Abstract
Base excision repair (BER) enzymes maintain the integrity of the genome, and in humans, BER mutations are associated with cancer. Given the remarkable sensitivity of DNA-mediated charge transport (CT) to mismatched and damaged base pairs, we have proposed that DNA repair glycosylases (EndoIII and MutY) containing a redox-active [4Fe4S] cluster could use DNA CT in signaling one another to search cooperatively for damage in the genome. Here, we examine this model, where we estimate that electron transfers over a few hundred base pairs are sufficient for rapid interrogation of the full genome. Using atomic force microscopy, we found a redistribution of repair proteins onto DNA strands containing a single base mismatch, consistent with our model for CT scanning. We also demonstrated in Escherichia coli a cooperativity between EndoIII and MutY that is predicted by the CT scanning model. This relationship does not require the enzymatic activity of the glycosylase. Y82A EndoIII, a mutation that renders the protein deficient in DNA-mediated CT, however, inhibits cooperativity between MutY and EndoIII. These results illustrate how repair proteins might efficiently locate DNA lesions and point to a biological role for DNA-mediated CT within the cell.
Keywords: DNA charge transport, DNA damage, iron–sulfur proteins, oxidative stress
Base excision repair (BER) proteins, from bacteria to humans, are challenged with combing the genome for DNA base lesions to maintain the integrity of our genetic material (1, 2). This challenge is remarkable, given the low copy number of these proteins and that they must discriminate among small differences between modified and natural bases. For MutY, a BER repair protein in Escherichia coli with a human homolog, there are ≤30 proteins in the E. coli cell (1) to interrogate 4.6 million bases; the ratio of binding affinities for the target lesion, an 8-oxoguanine:adenine mismatch, versus well-matched native base pairs is ≤1,000 (3). Endonuclease III (EndoIII) recognizes a less-prevalent lesion, hydroxylated pyrimidines, with equally low specificity; the copy number of EndoIII within E. coli is ≈500 (1). How these glycosylases fix their substrate lesions, once found, has been well characterized (1, 2), as are the structures of MutY and EndoIII bound to DNA (4, 5). Yet, how these lesions are efficiently detected before excision is not established.
Location of damaged bases in the genome is likely the rate-limiting step in BER within the cell (6). Current models for genome scanning to detect lesions involve protein sliding along the DNA, squeezing the backbone, slipping bases out to allow for interrogation, or finding transiently opened sites (7, 8). However, given the low copy number of these proteins and their need to sift through the genome to find often subtle base lesions, the time required for this search is long.
A subset of these BER proteins contains [4Fe4S] clusters, common redox cofactors in proteins (1, 2). Increasingly, iron–sulfur clusters are found associated with varied DNA-binding proteins and located far from the enzymatic active site, with no apparent function. For BER proteins, [4Fe4S] clusters were first thought to play a structural role. When not bound to DNA, these proteins are found in the [4Fe4S]2+ state and are not easily oxidized or reduced under physiological conditions (9). However, for MutY and EndoIII, we have demonstrated by using DNA-modified electrodes that DNA binding shifts the 3+/2+ cluster potential into a physiological range, ≈100 mV vs. normal hydrogen electrode for each BER enzyme (10, 11); DNA binding stabilizes the protein in the +3 form.
Given the sensitivity of DNA-mediated charge transport (CT) to mismatched and damaged bases, we have proposed that DNA repair glycosylases containing a redox-active [4Fe4S] cluster, including EndoIII and MutY, use DNA CT as the first step in substrate detection by signaling one another to search cooperatively for damage in the genome (10, 11). DNA-mediated CT can proceed over long molecular distances on a short timescale (12). Oxidative damage to DNA has been demonstrated, with oxidants covalently tethered and spatially separated from damage sites at distances of >200 Å and with negligible loss in efficiency (13). Reductive CT has been shown to have an equally shallow distance dependence both in electrochemical studies (14) and in assemblies in solution (15). Previous studies establish that CT through DNA is possible in biological environments that include nucleosomes (16) and cell nuclei (17). DNA CT is, however, extremely sensitive to perturbations in the intervening base pair stack, such as DNA mismatches and lesions (18–20). DNA-mediated electrochemistry has therefore been used in the development of sensors for mutational analysis (19) and protein binding (21).
Because this chemistry occurs at a distance and is modulated by the structural integrity of the base pair stack, these reactions may be useful within the cell for long-range signaling to proteins. In that context, we have established previously the long-range oxidation of the DNA-bound BER enzymes in spectroscopic studies monitoring oxidation of the [4Fe4S] clusters by guanine radicals in the duplex (22). Importantly, we have also shown the injection of an electron into the base pair stack from the DNA-bound BER enzymes, with the electron trapped by a well-coupled modified base in the duplex (23). Both with respect to hole injection into the DNA-bound proteins and electron injection into the DNA from the DNA-bound proteins, EndoIII and MutY behave equivalently, as expected, given their similar DNA-bound redox potentials and structures. Here, we now explore whether DNA-mediated CT may provide a means to facilitate the detection of damage in vivo.
Results
A CT Model to Detect DNA Damage.
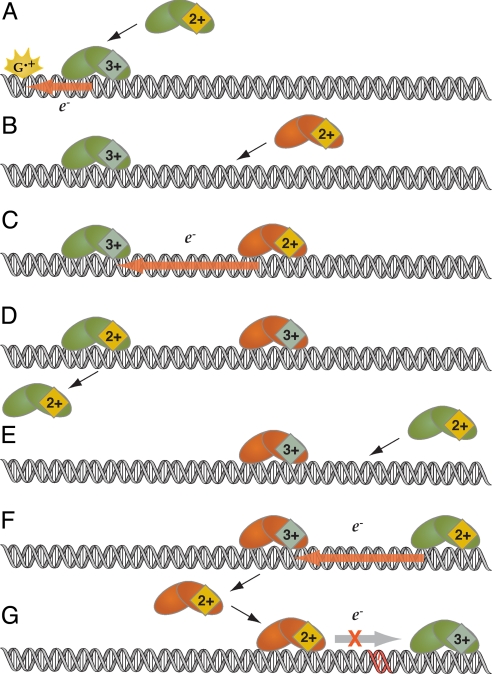
We have proposed that BER proteins bearing [4Fe4S] clusters exploit DNA-mediated CT as a fast, sensitive method to detect damage (Fig. 1). This redox signaling model is initiated when one 2+ protein (donor) binds DNA (Fig. 1 B and E), promoting electron transfer from the donor protein to a distal protein (acceptor; Fig. 1 C and F) already bound in the 3+ state. The newly oxidized donor protein remains DNA-bound while the reduced acceptor diffuses away (Fig. 1 D and F). Integral to this model is a differential DNA affinity for the [4Fe4S]3+ and [4Fe4S]2+ forms of the protein. We have demonstrated this differential affinity by measuring a −200-mV potential shift associated with DNA binding that corresponds thermodynamically to ≥1,000-fold difference in DNA affinity between the oxidized and reduced proteins (24).
Fig. 1.
A model for DNA-mediated CT in DNA repair. In this model, DNA repair proteins containing [4Fe4S]2+ clusters—for example, EndoIII (green) and MutY (orange)—bind DNA, activating them toward oxidation to the [4Fe4S]3+ state. The sequence of events is as follows: Guanine radical formation can oxidize a repair protein in a DNA-mediated reaction, stabilizing the oxidized protein bound to DNA (A). A second protein binds near the first protein (B and E). CT to a distally bound protein can occur if the intervening DNA is undamaged (C and F). The newly reduced protein has a diminished affinity for DNA and diffuses away (D). If a lesion site is present between the proteins (G), the DNA-mediated CT step is inhibited, and the oxidized protein remains bound. The sum of the DNA-mediated CT steps between proteins constitutes a full search of the genome.
Importantly, the DNA-mediated CT reaction between two repair proteins can be considered a scan of the integrity of the intervening DNA, because DNA-mediated CT can only proceed through a well-stacked duplex. As illustrated in Fig. 1G, when the repair protein, already oxidized, is bound near a base lesion, DNA-mediated CT does not provide a pathway for reduction and subsequent protein dissociation. The protein instead remains bound to the duplex, so that on a slower timescale, the protein can processively diffuse to the target site; now, however, sliding is needed only across a small region, and the low target specificity of the protein is sufficient for recognition (3, 25, 26). Essentially, then, our proposal for base lesion detection by using DNA CT yields a redistribution of the BER enzymes onto local regions of the genome that contain lesions. Critical to this mechanism is DNA-mediated signaling among proteins bound at long range so that the proteins, despite their low abundance, cooperate with one another in localizing onto target sites.
To exploit DNA-mediated CT, some proteins must exist in the oxidized state. There are many oxidants in the cellular milieu, and the level of oxidative stress will govern the proportion of oxidized protein. Indeed, we have shown that these proteins (22) and others (27) can be oxidized by guanine radicals, the first genomic signal of oxidative stress (28), via DNA-mediated CT.
A basic model of genome scanning involving only facilitated diffusion without CT but where interrogation is assumed to be instantaneous yields a genome-scanning time of at least 46 min for MutY (SI Text), wholly insufficient, given the doubling time in E. coli of 20 min. This calculated scanning time is based on measured values of protein diffusion constants, intracellular protein concentration (1), and genome size by using a model of 1D diffusion with short, localized hops (7). Recent calculations of global searches by low-copy number proteins suggest a still slower search time (29), and the nanomolar dissociation constant excludes 3D mechanisms. This estimate significantly understates the problem, moreover, because the actual interrogation time is not instantaneous, and protein traffic on the DNA necessarily interferes with sliding.
In our model, the DNA is essentially scanned by the electron, with the repair proteins facilitating electron migration. Thus, we calculate a genome-scanning time for MutY in E. coli that is significantly more efficient through DNA CT. Because an injected charge equilibrates on the nanosecond timescale (12), and protein diffusion occurs in microseconds to milliseconds (7), the rate-limiting step in this process is the 3D diffusion of this reduced protein within CT range of the oxidized DNA-bound protein. Hence, scanning can be modeled as a random walk of the electron (or hole) on the DNA, where the step time for the walk is the average time for a reduced protein to approach within range to carry out DNA-mediated CT to the oxidized protein. In our calculation, we conservatively use 3D diffusion of the reduced protein within the cellular volume to approach its target site on the DNA. We also use experimental values for protein concentrations (1), genome size, and protein-binding affinities (2).
Importantly, because this model involves cooperation among the repair proteins, we can use the total concentration of these proteins within the cell, rather than copy numbers for MutY or EndoIII individually. Thus, MutY, present in ≤30 copies, benefits from 500 copies of EndoIII (1). We do, however, neglect contributions from any other proteins that might participate in DNA-mediated signaling; other DNA-bound proteins that are redox-active may exhibit similar potentials (e.g., SoxR, OxyR, IscR), and CT reactions involving these proteins too would substantially speed the search process.
Our model relies on the fact that DNA-mediated interprotein CT is much faster than protein diffusion, and that the oxidized repair proteins have higher nonspecific DNA affinity than the reduced proteins; both assumptions have experimental support (12, 24). As we did for facilitated diffusion, we assume here also that intervening DNA-binding proteins do not inhibit scanning. In fact, one advantage of DNA CT over other search mechanisms is that the electron travels through the DNA base pairs, and no proteins need to be displaced (16).
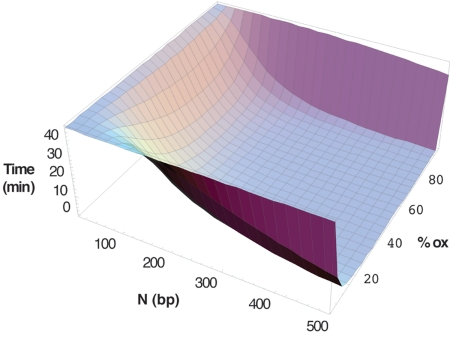
Fig. 2 shows how the interrogation time varies as a function of N, the maximum distance over which DNA-mediated CT proceeds, and ox, the percentage of proteins oxidized. Remarkably, with 20% oxidized protein, permitting DNA CT over 200 bp yielded an interrogation time of 8 min, and over several hundred base pairs it yielded scan times of less than a minute. These values are well within the 20-min doubling time of E. coli.
Fig. 2.
Scanning time as a function of maximum distance of DNA-mediated interprotein CT (N) and the fraction of repair proteins that are in the 3+ state (% ox) by using the CT scanning model. At 10% oxidized protein with a maximum CT distance of 500 bp, the time required to interrogate the genome is ≈5 min.
The dependence of interrogation time on the percentage of proteins oxidized is also noteworthy (Fig. 2). The scanning efficiency resembles a switch that is turned on at low levels of oxidation, when DNA repair is needed. Activation of this switch depends on the redox buffering capacity of the cell and the level of oxidative stress.
An Atomic Force Microscopy (AFM) Assay to Measure Protein Redistribution onto Mismatched DNA.
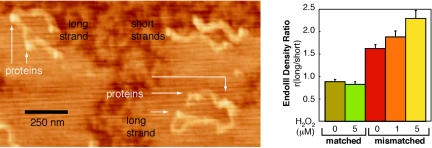
Although we have previously carried out studies establishing hole and electron injection across the protein–DNA interface (23, 24), our model also predicts that DNA–protein CT would promote the redistribution of repair proteins in the vicinity of base lesions or mismatches. We can assay for this redistribution by AFM. A mixture of DNAs, both long (3.8 kb) DNA duplexes containing a single CA mismatch and short (2.2 and 1.6 kb), well-matched duplexes of the same total sequence were prepared (30); the longer sequence was obtained by ligation of the two shorter sequences. This mixture of matched and mismatched DNA strands was incubated with EndoIII and examined by using established AFM techniques (Fig. 3) (31). Only clearly identifiable long or short strands were counted. Protein assignments were verified through analysis of their 4-nm heights in the images; without protein, features of this dimension are not observed, and still larger heights indicate salt precipitates. Although a CA mismatch effectively inhibits DNA CT (19), it is not a lesion that is preferentially bound by EndoIII; a gel-shift assay on 21-mers with and without a central CA mismatch showed no detectable difference in EndoIII binding. Thus, without DNA CT between bound EndoIII molecules, one might expect an equal density of proteins on the short and long strands.
Fig. 3.
Measurements of repair protein distributions on DNA by AFM. A zoomed-in view (Left) of representative AFM images of DNA strands incubated overnight with WT EndoIII. A higher density of proteins is apparent on the longer DNA strands containing the single CA mismatch. (Right) Quantitation of protein density ratios (<10% uncertainty). A CA mismatch is contained on the long strand except for the sample indicated by matched DNA, where both the long and the short strands are fully matched.
We found that EndoIII shows a significant preference for the longer strands containing the CA mismatch. Examination of the number of proteins bound to 300 long strands and 465 short strands revealed a greater density of proteins bound to the long strand: ratio of long to short was 1.6 (Fig. 3). If instead we examine the distribution of EndoIII on long versus short strands, where all strands are matched, we see a small preference for the short strands; the ratio of protein densities, long to short, was 0.9. When we calculate the strand preference based on DNA CT, this protein density ratio depends on the DNA CT length and/or the length of the DNA over which protein can diffuse before dissociating. By using a signaling/sliding length of 90 bp and allowing free sliding off the DNA ends, we calculated a protein density ratio of 1.6, where half of the protein population is near the mismatch.
AFM measurements as a function of oxidation of proteins bound to DNA, using H2O2 as oxidant, revealed an additional increase in the ratio of EndoIII bound to mismatch-containing strands. Examination of more than 250 long CA mismatch-containing strands and 300 shorter matched strands incubated with EndoIII and treated with 5 μM peroxide revealed a ratio of bound protein densities, long to short, of 2.4; when both long and short strands were matched, the ratio was 0.83 (Fig. 3).
These results are consistent with our model. DNA-mediated CT will drive the redistribution of repair proteins away from undamaged regions such that the proteins will cluster near damaged sites. As a result, we see the proteins redistribute preferentially onto the DNA strands containing the mismatch, even though a CA mismatch is not a substrate for EndoIII. Moreover, as predicted by the model, the redistribution of EndoIII is more pronounced in the presence of oxidative stress.
Cooperation Between EndoIII and MutY Inside the Cell.
This CT scanning model can also be tested in vivo by assaying for the cooperation among repair proteins facilitated by DNA-mediated signaling. If these proteins are able to help each other in their search for damage by using DNA CT, knocking out the gene for EndoIII or reducing its capability to carry out CT should lead to a decrease in MutY activity in vivo. Assays for MutY and EndoIII activity inside E. coli cells have already been developed (32). The assay for “helper function” used here employed engineered mutations in the lacZ gene to report the frequency of a particular base pair substitution. The strain that served as an assay for MutY activity, CC104, substitutes a cytosine for an adenine in the lacZ Glu-461 codon, which is essential for β-galactosidase activity. Because MutY prevents GC-to-TA transversions (33), reversion of this original mutation back to WT lacZ reflects a deficiency in MutY activity. Analogously, the CC102 strain (32) serves as an assay for EndoIII activity by monitoring GC-to-AT transitions (34).
In the CC104 MutY activity reporter strain (Table 1), 20 ± 9 lac+ revertants were observed per 109 cells, whereas inactivation of mutY in CC104 (CC104 mutY−) caused the number of lac+ revertants to increase 15 times (300 ± 33), as expected (32, 33). When the gene encoding EndoIII (nth) was inactivated in CC104 (CC104 nth−), the lac+ reversion frequency observed was 54 ± 5, more than a factor of two increase over CC104. Thus, loss of EndoIII does have a small but significant effect on MutY activity in vivo. This loss in activity is consistent with a loss in helper function by EndoIII, as predicted; the lower activity of MutY without EndoIII could reflect the lack of cooperative searching via DNA CT. An alternative explanation, however, is that MutY and EndoIII share some overlapping ability to repair lesions. In this case, the lac+ reversion frequency of the CC104 mutY−/nth− strain (270 ± 29) should be greater than that of CC104 mutY−, but they are, within error, equivalent.
Table 1.
Assay for DNA repair in E. coli by MutY (CC104)
| Strain | lac+ revertants* | Increase, x/CC104 |
|---|---|---|
| CC104† | 20 ± 9 | — |
| CC104 nth− | 54 ± 5 | 2.7 |
| CC104 mutY− | 300 ± 33 | 15 |
| CC104 mutY−/nth− | 270 ± 29 | 13.5 |
*The lac+ revertants are reported as the average number of lac+ colonies that arose per 109 cells plated on minimal lactose media. These data represent a single set of experiments, with 10 replicates per strain assayed concurrently. Values are reported as the mean ± SD
†CC104 strains reflect the rate of GC-to-TA transversion mutations and serve as a reporter for MutY activity in E. coli.
This in vivo relationship between EndoIII and MutY has been observed previously, although in different experimental contexts. Small increases in mutational frequency have been detected when mutY is inactivated in CC102 (SI Text) (32) or when nth is inactivated in CC104 (34). In the latter case, it was proposed that this could be due to some intrinsic ability of EndoIII to repair oxidatively damaged guanine residues. Reported EndoIII repair activities do not prevent GC-to-TA transversion mutations (34), and thus are not relevant to the CC104 assay.
We can furthermore test directly whether the loss of MutY activity in the CC104 assay is the result of overlapping glycosylase activities by determining whether the number of lac+ revertants is still suppressed by an EndoIII mutant that is biochemically incompetent to carry out the glycosylase reaction. A mutant of EndoIII (D138A) that is known to be deficient in glycosylase activity (35, 36) was introduced on a plasmid into both the CC102 and CC104 strains along with appropriate vector controls (Table S2). Because this mutant cannot perform the base excision reaction, D138A fails to reduce the high reversion frequency observed with CC102 nth−. However, D138A is able to complement the CC104 nth− strain. Thus, the glycosylase activity of EndoIII is not required for its helper function to aid MutY in repairing lesions inside the cell. Nonetheless, it appears that EndoIII lacking D138 can bind DNA and contains an intact [4Fe4S] cluster (37). Based on our model, D138A should be competent to carry out DNA-mediated CT, and thus serve as a helper to MutY, as we observe.
A Mutant Defective in DNA/Protein CT.
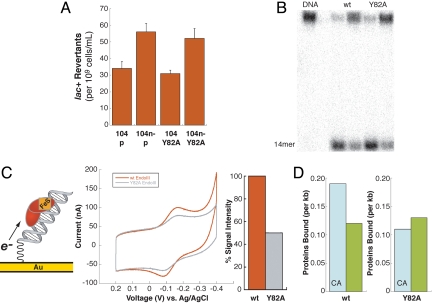
In our model, it is the ability to carry out DNA-mediated CT, not the glycosylase activity of EndoIII, that is critical to its helper function. Thus, perturbing the path for electron transfer to the DNA would interfere with this helper function. Aromatic tyrosine and tryptophan residues often facilitate long-range electron transfers in proteins (38), and EndoIII contains many of these residues. In particular, Y82 is conserved in most EndoIII and MutY homologs (39), and an analogous mutation (Y166S) in the human homolog of MutY is associated with cancer (37). In the crystal structure, Y82 is located very close to the DNA backbone (4). Y82A EndoIII was thus introduced on a plasmid into both reporter strains (CC102 and CC104) and their nth knockouts to explore whether this mutation attenuates helper function (SI Text). Significantly, Y82A in the CC104 nth− strain shows an increase in mutation rate versus the CC104/Y82A and CC104/p controls (Fig. 4). The number of lac+ revertants was found to increase by 53% ± 16% when comparing CC104 nth−/Y82A to CC104/p. When comparing CC104 nth−/Y82A to CC104/Y82A, the number of lac+ revertants increased by 68% ± 13%. Similarly, for these trials, the ratio of the number of lac+ revertants for CC104 nth−/p versus CC104/p was 165% ± 13%. These results clearly indicate that Y82A does not restore helper function.
Fig. 4.
Y82A EndoIII, a mutant in DNA-mediated CT capability. (A) Bar graph showing lac+ revertants for CC104/p, CC104 nth−/p, CC104/Y82A, and CC104 nth−/Y82A strains, where p denotes inclusion of an empty vector. Lac+ revertants are reported as the average number of lac+ colonies that arise per 109 cells plated on minimal lactose medium containing ampicillin. Data for the CC104 strains are shown based on five sets of independent experiments, each containing 10 replicates per strain. (B) Autoradiogram after denaturing PAGE of 32P-5′-TGTCAATAGCAAGXGGAGAAGTCAATCGTGAGTCT-3′ plus complementary strand, where X = 5-OH-dU base-paired with G. Protein samples (100 or 10 nM) were incubated with duplexes for 15 min at 37 °C and quenched with 1 M NaOH. Cleavage of the 32P-labeled strand at the lesion site (X) by EndoIII results in formation of a 14-mer. No significant difference in glycosylase activity (10% uncertainty) is observed between Y82A and WT EndoIII. (C) Cyclic voltammetry of Y82A EndoIII at an Au electrode modified with SH(CH2)2CONH(CH2)6NHOCO-5′-AGTACAGTCATCGCG-3′ plus complementary strand showing the reduction and reoxidation of the DNA-bound protein. DNA-modified surfaces were prepared and backfilled with mercaptohexanol, and WT or Y82A EndoIII was tested. Surfaces were then rinsed, and the other protein was analyzed on the same surface. Over several trials, the electrochemical signal associated with Y82A was 50% ± 13% smaller per [4Fe4S] cluster compared with WT EndoIII, reflecting poor electronic coupling of the mutant to the DNA-modified electrode. (D) Comparative densities for WT (Left) and Y82A (Right) EndoIII bound to matched versus mismatched (CA) strands measured by AFM. Although WT EndoIII preferentially redistributes onto the mismatched strand, Y82A shows no preference.
It is noteworthy that inclusion of Y82A EndoIII in CC102 nth− led to a diminished mutation rate, indicating that this mutant is competent for EndoIII activity inside the cell (SI Text). Interestingly, the observation that Y82A complements CC102 nth− but not CC104 nth− is consistent with the conclusion that the glycosylase activity of EndoIII is not a source of helper function. Moreover, the fact that Y82A complements CC102 nth− is understandable in the context of our model, because of the higher copy number of EndoIII in E. coli cells than MutY. In our model, without oxidative stress, we would predict that DNA CT is not essential for EndoIII repair activity inside the cell. We would therefore anticipate that the role of EndoIII in helping MutY search for lesions may be more important than the ability of EndoIII to find its own lesions.
To establish the biochemical characteristics of Y82A EndoIII, the protein was purified and its redox and glycosylase activities examined. Importantly, the mutant enzyme does contain the [4Fe4S] cluster, characterized by its distinctive absorbance spectrum (Fig. S3). Y82A EndoIII also maintained glycosylase activity against a 5-OH-dU lesion in a 32P-5′-end-labeled 35-mer duplex (Fig. 4); the activity of the mutant in this assay was equal to that of WT. Note that this experiment on a 35-mer duplex measured only the base excision reaction, not the search process. Similarly, in the E. coli EndoIII activity assay, where we expect that the search process is not rate-limiting, Y82A EndoIII activity was comparable to that of WT EndoIII. In contrast, D138A EndoIII, which instead inhibited the base excision reaction, failed to complement the nth knockout in the EndoIII activity reporter strain but did complement the nth knockout in the MutY activity reporter strain, where lesion detection was limiting.
To test for DNA-bound redox activity, Y82A was examined on a Au electrode modified with thiol-terminated DNA duplexes. Significantly, in the cyclic voltammogram, the potential for the DNA-bound mutant resembles that of the WT (11), but the signal intensity is diminished (Fig. 4). The protein concentrations were determined based on the 410-nm absorbance of the [4Fe4S] cluster; the smaller electrochemical signal observed with Y82A does not reflect a lower concentration of [4Fe4S] clusters. During several trials, Y82A EndoIII exhibited a signal that was 50% ± 13% smaller than that for WT EndoIII (per [4Fe4S] cluster). This signal intensity provides a reliable measurement of reduction/oxidation of the DNA-bound protein. Because the glycosylase activity on the 35-mer was equal for the mutant and WT, this diminished signal cannot reflect diminished binding of the mutant to the DNA. Instead, this lowered signal intensity would be expected with an attenuated efficiency of CT from the cluster to DNA and reflects poor electronic coupling of the mutant with the DNA duplex. These results therefore indicate that Y82A EndoIII is defective in DNA-mediated signaling.
Significantly, and consistent with these results, examination of the distribution of Y82A on mismatched and matched strands by AFM showed no preference for the mismatched strand; we observed 0.11 protein per kilobase long strand and 0.13 protein per kilobase short strand (Fig. 4). In fact, the ratio of protein densities on mismatched versus matched strands with Y82A, long to short, was 0.9, essentially equal to that of WT EndoIII bound to fully matched long versus short strands. Because the Y82A mutant, biochemically defective only in DNA CT, cannot redistribute to the vicinity of the lesion, DNA CT must play a role in finding the lesion both in the AFM experiment and in the helper function assay. These results together demonstrate a distinct connection between DNA-mediated CT to the [4Fe4S] cluster, the detection of DNA defects, and the in vivo relationship observed between MutY and EndoIII.
Discussion
These experiments indicate that MutY and EndoIII cooperate in their search for damage in the genome and redistribute in the vicinity of lesions consistent with CT scanning. This cooperation, or helper function, does not involve the glycosylase reaction. Furthermore, based on their chromosomal arrangement, the expressions of MutY and EndoIII do not appear to be linked (40). There is also no chemical evidence that the proteins physically bind to one another, and their low abundance within the cell makes random associations improbable. This cooperation thus arises from a distance. Importantly, what does appear to be required for helper function is an intact [4Fe4S] cluster as well as an electroactive protein–DNA interface. Mutation of an aromatic amino acid residue near the DNA-binding site, Y82A, leads to a decrease in CT efficiency in vitro, the inability of the protein to redistribute near lesions by AFM, and diminished helper function in vivo. These experiments thus establish a link between DNA-mediated CT and the cooperative search for damage by these repair proteins both in vitro and in vivo.
BER glycosylases are known to prevent mutations inside the cell, yet in most organisms, these enzymes are not required for normal growth and development (2). Recently, it was discovered that germ-line mutations in human BER homologs result in genetic predisposition to cancer (37). Specifically, the human homolog of mutY (MUTYH) is found mutated in a subset of patients predisposed to colorectal cancer. Many of the cancer-associated mutations in MUTYH are missense, or single-amino acid, mutations. Although several of the most common mutants have been characterized biochemically, it remains unclear exactly how these variants lead to disease. Given that initial detection of lesions is likely the rate-limiting step in BER (6), it is possible that mutants with defects in protein–DNA CT would be associated with cancer. Indeed, many of these MUTYH missense mutations found in colorectal cancer patients result in loss or gain of aromatic residues near predicted protein–DNA interfaces (37). Significantly, MUTYH contains two adjacent tyrosine residues (Y165 and Y166) that closely align with Y82 in E. coli EndoIII, and inherited mutations in these MUTYH residues (Y165C and, less commonly, Y166S) are clinically relevant in cancer. These results thus provide tantalizing evidence for association between defects in lesion detection via DNA-mediated CT by BER enzymes and human disease.
Iron–sulfur clusters are becoming increasingly recognized as a motif in proteins that repair, replicate, and transcribe DNA (41, 42). Recent characterizations of archaeal DNA primase, RNA polymerase, and nucleotide excision repair helicase (XPD) homologs reveal an iron–sulfur cluster required for normal enzyme function. Although the precise role of the cluster in these proteins is unclear, the cysteine residues ligating the cluster are conserved in eukaryotic homologs of these proteins. It is interesting to consider whether in these proteins, as in BER enzymes, the iron–sulfur cluster is poised to send and receive redox signals mediated by the DNA helix. Such long-range signaling among proteins bound to DNA would make searching for lesions much more efficient and may generally provide a means of genome-wide communication to monitor cellular stresses.
DNA-mediated CT serves as a fast and efficient reaction that is exquisitely sensitive to lesions in the base pair stack. This chemistry helps explain how these repair glycosylases may locate their lesions efficiently in the cell, a key function because mutations in these enzymes in humans are implicated in colorectal cancer (37). This mechanism furthermore provides a rationale for iron–sulfur clusters in DNA repair proteins. Other roles for DNA-mediated CT in biological signaling must now be considered.
Materials and Methods
Genome Scanning Calculations.
Methods used to calculate the genome scanning time for lesion detection by DNA repair proteins via DNA CT or facilitated diffusion may be found in the SI Text.
AFM Experiments.
Strands containing single-base mismatches were constructed by ligating together duplex strands with a single-strand overhang (to generate the mismatch); strands containing the single-strand overhang were generated by PCR using primers with a 2′-O-methyl-ribonucleotide to pause the polymerase and leave the overhang (30). Details regarding experimental procedures are described in the SI Text. For each AFM experiment, at least six images and >200 strands were counted by using several preparations of protein/DNA samples.
MutY Activity Assays.
Strain and plasmid construction for the genetics assays is provided in the SI Text. For the experiments, all strains were first streaked to selective media. Ten independent colonies of each strain were grown to a density of 109 cells per milliliter in minimal medium (NCE) supplemented with glucose. These cells were then plated on NCE medium supplemented with lactose and incubated for 36 h at 37 °C. The resulting lac+ revertants are reported as the average ± SD per 109 cells per milliliter plated (two highest and lowest values removed). In experiments where plasmids were used, all media were supplemented with ampicillin (40 μg/mL in NCE; 100 μg/mL in LB).
Y82A EndoIII Characterization.
Y82A EndoIII was purified as described in the SI Text. DNA-modified electrodes were also prepared as described previously (11). Protein solution was introduced to the electrode surface and allowed to incubate for ≈20 min until signal reached full intensity. Cyclic voltammetry experiments were performed with a 50-mV/s scan rate, Ag/AgCl reference electrode, and Pt wire auxiliary electrode in an electrochemical cell modified for protein experiments.
Supplementary Material
Acknowledgments.
We thank Sheila David (University of California, Davis), Jeffrey Miller (University of California, Los Angeles), Timothy O'Connor (City of Hope, Duarte, CA), and the Coli Genetic Stock Center (Yale University, New Haven, CT) for their generous donation of bacterial strains and plasmids. We are grateful to the National Institutes of Health for Grant GM49216 (to J.K.B.) and the Howard Hughes Medical Institute (to D.K.N.). A.K.B. thanks the Parsons Foundation for fellowship support.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908059106/DCSupplemental.
References
- 1.Demple B, Harrison L. Repair of oxidative damage to DNA: Enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 2.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francis AW, Helquist SA, Kool ET, David SS. Probing the requirements for recognition and catalysis in Fpg and MutY with nonpolar adenine isosteres. J Am Chem Soc. 2003;125:16235–16242. doi: 10.1021/ja0374426. [DOI] [PubMed] [Google Scholar]
- 4.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 6.Livingston AL, et al. Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY. Nat Chem Biol. 2008;4:51–58. doi: 10.1038/nchembio.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 8.Parker JB, et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham RP, et al. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989;28:4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- 10.Boon EM, et al. DNA-mediated charge transport for DNA repair. Proc Natl Acad Sci USA. 2003;100:12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boal AK, et al. DNA-bound redox activity of DNA repair glycosylases containing [4Fe4S] clusters. Biochemistry. 2005;44:8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- 12.Wagenknecht HA, editor. Charge Transfer in DNA: From Mechanism to Application. Weinheim, Germany: Wiley-VCH; 2005. [Google Scholar]
- 13.Nunez ME, Hall DB, Barton JK. Long-range oxidative damage to DNA: Effects of distance and sequence. Chem Biol. 1999;6:85–97. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 14.Kelley SO, Jackson NM, Hill MG, Barton JK. Long-range electron transfer through DNA films. Angew Chem Int Ed. 1999;38:941–945. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Elias B, Shao F, Barton JK. Charge migration along the DNA duplex: Hole versus electron transport. J Am Chem Soc. 2008;130:1152–1153. doi: 10.1021/ja710358p. [DOI] [PubMed] [Google Scholar]
- 16.Nunez ME, Noyes KT, Barton JK. Oxidative charge transport through DNA in nucleosome core particles. Chem Biol. 2002;9:403–415. doi: 10.1016/s1074-5521(02)00121-7. [DOI] [PubMed] [Google Scholar]
- 17.Nunez ME, Holmquist GP, Barton JK. Evidence for DNA charge transport in the nucleus. Biochemistry. 2001;40:12465–12471. doi: 10.1021/bi011560t. [DOI] [PubMed] [Google Scholar]
- 18.Boal AK, Barton JK. Electrochemical detection of lesions in DNA. Bioconjug Chem. 2005;16:312–321. doi: 10.1021/bc0497362. [DOI] [PubMed] [Google Scholar]
- 19.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Mutation detection by electrocatalysis at DNA-modified electrodes. Nat Biotechnol. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 20.Guo XF, Gorodetsky AA, Hone J, Barton JK, Nuckolls C. Conductivity of a single DNA duplex bridging a carbon nanotube gap. Nat Nanotechnol. 2008;3:163–167. doi: 10.1038/nnano.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boon EM, Salas JE, Barton JK. An electrical probe of protein-DNA interactions on DNA-modified surfaces. Nat Biotechnol. 2002;20:282–286. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 22.Yavin E, et al. Protein-DNA charge transport: Redox activation of a DNA repair protein by guanine radical. Proc Natl Acad Sci USA. 2005;102:3546–3551. doi: 10.1073/pnas.0409410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yavin E, et al. Electron trap for DNA-bound repair enzymes: A strategy for DNA-mediated signaling. Proc Natl Acad Sci USA. 2006;103:3610–3614. doi: 10.1073/pnas.0600239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorodetsky AA, Boal AK, Barton JK. Direct electrochemistry of endonuclease III in the presence and absence of DNA. J Am Chem Soc. 2006;128:12082–12083. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 25.O'Handley S, Scholes CP, Cunningham RP. Endonuclease III interactions with DNA substrates. Biochemistry. 1995;34:2528–2536. doi: 10.1021/bi00008a017. [DOI] [PubMed] [Google Scholar]
- 26.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Augustyn KE, Merino EJ, Barton JK. A role for DNA-mediated charge transport in regulating p53: Oxidation of the DNA-bound protein from a distance. Proc Natl Acad Sci USA. 2007;104:18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 29.Wunderlich Z, Mirny LA. Spatial effects on the speed and reliability of protein-DNA search. Nucleic Acids Res. 2008;36:3570–3578. doi: 10.1093/nar/gkn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donahue WF, Turczyk BM, Jarrell KA. Rapid gene cloning using terminator primers and modular vectors. Nucleic Acids Res. 2002;30:e95. doi: 10.1093/nar/gnf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Haushalter KA, Lieber CM, Verdine GL. Direct visualization of a DNA glycosylase searching for damage. Chem Biol. 2002;9:345–350. doi: 10.1016/s1074-5521(02)00120-5. [DOI] [PubMed] [Google Scholar]
- 32.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: A mutator locus in Escherichia coli that generates G·C to T·A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C to T transitions in vivo. Proc Natl Acad Sci USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thayer MM, et al. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo CF, et al. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1995;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 37.Cheadle JP, Sampson JR. MUTYH-associated polyposis—from defect in base excision repair to clinical genetic testing. DNA Repair. 2007;6:274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Shih C, et al. Tryptophan-accelerated electron flow through proteins. Science. 2008;320:1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Blaisdell JO, Wallace SS, Bond JP. Engineering functional changes in Escherichia coli endonuclease III based on phylogenetic and structural analyses. J Biol Chem. 2005;280:34378–34384. doi: 10.1074/jbc.M504916200. [DOI] [PubMed] [Google Scholar]
- 40.Gifford CM, Wallace SS. The genes encoding endonuclease VIII and endonuclease III in Escherichia coli are transcribed as the terminal genes in operons. Nucleic Acids Res. 2000;28:762–769. doi: 10.1093/nar/28.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan L, et al. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–900. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from archaea. Nature. 2008;451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.