B, upper panel) was incubated with a full-length GST-MTH2 fusion protein (Fig. 1B, middle panel), and Western blotting was then performed with anti-PCNA. As shown in Fig. 1B (lower panel), PCNA formed a complex with GST-MTH2 fusion protein in vitro. To further map regions of interaction, PCNA and MTH2 were divided into three fragments to perform GST pull-down in A549 cells. Results showed that the 76–165-amino acid region of MTH2 (Fig. 1C) and 7–100-amino acid region of PCNA (Fig. 1D) are the fragments involved in the interaction of PCNA and MTH2. Although the antibody of MTH2 was produced from mouse MTH2 cDNA, it can effectively detect human MTH2 assayed with purified protein in vitro or by RNAi in vivo (supplemental Fig. S1, A–C). These results suggested that PCNA physiologically interacts with MTH2 in cells.
FIGURE 1.
PCNA and MTH2 form a complex in vivo. A, proteins were extracted from A549, H1299, H719, 2BS, or HEK293T cells for immunoprecipitation using anti-MTH2, followed by Western blotting with anti-PCNA. B, PCNA was translated in vitro using pHPCNA15 and was detected by Western blotting with anti-PCNA (upper panel). Endogenous PCNA was used as a positive control, and a sample without plasmid was used as a negative control. GST or GST-MTH2 fusion protein was purified from bacteria and was detected by Western blotting with anti-GST (middle panel). GST or GST-MTH2 fusion protein was incubated with PCNA (translated in vitro), and Western blotting was performed to detect interaction with anti-PCNA. C and D, schematic diagrams show different fragments of GST-MTH2 (upper panel of C) and GST-PCNA (upper panel of D) used in GST pull-down assays. Both GST-MTH2 and GST-PCNA fusion proteins were purified from bacteria and incubated with total extracts of A549 cells. The interaction of PCNA and different fragments of GST-MTH2 (lower panel of C) or MTH2 and different fragments of GST-PCNA (lower panel of D) was evaluated with Western blotting using anti-PCNA or anti-MTH2, respectively.
UV Irradiation Induces Dissociation of PCNA and MTH2 in A549 Cells
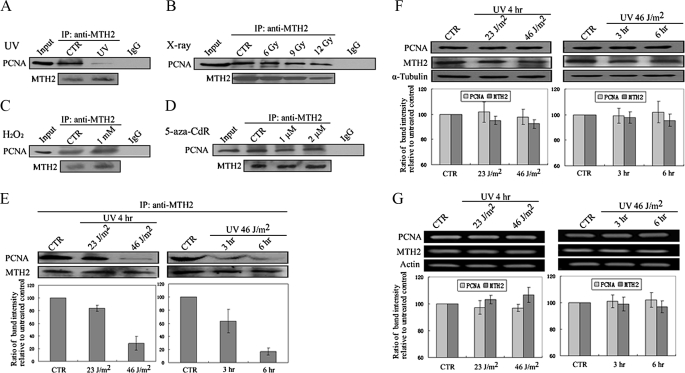
To investigate the effect of DNA-damaging stresses on the PCNA-MTH2 complex, A549 cells were treated with UV, x-ray, H2O2, or 5-aza-2′-deoxycytidine (5-aza-CdR, a DNA demethylating agent that causes DNA damage) (35, 36), and co-immunoprecipitation assays were then performed. It was a surprise to find that PCNA co-immunoprecipitated with anti-MTH2 was markedly decreased in UV-treated cells (Fig. 2A), and this result was dependent on UV dose or UV treatment time (Fig. 2E). For example, compared with untreated cells, the interaction of these proteins was decreased 1.3-fold at a dose of 23 J/m2 and 3.7-fold at a dose of 46 J/m2 in UV-irradiated cells as determined by Western blot band intensity measurement (Fig. 2E, left lower column diagram). In addition, at 3 h after UV irradiation the interaction of these proteins was decreased 2.3-fold, and at 6 h 5.5-fold (Fig. 2E, right lower column diagram). It is interesting that there were no obvious changes in the interaction of these proteins in cells treated with x-ray, H2O2, or 5-aza-CdR (Fig. 2, B–D), which shows that UV treatment may have a unique capacity to induce a decrease in the interaction of PCNA-MTH2.
FIGURE 2.
Dissociation of PCNA and MTH2 in A549 cells treated with UV. A–D, A549 cells were treated with UV at 46 J/m2 and then incubated for 6 h or with X-rays at different doses (6Gy, 9Gy,12Gy) and then incubated for 12 h, or with H2O2 (1 mm for 12 h) or with 5-aza-CdR (1 μm for 72 h). Protein was extracted for immunoprecipitation using anti-MTH2, followed by Western blotting using anti-PCNA or anti-MTH2. E, A549 cells were irradiated with UV at different doses and then incubated for 4 h or incubated for different times after UV irradiation at 46 J/m2. Protein was extracted for immunoprecipitation using anti-MTH2, followed by Western blotting using anti-PCNA or anti-MTH2. PCNA bands were scanned, and relative band intensities were normalized for each MTH2 band. The band intensity of CTR was set as 100, and the numerical value of the intensity of each band represents the percentage of band intensity with respect to that of CTR. The representative value in the lower column diagram is an average relative band intensity of PCNA, with standard error from several independent experiments. F and G, both PCNA and MTH2 were detected by Western blotting (F) or RT-PCR (G) in A549 cells irradiated with UV at different doses or for different incubation times after irradiation. The α-tubulin loading control is shown in the lower panel. Protein and mRNA bands were scanned, and relative band intensities were normalized for each α-tubulin band. The lower column diagrams represent an average relative band intensity of PCNA or MTH2 with standard error from several independent experiments.
To investigate whether the decrease in interaction between PCNA and MTH2 is due to dissociation of these proteins or down-regulation of individual protein expression, Western blotting and RT-PCR were performed to evaluate expression of PCNA and MTH2 (respectively). Results showed that levels of both proteins and mRNA were sustained almost without change within a period of 6 h after UV irradiation (Fig. 2, F and G), indicating that UV irradiation led to reduced MTH2/PCNA association and not to a decrease in the level of either protein.
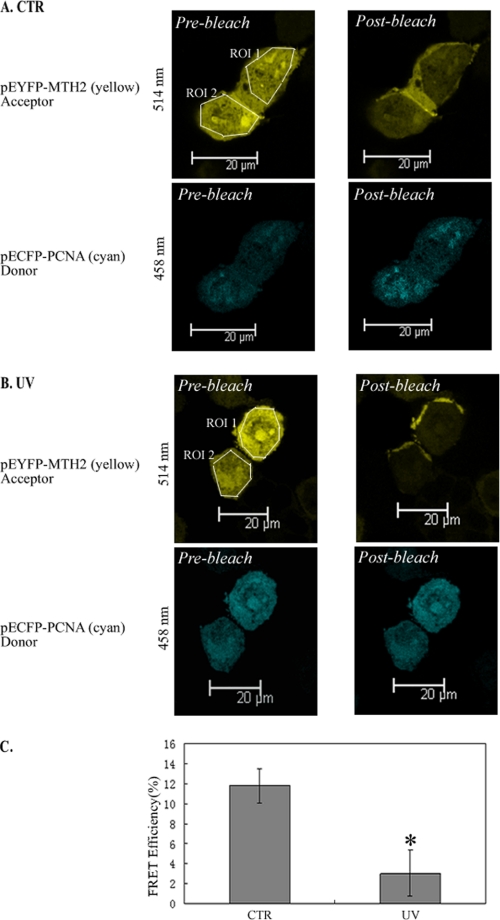
In addition, the FRET assay was performed to confirm the occurrence of dissociation of PCNA and MTH2 in response to UV treatment. HEK293T cells were transfected with plasmids expressing ECFP-PCNA and EYFP-MTH2. In this assay, the acceptor photobleaching technique was used to compare the emission of the donor fluorophore (ECFP-PCNA) before and after photobleaching of the acceptor fluorophore (EYFP-MTH2) within both ROI 1 and ROI 2 (ROI, region of interest). The images on the left panels of Fig. 3, A and B show the fluorescence of EYFP-MTH2 and ECFP-PCNA before photobleaching. In unstressed cells, after photobleaching of both ROI 1 and ROI 2 of the acceptor (upper right panel, Fig. 3A), the fluorescence of the donor in the ROIs was markedly enhanced (lower right panel, Fig. 3A), demonstrating that FRET had occurred between PCNA and MTH2. However, in UV-treated cells, after both ROI 1 and ROI 2 of the acceptor were photobleached (upper right panel, Fig. 3B), the fluorescence of the donor in the ROIs showed no obvious change (lower right panel, Fig. 3B), suggesting that FRET was weakened between these two proteins. Statistically, the energy transfer efficiency was obviously decreased in UV-irradiated HEK293T cells as compared with that in untreated cells (Fig. 3C). These data clearly demonstrate that the interaction between PCNA and MTH2 was disrupted in response to UV irradiation.
FIGURE 3.
FRET analysis of PCNA-MTH2 interaction. A and B, FRET assay was performed to detect the energy transfer efficiency between PCNA and MTH2 before (A) or after (B) UV treatment. pEYFP-MTH2 and pECFP-PCNA were transfected into HEK293T cells and visualized by excitation at 514 nm for EYFP-MTH2 (upper panel of A and B) or at 458 nm for ECFP-PCNA (lower panel of A and B). In the FRET assay, EYFP-MTH2 acted as the acceptor, and ECFP-PCNA as the donor. In the upper left panel (A and B), the regions chosen for photobleaching (ROI) are bounded by a white line. C, FRET efficiency was shown in the column diagram, and the representative value is an average FRET efficiency of 25 cells with standard error (*, p < 0.05).
UV Induces Acetylation of PCNA
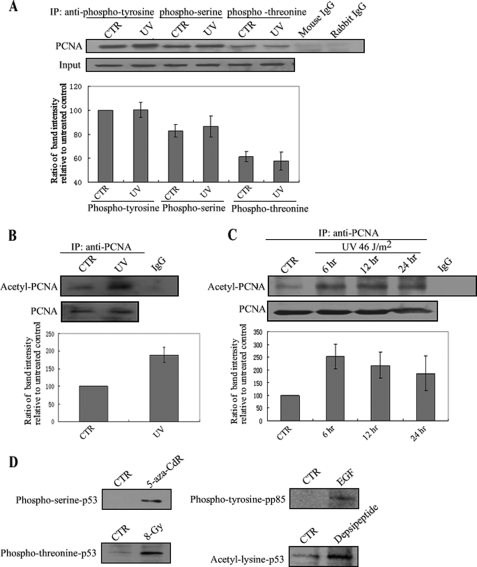
Both acetylation and phosphorylation have been reported to be related to the PCNA function (37, 38), which raises a question as to whether post-translational modification of PCNA is involved in UV-induced dissociation. To evaluate this question, cell lysate from A549 cells was first co-immunoprecipitated with anti-phosphotyrosine, anti-phosphoserine, or anti-phosphothreonine, and Western blotting was carried out with anti-PCNA. As shown in Fig. 4A, we did not observe an obvious change in PCNA phosphorylation at tyrosine, serine, or threonine sites. However, UV led to significant acetylation of PCNA (Fig. 4B), and the acetylation was sustained for up to 24 h after UV irradiation (Fig. 4C). In addition, to test the specificity of these antibodies, A549 cells and A431 cells were treated with various stimuli. 5-aza-CdR and x-ray were shown to induce phosphorylation of p53 serine or phosphorylation of p53 threonine in A549 cells (35). Epidermal growth factor (EGF) was shown to stimulate the phosphotyrosine of pp85 (39). Depsipeptide (a HDAC inhibitor) was shown to induce the acetylation of p53 lysine (34). As shown in Fig. 4D, these stimuli significantly induced post-translational modifications of p53 at these sites which were tested, demonstrating that acetylation is a unique modification of PCNA in response to UV treatment.
FIGURE 4.
UV irradiation induces PCNA acetylation in A549 cells. A, A549 cells were irradiated with UV at 46 J/m2 and incubated for 6 h. Protein was extracted and then immunoprecipitated using anti-phosphotyrosine, phosphoserine, or phosphothreonine, followed by Western blotting using anti-PCNA. Mouse or rabbit IgG was used as a negative control. PCNA bands were scanned, and relative band intensities were normalized for each input band. The lower column diagram represents the average relative band intensity of PCNA with standard error from several independent experiments. B and C, A549 cells were irradiated with UV at 46 J/m2 and incubated for 6 h (B), or at 23 J/m2 for different incubation times (6, 12, or 24 h) (C). Protein was extracted and then immunoprecipitated using anti-PCNA followed by Western blotting using anti-acetyl-lysine. IgG was used as a negative control. The representative value of the lower column diagram is the average relative band intensity of PCNA with standard error from several independent experiments. D, epidermal growth factor (EGF) was added to human A431 carcinoma cells to identify phosphotyrosine in pp85. A549 cells were treated with 5-aza-CdR (0.1 μm, 48 h), x-ray (8-Gy), or depsipeptide (0.1 μm, 6 h) to test for phosphoserine, phosphothreonine, or acetyl-lysine of p53, respectively.
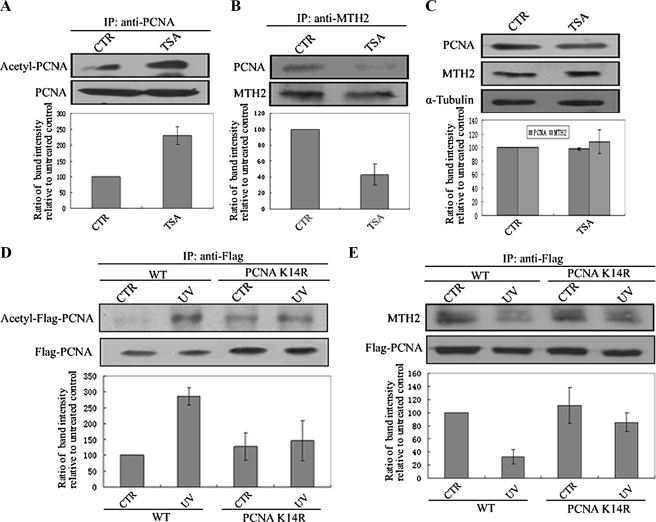
To test whether modification of PCNA by acetylation is related to dissociation of these proteins, A549 cells were treated with TSA (Trichostatin A, a HDAC inhibitor) to mimic protein acetylation status, and a co-immunoprecipitation assay was carried out. Results showed that TSA also induced acetylation of PCNA (Fig. 5A), and dissociation of these proteins followed (Fig. 5B). However, the level of the two proteins was sustained in TSA-treated cells (Fig. 5C).
FIGURE 5.
PCNA acetylation at Lys-14 is required for the dissociation of PCNA and MTH2. A and B, A549 cells were treated with TSA (1 μm, 12 h), and protein was then extracted for immunoprecipitation using anti-PCNA (A) or anti-MTH2 (B), followed by Western blotting using anti-acetyl-lysine or anti-PCNA. The lower column diagram represents the average relative band intensity of PCNA with standard error from several independent experiments. C, protein was extracted from TSA-treated cells (1 μm, 12 h), and Western blotting was performed using anti-PCNA, anti-MTH2, or anti-α-tubulin. The lower column represents the average band intensity of PCNA or MTH2 with standard error from several independent experiments. D and E, wild-type (WT) or K14R Flag-PCNA was transfected into HEK293T cells for 48 h. Cells were then irradiated with UV at 46 J/m2 and incubated for 6 h. Protein was extracted for immunoprecipitation using anti-Flag, followed by Western blotting using anti-acetyl-lysine (D) or anti-MTH2 (E). The bands representing acetyl-Flag-PCNA (D) or MTH2 (E) were scanned, and relative band intensities were normalized for each Flag-PCNA band. The lower column diagram represents the average relative band intensity of acetyl-Flag-PCNA (D) or MTH2 (E) with standard error from several independent experiments.
In addition, to identify which acetylation site is critical for the interaction of these two proteins, several site-directed mutants of PCNA were established. As PCNA binds to MTH2 via its N terminus (including Lys-13, Lys-14, Lys-20, Lys-77, Lys-80), Flag-tagged wild-type and various mutants of PCNA (mutated Lys-13, Lys-14, Lys-20, Lys-77, or Lys-80) was generated and transfected in HEK293T cells. However, it was found that UV irradiation did not induce an obvious increase in PCNAK14R acetylation as compared with that of wild-type PCNA (Fig. 5D). Consequently, UV could not lead to the dissociation of PCNAK14R and MTH2 (Fig. 5E), indicating that acetylation of PCNA at Lys-14 may play a key role in regulating interaction of MTH2 and PCNA.
Dissociated PCNA Has a Shorter Half-life and Is More Readily Degraded
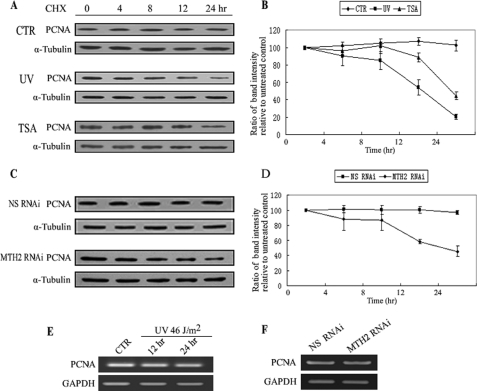
PCNA is a highly conserved protein and its half-life is comparatively long. For example, in NIH/3T3 cells, the reported half-life of PCNA is 20 h (40). When cycloheximide (CHX), an inhibitor of protein synthesis, was added into the medium at 25 μg/ml for up to 24 h, the expression levels of PCNA remained unchanged in A549 cells (Fig. 6A, upper panel). However, in response to UV irradiation, the half-life of PCNA was obviously shortened after a lag time (Fig. 6A, middle panel). For example, the level of PCNA did not obviously change in the initial time period (∼6 h) after UV irradiation. However, the PCNA level was decreased to 58.7% at 12 h and further decreased to 21.4% at 24 h as compared with the level of PCNA immediately after UV irradiation (0 h) (Fig. 6B). Because the PCNA mRNA level was not changed in response to UV irradiation (Fig. 6E), it is likely that UV irradiation accelerates degradation of PCNA. To determine whether there is a link between UV-induced dissociation of these proteins and UV-induced degradation of PCNA, the half-life of PCNA in TSA-treated or MTH2-knocked down A549 cells was also evaluated. A similar result was obtained in the TSA-treated cells, in which the half-life of PCNA was shortened to 12–24 h (Fig. 6, A, lower panel and B). Subsequently, knockdown of MTH2 was used to simulate the dissociated status of PCNA and MTH2. The efficiency of MTH2 RNAi is shown in supplemental Fig. S1C. The level of PCNA was sustained for up to 24 h after treatment with CHX in nonspecific RNAi A549 cells (Fig. 6, C, upper panel and D). However, the half-life of PCNA in MTH2-knocked down cells was shortened to a period of between 12–24 h (Fig. 6, C, lower panel and D), and the mRNA level of PCNA was sustained in MTH2-knocked down cells as compared with that in nonspecific RNAi cells (Fig. 6F). These data suggest that PCNA dissociated from its complex with MTH2 has a shorter half-life and is more easily degraded.
FIGURE 6.
Half-life of PCNA is shortened after dissociation of PCNA and MTH2. A and B, CHX was added into untreated, UV-treated, or TSA-treated A549 cells at 25 μg/ml for different times as indicated. Western blotting was performed from whole cell extracts with anti-PCNA (A). The hyphenated line graph shows the change in PCNA level at different times. The representative value is the average band intensity of PCNA with standard error from several independent experiments (B). C and D, 24 h after NS-siRNA or MTH2-siRNA was transfected into A549 cells, CHX was added to the medium at 25 μg/ml for various lengths of time as indicated. NS-siRNA was used as negative control for the RNAi assay. The hyphenated line graph shows the change in PCNA level at different times. The representative value is the average band intensity of PCNA with standard error from several independent experiments (D). E and F, A549 cells were irradiated with UV at 46 J/m2 and then incubated for 12 h or 24 h (F). NS-siRNA or MTH2-siRNA was transfected into A549 cells for 48 h (G). mRNA was extracted for RT-PCR.
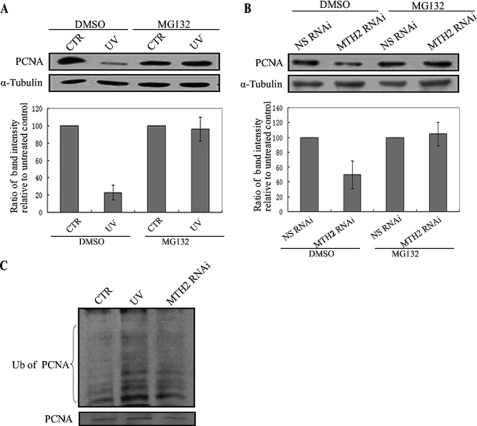
To further analyze the pathway of PCNA degradation, UV-irradiated or MTH2-knocked down A549 cells were investigated in the presence of the proteosome inhibitor MG132. Down-regulation of PCNA induced by UV irradiation or MTH2-siRNA treatment was reversed in cells in the presence of MG132 (Fig. 7, A and B). Moreover, PCNA-conjugated ubiquitin in UV or MTH2 siRNA-treated cells was increased compared with that in untreated cells (Fig. 7C). These results demonstrate that the degradation of PCNA likely occurs mainly through a proteosome pathway.
FIGURE 7.
Degradation of PCNA is proteosome-dependent. A and B, A549 cells were UV-irradiated at 46 J/m2 and then incubated for 24 h with DMSO or MG132 at 25 μm, or NS-siRNA or MTH2-siRNA was transfected into A549 cells for 48 h with DMSO or MG132 at 25 μm. Proteins were extracted for Western blotting with anti-PCNA. The lower column diagram represents the average band intensity of PCNA with standard error from several independent experiments. C, A549 cells were treated with UV (46 J/m2, 24 h) or MTH2-siRNA in the presence of proteasome inhibitors (25 μm MG132). The treated cells were then lysed and immunoprecipitated with anti-PCNA, followed by Western blotting with anti-ubiquitin.
The Degradation of PCNA Is Related to Inhibitory Effects of UV on DNA Synthesis and Cell Cycle Progression
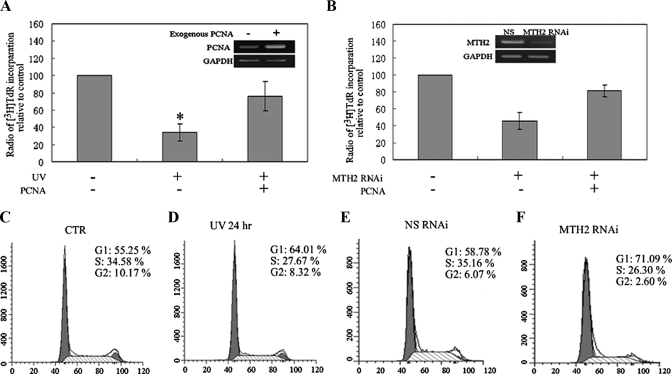
It is known that PCNA is involved in many metabolic pathways and is in particular involved in regulating DNA synthesis. This led to the supposition that PCNA-mediated DNA synthesis may be influenced by MTH2 in response to UV treatment. Thus, an [3H]TdR incorporation assay was used to evaluate DNA synthesis rate and cell proliferation. As shown in Fig. 8A, a significant decrease in [3H]TdR incorporation was detected in UV-treated cells, and this decrease was reversed by expression of exogenous PCNA. To further investigate whether knockdown of MTH2 inhibits DNA synthesis by promoting PCNA degradation, [3H]TdR was added into the medium for 12 h after MTH2-siRNA treatment. Results showed that radioactivity from [3H]TdR was obviously reduced in MTH2-knocked down cells, and this reduction was also reversed by expression of exogenous PCNA (Fig. 8B). These data suggest that knockdown of MTH2 was at least partly associated with inhibition of DNA synthesis through promoting the degradation of PCNA.
FIGURE 8.
PCNA degradation inhibits DNA synthesis and cell cycle progression. A and B, after A549 cells were irradiated with UV (46 J/m2), or MTH2 was knocked down from A549 cells, exogenous PCNA was transfected into the cells. [3H]TdR was then added to the medium at 1 μCi/ml for 12 h, and DNA was precipitated with cold trichloroacetic acid. The radioactivity of three separate experiments was measured, and relative radioactivity per μg of DNA was counted. Statistical analysis was performed by comparing the radioactivity of UV to CTR or MTH2 RNAi to NS RNAi (*, p < 0.05). The results of RT-PCR in the top panels showed the efficiency of expression of exogenous PCNA by transfecting pHPCNA15 into A549 cells (A) and the efficiency of knocking down MTH2 by transfecting MTH2 siRNA into A549 cells (B). C–F, representative flow cytometry histograms at 24 h after irradiating A549 cells with UV at 46 J/m2, or at 48 h after NS-siRNA or MTH2-siRNA was transfected into A549 cells. The y axis represents the cell count, and the x axis represents DNA content. These experiments were conducted four times. Differences between groups were statistically analyzed using the unpaired Student's t test (*, p < 0.05).
In addition to DNA synthesis, PCNA is also required for G1-phase progression by interacting with the cyclin D-CDK4 complex (41). We therefore further analyzed changes in the cell cycle in UV-treated or MTH2-knock down A549 cells. By flow cytometry analysis, an obvious G1-phase arrest was observed in both UV-treated A549 cells and MTH2-knocked down A549 cells when compared with the untreated controls (Fig. 8, C–F). For example, after the treatment of UV or RNAi against MTH2, the cells accumulated in G1-phase increased to 64.01% from 55.25% and to 71.09% from 58.78%, respectively. These results suggested that UV irradiation induced changes in the cell cycle may be related to PCNA degradation.
An additional question of interest raised in this study is the fate of MTH2 after dissociation from PCNA in response to UV treatment. To explore the effect of PCNA on MTH2 activity, A549 cells were treated with TSA to induce the dissociation of PCNA and MTH2, and the cellular 8-oxo-dG was then detected with anti-8-oxo-dG. MTH2, as a 8-oxo-dGTPase, could degrade 8-oxo-dGTP to 8-oxo-dGMP, and prevent the misincorporation of 8-oxo-dG into DNA. Therefore, there was a correlation between the activity of MTH2 and the amount of cellular 8-oxo-dG. As a positive control, H2O2-treated cells showed a significant increase in 8-oxo-dG. Comparably, as a negative control, N-acetyl-l-cysteine, a ROS inhibitor, showed an obvious decrease in 8-oxo-dG. The amount of 8-oxo-dG was decreased in TSA-treated cells, compared with untreated control (supplemental Fig. S3). However, the difference between two groups was not significant by a statistic analysis. It suggests that PCNA may be not a key factor to regulate the activity of MTH2.
DISCUSSION
The data presented here provide evidence that the newly identified molecule MTH2 is of potential importance in its functions in association with PCNA. In particular, our findings demonstrate that the interaction between PCNA and MTH2 may protect PCNA from degradation. When these proteins were dissociated in response to UV irradiation, degradation of PCNA was accelerated, and DNA synthesis decelerated.
UV irradiation has been reported to induce apoptosis in many cell lines. For example, human keratinocyte cell line HaCaT and human leukemia cell line HL-60 were sensitive to UV-induced apoptosis (42–44). In this study, to avoid apoptosis induced by UV irradiation, we used A549 cells to observe the PCNA/MTH2 association, as A549 cells were resistant to UV irradiation-induced apoptosis (45, 46). In addition, LKB1, a tumor suppressor, is frequently inactivated in non-small cell lung cancer (47). Therefore, in addition to non-small cell lung cancer cell lines (A549, H1299, and H719), we also selected human epithelial cell line 2BS (LKB1-proficient), and human embryonic kidney 293T cell (LKB1-proficient) to further evaluate the interaction of MTH2-PCNA.
Many PCNA-binding proteins, such as p21, polymerase β, specific endonuclease Fen1, and XPG contain a consensus PCNA-interacting motif, referred to as a PIP-box [Q-XX-(h)- X-X-(a)-(a)] (48–51). Recently, a novel PCNA binding motif has been identified, which is called a KA-box [KA-(A/L/I)-(A/L/Q)-X-X-(L/V)] (52). This motif is also present in several PCNA-binding proteins, such as polymerase δ, polymerase ϵ, and RFC (52). However, the PCNA-binding domain of MTH2 contains neither the PIP-box nor the KA-box motif. Although MTH2 lacks a consensus motif for PCNA binding, the C-terminal region of MTH2 was demonstrated in this study to bind to PCNA (Fig. 1C). Consistent with our results, GADD45 has been reported to bind to PCNA in its C terminus, although there is no consensus motif in its C-terminal regions (53).
PCNA has previously been found to interact with multiple different molecules, which are involved in several important metabolic pathways (2, 54, 55). In addition, the interaction of GADD45 with PCNA is involved in nucleotide excision repair (NER) (56, 57). DNA methyltransferase (58, 59), histone deacetylases (HDAC) (60), and p300 (61–63) also bind to PCNA to regulate DNA methylation and chromatin remodeling. More importantly, PCNA is able to mediate switching of different pathways by interacting with different molecules. For example, PCNA interacts with different DNA polymerases resulting in a switch from accurate DNA synthesis to error-prone translesion synthesis (TLS) (64–66). Moreover, monoubiquitination of PCNA triggers a switching of interactions between PCNA and different polymerases (67–69). After DNA damage, translesion DNA synthesis (TLS) occurs to prevent stalling of DNA synthesis, and DNA repair machinery is activated to maintain DNA integrity. Therefore, after UV irradiation, TLS polymerase or GADD45 binds to PCNA to perform TLS or nucleotide excision repair (NER). However, PCNA involved in TLS or NER is chromatin-bound PCNA, whereas PCNA associated with MTH2 is chromatin-unbound (supplemental Fig. S2). Therefore, it seems likely that the dissociation of MTH2 and PCNA is independent of TLS polymerase or GADD45.
It has been reported that UV irradiation can induce expression of PCNA, wherein the mRNA level of PCNA was increased by 2-fold over the initial 1–2 h after irradiation as assayed with Northern blotting (70). However, over a period of up to 4–6 h after UV irradiation, PCNA gradually returned its original level. Therefore, the up-regulation of PCNA is a relatively acute response to UV irradiation. This is consistent with our result that levels of PCNA did not obviously change over the 3–6-h period after UV irradiation (Fig. 2, F and G), although we did not detect changes in PCNA in the initial 1–2 h after irradiation.
It has been reported that mammalian cells contain three different PCNA isoforms which differ in their acetylation status, and these differences are associated with the subcellular localization and functions of these isoforms (37). In this study, we observed that acetylation of PCNA at Lys-14 was induced by UV irradiation, and followed the dissociation of PCNA and MTH2 (Fig. 5, D and E). This therefore suggests that acetylation of PCNA at Lys-14 is a critical modification, which may bring about a change in PCNA configuration and in turn disrupt the interaction of MTH2 and PCNA.
Although PCNA functions have been well studied, the mechanisms by which its functions are controlled remains to be investigated. In this study, we observed that the half-life of PCNA was shorter in UV-treated and MTH2-knocked down cells (Fig. 6, A–D), and observed that the loss of PCNA was prevented by the proteosome inhibitor MG132 (Fig. 7, A and B). However, PCNA did not accumulate in MG132-treated cells as compared with untreated cells, which is consistent with evidence that has been obtained by other researchers (38, 71). This lack of PCNA accumulation may result from the fact that although PCNA degradation is proteosome-dependent, the degradation level of PCNA is usually very low in nonstressed cells (72) and an obvious accumulation of PCNA in cells in the presence of MG132 is thus not observed. Nevertheless, these results indicated that the dissociation of PCNA and MTH2 is a trigger for degradation of PCNA and further suggested that MTH2 forms a complex with PCNA to protect PCNA from degradation. The region of PCNA cleaved by proteosomes lies in its N-terminal region (72), which is precisely the MTH2-binding region of PCNA, and thus suggesting that the binding of MTH2 to PCNA overlies the N-degron of PCNA. As such, after the association of PCNA and MTH2 is disrupted by UV, the degron of PCNA is exposed, and PCNA degradation is triggered. Therefore, the association of MTH2 and PCNA stabilizes PCNA, but involves mainly the chromatin-unbound, free PCNA.
Because PCNA has been used as a proliferation index to reflect tumor growth because of its high levels in proliferating cells (55, 73–75), reduction of PCNA stability or acceleration of PCNA degradation might offer an effective strategy for regulating tumor growth. UV irradiation is a universal DNA-damaging factor, which acts mainly by inducing formation of DNA photoproducts between adjacent pyrimidine bases on the same strand such as cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts (6–4PPs) (76). UV-induced DNA photoproducts are the main reason that UV leads to stalled DNA synthesis. Here our results showed that the expression of exogenous PCNA could partly reverse the inhibitory effect of UV on DNA synthesis (Fig. 8A), suggesting that degradation of PCNA is one of the reasons that UV particularly inhibits DNA synthesis.
In addition, UV-induced cell cycle arrest is a dynamic process, in which there is a redistribution of cells from G1-phase to S-phase (77). For instance, UV-irradiated U343 cells initially accumulated in G1-phase, and reached a peak between 24 and 36 h, followed by an S-phase arrest between 36 and 48 h. This is consistent with our results wherein an obvious G1-phase arrest was induced at 24 h after UV irradiation (Fig. 8D).
In conclusion, this study demonstrated that PCNA and MTH2 form a complex to maintain the stability of PCNA in unstressed cells. However, UV irradiation induces dissociation of PCNA and MTH2, which leads to degradation of PCNA. Therefore, disruption of this specific interaction between PCNA and MTH2 may prove to be a worthwhile strategy for inhibition of development of malignant tumors.
Supplementary Material
Acknowledgments
We thank our colleagues Donglai Wang, Zhixing Zheng, Jing Yang, Wen Zhou, and Xi Wang for technology used in this work and useful discussions.
This study was supported by the National Natural Science Foundation of China (No. 90919030, No.30425017, 30670417, and 30621002) and Grants 2005CB522403, 2006CB910300, and B07001 from the Ministry of Science and Technology of China.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PCNA
- proliferating cell nuclear antigen
- MTH2
- MutT homolog2
- 8-oxo-dGTP
- 8-oxo-7,8-dihydrodeoxyguanosine triphosphate
- GST
- glutathione S-transferase
- PMSF
- phenylmethylsulfonyl fluoride
- TSA
- trichostatin A
- ROI
- region of interest
- FRET
- fluorescence resonance energy transfer
- PIPES
- 1,4-piperazinediethanesulfonic acid
- RNAi
- RNA interference
- CHX
- cycloheximide
- TLS
- translesion synthesis
- RT
- reverse transcription.
REFERENCES
- 1.Indiani C., O'Donnell M. (2006) Nat. Rev. Mol. Cell Biol. 7, 751–761 [DOI] [PubMed] [Google Scholar]
- 2.Moldovan G. L., Pfander B., Jentsch S. (2007) Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 3.Warbrick E. (2000) Bioessays 22, 997–1006 [DOI] [PubMed] [Google Scholar]
- 4.Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 5.Kong X. P., Onrust R., O'Donnell M., Kuriyan J. (1992) Cell 69, 425–437 [DOI] [PubMed] [Google Scholar]
- 6.Bravo R., Frank R., Blundell P. A., Macdonald-Bravo H. (1987) Nature 326, 515–517 [DOI] [PubMed] [Google Scholar]
- 7.Prelich G., Tan C. K., Kostura M., Mathews M. B., So A. G., Downey K. M., Stillman B. (1987) Nature 326, 517–520 [DOI] [PubMed] [Google Scholar]
- 8.Jaskulski D., deRiel J. K., Mercer W. E., Calabretta B., Baserga R. (1988) Science 240, 1544–1546 [DOI] [PubMed] [Google Scholar]
- 9.Prelich G., Kostura M., Marshak D. R., Mathews M. B., Stillman B. (1987) Nature 326, 471–475 [DOI] [PubMed] [Google Scholar]
- 10.Levin D. S., Bai W., Yao N., O'Donnell M., Tomkinson A. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12863–12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosfield D. J., Mol C. D., Shen B., Tainer J. A. (1998) Cell 95, 135–146 [DOI] [PubMed] [Google Scholar]
- 12.Wu X., Li J., Li X., Hsieh C. L., Burgers P. M., Lieber M. R. (1996) Nucleic Acids Res. 24, 2036–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prelich G., Stillman B. (1988) Cell 53, 117–126 [DOI] [PubMed] [Google Scholar]
- 14.Shivji K. K., Kenny M. K., Wood R. D. (1992) Cell 69, 367–374 [DOI] [PubMed] [Google Scholar]
- 15.Clark A. B., Valle F., Drotschmann K., Gary R. K., Kunkel T. A. (2000) J. Biol. Chem. 275, 36498–36501 [DOI] [PubMed] [Google Scholar]
- 16.Masih P. J., Kunnev D., Melendy T. (2008) Nucleic Acids Res. 36, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou H., Theriot C. A., Das A., Hegde M. L., Matsumoto Y., Boldogh I., Hazra T. K., Bhakat K. K., Mitra S. (2008) J. Biol. Chem. 283, 3130–3140 [DOI] [PubMed] [Google Scholar]
- 18.Matsuoka S., Yamaguchi M., Matsukage A. (1994) J. Biol. Chem. 269, 11030–11036 [PubMed] [Google Scholar]
- 19.Scorah J., Dong M. Q., Yates J. R., 3rd, Scott M., Gillespie D., McGowan C. H. (2008) J. Biol. Chem. 283, 17250–17259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arias E. E., Walter J. C. (2006) Nat Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 21.Engel F. B., Hauck L., Boehm M., Nabel E. G., Dietz R., von Harsdorf R. (2003) Mol. Cell. Biol. 23, 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maki H., Sekiguchi M. (1992) Nature 355, 273–275 [DOI] [PubMed] [Google Scholar]
- 23.Shibutani S., Takeshita M., Grollman A. P. (1991) Nature 349, 431–434 [DOI] [PubMed] [Google Scholar]
- 24.Cai J. P., Kawate H., Ihara K., Yakushiji H., Nakabeppu Y., Tsuzuki T., Sekiguchi M. (1997) Nucleic Acids Res. 25, 1170–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tajiri T., Maki H., Sekiguchi M. (1995) Mutat. Res. 336, 257–267 [DOI] [PubMed] [Google Scholar]
- 26.Akiyama M., Maki H., Sekiguchi M., Horiuchi T. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 3949–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishibashi T., Hayakawa H., Sekiguchi M. (2003) EMBO Rep. 4, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekiguchi M., Tsuzuki T. (2002) Oncogene 21, 8895–8904 [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi M., Ohara-Nemoto Y., Kaneko M., Hayakawa H., Sekiguchi M., Yamamoto K. (1998) J. Biol. Chem. 273, 26394–26399 [DOI] [PubMed] [Google Scholar]
- 30.Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M. (1993) J. Biol. Chem. 268, 23524–23530 [PubMed] [Google Scholar]
- 31.Tsuzuki T., Egashira A., Igarashi H., Iwakuma T., Nakatsuru Y., Tominaga Y., Kawate H., Nakao K., Nakamura K., Ide F., Kura S., Nakabeppu Y., Katsuki M., Ishikawa T., Sekiguchi M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11456–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai J. P., Ishibashi T., Takagi Y., Hayakawa H., Sekiguchi M. (2003) Biochem. Biophys. Res. Commun. 305, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 33.Zhu W. G., Lakshmanan R. R., Beal M. D., Otterson G. A. (2001) Cancer Res. 61, 1327–1333 [PubMed] [Google Scholar]
- 34.Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., Zhu W. G. (2006) Mol. Cell. Biol. 26, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H., Zhao Y., Li L., McNutt M. A., Wu L., Lu S., Yu Y., Zhou W., Feng J., Chai G., Yang Y., Zhu W. G. (2008) J. Biol. Chem. 283, 2564–2574 [DOI] [PubMed] [Google Scholar]
- 36.Zhu W. G., Hileman T., Ke Y., Wang P., Lu S., Duan W., Dai Z., Tong T., Villalona-Calero M. A., Plass C., Otterson G. A. (2004) J. Biol. Chem. 279, 15161–15166 [DOI] [PubMed] [Google Scholar]
- 37.Naryzhny S. N., Lee H. (2004) J. Biol. Chem. 279, 20194–20199 [DOI] [PubMed] [Google Scholar]
- 38.Wang S. C., Nakajima Y., Yu Y. L., Xia W., Chen C. T., Yang C. C., McIntush E. W., Li L. Y., Hawke D. H., Kobayashi R., Hung M. C. (2006) Nat. Cell Biol. 8, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 39.Cohen B., Yoakim M., Piwnica-Worms H., Roberts T. M., Schaffhausen B. S. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4458–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bravo R., Macdonald-Bravo H. (1987) J. Cell Biol. 105, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H., Xiong Y., Beach D. (1993) Mol. Biol. Cell 4, 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gniadecki R., Hansen M., Wulf H. C. (1997) J. Invest. Dermatol. 109, 163–169 [DOI] [PubMed] [Google Scholar]
- 43.Schwarz A., Bhardwaj R., Aragane Y., Mahnke K., Riemann H., Metze D., Luger T. A., Schwarz T. (1995) J. Invest. Dermatol. 104, 922–927 [DOI] [PubMed] [Google Scholar]
- 44.Martin S. J., Cotter T. G. (1991) Int. J. Radiat. Biol. 59, 1001–1016 [DOI] [PubMed] [Google Scholar]
- 45.Magrini R., Russo D., Ottaggio L., Fronza G., Inga A., Menichini P. (2008) J. Cell. Biochem. 104, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 46.Volm M., Rittgen W. (2000) Anticancer Res. 20, 3449–3458 [PubMed] [Google Scholar]
- 47.Ji H., Ramsey M. R., Hayes D. N., Fan C., McNamara K., Kozlowski P., Torrice C., Wu M. C., Shimamura T., Perera S. A., Liang M. C., Cai D., Naumov G. N., Bao L., Contreras C. M., Li D., Chen L., Krishnamurthy J., Koivunen J., Chirieac L. R., Padera R. F., Bronson R. T., Lindeman N. I., Christiani D. C., Lin X., Shapiro G. I., Jänne P. A., Johnson B. E., Meyerson M., Kwiatkowski D. J., Castrillon D. H., Bardeesy N., Sharpless N. E., Wong K. K. (2007) Nature 448, 807–810 [DOI] [PubMed] [Google Scholar]
- 48.Kedar P. S., Kim S. J., Robertson A., Hou E., Prasad R., Horton J. K., Wilson S. H. (2002) J. Biol. Chem. 277, 31115–31123 [DOI] [PubMed] [Google Scholar]
- 49.Waga S., Hannon G. J., Beach D., Stillman B. (1994) Nature 369, 574–578 [DOI] [PubMed] [Google Scholar]
- 50.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. (2005) EMBO J. 24, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gary R., Ludwig D. L., Cornelius H. L., MacInnes M. A., Park M. S. (1997) J. Biol. Chem. 272, 24522–24529 [DOI] [PubMed] [Google Scholar]
- 52.Xu H., Zhang P., Liu L., Lee M. Y. (2001) Biochemistry 40, 4512–4520 [DOI] [PubMed] [Google Scholar]
- 53.Vairapandi M., Azam N., Balliet A. G., Hoffman B., Liebermann D. A. (2000) J. Biol. Chem. 275, 16810–16819 [DOI] [PubMed] [Google Scholar]
- 54.Maga G., Hubscher U. (2003) J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 55.Waga S., Stillman B. (1998) Annu. Rev. Biochem. 67, 721–751 [DOI] [PubMed] [Google Scholar]
- 56.Liebermann D. A., Hoffman B. (2007) Blood Cells Mol. Dis. 39, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liebermann D. A., Hoffman B. (2008) J. Mol. Signal 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huen M. S., Sy S. M., van Deursen J. M., Chen J. (2008) J. Biol. Chem. 283, 11073–11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F. (1997) Science 277, 1996–2000 [DOI] [PubMed] [Google Scholar]
- 60.Milutinovic S., Zhuang Q., Szyf M. (2002) J. Biol. Chem. 277, 20974–20978 [DOI] [PubMed] [Google Scholar]
- 61.Hasan S., Hassa P. O., Imhof R., Hottiger M. O. (2001) Nature 410, 387–391 [DOI] [PubMed] [Google Scholar]
- 62.Hong R., Chakravarti D. (2003) J. Biol. Chem. 278, 44505–44513 [DOI] [PubMed] [Google Scholar]
- 63.Cazzalini O., Perucca P., Savio M., Necchi D., Bianchi L., Stivala L. A., Ducommun B., Scovassi A. I., Prosperi E. (2008) Nucleic Acids Res. 36, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haracska L., Johnson R. E., Unk I., Phillips B., Hurwitz J., Prakash L., Prakash S. (2001) Mol. Cell. Biol. 21, 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maga G., Villani G., Ramadan K., Shevelev I., Tanguy Le Gac N., Blanco L., Blanca G., Spadari S., U. (2002) J. Biol. Chem. 277, 48434–48440 [DOI] [PubMed] [Google Scholar]
- 66.Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 67.Kannouche P. L., Wing J., Lehmann A. R. (2004) Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 68.Matunis M. J. (2002) Mol. Cell 10, 441–442 [DOI] [PubMed] [Google Scholar]
- 69.Niimi A., Brown S., Sabbioneda S., Kannouche P. L., Scott A., Yasui A., Green C. M., Lehmann A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng X. R., Hao H., Jiang Y., Lee M. Y. (1994) J. Biol. Chem. 269, 24027–24033 [PubMed] [Google Scholar]
- 71.Izumi M., Yatagai F., Hanaoka F. (2001) J. Biol. Chem. 276, 48526–48531 [DOI] [PubMed] [Google Scholar]
- 72.Naryzhny S. N., Lee H. (2003) Proteomics 3, 930–936 [DOI] [PubMed] [Google Scholar]
- 73.Kamat A. A., Kim T. J., Landen C. N., Jr., Lu C., Han L. Y., Lin Y. G., Merritt W. M., Thaker P. H., Gershenson D. M., Bischoff F. Z., Heymach J. V., Jaffe R. B., Coleman R. L., Sood A. K. (2007) Cancer Res. 67, 281–288 [DOI] [PubMed] [Google Scholar]
- 74.Han L. Y., Landen C. N., Trevino J. G., Halder J., Lin Y. G., Kamat A. A., Kim T. J., Merritt W. M., Coleman R. L., Gershenson D. M., Shakespeare W. C., Wang Y., Sundaramoorth R., Metcalf C. A., 3rd, Dalgarno D. C., Sawyer T. K., Gallick G. E., Sood A. K. (2006) Cancer Res. 66, 8633–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chodon D., Banu S. M., Padmavathi R., Sakthisekaran D. (2007) Mol. Cell Biochem. 297, 73–80 [DOI] [PubMed] [Google Scholar]
- 76.Friedberg E. C. (2003) Nature 421, 436–440 [DOI] [PubMed] [Google Scholar]
- 77.Hiyama H., Reeves S. A. (1999) Cell Death Differ. 6, 565–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.