Abstract
The pursuit of small molecules that bind to DNA has led to the discovery of selective and potent antitrypanosomal agents, specifically 4,4′-bis(imidazolinylamino) and 4,4′-bis(guanidino) diphenylamine compounds, CD27 and CD25, respectively. Although the antitrypanosomal properties of these compounds have been characterized, further development of this series of compounds requires assessment of their DNA site-selectivities and affinities. Toward this end, both compounds have been analyzed and found to selectively bind AT sequences. However, CD27 was found to bind with higher affinity to 5′-AATT than 5′-ATAT while CD25 bound more weakly but equally well to either sequence. To detail the nature of its interactions with DNA, the crystal structure of CD27, bound to its preferred DNA binding site 5′-AATT within a self-complementary oligonucleotide, 5′-d(CTTAATTCGAATTAAG), was determined at 1.75 Å using a host-guest approach. Although CD27 is predicted to be highly twisted in its energy minimized state, it adopts a more planar crescent shape when bound in the minor groove of the DNA. Interactions of CD27 with 5′-AATT include bifurcated hydrogen bonds, providing a basis for selectivity of this site, and favorable van der Waals interactions in a slightly widened minor groove. Thus, an induced fit results from conformational changes in both the ligand and the DNA. Our studies suggest a basis for understanding the mechanism of the antitrypanosomal activity of these symmetric diphenylamine compounds.
Trypanosomiasis is a potentially deadly disease that affects 50,000 to 70,000 people in sub-Saharan Africa (1, 2). Currently, human African trypanosomiasis (HAT), commonly referred to as African sleeping sickness, belongs to the group of most neglected diseases identified by a World Health Organization Industry working group (1). Treatments available for late stages of the disease, an arsenical drug (melarsoprol) and an ornithine decarboxylase irreversible inhibitor (eflornithine, α-difluoromethylornithine), are highly toxic and must be delivered intravenously, a mode unsuitable for patients in these underdeveloped regions (3). Therefore, research dedicated to finding alternative drugs has led to the discovery of two potent anti-trypanosomal compounds, bisaminoimidazoline and bisguanidine derivatives of diphenylamine (CD27 and CD25, respectively, Figure 1) (4). These compounds are related to the diamidine DB75, retaining the dicationic ends and a diphenyl core with a spacer unit; in DB75, the spacer is a furan rather than an amine linking the two phenyl units. The methoxime prodrug of DB75, DB289, has reached Phase III clinical trials as a trypanocidal compound (5-7),
Figure 1.
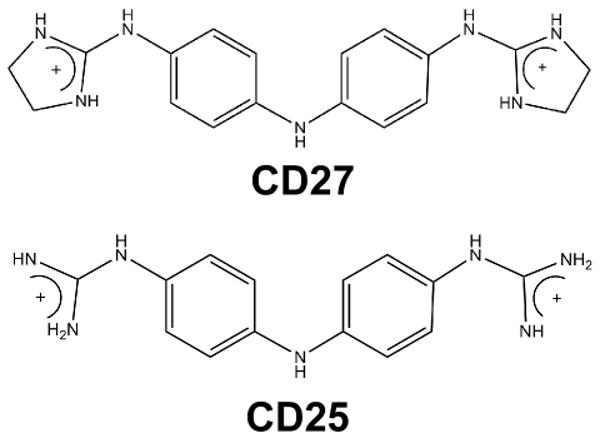
Chemical structures of CD27 and CD25 shown as dicationic species.
CD27 and CD25 have demonstrated in vitro activity against Trypanosomal parasites (T.b. rhodesiense STIB900), the bisaminoimidazoline CD27, in particular, has very low cytotoxicity (4). Trypanosomes have extensive AT tracts in their mitochondrial genome and are very sensitive to guanidine- and aminoimidazoline-containing compounds that are often AT selective DNA binders (8). The bulk DNA-binding properties of both CD27 and CD25 have been determined by measuring the thermal melting temperature for poly(dA•dT)2, which increases by 38.5° C and 29.6° C, respectively, consistent with significant binding affinity for AT-containing sequences (8). However, the selectivity and affinity of DNA binding for these compounds had not been characterized further. In addition to their ability to target DNA, the effectiveness of these compounds can also be accredited to their robust cellular uptake even in the absence of a functional P2-aminopurine transporter, which is closely associated with drug resistance (8). Symmetrical molecules of aminoimidazolines or guanidines were found to be the most potent compounds (9). Interestingly, these same molecules have been tested for binding to α2-adrenoceptor in a search for new antidepressants (9, 10), and derivatives of the half molecules, phenyl aminoimidazoline or guanidine compounds, rather than the symmetric compounds were found to bind with higher affinity to α2-adrenoceptor (10).
Here, we report the crystal structure of a CD27-DNA complex as well as evaluations of DNA binding selectivity and affinity for both CD27 and CD25. Both fluorescent intercalation displacement (FID) and surface plasmon resonance (SPR) studies were performed in assessing the DNA binding properties of CD27 and CD25. Crystallographic studies employed a host-guest approach with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase serving as the host and the DNA with bound ligand as the guest. This approach has been used previously to study the binding interactions of DNA with ligands such as netropsin (11), RT29 (12), and bleomycin (13) and was used in this study to determine the crystal structure of CD27 bound to 5′-AATT containing oligonucleotide at 1.75 Å resolution. To our knowledge, this is the first crystal structure reported for a bisaminoimidazoline diphenylamine compound bound to DNA. Although we refer to these compounds as bisaminoimidazoline and bisguanidine diphenylamines in keeping with previous reports, they are in fact expected to be dicationic molecules under physiological conditions (Figure 1). Our comparative structural analysis of CD27 with related compounds, DB75 and berenil, along with the DNA-binding analyses provide a basis for understanding the selectivity of CD27 as well as providing a platform for future antitrypanosomal drug design.
Materials and Methods
Fluorescent Intercalator Displacement Analyses
A DNA library of 136 unique oligonucleotide hairpins was purchased from Trilink Biotechnologies, Inc. as individual lyophilized solids. Concentrations of the hairpin deoxyoligonucleotides were determined spectrophotometrically at 90° C by the method described by Boger et al. (14) using single strand extinction coefficients to ensure accurate quantitation. To carry out the assay, each well of a Costar black 96-well plate was loaded with 150 μL of a solution containing 0.012 M Tris pH 7.4, 0.12 M NaCl, and 5.5 μM ethidium bromide (EtBr) followed by the addition of each hairpin oligonucleotide (30 μL of 9 μM hairpin solution, 64 μM base pairs, in H2O) to the corresponding well. Final concentrations in each well were 1.5 μM DNA hairpin and 4.5 μM EtBr, giving a 1 to 3 ratio, with final buffer concentrations at 10 mM Tris pH 7.4 and 100 mM NaCl. Upon addition of binding agent, each well was incubated for 30 min at 20º C prior to the measurement of fluorescence. Fluorescence measurements were obtained on a Varian Cary Eclipse fluorescence plate reader using an excitation wavelength of 545 nm and emission wavelength of 595 nm. Assessment of compounds was carried out over a range of concentrations (0 –30 μM) with each well acting as its own control well (no binding agent corresponds to 100% fluorescence and no DNA to 0% fluorescence). Concentrations of the compounds CD27 and CD25 were determined by weight. All fluorescence readings are given as a percent decrease relative to control wells.
Surface Plasmon Resonance Analyses
Biosensor experiments were conducted in degassed MES buffer (100 mM Na+, 10 mM [2-(N-morpholino)ethanesulfonic acid], 1 mM EDTA, 0.0005% v/v of surfactant P20, pH 6.25) at 25º C. The 5′-biotin-labeled DNA hairpins were purchased from Midland Certified Reagent Company, Inc. (Midland, TX) with HPLC purification. The DNA hairpin sequences examined included: 5′-biotin-CGAATTCGTCTCCGAATTCG-3′, 5′-biotin-CATATATATCCCCATATATATG-3′, and 5′-biotin-CGCGCGCGTTTTCGCGCGCG-3′, referred to in the text as AATT, (AT)4, and (CG)4, respectively. The experiments were conducted with a BIACORE 2000 instrument (Biacore AB). The DNA hairpins were immobilized on a streptavidin-derivatized gold chip (SA chip from BIAcore) by manual injection of 25 nM hairpin DNA solution with flow rate of 1 μL/min until the response units reach about 375-415 RUs. Flow cell 1 was left blank while flow cells 2, 3, and 4 were immobilized with three different DNA hairpins. Typically, a series of different concentrations of ligand was injected onto the chip at 25º C with a flow rate of 20 μL/min for a period of 5 min followed by a dissociation period of 5 min. After the dissociation process, the chip surface was regenerated with a 20 μL injection of 200 mM NaCl, 10 mM NaOH solution, injection tube rinsing, and multiple 1 min buffer injections. The observed steady-state responses, RUobs, were divided by predicted maximum response per ligand, RUmax (15, 16), and plotted against the free ligand concentrations, L, and fitted with a 2:1 model. r = (RUobs/RUmax) = (K1L + 2K1K2L2) / (1 + K1L + K1K2L2) L: the ligand concentrations in the flow solution.
Purification and Crystallization
The DNA oligonucleotide 5′-CTTAATTCGAATTAAG (TriLink Biotechnologies, Inc.) was purified by standard reverse phase HPLC methods and stored at a concentration of 2.5 mM duplex in 10 mM HEPES pH 7.0/10 mM MgCl2 (17). The N-terminal fragment of Moloney murine leukemia virus reverse transcriptase (MMLV RT) including residues 24-278 was purified as described previously (17). Briefly, the N-terminal-6XHis-tagged protein was overexpressed in E. coli and purified using Ni-NTA affinity chromatography, followed by S-sepharose ion-exchange chromatography, removal of the affinity tag using thrombin, and a final S-sepharose ion-exchange chromatography yielding about 5-10 mg per 1 L of culture. The protein was then concentrated to approximately 1.8 mM in 0.3 M NaCl/100 mM HEPES pH 7.5 for use in crystallization experiments.
RT-DNA-CD27 crystals were microseeded using crystals of an RT-DNA complex of the same DNA sequence and grown by hanging drop vapor diffusion crystallization at 20º C (17-19). Microseeding allows the rapid growth of crystals (1-2 days) of any DNA sequence co-crystallized (with or without drug complexes) and the RT protein. Crystals were obtained from a reservoir solution containing 7% PEG 40000, 5 mM magnesium acetate, 50 mM ADA pH 6.5. Hanging drops contained a 1:1 ratio of reservoir solution and complex, which included a ratio of DNA/protein of 1:1.8 and a DNA/drug ratio of 1:3. Stock concentrations were 1.8 mM RT in 50 mM MES pH 6.0/0.3 M NaCl, 1.6 mM oligonucleotide in 10 mM HEPES pH 7.0/10mM MgCl2, and 25 mM CD27 in 25% DMSO. The RT-DNA-CD27 crystals were soaked in cryosolutions including 9% PEG 4000, 5 mM magnesium acetate, 100 mM HEPES pH 8.0, 2 mM CD27, and increasing concentrations of ethylene glycol beginning with 2 % and concluding with 20%. The data for the crystal were collected at beamline 19-BM of the Advanced Photon Source (APS) Argonne, IL (λ=1.008001 Å) to Bragg spacings of 1.75 Å for a crystal that was ∼ 50 × 60 × 160 μm3. The space group was P21212 with unit cell parameters of a = 54.93 Å, b = 146.75 Å, c = 46.85 Å. Data were processed using the HKL 2000 package (20). See Table 1 for data processing statistics.
Table1. Summary of Crystallographic and Refinement Data.
| Cell Parameters | RT:AATT:CD27 |
|---|---|
| Cell constants (Å) | a=53.74 |
| b=145.6 | |
| c=46.57 | |
| Space Group | P21212 |
| Statistics | |
| Maximum Resolution | 1.75 Å |
| Reflections (unique) | 35,935 |
| (total) | 165,567 |
| Completeness (%) | 95% (94.6%) |
| Rsym (%) | 0.045 (0.355) |
| I/ σ | 24.7 (4.1) |
| Refinement | |
| Resolution range (Å) | 50-1.75 |
| Number of waters | 257 |
| Average B-factor | |
| (ptn) | 28.7 |
| (waters) | 36.1 |
| (DNA) | 47.3 |
| (CD27) | 51.0 |
| Rvalue (%) | 23.1 |
| Rfree (%) | 25.4 |
Structure Determination and Refinement
The crystal structure of RT-DNA-CD27 was phased by molecular replacement in AMoRe using the refined model of the N-terminal fragment of MMLV-RT as the search model (PDB ID code 1ZTW) (21). The initial model was subjected to rigid-body, positional, and B-factor refinement in CNS (22), and resulting difference maps included unbiased electron density for the DNA and CD27. The B-form DNA model for our desired sequence was generated using Nucleic Acid Builder (23) and adjusted manually to fit the electron density in O (See Supporting Information, Figure S-1) (24). This was followed by subsequent additions of water molecules, excluding those associated with the DNA, and several rounds of positional and B-factor refinement in CNS. Although CD27 was evident in initial Fo-Fc electron density maps, it was not positioned in the minor groove until the DNA model had been refined. Inspection of Fo-Fc maps at this stage of the refinement revealed clear density indicating the position and orientation of the CD27 molecule. The aminoimidazoline end closest to the 5′ end in the base of the groove had well-defined density and was used as a guide to fit the remainder of the symmetrical molecule. The phenyl ring adjacent to this first aminoimidazoline also had well-ordered density leading up to the nitrogen bridge that points out away from the minor groove. The second phenyl had sufficient density to deduce that this phenyl moiety is planar relative to the first. Upon additional rounds of refinement, density improved for the bound ligand and DNA revealing the position of the second aminoimidazoline end. This end is relatively planar allowing the molecule to adopt a nearly crescent shape characteristic of minor groove binding compounds. Additional refinement calculations using CNS (22) were done until convergence was achieved (see Table 1 for refinement statistics), and no large peaks remained in the Fo-Fc electron density maps.
Calculated Equilibrium Geometry
The molecular modeling calculations with CD27 were carried out with Spartan 4.0 software (Wavefunction, Inc.) in the dicationic state. Equilibrium geometry calculations were initiated from both the coordinates of CD27 when bound to DNA and from a molecular mechanics energy minimized structure built in Spartan. Calculations were carried out using the Hartree-Fock method at the 6-31G(d,p) level. Calculations from both starting structures minimized to a very similar final equilibrium geometry conformation.
Calculated Solvent Accessible Surface Areas
Solvent accessible surface areas (SASA) of the CD27-DNA complex were calculated using NACCESS V2.1.1. This program outlines the accessible surface of a molecule using a path generated by the center of a 1.4 Å radius-probe rolled around the van der Waals surface of the molecule based on the Lee & Richards method (25, 26). The surface area is calculated by slicing the three-dimensional molecular volume to determine the accessible surface of individual atoms. SASAs for unbound DNA and CD27 were calculated using only the coordinates for DNA or CD27, respectively. SASAtotal = SASAcomplex – (SASAfree DNA + SASAfree CD27). Coordinates have been deposited with the PDB, 3FSI.
Results
DNA Sequence Selectivity of CD27 vs. CD25
A fluorescent intercalator displacement (FID) assay (14) was employed to verify the A/T site-selectivity of CD27 and CD25 (Figure 1) using a library of 136 hairpin oligonucleotides containing four base pair cassettes comprising all possible four base pair sites within DNA. The FID assay relies upon competitive binding and/or displacement of intercalated ethidium bromide from hairpin oligonucleotides containing preferred binding sites by the compound of interest resulting in a decrease in fluorescence. We have used this assay previously to determine the selectivities of DNA-binding compounds including RT29, bleomycin, actinomycin, and netropsin (12, 27). As expected based on their structural similarities to other known A/T-rich DNA binders, CD27 and CD25 were found to preferentially bind to A/T-rich sites in the FID assay (Figure 2). The histogram for CD27 displayed slightly steeper plot curvature than CD25 at the 30 μM concentration (60% decrease in F) consistent with higher affinity binding and greater selectivity for the highest-ranked sequences: 5′-AAAA, 5′-AATT, and 5′-AAAT. In comparison, CD25 only promoted a 40% decrease in F at the highest-ranked end of the histogram consistent with lower affinity binding and less selectivity among the top ranked sites.
Figure 2.
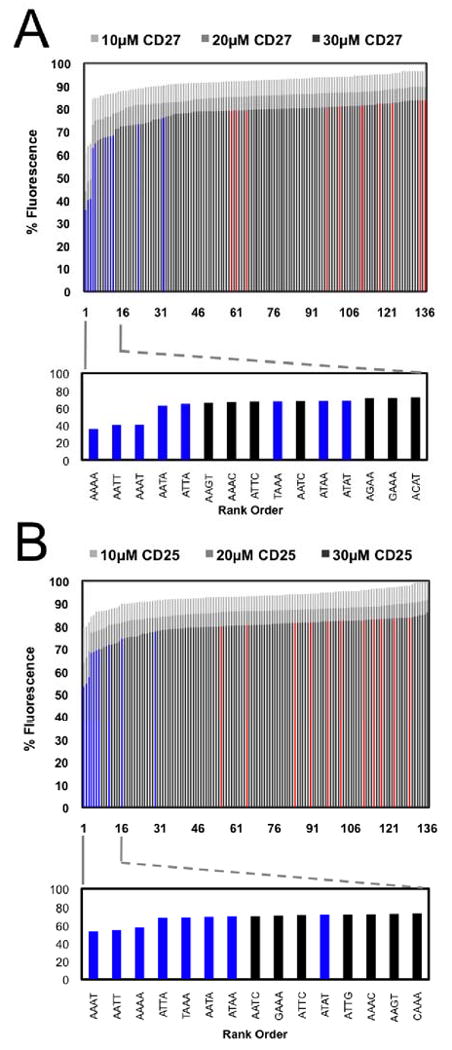
FID results cast as merged-bar histograms and identification (lower expansion) of the top 15 four base pair cassettes selected (red bars depict GC-only four base pair cassettes, blue bars depict AT-only four base pair cassettes, and black bars depict mixed sequence four base pair cassettes). (A) CD27 analyzed at 10, 20, and 30 μM concentrations and (B) CD25 analyzed at 10, 20, and 30 μM concentrations.
To further investigate CD25 and CD27 DNA binding quantitatively, surface plasmon resonance (SPR) experiments were performed to determine the binding affinity of these compounds to DNA. Representative sensorgrams for CD27 and CD25 binding to AATT, (AT)4 or (CG)4-containing oligonucleotides are shown in Figure 3. The primary binding constant K1 (see Materials and Methods and Table 5) for CD27 was determined to be 9.2 × 105 M-1 for a hairpin oligonucleotide containing a 5′-AATT site and 4.6 × 105 M-1 (fitting uncertainties of less than 10%) for an 5′-ATAT-containing hairpin DNA, a two-fold difference in binding affinity. In contrast, the primary binding constant for CD25 was determined to be 3.2 × 105 M-1 (fitting uncertainty less than 10%) for both 5′-AATT and 5′-ATAT containing hairpin oligonucleotides. CD25 and CD27 binding to a (CG)4- containing hairpin oligonucleotide was found to be too weak for accurate fitting of a binding constant.
Figure 3.
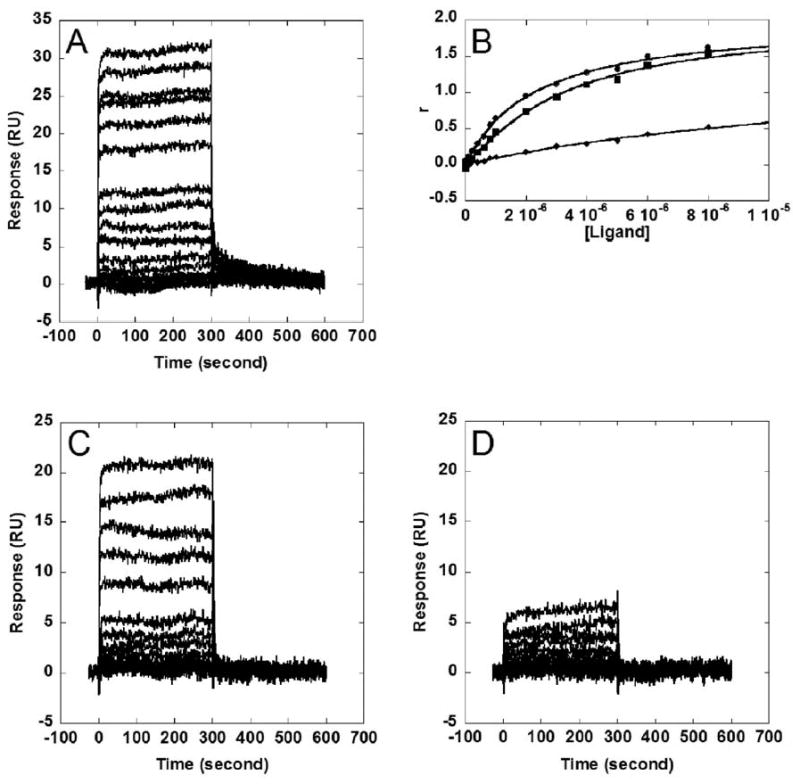
(A) Sensorgrams for the interaction of CD27 with the AATT hairpin. (B) Binding curves of CD27 to three different DNA hairpins; AATT (circles), alternating AT (squares), and alternating CG (diamonds). The strongest binding was observed with AATT hairpin. (C) Sensorgrams for the interaction of CD25 with (AT)4 hairpin and (D) with (CG)4 hairpin. The plots illustrate preferential binding to the AT sequences. Both plots have the same concentration range from 1 nM (lowest sensorgram) to 8 μM (highest sensorgram).
Table 5. Primary binding constants (K1(M-1), fitting errors (<10%)).
| AATT | (AT)4 | (CG)4 | |
|---|---|---|---|
| CD25 | 3.2×105 | 3.2×105 | ND |
| CD27 | 9.2×105 | 4.6×105 | ND |
Secondary binding (K2) to AATT and (AT)4 is 4-5 times weaker. ND: Too weak for accurate fitting
Both SPR and FID binding measurements confirm the preferential binding of CD25 and CD27 to A/T-containing oligonucleotides. Both methods also suggest greater selectivity and higher affinity binding of CD27 to DNA as compared to CD25. By SPR, CD27 binds with higher affinity to 5′-AATT vs. 5′-ATAT sites while CD25 binds with the same affinity to both 5′-AATT and 5′-(AT)4 containing oligonucleotides, thus displaying a lack of selectivity for these two sequences.
Description of the Crystal Structure
The host-guest crystallographic approach (11, 17, 19) was used to analyze the molecular basis of the binding interactions of CD27 in the minor groove of its preferred 5′-AATT site in atomic detail. The N-terminal fragment of Moloney Murine leukemia virus reverse transcriptase (MMLV RT) serves as the host, and DNA in the presence or absence of a ligand as the guest (Figure 4). As previously reported, this method allows for the analysis of novel DNA-binding compounds and a comparative analysis of the same oligonucleotide free of ligand (11). Advantages of the method include the ability to readily crystallize 16 base pair oligonucleotide duplexes of any sequence, to phase the structure by molecular replacement using the protein model, to interpret unbiased electron density for both the DNA and bound ligand, and to characterize simple minor groove binding compounds as well as ligands with more complex interactions such as bleomycin (11-13).
Figure 4.
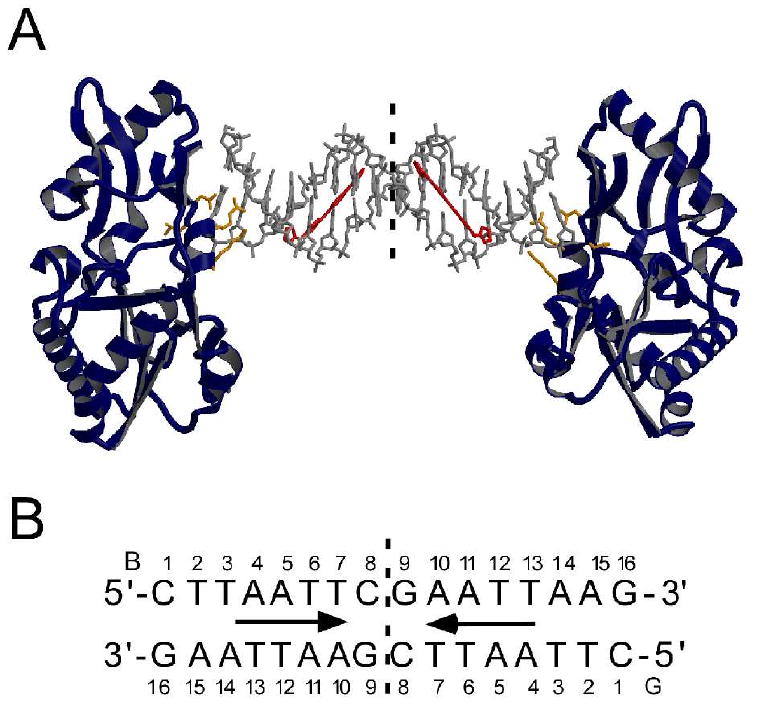
(A) The crystal structure of the RT-DNA-CD27 complex. The asymmetric unit consists of one protein molecule, 8 base pairs of the symmetric 16 base pair oligonucleotide duplex, and one CD27 molecule representing half of the symmetric complex. The dashed vertical line represents the dyad. The DNA oligonucleotide is shown as a gray stick model and CD27 is shown in a red stick model. RT is shown as a ribbon rendering with beta strands, alpha-helices and coils in navy. Residues Tyr64, Asp114, Lue115, Arg116, and Gly191 make contacts with the DNA and are shown as an orange ball-and-stick model. (B) Schematic of the oligonucleotide duplex with the two complementary strands and numbering scheme referred to in the text. Arrow denotes span of CD27 binding to oligonucleotide.
The complex analyzed includes two protein molecules bound to either end of a symmetric 16 base pair duplex oligonucleotide (5′-CTTAATTCGAATTAAG) including two CD27 binding sites as shown in Figure 4, while the asymmetric unit within the crystal includes one RT protein, 8 base pairs of duplex DNA, and one CD27 molecule. The structure was determined by molecular replacement using the protein as the search model (See Materials and Methods and Table 1). Difference electron density maps following refinement of the DNA and protein model (Figure 5) revealed the presence of CD27 bound in the minor groove of the 5′-AATT site as shown in Figures 4 and 6. Interactions of the protein with the DNA include 7 hydrogen bonds with the terminal three base pairs of the duplex DNA as summarized in Table 2. As a consequence, the intervening ten base pairs are relatively flexible as reflected in the higher B factors observed for the DNA than the protein in this complex (Table 1, Supporting Information Figure S-1). A critical design feature of the oligonucleotide is the placement of the 5′-AATT site beginning at position 4 from the end of the duplex to allow interactions of the oligonucleotide with the protein required to form the crystal lattice and yet allow CD27 to interact freely with the DNA (Figure 4). Inherent in the flexibility of the DNA is the ability to accommodate structural changes that allow for ligand binding to the minor groove as discussed below for CD27.
Figure 5.
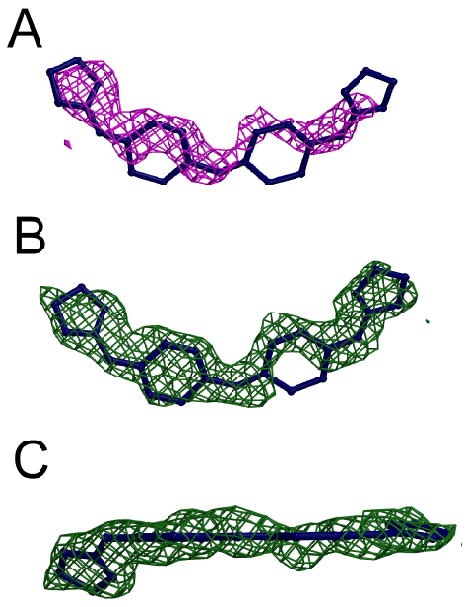
(A) Initial Fo-Fc map of CD27 density contoured at 2.5 σ shown in a magenta cage rendering superimposed on the final CD27 model. (B) Final 2Fo-Fc map of CD27 density contoured at 1 σ shown in green cage rendering and the final CD27 model superimposed in blue. The view in (C) is rotated ∼90° from that shown in (B).
Figure 6.
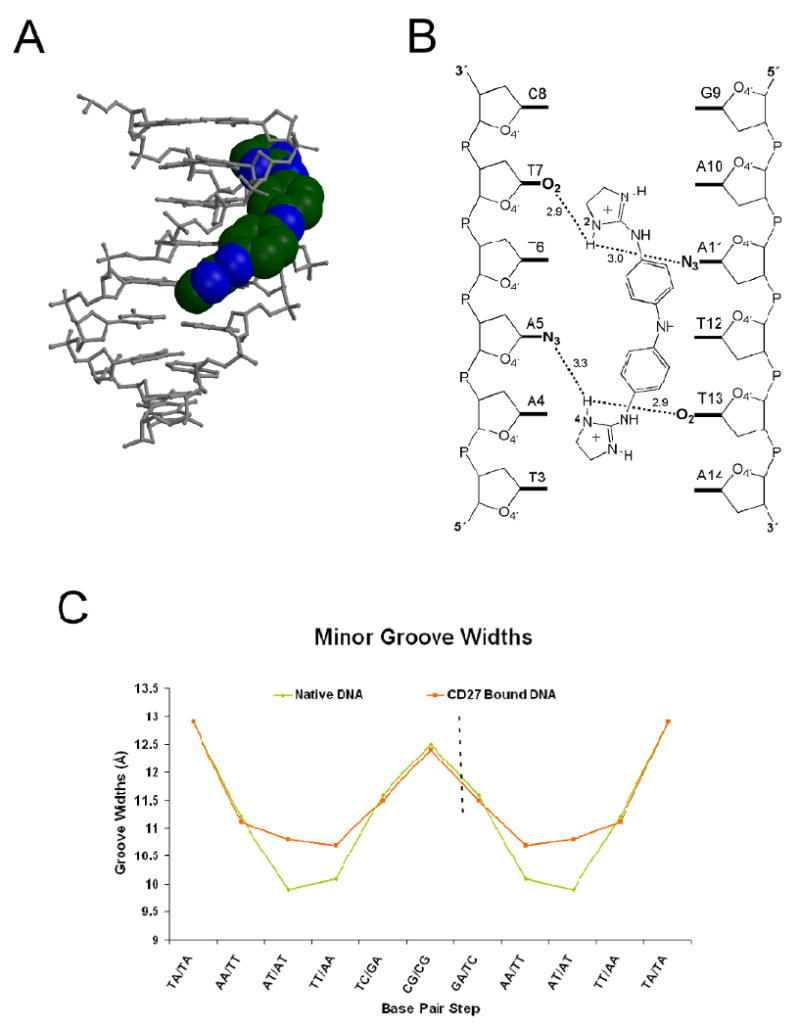
(A) Space-filling CPK model of CD27 bound in the minor groove of the DNA represented as a gray stick model, nitrogen in blue and carbon atoms in green. (B) Schematic of bifurcated hydrogen-bonding interactions between CD27 and the minor groove of the DNA. (C) Minor groove widths in Å calculated in 3DNA based on the cross-strand distances of phosphate groups is shown in orange for the CD27-DNA complex and green for the same DNA in the absence of CD27 for the entire 16 base pair oligonucleotide. The dashed line indicates the axis of symmetry present within DNA molecule, with one half representing the asymmetric unit within the crystal.
Table 2. Hydrogen bonding interactions between the protein and DNA (Å).
| Residue | Atom | Base pair | Atom | Distance |
|---|---|---|---|---|
| Asp 114 | OD2 | Gua 16 | N2 | 3.1 |
| Leu 115 | N | Gua 16 | O3′ | 3.0 |
| Arg 116 | Thy 3 | O4′ | 2.8 | |
| Arg 116 | NH2 | Gua 16 | N2 | 3.4 |
| Gly 191 | O | Gua 16 | O3′ | 3.0 |
| Arg 116 | NH2 | Thy 2 | O2 | 2.7 |
Attempts were also made to obtain a complex of CD25 bound to DNA using the same host-guest approach. CD25 was co-crystallized with the DNA and protein yielding suitable crystals as described for CD27 in the Materials and Methods; the crystals were then soaked in cryosolvents that included CD25. However, in contrast to CD27, the crystals cracked rendering them unsuitable for data collection. Alternatively, crystals were soaked in cryosolvents lacking CD25. Diffraction data were then collected, and the structures were determined. In these structures, there was no evidence of CD25 bound to the DNA in the electron density maps. Thus, efforts to date to determine a crystal structure of CD25 bound to DNA have been unsuccessful.
Interactions of CD27 with the AATT Site
CD27 forms two bifurcated hydrogen bonds within the 5′-AATT site: N4 of CD27 hydrogen bonds to O2 of T13 and N3 of A5 (2.9 Å and 3.3 Å, respectively), and, similarly, N2 of CD27 with O2 of T7 and N3 of A11 (2.9 Å and 3.0 Å, respectively) as shown in Figures 6 and 7, and Table 3. Thus, interactions of CD27 with the DNA involve direct hydrogen bonding and van der Waals interactions but do not involve ordered water molecules as have been observed in the structures of berenil and DB75 (28). In fact, interactions of ordered solvent with the DNA are somewhat limited in this structure including only 5 water molecules, 4 of which are hydrogen-bonded to the phosphodiester backbone and one in the major groove (See Table 4 and Supporting Information, Figure S-2), a finding not atypical of what we have observed in other host-guest complex structures that we have determined (11, 12, 29, 30).
Figure 7.
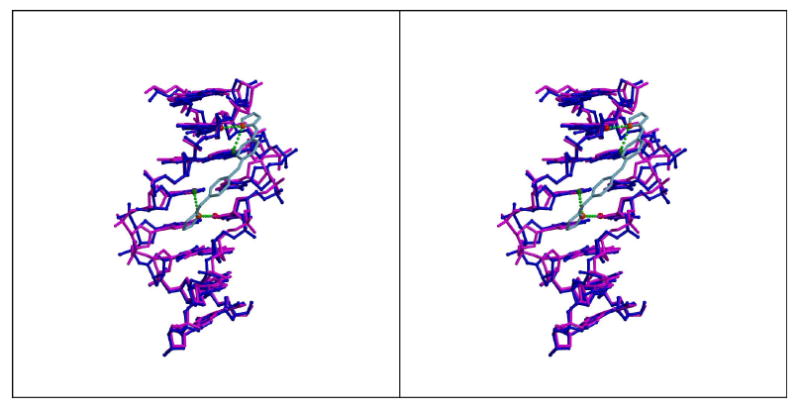
(A) Stereodiagram of 5′-AATT DNA in the presence (blue) and absence (magenta) of CD27 (sky blue) bound to DNA. Final models were superimposed (rmsd = 0.6 Å) using CCP4 based on all atoms within the 8 base pairs. Hydrogen bonds between DNA and CD27 are shown in light green dashed lines. N3 atoms of A5 and A11 shown in green, O2 atoms of T7 and T13 shown in red, and N2 and N4 of CD27 in orange.
Table 3. Hydrogen bonding interactions between the DNA and CD27 (Å).
| Base | Atom | Ligand | Atom | Distance |
|---|---|---|---|---|
| Ade 5 | N3 | CD27 | N4 | 3.3 |
| Thy 7 | O2 | CD27 | N2 | 2.9 |
| Ade 11 | N3 | CD27 | N2 | 3.0 |
| Thy 13 | O2 | CD27 | N4 | 2.9 |
Table 4. Hydrogen bonding interactions between the DNA and water (Å).
| Base | Atom | Number | Distance | |
|---|---|---|---|---|
| Cyt 1 | O2 | H2O | 257 | 3.0 |
| Thy 3 | O4 | H2O | 177 | 2.7 |
| Ade 15 | O1P | H2O | 243 | 2.7 |
| Gua 16 | O1P | H2O | 154 | 2.9 |
| Gua 16 | N7 | H2O | 185 | 2.9 |
Despite the symmetry of CD27 and the sequence of its recognition site, 5′-AATT, the electron density for the ligand is not symmetric. As shown in Figure 5, the density for the aminoimidazoline phenyl bound to A5 and T13 is better ordered than that bound to T7 and A11. Within the aminoimidazoline phenyl bound to T7 and A11, the phenyl ring is partially disordered lacking electron density for the two carbon atoms farthest from the floor of the minor groove. Thus, the better ordered electron density occurs within the narrower part of the groove for the 5′-AATT site. As previously noted (11), despite the chemical symmetry of the 5′-AATT site, it is in fact structurally asymmetric with regard to groove width (Figure 6C). Although CD27 does form symmetric bifurcated hydrogen bonds within the 5′-AATT site (Figures 3 and 4), the symmetry does not extend to the length of the hydrogen bonds. The hydrogen bonds of N2 or N4 to O2 of T7 and T13 are of the same length (2.9 Å), while the hydrogen bond of N4 with N3 of A5 in the narrower part of the groove is somewhat longer with a less favorable angle than the corresponding hydrogen bond of N2 with N3 of A11 (3.3 vs. 3.0 Å, respectively). This finding suggests that the better ordered density within the narrower part of the groove potentially results from more favorable van der Waals interactions rather than from stronger hydrogen bonds. In fact, the carbon adjacent to N4 within the ethyl bridge linking N4 and the other N atom in the imidazoline (C17 in the coordinate file) makes van der Waals contacts with O4′ of A5, O4′ of A14, and O2 of T13 (See Supporting Information, Figure S-3). The other C atoms (C18, C15, and C16) within the ethyl bridge of either imidazoline make only a single van der Waals contact. Interactions of CD27 with the 5′-AATT bury a total surface area of 729.6 Å2.
Binding of CD27 Results in Changes in the DNA Structure
Differences in the DNA in the presence and absence of CD27 (rmsd of 0.6 Å) have been evaluated by comparison of the corresponding structures, which were determined in the same crystal lattice, thereby limiting artifactual crystallographic factors (Figure 4). Binding of CD27 to the 5′-AATT site, base pairs 4-7 and 10-13, results in a significant widening of the minor groove. Specifically, the groove is widened by 0.9 Å within the A5T6/T12A11 dinucleotide step and 0.6 Å for the T6T7/A11A10 dinucleotide step as compared to the unbound structure (See Figure 1 for oligonucleotide numbering scheme). A further point of interest is that the narrowest point within the groove is shifted from A5T6/T12A11 in the unbound structure to the T6T7/A11A10 dinucleotide step, which differ by 0.1 Å, 10.7 Å vs. 10.8 Å, respectively, as calculated using 3DNA (31). Overall, the groove width varies less across the 5′-AATT site in the CD27-DNA complex than in the unbound structure as shown in Figure 6.
In addition to changes in the width of the minor groove, the binding of CD27 also affects the local nucleobase structure particularly in the regions of direct contact including A5, T13, T7, and A11 as shown in Figure 7. Upon binding of CD27, buckling within the A5-T12 base pair is decreased by 7.8° while propeller twist is increased by 3.9°. Propeller twist increases of 5.2° and 3.9° in base pairs T6-A11 and T7-A10 were also observed upon CD27 binding. Base pair opening is increased in all of the base pairs within the 5′-AATT site in the RT-DNA-CD27 structure as compared to the unbound structure, the most dramatic being an increase of 13.7° for the T7-A10 base pair. Increases in base pair opening for A4-T13, A5-T12, and T6-A11 are 3.7°, 5.2°, and 2.9°, respectively. Differences were also observed in local step parameters with the tilt of T6T7/A10A11 dinucleotide step increasing by 5.9° while that of the T7C8/G9A10 step decreased by 6.4°. Also noteworthy, the roll parameter at T7C8/G9A10 increased by 7.2° but decreased by 5.2° for the CG/CG dinucleotide step at the crystallographic dyad axis. These changes in local base and step parameters likely occur as a direct consequence of specific hydrogen-bonding and van der Waals interactions of CD27 with the DNA.
Comparison of DNA-Bound vs. Free CD27 Structure
The calculated, energy-minimized structure predicts that CD27 will adopt a staggered conformation with a dihedral angle of 43° between the two phenyl rings, and this reduces electrostatic and steric repulsion. The two aminoimidazoline ends are likewise staggered relative to the phenyl rings with dihedral angles of 85° in the energy minimized conformation. However, upon binding to the DNA, CD27 adopts a more classical crescent shape as shown in Figure 8 with a dihedral angle of near 0º between the phenyl rings. As the structure flattens, it should become more energetically favored due to enhanced conjugation; however, this aspect is attenuated by accompanying steric clashes and charge repulsion. As shown in Figure 8, the variations in electrostatic potential are greater in the DNA-bound than in the equilibrium structure. Thus, as observed for other minor groove binding compounds such as RT29 (12), the interaction of ligand with DNA appears to involve an induced fit of both the ligand and DNA in order to maximize favorable interactions.
Figure 8.
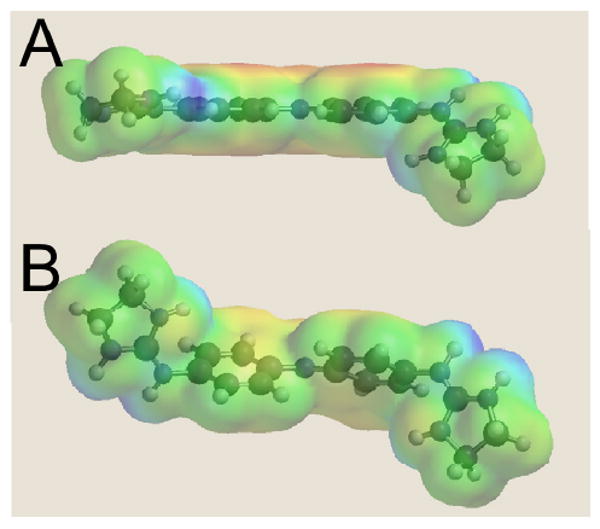
(A) Representation of CD27 as bound to DNA in the crystal structure shown within the electrostatic potential molecular surface. (B) The energy minimized structure of CD27, as calculated using Hartree-Fock method in Spartan 2004 and shown within the electrostatic potential molecular surface for the molecule, is highly twisted in contrast to the DNA-bound form in (A) which has a relatively planar crescent shape.
Discussion
As previously reported, dicationic compounds such as CD27 and CD25 bind DNA with relatively high affinity as indicated by the ΔTm values observed upon the melting of poly (dA•dT)2 in their presence: 38.5 °C and 29.6 °C, respectively (8). The compounds differ only in the nature of the dicationic moieties comprising either end of the molecule, a bisguanidine in the case of CD25 and bisaminoimidazoline in CD27. Through our structural analysis of CD27 bound to DNA and further characterization of the DNA binding properties of CD27 and CD25, we provide new insights with regard to understanding how these diphenylamines bind and recognize DNA.
CD27 binds to the minor groove floor via bifurcated hydrogen bonding interactions between the N2 and N4 imidazoline atoms and the O2 and N3 atoms of T and A, respectively, of adjacent base pairs (See Figures 3B and 4). Recognition of the 5′-AATT site within the DNA appears to result directly from the ability of CD27 to form these hydrogen bonds, which specifically involve T of one strand and A from adjacent base pair of the complementary strand within the site (Figure 7). Consistent with the somewhat lower binding affinity observed for 5′-ATAT, we predict that CD27, like netropsin (32), would not form bifurcated hydrogen bonds with a 5′-ATAT site. In fact, netropsin was found to be disordered within a 5′-ATAT site (32) and has also been shown to bind 5′-ATAT approximately 2.5 times less well than 5′-AATT as assessed by DNase I footprinting (33). Thus, we suggest that hydrogen-bonding along with favorable van der Waals interactions resulting from interactions with the internal adenines provides a basis for the selectivity of CD27 for 5′-AATT sites. Binding of CD27 induces significant changes both in itself and the DNA, specifically the properties of the minor groove and local base and step parameters within the DNA. The nitrogen bridge within CD27 forms no hydrogen bonding interactions with the DNA but may confer desirable properties as bioisosteric substitution (e.g., CH2, S, O) at this position results in a decrease in trypanocidal activity in vitro (8, 9). Interestingly, neither of the closely related compounds, diamidine diphenylfuran (or 2,5-bis(4-guanylphenyl)furan, DB75) or berenil, form bifurcated hydrogen bonds involving O2 and N3 of adjacent base pairs in the 5′-AATT site, rather the amidines hydrogen bond directly to O2 atoms of T or N3 atoms of A on either end of the 5′ -AATT site (28). In addition, water mediated interactions are involved in binding of both DB75 and berenil to DNA but not for CD27. Both DB75 and berenil were designed to have a preformed planar, crescent shape through the inclusion of a furan spacer linking the phenyl units in the case of DB75 and a triazene in the case of berenil and therefore form very favorable van der Waals interactions with the DNA. In energy-minimized structures, both compounds have planar diphenyl furan or triazene core structures as shown in Supporting Information, Figure S-4. Thus, DB75 and berenil differ significantly from CD27, which includes a single NH spacer allowing the molecule to adopt a much more twisted conformation in its unbound state. Both DB75 and berenil are potent antitrypanosomal compounds but show little sequence selectivity among A/T-rich sites (7).
In contrast to CD27 which binds 5′-AATT with two-fold higher affinity than 5′-ATAT sites as measured by SPR, CD25 shows no difference in its primary binding constant for 5′-ATAT vs. 5′-AATT. The decreased relative selectivity displayed by CD25 may have contributed therefore to difficulties encountered in attempting to obtain a crystal structure of the CD25-DNA complex. Regarding H-bonding interactions, although it is not possible to ascertain definitively whether or not CD25 forms bifurcated hydrogen bonds with the DNA, we speculate that by comparison with DNA-bound DB75, berenil, RT29, and netropsin that it may not. DB75, berenil, and RT29 are diamidines, which hydrogen bond directly to only one strand of the DNA within the minor groove (12, 28). Netropsin is also dicationic including both an amidine and guanidine, neither of which forms a bifurcated hydrogen bond to the DNA in any of the reported crystal structures (11, 34, 35). Thus, although CD27 and CD25 are similar, their interactions with the DNA may differ in detail. The aminoimidazoline ring in CD27 offers the same hydrogen bonding potential as found in the guanidine but may form more favorable van der Waals interactions with the minor groove of 5′-AATT while the more flexible guanidine may bind more indiscriminately with alternative AT sites. In turn, the more favorable van der Waals interactions with the aminoimidazoline may promote bifurcated hydrogen bonding thereby facilitating recognition of both strands of the DNA within the minor groove.
Both CD27 and CD25 are documented as having excellent in vivo anti-trypanosomal activity; however, previous studies have shown that the aminoimidazoline has a greater selectivity for the parasite of interest as compared to guanidine analogues and is a more potent trypanocide (8). Based on our comparative binding studies, we conclude that CD27 binds DNA more selectively and with higher affinity to 5′-AATT sites than does CD25. Our structural and DNA-binding results taken together with the in vitro findings that CD27 has greater selectivity for parasites of interest suggest that selectivity is an important property for trypanocidal compounds and that the aminoimidazoline moieties likely contribute to selectivity through favorable van der Waals interactions. The crystal structure of CD27 bound to DNA provides a basis for understanding in atomic detail the interactions between CD27 and DNA as well as a basis for future design of trypanocidal compounds.
Supplementary Material
Acknowledgments
We thank Marianne Cuff, Steve Ginell and Andrzej Joachimiak from the Structural Biology Center (SBC) Collaborative Access Team (CAT) at the Advanced Photon Source (APS). Use of the Argonne National Laboratory, SBC beamline at the APS was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
Footnotes
This work was supported by a National Institutes of Health Grant AI064200 (to W.D.W.) and the Bill and Melinda Gates Foundation (to W.D.W.). SPR instrumentation was funded in part by the Georgia Research Alliance. The work was also supported by the Spanish Ministerio de Educación y Ciencia grant SAF2006-04698 and the CSIC-I3 program PIE200680I121 (to C. D.).
Supporting Information Available: Unbiased difference electron density maps for the DNA, 2Fo-Fc electron density maps for the final structure, a stereoimage of the DNA structure with associated water, a 2-D van der Waals contact map for one of the two amidazoline groups, and energy minimized structures of berenil and DB75 are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.WHO-IFPMA. Working Paper on Priority Infectious Diseases Requiring Additional R & D, Round Table; Geneva, Switzerland. 2001. [Google Scholar]
- 2.WHO. Human African trypanosomiasis (sleeping sickness): epidemiological update. Wkly Epidemiol Rec. 2006;81:71–80. [PubMed] [Google Scholar]
- 3.Brun R, Schumacher R, Schmid C, Kunz C, Burri C. The phenomenon of treatment failures in Human African Trypanosomiasis. Trop Med Int Health. 2001;6:906–914. doi: 10.1046/j.1365-3156.2001.00775.x. [DOI] [PubMed] [Google Scholar]
- 4.Dardonville C, Brun R. Bisguanidine, bis(2-aminoimidazoline), and polyamine derivatives as potent and selective chemotherapeutic agents against Trypanosoma brucei rhodesiense. Synthesis and in vitro evaluation. J Med Chem. 2004;47:2296–2307. doi: 10.1021/jm031024u. [DOI] [PubMed] [Google Scholar]
- 5.Tidwell RR, Boykin DW. In: Dicationic DNA minor-groove binders as antimicrobial agents, in DNA and RNA Binders: From Small Molecules to Drugs. Demeunynck M, Bailly C, Wilson WD, editors. Wiley-VCH; Weinheim, Germany: 2003. pp. 415–460. [Google Scholar]
- 6.Mathis AM, Holman JL, Sturk LM, Ismail MA, Boykin DW, Tidwell RR, Hall JE. Accumulation and intracellular distribution of antitrypanosomal diamidine compounds DB75 and DB820 in African trypanosomes. Antimicrob Agents Chemother. 2006;50:2185–2191. doi: 10.1128/AAC.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson WD, Nguyen B, Tanious FA, Mathis A, Hall JE, Stephens CE, Boykin DW. Dications that target the DNA minor groove: compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr Med Chem Anticancer Agents. 2005;5:389–408. doi: 10.2174/1568011054222319. [DOI] [PubMed] [Google Scholar]
- 8.Dardonville C, Barrett MP, Brun R, Kaiser M, Tanious F, Wilson WD. DNA binding affinity of bisguanidine and bis(2-aminoimidazoline) derivatives with in vivo antitrypanosomal activity. J Med Chem. 2006;49:3748–3752. doi: 10.1021/jm060295c. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez F, Rozas I, Kaiser M, Brun R, Nguyen B, Wilson WD, Garcia RN, Dardonville C. New bis(2-aminoimidazoline) and bisguanidine DNA minor groove binders with potent in vivo antitrypanosomal and antiplasmodial activity. J Med Chem. 2008;51:909–923. doi: 10.1021/jm7013088. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez F, Rozas I, Ortega JE, Meana JJ, Callado LF. Guanidine and 2-aminoimidazoline aromatic derivatives as alpha(2)-adrenoceptor antagonists, 1: toward new antidepressants with heteroatomic linkers. J Med Chem. 2007;50:4516–4527. doi: 10.1021/jm070229q. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin KD, Long EC, Georgiadis MM. A host-guest approach for determining drug-DNA interactions: an example using netropsin. Nucleic Acids Res. 2005;33:4106–4116. doi: 10.1093/nar/gki717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin KD, Lewis MA, Tanious FA, Tidwell RR, Wilson WD, Georgiadis MM, Long EC. A high-throughput, high-resolution strategy for the study of site-selective DNA binding agents: Analysis of a “highly twisted” benzimidazole-diamidine. J Am Chem Soc. 2006;128:7846–7854. doi: 10.1021/ja0600936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin KD, Lewis MA, Long EC, Georgiadis MM. Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding. Proc Natl Acad Sci U S A. 2008;105:5052–5056. doi: 10.1073/pnas.0708143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse WC, Boger DL. A fluorescent intercalator displacement assay for establishing DNA binding selectivity and affinity. Acc Chem Res. 2004;37:61–69. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]
- 15.Davis TM, Wilson WD. Determination of the refractive index increments of small molecules for correction of surface plasmon resonance data. Anal Biochem. 2000;284:348–353. doi: 10.1006/abio.2000.4726. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Sun D, Jessen S, Liu C, Liu X, Najmudin S, Georgiadis MM. Cloning, expression, and purification of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase: crystallization of nucleic acid complexes. Protein Sci. 1998;7:1575–1582. doi: 10.1002/pro.5560070711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cote ML, Pflomm M, Georgiadis MM. Staying straight with A-tracts: a DNA analog of the HIV-1 polypurine tract. J Mol Biol. 2003;330:57–74. doi: 10.1016/s0022-2836(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 19.Cote ML, Yohannan SJ, Georgiadis MM. Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 9):1120–1131. doi: 10.1107/s0907444900008246. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. In: Processing of X-ray diffraction data collected in oscillation mode, in Methods in Enzymology. Carter J CW, Sweet RM, editors. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 21.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Macke T, Case DA. In: Modeling unusual nucleic acid structures, in Molecular Modeling of Nucleic Acids. Leontes NB, SantaLucia JJ, editors. Vol. 1. American Chemical Society; Washington, D.C.: 1998. pp. 379–393. [Google Scholar]
- 24.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 26.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 27.Lewis MA, Long EC. Fluorescent intercalator displacement analyses of DNA binding by the peptide-derived natural products netropsin, actinomycin, and bleomycin. Bioorg Med Chem. 2006;14:3481–3490. doi: 10.1016/j.bmc.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Laughton CA, Tanious F, Nunn CM, Boykin DW, Wilson WD, Neidle S. A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity. Biochemistry. 1996;35:5655–5661. doi: 10.1021/bi952162r. [DOI] [PubMed] [Google Scholar]
- 29.Cote ML, Georgiadis MM. Structure of a pseudo-16-mer DNA with stacked guanines and two G-A mispairs complexed with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase. Acta Crystallogr D Biol Crystallogr. 2001;57:1238–1250. doi: 10.1107/s090744490100943x. [DOI] [PubMed] [Google Scholar]
- 30.Montano SP, Cote ML, Roth MJ, Georgiadis MM. Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus. Nucleic Acids Res. 2006;34:5353–5360. doi: 10.1093/nar/gkl693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coll M, Aymami J, van der Marel GA, van Boom JH, Rich A, Wang AH. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989;28:310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- 33.Abu-Daya A, Brown PM, Fox KR. DNA sequence preferences of several AT-selective minor groove binding ligands. Nucleic Acids Res. 1995;23:3385–3392. doi: 10.1093/nar/23.17.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985;82:1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nunn CM, Garman E, Neidle S. Crystal structure of the DNA decamer d(CGCAATTGCG) complexed with the minor groove binding drug netropsin. Biochemistry. 1997;36:4792–4799. doi: 10.1021/bi9628228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


