Abstract
Human cytomegalovirus (HCMV) lytic DNA replication is initiated at the cis-acting oriLyt region and requires six core replication proteins along with UL84 and IE2. Although UL84 is thought to be the replication initiator protein, little is known about its interaction with oriLyt. We have now performed chromatin immunoprecipitation assays (ChIP) using antibodies specific to UL84, IE2, UL44, CCAAT/enhancer binding protein (C/EBPα) and PCR primers that span the entire oriLyt region to reveal an evaluation of specific protein binding across oriLyt. UL84 interacted with several regions of oriLyt that contain C/EBPα transcription factor binding sites. Mutation of either of one of C/EBPα (92,526 or 92535) sites inactivated oriLyt and resulted in the loss of binding of UL84. These data reveal the regions of interaction within oriLyt for several key replication proteins and show that the interaction between UL84 and C/EBPα sites within oriLyt is essential for lytic DNA replication.
Introduction
Human cytomegalovirus (HCMV) lytic replication is mediated by the cis acting element oriLyt (Anders et al., 1992a; Anders and Punturieri, 1991; Hamzeh et al., 1990; Masse et al., 1992). OriLyt is composed of a “core” domain located between nts 91751 and 93299 containing two essential regions (I and II) (Xu et al., 2004b; Zhu, Huang, and Anders, 1998). Essential region II contains an RNA-DNA hybrid structure that can form a stemloop composed of an RNA strand. This RNA stemloop interacts with the virus-encoded protein UL84 in vitro as well as in the infected cell environment and in packaged virions (Colletti et al., 2007; Prichard et al., 1998). Part of essential region I contains an IE2/UL84-responsive bidirectional promoter suggesting that transcription plays a major role in oriLyt activation (Xu et al., 2004b). Although a functional non-conventional IE2 binding site was identified within in the oriLyt promoter region, other cis acting elements within oriLyt that participate in activation/repression of initiation of lytic DNA synthesis are unknown. Essential region I also contains several transcription factor binding sites as well as a oligopyrimidine rich region referred to as the Y-BLOCK (Zhu, Huang, and Anders, 1998).
HCMV UL84 is a multifunctional factor required for lytic DNA replication and the production of infectious virus (Xu et al., 2004a). UL84 has the ability to activate as well as repress transcriptional activation (Gebert et al., 1997a). In addition, UL84 can interact with RNA and shuttle between the nucleus and the cytoplasm (Lischka et al., 2006). Many of these activities and the presence of specific protein sequence domains, point to UL84 as being a member of the DExD/H box family of proteins (Colletti et al., 2005). UL84 is associated with IE2 in infected cells (Spector and Tevethia, 1994) and although the exact nature of this association is unknown, this interaction apparently leads to a repression of transactivation of at least one HCMV encoded gene in transient assays (Gebert et al., 1997b). Additionally, an IE2/UL84 interaction serves to activate the oriLyt promoter and the binding of the two proteins is essential for oriLyt-dependent DNA replication (Xu et al., 2004b). Recently, UL84 was shown to interact with two other viral encoded factors: UL44, the viral polymerase processivity factor and pp65, a tegument protein (Gao, Colletti, and Pari, 2008).
Although UL84 is presumed to be the oriLyt initiation protein little is known about the interaction of this protein with oriLyt. One study predicted that UL84 is a dUTPase, however no experimental evidence exists to show that the protein has this activity (Davison and Stow, 2005). In an effort to define the role of UL84 in lytic replication we investigated the DNA binding profile of UL84 and two other viral encoded proteins, UL44 and IE2, within the lytic origin in an infected cell environment and, in the case of UL84, in the packaged virion. In this report we identify UL84, UL44 and IE2 interaction domains within oriLyt using the chromatin immunoprecipitation assay (ChIP). We show that UL84 interacts with DNA sequences in oriLyt that contain several CCAAT/enhancer binding protein (C/EBPα) transcription factor binding sites. A 3-nucleotide mutation introduced into the C/EBPα consensus sequences within HCMV oriLyt resulted in the inability of UL84 to interact with these sites in transfected cells and the inactivation of oriLyt in the transient replication assay. It also appears that UL84 interacts with these elements independent of binding with C/EBPα in that co-immunoprecipitations failed to detect a UL84-C/EBPα interaction in infected or cotransfected cells. These results strongly suggest that UL84 interacts with specific transcription factor binding sites within oriLyt and imparts an as yet unidentified function that is essential for oriLyt amplification.
Results
Interaction of UL84, IE2, UL44 and C/EBPα with oriLyt
Although we previously demonstrated that UL84 interacts with a specific stem loop structure within oriLyt, we wanted to identify other regions of oriLyt that interact with UL84. Since UL84 was shown to associate with UL44 and IE2 we were also interested in regions of oriLyt that interacted with these proteins and if their interaction with oriLyt overlapped with UL84 (Gao, Colletti, and Pari, 2008; Spector and Tevethia, 1994). Lastly, HCMV oriLyt contains several C/EBP-binding sites and we wanted to investigate if UL84 interacted with regions of oriLyt that contained these sites (Fig. 1A). C/EBP binding sites as well as other transcription factor binding sites are found in other herpesvirus lytic origins and were shown to be substrates for viral replication proteins (Lieberman et al., 1990; Wang et al., 2003a; Wang et al., 2003b). In order to identify regions of interaction we employed the ChIP assay using primers that spanned most of oriLyt (Fig. 1). We previously used the ChIP assay to identify IE2 and UL84 binding sites within oriLyt in infected cells and, for UL84, in packaged virions at specific region within oriLyt (Colletti et al., 2007; Xu et al., 2004b). We were interested in expanding those studies to include the entire oriLyt region. For our ChIP assays we used a C/EBPα-specific antibody in addition to IE2, UL44 and UL84 specific antibodies. By examining the binding domains for all of these proteins we sought to assemble a picture of oriLyt interaction domains for UL84 and it's identified binding partners. Additionally, we wanted to determine if C/EBPα interacted with oriLyt in an effort to identify a possible connection between this protein and UL84.
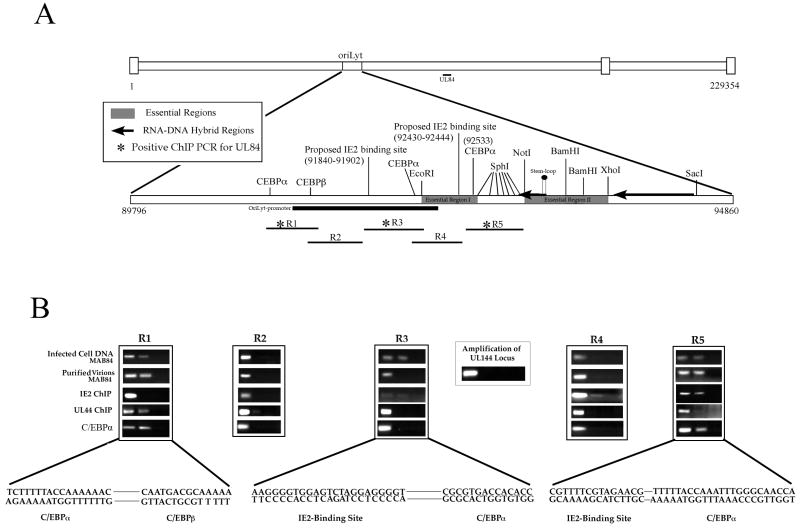
Figure 1. Interaction of UL84, UL44, IE2 and C/EBPα with oriLyt.
(A) Schematic of HCMV genome showing the location of oriLyt and the positions of three essential regions, various restriction enzyme recognition sites, RNA/DNA hybrids, C/EBPα/β, IE2-binding sites, oriLyt promoter region and the RNA stem-loop structure. Also shown are the relative locations of PCR primers used for ChIP assays for DNA isolated from infected cells and purified virions. (B) ChIP assays showing the interaction of UL84, IE2, UL44 and C/EBPα with various regions of oriLyt. Infected cells were prepared as described and specific antibodies to UL84 (Virusys), IE2 (Vancouver Biotech), UL44 (Bill Britt) or C/EBPα (Santa Cruz) were used for immunoprecipitations. PCR primers from regions 1-5 are shown above each ChIP assay gel. The lanes of each ChIP assay are as follows: 1, PCR product from input DNA; 2, PCR product from immunoprecipitations using specific antibodies shown to the left of the figure; 3, PCR product from immunoprecipitations using an unrelated antibody that was the same isotype as the test antibody. Also shown is a PCR amplification of the HCMV UL144 loci from samples immunoprecipitated using the anti-UL84 antibody. Lanes: 1, PCR product from input DNA; 2, PCR product from immunoprecipitations using the anti-UL84 antibody; 3, PCR product from immunoprecipitations using an unrelated antibody that was the same isotype as the test antibody. Solid line shows sequences gaps.
We used infected cells or purified virus (for UL84) for ChIP assays as previously described (Colletti et al., 2007). Only UL84 immunoprecipitations were done from purified virions. DNA fragments were estimated within the 300-500 base pair (bp) range (data not shown). Figure 1A is a schematic of the HCMV genome showing the location of oriLyt and the specific primers used to amplify regions of the lytic origin (Fig. 1. Regions: R1-R5). Also shown is the location of three clusters of C/EBPα transcription factor binding sites and several key elements within oriLyt including the location of 2 essential regions and RNA-DNA hybrid structures. The ChIP assay revealed that UL84 interacts with various regions of oriLyt all of which contain C/EBPα transcription factor binding sites (Fig. 1B, R1, R3 and R5). UL84 protein was also found bound to DNA in purified virus localizing to regions R1 and R5 as shown by positive PCR products from these samples (Fig. 1B). Since UL84 is known to interact with IE2, we also investigated the binding of IE2 to specific regions of oriLyt using an IE2-specific antibody (G13-12E2, Vancouver Biotech). IE2 interacted with oriLyt regions R3, R4 and R5. The interaction of IE2 with R4 and R5 is located in a domain that contains a previously reported IE2-responsive promoter (Xu et al., 2004b). We also examined the binding of UL44 (polymerase accessory factor/UL84 binding partner) to oriLyt. UL44 was shown to interact with regions R1, R2 and R5 (Fig. 1B). Two of these regions R1 and R5 are also substrates for UL84, suggesting that the interaction of UL44 with these regions is part of an UL84-UL44 protein complex. Lastly we investigated the interaction of C/EBPα with oriLyt. C/EBPα interacted with oriLyt domains R1 and R5 (Fig. 1B). Interestingly, no PCR signal was detected in domain R4 which contains a proposed C/EBPα binding site (Fig. 1B). As controls for the ChIP assay we performed immunoprecipitations with an isotype specific unrelated antibody. We also used a PCR primer set complementary to a region outside of oriLyt, the UL144 locus. All of these control reactions failed to yield a detectable PCR product (Fig. 1B).
These results show specific protein binding to HCMV oriLyt. One interesting finding is that UL84 interacts with regions of oriLyt that contain C/EBPα transcription factor binding sites. These findings lead us to further investigate the functional significance of a possible interaction of UL84 with C/EBPα sites within oriLyt.
Predicted oriLyt C/EBPα binding motifs interact with C/EBPα in vitro
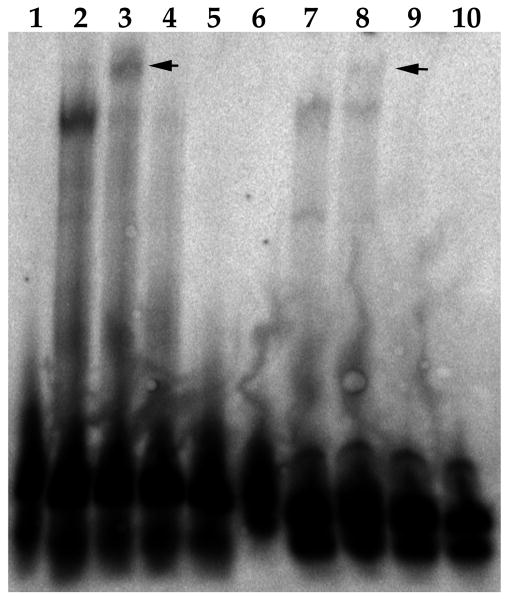
Since the ChIP assay showed that C/EBPα interacted with oriLyt, we evaluated the identified regions of oriLyt for the presence of possible C/EBPα binding motifs. We used the software program TRANSFAC to identify the sequence 5′- CCAAAT-3′ as predicted to interact with C/EBPα. In order to show this experimentally, we performed an EMSA using a double stranded (ds) oligonucleotide containing the oriLyt sequence 5′-TTGCCCAAATTTGGTAAAAATTTGC-3′ or a control ds oligonucleotide, 5′-ATCTGCTGATTGGCCCAGAGCGGGAACCAATCAGCG-3′, shown previously to interact with C/EBPα (Wu et al., 2003). Nuclear extracts were prepared form HEK293 cells transfected with a C/EBPα expression plasmid and incubated with the oriLyt proposed and control C/EBPα-containing oligonucleotides. Both the control ds oligonucleotide sequence as well as the oriLyt C/EBPα sequence interacted with C/EBPα as demonstrated by the super-shifted band in the presence of a C/EBPα-specific antibody (Fig. 2, arrows show supershift). This experiment indicated that the predicted oriLyt C/EBPα binding site interacted with C/EBPα and strongly suggested that this is the C/EBPα-interacting motif with in oriLyt identified in the ChIP assay.
Figure 2. C/EBPα interacts with the proposed oriLyt C/EBPα-binding site sequence in vitro.
Nuclear extract was prepared from HEK293 cells transfected with a C/EBPα expression plasmid. Three microliters of nuclear extract was incubated with ds oligonucleotides in the presence or absence of a C/EBPα specific antibody. Lanes: 1, control oligonucleotide; 2, control oligonucleotide plus nuclear extract from cells transfected with a C/EBPα expression plasmid; 3, control oligonucleotide plus nuclear extract from cells transfected with a C/EBPα expression plasmid plus a C/EBPα specific antibody; 4, control oligonucleotide plus nuclear extract from cells transfected with a C/EBPα expression plasmid plus 20X; unlabeled control oligonucleotide; 5, control oligonucleotide plus untrasfected nuclear extract; 6; oriLyt a C/EBPα oligonucleotide; 7, orilyt C/EBPα oligonucleotide plus nuclear extract from cells transfected with a C/EBPα expression plasmid; 8, oriLyt C/EBPα oligonucleotide plus nuclear extract from cells transfected with a C/EBPα expression plasmid plus a C/EBPα specific antibody; 9, oriLyt C/EBPα oligonucleotide plus nuclear extract from cells transfected with a C/EBPα expression plasmid plus 20X; unlabeled control oligonucleotide; 10, oriLyt C/EBPα oligonucleotide plus untrasfected nuclear extract; 6; oriLyt a C/EBPα oligonucleotide.
Intact C/EBPα sites are essential for amplification of oriLyt
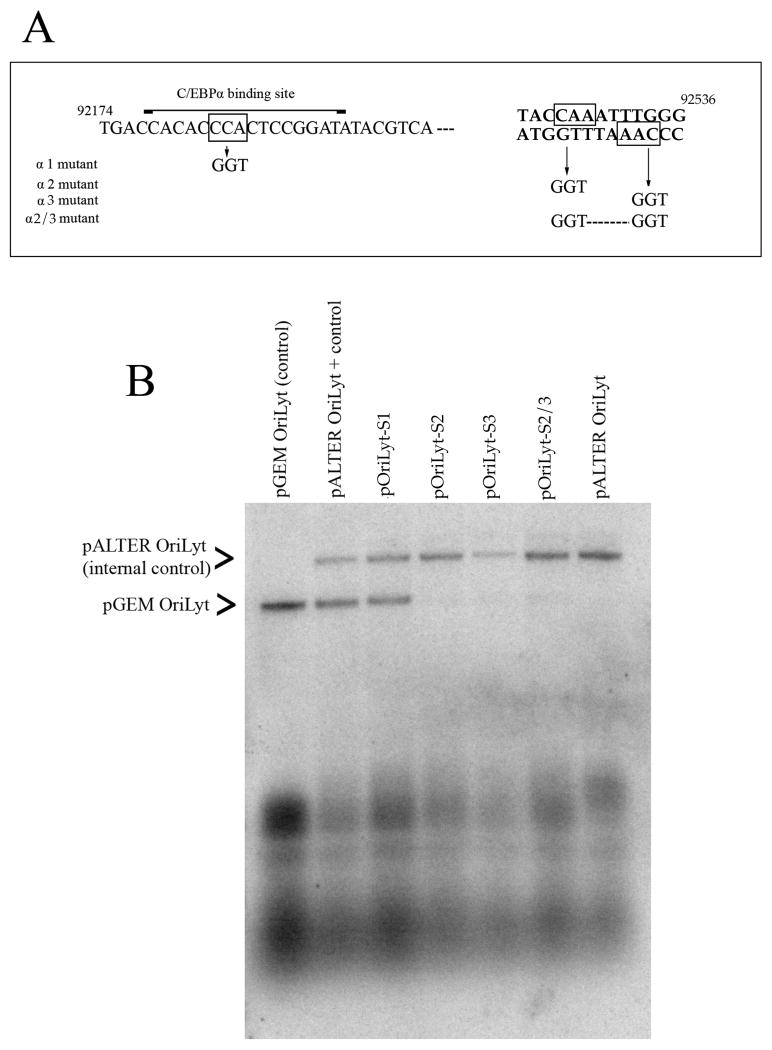
As a first step in indentifying a possible role for C/EBPα sites in oriLyt function and DNA replication we used the powerful transient DNA replication assay to determine if C/EBPα binding sites contribute to lytic DNA replication (Anders et al., 1992b). We generated several plasmids that contained mutations in one or more of the C/EBPα sites found within oriLyt. Figure 2A shows the location and nucleotide coordinates of three C/EBPα transcription factor-binding sites that were mutated within oriLyt. Site 1 is at nts 92,183, site 2 is located at 92,527 and site 3 is located at nts 92,534 (Fig. 3A). Four plasmids were generated: pOriLyt-S1 contains a mutation in C/EBPα binding site 1, pOriLyt-S2 contains a mutation in C/EBPα binding site 2, pOriLyt-S3 contains a mutation in C/EBP binding site 3 and pOriLyt-S2/3 contains mutations in C/EBPα binding sites 2 and C/EBPα 3 (Fig.3A). These plasmids were transfected into HF cells that were subsequently infected with HCMV. As an internal control for oriLyt amplification we also transfected the plasmid pALTER-oriLyt. This plasmid contains wt oriLyt, however the cloning vector is larger (pALTER) than the parent vector for wt oriLyt or the mutated oriLyt plasmids (pGEM7zf). This allowed for the detection of both mutated and wt oriLyt plasmids in the same cells and transfection sample. The internal control would also demonstrate that the replication assay in each case was functioning properly.
Figure 3. Intact C/EBPα sites are required for amplification of oriLyt.
(A) Schematic of oriLyt sequence from nucleotides 92,174-92,541 showing the positions of C/EBPα transcription factor binding sites and the mutations made within the oriLyt sequence. (B) Southern blot of a transient transfection replication assay. HF cells were transfected with various oriLyt containing plasmids plus the internal control plasmid pALTER-OriLyt. Cells were infected 24 h post transfection and total cell DNA was harvested 5 days post transfection and cleaved with DpnI and EcoRI. Following agarose gel separation and transfer to a nylon membrane, the blot was hybridized to a 32P-labbled-pGEM probe. Arrows to the left of the figure show the identification of each plasmid.
Total cellular DNA was harvested 5 days post infection and cleaved with EcoRI and DpnI. DNA samples were resolved using a 0.8% agarose gel that was subsequently transferred to a nylon membrane (ZetaProbe, BioRad) and hybridized with a 32P-labeled pGEM7zf (-) probe as described previously (Xu et al., 2004b). Replicated, DpnI-resistant, oriLyt plasmid is shown by the arrows at the left of Figure 3B. Mutation of C/EBPα binding site 1 had no effect on the efficiency of oriLyt amplification (Fig. 3B). However, mutations in either C/EBPα binding site 1 or 2 resulted in a complete eradication of oriLyt amplification (Fig. 3B). Amplification of the oriLyt internal control plasmid pALTER-oriLyt was unchanged and indicated that transfection efficiency and the assay itself was adequate (Fig. 3B). These results showed that C/EBPα binding sites 2 and 3 were essential for oriLyt replication and coupled with the results from the ChIP assay suggested that UL84 interacted with these sites in the infected cell environment.
UL84 does not interact with C/EBPα
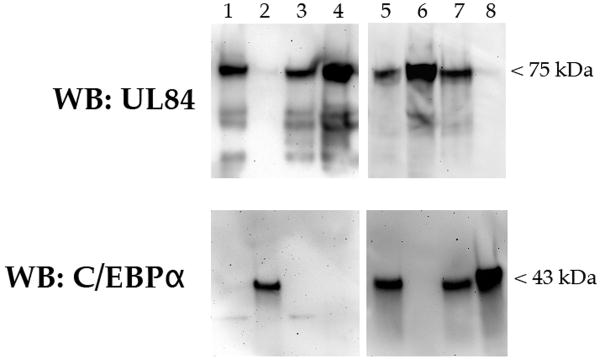
A recent proteomics study of cellular and viral factors interacting with UL84 in infected cells did not reveal C/EBPα (or C/EBPβ) as a binding partner of UL84 (Gao, Colletti, and Pari, 2008). In order to take a more exhaustive investigation of this we infected HF cells with HCMV and 5 days post infection protein lysates were prepared. Protein lysates were used to immunoprecipitate C/EBPα using a specific antibody followed by western blot analysis of immunoprecipitated protein for the presence of UL84. No specific protein band was detected using an antibody to UL84 on western blots (Fig. 4, lane 1, WB: UL84). We also did the reverse immunoprecipitation where we used a UL84-specific antibody and then analyzed western blots for the presence of C/EBPα using a specific antibody. This reverse experiment also showed that no C/EBPα was detected, indicating that these two proteins do not interact in infected cells (Fig. 4, lane 3, WB: C/EBPα). Due to the low abundance of C/EBPα in human fibroblasts we could not detect the protein in the lysate (Fig. 4, lane 2).
Figure 4. C/EBPα does not interact with UL84 in infected or transfected cells.
HF cells were infected with HCMV and protein lysates were prepared 5 days post infection. Proteins were immunoprecipitated using either a UL84 or C/EBPα - specific antibody. Immunoprecipitated protein was analyzed by western blot using the same antibodies. Lanes: 1, infected cell lysate, 2, infected cell protein lysate immunoprecipitated with anti- C/EBPα 3, Infected cell protein lysate; 4, infected cell protein lysate immunoprecipitated with anti- UL84; 5, UL84 and C/EBPα cotransfected cell protein lysate; 6, cotransfected protein lysate immunoprecipitated with anti-UL84; 7, UL84 and C/EBPα cotransfected cell protein lysate; 8, cotransfected protein lysate immunoprecipitated with anti-C/EBPα. The specific antibodies used to react with the Western blot are shown to the left of the figure.
As a further test for the possible interaction of UL84 with C/EBPα, we also cotransfected a UL84 expression plasmid along with a C/EBPα expression plasmid and performed the same immunoprecipitations that were done in infected cells. Again we could not detect C/EBPα when immunoprecipitations were performed using anti-UL84 antibody (Fig. 4, lane 5, WB: C/EBPα), likewise we could not detect UL84 when immunoprecipitations were performed using a C/EBPα-specific antibody (Fig. 4, lane 7, WB: UL84). Based on these findings we conclude that UL84 does not directly or indirectly interact with C/EBPα.
Mutation of oriLyt C/EBPα sites resulted in the loss of UL84 interaction with oriLyt
Our findings strongly suggest that UL84 interacts with C/EBPα transcription factor binding sites within oriLyt and UL84 binds independent of an interaction with C/EBPα. In order to analyze binding of UL84 to C/EBPα in a cellular environment we again performed a ChIP assay using the C/EBPα mutated versions of oriLyt. We cotransfected HF cells with a UL84 expression plasmid along either the wt oriLyt containing plasmid or pOriLyt-S2/3. In this plasmid based ChIP assay, the transfection of UL84 alone resulted in a positive PCR signal when DNA samples were prepared from cells transfected with wild-type oriLyt (Fig. 5, lane 1). This indicated that UL84 interacts with C/EBPα/β sites in the absence of any other viral protein. However no PCR product was detected from samples transfected with pOriLyt-S2/3, which has mutated C/EBPα sites (Fig. 5, lane 2). No PCR product was detected in control lanes where ChIP assays were preformed using an isotype specific but unrelated antibody (Fig. 5, lane 3). These experiments show that UL84 interacts with the C/EBPα sites within oriLyt and suggests that UL84 mediates oriLyt DNA synthesis through this interaction.
Figure 5. Mutation of C/EBPα site 2 results in the loss of UL84 binding in transfected cells.
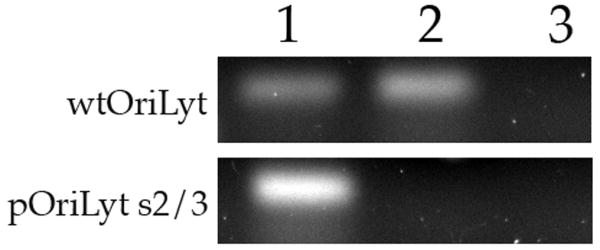
HF cells (1 × 107) were transfected by electroporation with either wt OriLyt (10μg) or pOriLyt-2/3 (10μg) and a UL84 (5μg) expression plasmid. Two days post transfection cells were prepared for ChIP assays using a UL84 specific antibody or, in the case of the control ChIP an unrelated antibody of the same isotype as the UL84 antibody. Lanes: 1, PCR product input DNA from cells cotransfected with wt OriLyt containing plasmid and a UL84 expression plasmid; 2, PCR product from a ChIP assay from cells transfected with an OriLyt containing plasmid and a UL84 expression plasmid; 3, PCR product from a ChIP assay from cells transfected with an OriLyt containing plasmid and a UL84 expression plasmid using an isotype control antibody. PCR reactions were performed using primers that flanked the C/EBPαβ transcription factor binding sites with in oriLyt: Forward 5′-ACTCGAGTCACCATCCCATAAT-3′ and reverse 5′-TTTTCGTAGAACGTTTCGTTAGAAG-3′. The plasmid used in the transfection experiments is shown at the left of the figure.
Discussion
Although it was shown previously that UL84 interacts with an RNA stemloop structure within oriLyt, we investigated if there were other binding sites within the lytic origin. As part of this endeavor we also explored the possible interactions of two other viral proteins that are known to bind with UL84, IE2 and UL44. Interestingly, UL84 was found to be in purified (packaged) virions bound to DNA in several loci as well as in the previously reported region containing the RNA stemloop structure. In the case of protein binding to R3 of oriLyt, we found UL84 interacting with this region in the infected cell environment but not in purified virions. This apparent discrepancy could be due to the binding of UL84 to actively replicating sites as well as interacting with DNA or RNA within the virion. The ChIP analysis of the entire oriLyt region revealed that IE2 interacts with regions of the lytic origin that contain IE2 binding sites and the region that is adjacent to the RNA-DNA hybrid. It was previously demonstrated that IE2 interacts with a IE2-UL84 responsive promoter within oriLyt (Xu et al., 2004b). In this report we show that IE2 interacts with region 5 (R5), which is also associated with UL84 and UL44 binding. UL44 also interacts with oriLyt in several regions associated with IE2 and UL84. Interestingly, UL44, IE2 and UL84 all interact with R5 of oriLyt. This is the region of oriLyt just adjacent to the RNA stemloop structure and the association of these proteins with this domain suggests that this area could be the point of initiation of DNA replication within oriLyt. At the very least, the interaction of all of these replication-associated proteins with this region suggests that R5 is a significant active site for protein binding within oriLyt.
One of the main findings of this report is that UL84 binds to C/EBPα transcription factor binding sites within oriLyt. These two sites located at 92,534 and 92,527, are approximately 300 nts upstream from the RNA stemloop within oriLyt. This region is within a previously described promoter region within oriLyt (Xu et al., 2004b). The oriLyt promoter is essential for activity and its function is not very well defined. The C/EBPα binding sites are also just down stream of the Y-BLOCK, another element known to contribute to oriLyt function (Zhu, Huang, and Anders, 1998). This report defines the C/EBPαβ sites as essential elements within this region. Interestingly, C/EBPα did not interact with R4 which contains a consensus C/EBPα-binding site. This site may not interact with C/EBPα for various reasons including that it may not be accessible by the protein. Mutation of either one of the oriLyt C/EBPα binding motifs that do interact with UL84 or C/EBPα resulted in a complete inactivation of oriLyt in the transient assay. Because of the close proximity of these two sites we acknowledge the possibility that the region identified could in fact be only one UL84 binding site. The transient replication assay is perfectly suited to evaluate the effect of mutations within oriLyt and the impact on DNA synthesis directly. Since the transient replication assay focuses only on DNA synthesis, the ability of this powerful assay to efficiently and quantitatively assess specific mutations has been invaluable in the herpesvirus field (Anders et al., 1992b; AuCoin et al., 2004; Dykes et al., 1997; Huang, Zhu, and Anders, 1996; Schepers et al., 1993; Sinigalia et al., 2008; Zhu, Huang, and Anders, 1998). The next step is to generate HCMV BACmid clones that have mutations within oriLyt, however the results presented here are the first report of UL84 interacting with specific elements within oriLyt. This observation points to a novel mechanism of activation for HCMV lytic replication that is dependent upon the recognition by UL84 of these two cis-acting sites and possibly the RNA stemloop structure. Also as mentioned above, given the richness of novel structures and protein binding within this region, R5, this suggests that the initiation of DNA synthesis may occur in this area. This is the first report to show an interaction of UL84 with a transcription factor-binding site within oriLyt.
Our attempts to show direct binding of UL84 to C/EBPα motifs using in vitro assays did not show any specific interaction, suggesting that other, as yet unidentified, cellular factors could be facilitating the interaction of UL84 with these elements. Although we have not explored the possibilities in this report we acknowledge that UL84 could be interacting with oriLyt through other forms of C/EBPα or a Jun-C/EBPα complex. Although the motifs we identified within oriLyt are consensus C/EBPα binding motifs we acknowledge that other as yet unidentified factors could be interacting with these sequences. However, our data strongly suggests that these motifs are essential for oriLyt amplification and the interaction of UL84 with these sequences is required in the cellular environment.
The initial ChIP assay on infected cell DNA did identify C/EBPα binding to regions of oriLyt that also interacted with UL84 (Fig.1). This result suggested that UL84 is interacting with C/EBPα and both proteins are binding to DNA. However, our evidence did not confirm this hypothesis and suggests that UL84 interacts with C/EBPα binding sites in oriLyt independent of a C/EBPα-UL84 protein-protein interaction. This is based on the observations that a proteomic analysis of UL84 binding partners failed to show an interaction with C/EBPα (Gao, Colletti, and Pari, 2008) and we failed to immunoprecipitate C/EBPα when using specific UL84 antibodies in pull down assays in transfected as well as infected cells. We were also unable to immunoprecipitate UL84 when using a C/EBPα -specific antibody in similar pull down assays.
Interaction of viral initiator proteins with transcription factor binding sites is observed in other herpesvirus systems. For example, Epstein-Barr Virus (EBV) lytic replication initiator protein Zta can interact with AP-1 sites within oriLyt in the absence of AP-1 binding (Lieberman et al., 1990). Zta is a transcriptional activator and appears to activate lytic replication by a dual mechanism (Sarisky et al., 1996). UL84 alone is not associated with any known transactivation function; although in conjunction with IE2 can activate the oriLyt promoter. UL84 could play a dual role where binding to RNA within the lytic origin could facilitate DNA synthesis and interaction with transcription factor bindings sites triggers transcription that also modulates initiation of DNA replication. Further analysis is needed to determine if the interaction of UL84 with C/EBPα binding motifs results in transcriptional activation.
Materials and Methods
Cells and Virus
Human fibroblasts. HEK293 cells and virus (AD169) were maintained as previous described (Colletti et al., 2007; Colletti et al., 2005; Xu, Colletti, and Pari, 2002).
Co-immunoprecipitation
Immunoprecipitations were performed as previously described (Colletti et al., 2004) with the following minor changes. 293FT cells were plated to 70 to 90% confluency in 100-mm dishes. Cells were transfected with 10 μg of the appropriate plasmids by using Trans-IT LT (Mirius) per manufacturer's recommendations. At 48 h posttransfection cells were washed twice with PBS (pH 7.4) and lysed by using 1 ml of lysis buffer A by shaking for 30 min at room temperature. Cells were scraped from the plate and passed through a 22-gauge needle three times. Cellular debris was removed by centrifugation at 1,500 × g for 10 min. Lysates were mixed incubated overnight at 4°C specific antibodies. Coimmunoprecipitations were carried out by using a nonconjugated immunoprecipitation system.
Nuclear extract preparation and EMSA
HEK 293FT cells were transfected (TransIT-LT, Mirrus) with 5μg of the plasmid pUC-hC/EBPα (gift from Dr. Darlington, Baylor College of Medicine) expressing full-length human C/EBPα (Harris et al., 2001). Two days post transfection cells were harvested and nuclear extract was prepared using Sigma Nxtract CellLYTIC NuCLEAR extraction kit according to the manufacturer's protocol. 5μL of the nuclear extract was subjected to SDS-PAGE and western blot analysis to verify protein expression.
For electromobility shift assays, 3μL of nuclear extract was added to a 20μl reaction mixture containing 1× binding buffer (10 mM HEPES [pH 7.5], 50 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 5% glycerol) and incubated with 1μL of [32P]dCTP-labeled oligonucleotides and incubated at 25°C for 30 minutes. For supershift assays 4μL of C/EBPα polyclonal antibody (Active Motif catalog # 39306) was incubated with the reaction mixture for an additional 30 minutes. Samples were resolved through a 4.5 % non-denaturing polyacrylamide gel using running buffer (10mM HEPES, 1mM EDTA, 0.5mM EGTA; pH 7.5) at 25°C. Gels were dried and shifted bands were detected using a phosphoimager (GE).
Chromatin Immunoprecipitation assay (ChIP)
Infected cells were treated as described previously (Xu et al., 2004b). Protein-DNA complexes were immunoprecipitated using antibodies specific for UL44 (gift from William Britt), UL84 (Virusys), IE2 (Vancouver Biotech) and C/EBPα (Santa Cruz). PCR primer sequences are as follows: R1: Forward 5′-AAAGATCCGAACTTTAAAATTGTGTGTTTTT - 3′ and Reverse 5′- TGCTCACCGCCTCGCCGGCCACGGGGTTGA - 3′; R2: Forward 5′- CCTGCGCTGATGGTCACGTGACCAACAA- 3′ Reverse 5′- CGTCGTGTATACATAACGGTGCCCGGTG- 3′ R3: Forward 5′- CCGTTAGGGTTCCACCGTCCTCGGTGTACG- 3′ R4: Forward 5- CGTCATCCTGTGGAATTCCGGACATACGGT - 3′ and reverse 5′- TTTGCCCCCCCCGGTTCCGGAGG - 3′; R5: Forward 5′- ACGGCGCACATCTAGTGGAATTTTACCG - 3′ and reverse, 5′- CTCCGGAACCGGGGGGGGCAAATTTTTA -3′.
Site-directed mutagenesis
The C/EBPα/β sites within oriLyt were mutated using QuickChange II (Stratagene) and primers: For pOriLyt-S1: Forward 5′-
CCTACGTCACACTCGCGTGAGGTCACCCACTCCGGATATACG-3′ and Reverse: 5′- CGTATATCCGGAGTGGGTGACCTCACGCGAGTGTGACGTAGG-3′; for pOriLyt-S2: Forward: 5′-GGGCAAATTTTTAGGTAATTTGGGCAACCAT-3′ and Reverse: 5- ATGGTTGCCCAAATTACCTAAAAATTTGCCC-3′; for pOriLyt-S3: Forward: 5′- AATTTTTACCAAATCCAGGCAACCATGATTT-3′ and Reverse 5′- AAATCATGGTTGCCTGGATTTGGTAAAAATT-3′; For pOriLyt-S2/3: Forward, 5′- GGGCAAATTTTTAGGTAATCCAGGCAACCATGATTTCCAATGG-3′ and Reverse, 5′- CCATTGGAAATCATGGTTGCCTGGATTACCTAAAAATTTGCCC-3′
Cotransfection Replication assay
The cotransfection-replication assay was performed using HCMV replication proteins as described previously (Pari and Anders, 1993; Xu et al., 2004b) and wt UL84 expression vectors or plasmids pOriLyt-S1, pOriLyt-S2, pOriLyt-S3 or pOriLyt-S2/3. Transfections also contained pALTER-oriLyt which was generated by subcloning oriLyt into the pALTER vector (Promega).
Table 1. oriLyt ChIP assay regions.
| Region | Nucleotide coordinates | Protein binding | Primers |
|---|---|---|---|
| R1 | 91150-91597 | UL84 (v/c), UL44, C/EBPα | F: AAAGATCCGAACTTTAAAATTGTGTGTTTTT R:TGCTCACCGCCTCGCCGGCCACGGGGTTGA |
| R2 | 91317-91627 | UL44 | F: ACGTCGTGTATACATAACGGTGCCCGGTGT R:TGCTCACCGCCTCGCCGGCCACGGGGTTGA |
| R3 | 91627-92228 | UL84 (c), IE2 | F:CTTTTGTTGGTCACGTGACCATCAGCGCAGG R: ACCGTATGTCCGGAATTCCACAGGATGACG |
| R4 | 92110-92519 | IE2 | F: AAACTTAACGCCGCCGCTTCTCAC R:TTTGCCCCCCCCGGTTCCGGAGG |
| R5 | 92497-92830 | UL84 (v/c), UL44, IE2, C/EBPα | F: CTCCGGAACCGGGGGGGGCAAATTTTTA R: ACGGCGCACATCTAGTGGAATTTTACCG |
v = bound in virion
c = infected cells
Acknowledgments
This work was funded by NIH Public Health Service AI45096. We thank Bill Britt for the UL44 specific antibody.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Dominique Kagele, University of Nevada, Reno School of Medicine, Department of Microbiology & Immunology and the Cell and Molecular Biology Graduate Program, Reno NV 89557.
Yang Gao, University of Nevada, Reno School of Medicine, Department of Microbiology & Immunology and the Cell and Molecular Biology Graduate Program, Reno NV 89557.
Kate Smallenburg, University of California Berkeley.
Gregory S Pari, University of Nevada, Reno School of Medicine, Department of Microbiology & Immunology and the Cell and Molecular Biology Graduate Program, Reno NV 89557.
References
- Anders DG, Kacica MA, Pari G, Punturieri SM. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J Virol. 1992a;66(6):3373–84. doi: 10.1128/jvi.66.6.3373-3384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders DG, Kacica MA, Pari G, Punturieri SM. Boundaries and structure of human cytomegalovirus oriLyt, a complex origin for lytic-phase DNA replication. J Virol. 1992b;66(6):3373–84. doi: 10.1128/jvi.66.6.3373-3384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders DG, Punturieri SM. Multicomponent origin of cytomegalovirus lytic-phase DNA replication. J Virol. 1991;65:931–937. doi: 10.1128/jvi.65.2.931-937.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AuCoin DP, Colletti KS, Cei SA, Papouskova I, Tarrant M, Pari GS. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP) Virology. 2004;318(2):542–55. doi: 10.1016/j.virol.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Colletti KS, Smallenburg KE, Xu Y, Pari GS. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J Virol. 2007;81(13):7077–85. doi: 10.1128/JVI.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti KS, Xu Y, Cei SA, Tarrant M, Pari GS. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J Virol. 2004;78(17):9203–14. doi: 10.1128/JVI.78.17.9203-9214.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti KS, Xu Y, Yamboliev I, Pari GS. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J Biol Chem. 2005;280(12):11955–60. doi: 10.1074/jbc.C400603200. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Stow ND. New genes from old: redeployment of dUTPase by herpesviruses. J Virol. 2005;79(20):12880–92. doi: 10.1128/JVI.79.20.12880-12892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes C, Chan H, Krenitsky DM, Dewhurst S. Stringent structural and sequence requirements of the human herpesvirus 6B lytic-phase origin of DNA replication. J Gen Virol. 1997;78(Pt 5):1125–9. doi: 10.1099/0022-1317-78-5-1125. [DOI] [PubMed] [Google Scholar]
- Gao Y, Colletti K, Pari GS. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J Virol. 2008;82(1):96–104. doi: 10.1128/JVI.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J Virol. 1997a;71(9):7048–60. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J Virol. 1997b;71(9):7048–60. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzeh FM, Lietman PS, Gibson W, Hayward GS. Identification of the lytic origin of DNA replication in human cytomegalovirus by a novel approach utilizing ganciclovir-induced chain termination. J Virol. 1990;64:6184–9195. doi: 10.1128/jvi.64.12.6184-6195.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Albrecht JH, Nakanishi M, Darlington GJ. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J Biol Chem. 2001;276(31):29200–9. doi: 10.1074/jbc.M011587200. [DOI] [PubMed] [Google Scholar]
- Huang L, Zhu Y, Anders DG. The variable 3′ ends of a human cytomegalovirus oriLyt transcript (SRT) overlap an essential, conserved replicator element. J Virol. 1996;70(8):5272–81. doi: 10.1128/jvi.70.8.5272-5281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP- 1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64(3):1143–55. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P, Rauh C, Mueller R, Stamminger T. Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J Virol. 2006;80(20):10274–80. doi: 10.1128/JVI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse MJO, Karlin S, Schachtel GA, Mocarski ES. Human cytomegalovirus origin of replication (oriLyt) resides within a highly complex repetitive region. Proc Natl Acad Sci. 1992;89:5246–5250. doi: 10.1073/pnas.89.12.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pari GS, Anders DG. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67(12):6979–88. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Jairath S, Penfold ME, Jeor S, Bohlman MC, Pari GS. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J Virol. 1998;72(9):6997–7004. doi: 10.1128/jvi.72.9.6997-7004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarisky RT, Gao Z, Lieberman PM, Fixman ED, Hayward GS, Hayward SD. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70(12):8340–7. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AD, Pich D, Mankertz J, Hammerschmidt W. Cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigalia E, Alvisi G, Mercorelli B, Coen DM, Pari GS, Jans DA, Ripalti A, Palu G, Loregian A. Role of Homodimerization of Human Cytomegalovirus DNA Polymerase Accessory Protein UL44 in Origin-Dependent DNA Replication in Cells. J Virol. 2008;82(24):12574–9. doi: 10.1128/JVI.01193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DJ, Tevethia MJ. Protein-protein interactions between human cytomegalovirus IE2-5f80aa and pUL84 in lytically infected cells. J Virol. 1994;68:7549–7553. doi: 10.1128/jvi.68.11.7549-7553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, Wu FY, Fujimuro M, Zong J, Hayward SD, Hayward GS. Role of CCAAT/Enhancer-Binding Protein Alpha (C/EBPalpha) in Activation of the Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic-Cycle Replication-Associated Protein (RAP) Promoter in Cooperation with the KSHV Replication and Transcription Activator (RTA) and RAP. J Virol. 2003a;77(1):600–23. doi: 10.1128/JVI.77.1.600-623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, Wu FY, Yu Y, Hayward GS. CCAAT/Enhancer-Binding Protein-alpha Is Induced during the Early Stages of Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic Cycle Reactivation and Together with the KSHV Replication and Transcription Activator (RTA) Cooperatively Stimulates the Viral RTA, MTA, and PAN Promoters. J Virol. 2003b;77(17):9590–612. doi: 10.1128/JVI.77.17.9590-9612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FY, Wang SE, Tang QQ, Fujimuro M, Chiou CJ, Zheng Q, Chen H, Hayward SD, Lane MD, Hayward GS. Cell Cycle Arrest by Kaposi's Sarcoma-Associated Herpesvirus Replication-Associated Protein Is Mediated at both the Transcriptional and Posttranslational Levels by Binding to CCAAT/Enhancer-Binding Protein alpha and p21(CIP-1) J Virol. 2003;77(16):8893–8914. doi: 10.1128/JVI.77.16.8893-8914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Cei SA, Huete AR, Pari GS. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J Virol. 2004a;78(19):10360–9. doi: 10.1128/JVI.78.19.10360-10369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Cei SA, Rodriguez Huete A, Colletti KS, Pari GS. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J Virol. 2004b;78(21):11664–77. doi: 10.1128/JVI.78.21.11664-11677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Colletti KS, Pari GS. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J Virol. 2002;76(17):8931–8. doi: 10.1128/JVI.76.17.8931-8938.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang L, Anders DG. Human cytomegalovirus oriLyt sequence requirements. J Virol. 1998;72(6):4989–96. doi: 10.1128/jvi.72.6.4989-4996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]