Abstract
Fusion between the membrane of HIV and the membrane of a host cell is a crucial step in HIV infection and is catalyzed by the binding of the fusion peptide domain (HFP) of the HIV gp41 protein to the host cell membrane. The HFP by itself induces vesicle fusion and is a useful model system to understand the fusion peptide/host cell membrane interaction. This article reports an experimental correlation between the membrane locations of different HFP constructs and their fusogenicities. The constructs were the HFP monomer with Val-2 to Glu-2 mutation (HFPmn_mut), wild type HFP monomer (HFPmn), and wild type HFP trimer (HFPtr). All constructs have predominant β sheet structure in membranes with physiologically relevant cholesterol content. HFPmn_mut does not fuse vesicles, HFPmn has moderate fusion rate, and HFPtr has the putative oligomerization state of HIV gp41 and a very rapid fusion rate. The HFP membrane locations were probed with solid-state NMR measurements of distances between labeled carbonyl (13CO) nuclei in the HFP backbone and lipid nuclei in the surface or interior regions of the membrane bilayer. HFPmn_mut is located at the membrane surface, HFPmn is inserted into a single membrane leaflet, and HFPtr is the most deeply inserted construct with contact with the center of the membrane. These results show a clear positive correlation between the insertion depths and the fusion activities of the HFP constructs. Other disease-causing enveloped viruses contain fusion peptides and this correlation may be a general structure-function model for these peptides.
Keywords: NMR, enveloped virus, AIDS, trimer, location
Like many viruses that cause human disease, HIV is enveloped by a membrane obtained during virus budding from a previously infected host cell and infection of a new cell requires fusion between the viral membrane and the cell membrane. Fusion is catalyzed by the HIV fusion protein gp41, which has ≈170 ectodomain residues outside the virus including a ≈20-residue N-terminal fusion peptide (HFP) that binds to target cell membranes (1). Studies of HIV with a truncated or mutated HFP showed that the HFP is crucial in the fusion process (2, 3). Functionally critical fusion peptides are also found in fusion proteins of other enveloped viruses such as influenza and Ebola (1). Chemically synthesized peptides with HFP sequences have been developed as fusion model systems and provide information about HFP perturbation of target membranes. Free HFPs induce vesicle fusion and there are strong correlations between the mutation/activity relationships of HFP-induced fusion and HIV/host cell fusion (3).
There have been some number of HFP structural studies, but in our view, there have not yet been clear correlations between HFP structure and fusogenic function. For example, membrane-associated HFP can adopt either helical or β strand conformation and there has been effort to determine a correlation between conformation and fusogenicity. However, this work has resulted in conflicting models such as: (i) the helical conformation is fusogenic and the β strand conformation is nonfusogenic (4); (ii) the β strand conformation is fusogenic and the helical conformation is nonfusogenic (5); (iii) a transient random coil conformation is fusogenic (6); and (iv) both the helical and β strand conformations are fusogenic (7). In the present study, the structural focus is on HFP membrane location rather than conformation and a clear correlation is observed between depth of membrane insertion and fusogenicity. In addition, this article describes a general solid-state NMR method for determining the membrane location of a peptide or protein that is an alternative to existing EPR or fluorescence membrane location methods (8–10).
Table 1 displays the sequences of the three HFP constructs of the present study. There are large differences in the rates and extents of vesicle fusion induced by these constructs. The wild-type HFP monomer (HFPmn) induces fusion with moderate rate (7). HFPmn with the V2E mutation denoted HFPmn_mut does not induce vesicle fusion and was chosen because viruses and cells expressing gp41 V2E mutant have greatly impaired fusion and infectivity (2, 11). Relative to all wild-type gp41, a 10:1 mixture of wild-type:V2E gp41 shows only 40% fusion which suggests that fusion requires assembly of many gp41s with wild-type fusion peptides. The high-resolution structures of the soluble ectodomain of gp41 which lacks the HFP are trimeric and suggest that HFP trimers insert into the target cell membrane (1). The putative functional significance of trimers is supported by the 15–40-fold higher vesicle fusion rates of the chemically cross-linked HFP trimer (HFPtr) relative to HFPmn (7). Thus, the fusion rates are ordered HFPmn_mut < HFPmn < HFPtr and the present study examines the structures and membrane locations of these constructs with correlation to their very different fusogenicities.
Table 1.
Names and sequences of the HIV fusion peptide constructs
| Name | Sequence |
|---|---|
| HFPmn | AVGIGALFLGFLGAAGSTMGARSWKKKKKA |
| HFPmn_mut | AEGIGALFLGFLGAAGSTMGARSWKKKKKA |
| HFPtr | AVGIGALFLGFLGAAGSTMGARSWKKKKKA |
| AVGIGALFLGFLGAAGSTMGARSWKKKKC̍ | |
| AVGIGALFLGFLGAAGSTMGARSWKKKKC̍G |
HFPmn structure has been studied in detergent micelles by liquid-state NMR and for HFPmn:detergent mol ratio ≤0.02, helical conformation has been observed (4, 12). The conformation of HFPs in membranes is modulated by the HFP:lipid mol ratio and the membrane composition (3, 8, 10). In the present HFP study, the membranes contained ≈30 mol% cholesterol which correlated with the ≈30 and ≈45 mol% cholesterol in membranes of host cells of HIV and in membranes of HIV, respectively (13). In this composition, solid-state NMR data supported a fully extended β sheet conformation for the Ala-1 to Gly-16 region of HFPmn with crossing of adjacent hydrogen bonded HFPmns near Phe-8 and Leu-9 (14).
The membrane locations of HFPs have been studied by using both experimental and computational methods. Fluorescence studies of HFPmn suggested that the indole group of a HFPmn F8W mutant is inserted in the membrane and located approximately midway along the bilayer longitude between the bilayer center and the phosphate headgroups (9, 10). In addition, an EPR study of HFPmn was consistent with an Ala-1 location close to the aqueous phase (8). A solid-state NMR study of HFPmn and HFPtr showed that for a large fraction of both constructs, the Ala-15 carbonyl (13CO) nuclei are 5–6 Å from 31P nuclei in the lipid headgroups (15). This result was obtained both for helical HFPs in membranes which lacked cholesterol and for β strand HFPs in membranes with cholesterol. Simulations have been carried out for a single HFPmn molecule in a membrane and showed predominant α helical conformation. There was disagreement among the simulation studies about the membrane insertion depth of HFPmn with reports of either shallow insertion into a single leaflet or traversal across both membrane leaflets (16, 17).
In the present work, a systematic study of membrane location as a function of residue position has been carried out for HFPmn_mut, HFPmn, and HFPtr. Solid-state NMR 13CO-31P rotational-echo double resonance (REDOR) measurements detected distances between labeled 13CO nuclei in the HFP backbone and 31P nuclei in the lipid headgroups (18). In addition, 13CO-19F REDOR experiments probed distances between the backbone 13COs and 19F substituted for a 1H at either the terminal C16 or the C5 position of the sn2 acyl chain of DPPC lipid. These 16-19F or 5-19F nuclei are respectively positioned at either the bilayer center or along the bilayer longitude midway between the bilayer center and the 31P nuclei near the bilayer surface. The 13CO-31P and 13CO-19F distance measurements are combined to determine the membrane location of a particular residue's 13CO nucleus.
Results
13CO-31P and 13CO-(16-19F) REDOR Reveal Different Membrane Locations for Different HFP Constructs.
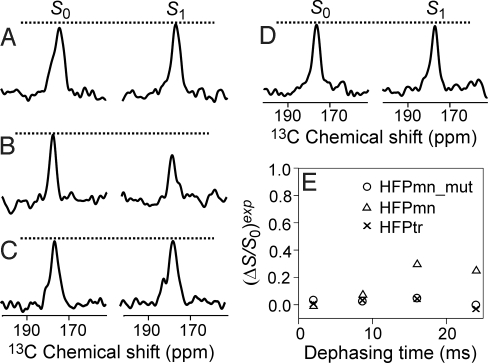
Fig. 1 displays 13CO REDOR NMR spectra for some of the membrane-associated HFP samples. The membrane composition was DTPC:DTPG:16-19F-DPPC:cholesterol in a 8:2:1:5 mol ratio. The ratios HFPmn_mut or HFPmn:total lipid = 0.036 and HFPtr:total lipid = 0.012 where total lipid = (DTPC + DTPG + 16-19F-DPPC). Thus, all samples had the same peptide strand:lipid mol ratio. The ether-linked DTPC and DTPG lipids were used because they lacked ester 13COs whose signals would overlap with the HFP 13CO signals. Use of ether-linked rather than ester-linked lipids does not affect HFP structure (14).
Fig. 1.
REDOR 13C S0 and S1 NMR spectra for samples with different labeled HFPs. The dephasing time for each 13CO-31P spectrum was 32 ms and for each 13CO-19F spectrum was 24 ms. The membranes included 9 mol% 16-19F-DPPC lipid. Each spectrum was the sum of ≈30,000 scans for 13CO-31P experiments and ≈20,000 scans for 13CO-19F experiments.
Each HFP construct contained a single 13CO label at either Ala-1, Ile-4, Ala-6, Leu-9, Leu-12, Ala-14, or Ala-21, and a construct is denoted HFPmn_mut-A1, HFPmn-A1, HFPtr-A1, etc. Residues 1–16 are in the apolar region of the HFP sequence whereas Ala-21 is in the more polar C-terminal region. The local conformation at each labeled residue in each construct was probed through comparison of the peak 13CO chemical shift with characteristic chemical shift distributions in helical or β strand conformations (supporting information (SI) Table S1). All of the peak shifts agree better with β strand than with helical conformation. There may be a ≈30% population of helical HFPmn_mut as evidenced by a shoulder at ≈180 ppm for samples labeled at Ala-6, Leu-9, or Leu-12. The typical linewidth for samples labeled between Ala-1 and Ala-14 is 3–5 ppm whereas the linewidth for samples labeled at Ala-21 is ≈8 ppm (SI Text). The overall conclusions of the analysis of the chemical shifts and linewidths are predominant β strand conformation for the N-terminal apolar regions of all three constructs and more disordered structure in the C-terminal polar regions.
The REDOR NMR spectra also provided information about the proximity of the labeled 13CO nucleus in the HFP to the 31P or 19F nuclei in the membrane lipids. In particular, “d ” or the magnitude of the 13CO-31P or 13CO-19F dipolar coupling was probed and is structurally significant because d for a single spin pair is quantitatively related to the internuclear separation (SI Text). During acquisition of the “S1” REDOR spectrum, the 13C magnetization evolved for the dephasing time “τ ” under the effect of the dipolar coupling. The “S0” spectrum served as a reference for which the time average of the dipolar evolution was zero. The effect of the dipolar coupling was observable in the reduced intensity of the S1 spectrum relative to the S0 spectrum (Fig. 1). For each S0 and S1 spectrum, a 1 ppm interval around the peak was integrated and the integrals were denoted as “S0exp” and “S1exp,” respectively. The experimental dephasing (ΔS/S0)exp = (S0exp − S1exp)/S0exp was then calculated and d determined from analysis of (ΔS/S0)exp vs. τ. Subsequent sections of this article describe this quantitative analysis with accompanying detailed description of HFP membrane location. However, a qualitative picture of membrane location is first obtained based on the (ΔS/S0)exp of the 13CO-31P and 13CO-(16-19F) experiments at large τ (Fig. 2). The 31P and 16-19F lipid nuclei are respectively located near the surface and the center of the membrane. The analysis of membrane location assumes that all of the constructs have the known HFPmn structure in which the Ala-1 to Gly-16 region is fully extended and a large fraction of HFPs assemble into an antiparallel β sheet structure with adjacent strand crossing near Phe-8 and Leu-9 (14). The β sheet structure is supported by the similar 13CO shifts for a given residue among the different constructs (Table S1).
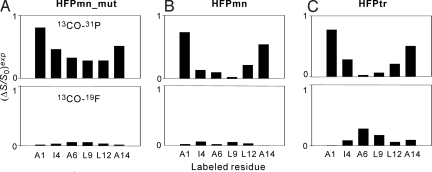
Fig. 2.
Summary of experimental REDOR dephasing (ΔS/S0)exp for the spectra displayed in Fig. 1. The top box in each image is the 13CO-31P data and the bottom box is the 13CO-(16-19F) data. The (ΔS/S0)exp values are shown as bars and a typical uncertainty is ±0.04.
In Fig. 2, one example observation is that all constructs labeled at Ala-1 have 13CO-31P (ΔS/S0)exp ≈ 0.8. This observation is interpreted to mean that a major fraction of Ala-1 13COs are 5–6 Å from a 31P (19). In some contrast, the HFPmn-L9 and HFPtr-L9 samples displayed 13CO-31P (ΔS/S0)exp ≈ 0 which is interpreted to mean that most of these Leu-9 13COs are >10 Å from a 31P. This type of analysis of Fig. 2 leads to the following general conclusions about the HFP membrane locations: (i) All HFPmn_mut samples have 13CO-31P (ΔS/S0)exp > 0.3 and 13CO-(16-19F) (ΔS/S0)exp ≈ 0 which indicates that HFPmn_mut lies on the membrane surface near the phosphate headgroups and is far from the bilayer center. (ii) The HFPmn and HFPtr samples with Ala-1 or Ala-14 labeling show 13CO-31P (ΔS/S0)exp > 0.5 and 13CO-(16-19F) (ΔS/S0)exp ≈ 0. These residues are near the ends of the β sheet structure and are close to the phosphate headgroups. (iii) The HFPtr-A6 and HFPtr-L9 samples have 13CO-31P (ΔS/S0)exp ≈ 0 and 13CO-(16-19F) (ΔS/S0)exp significantly >0. This suggests that the interior residues of the HFPtr β sheet are close to the bilayer center; i.e., a significant fraction of HFPtr is deeply inserted in the membrane. (iv) HFPmn samples labeled at Ile-4, Ala-6, or Leu-9 have 13CO-31P (ΔS/S0)exp and 13CO-(16-19F) (ΔS/S0)exp ≈ 0 which suggests that these interior β sheet residues are neither close to the headgroups nor to the bilayer center and are instead located midway between these two regions. Fig. 3 displays the membrane location models based on these analyses.
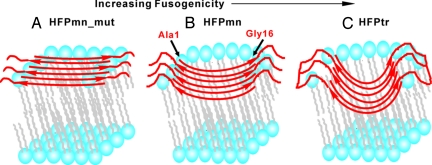
Fig. 3.
Membrane location models of β sheet (A) HFPmn_mut, (B) HFPmn, and (C) HFPtr. Lipid headgroups are drawn as blue balls, lipid alkyl chains are drawn in gray, and peptides are drawn in red. The lines at the C terminus of HFPtr represent the chemical cross-linking of the HFPtr construct. In all models, peptides are represented as oligomers with either six (HFPmn and HFPmn_mut) or two (HFPtr) molecules. The strands are in antiparallel β sheet structure with adjacent strand crossing near Phe-8 and Leu-9. This is the known structure for a large fraction of HFPmn peptides (14). The number of strands in a sheet is not known but is likely a small number (19). For clarity, not all lipid molecules are shown near the HFP.
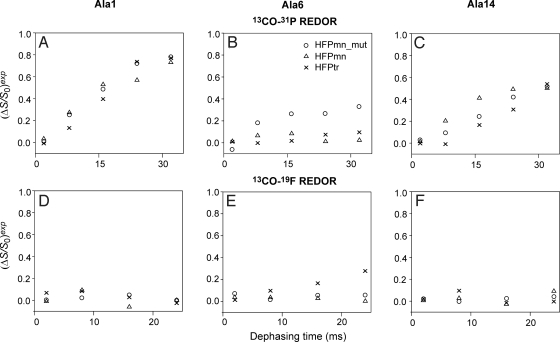
Fig. 4 displays plots of (ΔS/S0)exp vs. τ for samples labeled at Ala-1, Ala-6, or Ala-14. These residues were selected to represent the N-terminal, middle and C-terminal parts of the apolar region of the HFP sequence. These plots support the qualitative discussion of membrane location of the previous paragraph. The samples labeled at Ala-1 (Fig. 4 A and D) have 13CO-31P (ΔS/S0)exp that increase rapidly with τ whereas the 13CO-(16-19F) (ΔS/S0)exp ≈ 0 for all τ. After removal of natural abundance 13C contributions from the 13CO-31P (ΔS/S0)exp (SI Text), the remaining (ΔS/S0)lab reach ≈1 at large τ. The (ΔS/S0)lab represent only the Ala-1 13CO signals and the value of ≈1 indicates that the N termini of all constructs are close to the phosphate headgroups (18, 20). These N termini are likely positively charged and are therefore attracted to the negatively charged phosphates. Samples labeled at Ala-14 (Fig. 4 C and F) show similar large 13CO-31P (ΔS/S0)exp and 13CO-(16-19F) (ΔS/S0)exp ≈ 0 indicating proximity to the phosphate headgroups. However, at large τ, the 13CO-31P (ΔS/S0)lab < 1 which might be explained by a fraction of 13COs in the interior strands of the β sheet which are far from any lipid molecule and which have (ΔS/S0) = 0. For samples labeled at Ala-6, the CO-31P (ΔS/S0)exp vary with HFP construct and the 13CO-(16-19F) (ΔS/S0)exp also display significant variation (Fig. 4 B and E). For example, HFPmn_mut was the only construct with 13CO-31P (ΔS/S0)exp significantly greater than zero and HFPtr was the only one with 13CO-(16-19F) (ΔS/S0)exp significantly greater than zero. These results revealed that the middle regions of different HFP constructs had different membrane locations. HFPtr had the deepest insertion and induced the most rapid vesicle fusion whereas HFPmn_mut was located on the membrane surface and was the least fusogenic construct.
Fig. 4.
13CO-31P and 13CO-19F REDOR dephasing curves for different samples containing 16-19F-DPPC lipid and labeled at (A and D) Ala-1, (B and E) Ala-6 or (C and F) Ala-14. The symbol for each construct is given in the legend of B. For 2 ms dephasing time, the typical uncertainty in (ΔS/S0)exp is ±0.02 and for the other dephasing times, the typical uncertainty is ±0.04.
13CO-(5-19F) REDOR Confirms the HFPmn Membrane Location.
The previous paragraphs suggested that the interior β sheet region of HFPmn was located midway between the headgroups and the bilayer center and more direct evidence was provided by experiments on samples containing 5-19F DPPC lipid. The location of this 19F along the bilayer longitude should be close to the position of the C5 carbon to which it is directly bonded. For gel phase DPPC bilayers, C5 is ≈10 Å from the 31P latitude and ≈12 Å from the bilayer center, i.e., C5 is located approximately midway across a single membrane leaflet (21). Although the cholesterol-rich membranes of the present study form a liquid-ordered rather than a gel phase bilayer, the acyl chain conformation is ordered in both phases with the C5 carbon midway between the 31P and the bilayer center (22). Fig. 5 shows the REDOR spectra at τ = 24 ms for samples labeled at Ala-1 and Ala-6 and plots of (ΔS/S0)exp vs. τ for Ala-6 samples. As a negative control, the sample with HFPmn-A1 (Fig. 5A) had (ΔS/S0)exp ≈ 0 which was consistent with the proximity of Ala-1 to the lipid phosphate groups deduced from the 13CO-31P REDOR data. Fig. 5E shows that among constructs labeled at Ala-6, HFPmn-A6 was the only one with nonzero (ΔS/S0)exp. Similar results were obtained for HFPmn-L9 (SI Text). These data along with (ΔS/S0)exp ≈ 0 for the 13CO-31P and 13CO-(16-19F) experiments support a membrane location for interior residues of HFPmn which is midway between the headgroups and the bilayer center, i.e., intermediate between HFPmn_mut and HFPtr (Fig. 3). This location correlates with the intermediate fusion rate of HFPmn.
Fig. 5.
13C S0 and S1 NMR spectra from 13CO-19F REDOR experiments of samples containing 5-19F-DPPC lipid. For A–D, the samples were respectively made with HFPmn-A1, HFPmn-A6, HFPmn_mut-A6, and HFPtr-A6. The dephasing time was 24 ms and each S0 or S1 spectrum was the sum of ≈20,000 scans. 13CO-19F REDOR dephasing curves for different HFP-A6 constructs are plotted in E and the construct symbols are shown in the legend. For 2 ms dephasing time, the typical uncertainty in (ΔS/S0)exp is ±0.02 and for the other dephasing times, the typical uncertainty is ±0.04.
Insertion Models for HFP Constructs and Quantitative Analysis of HFP Membrane Locations.
Fig. 3 shows experimentally-based membrane insertion models for HFPmn_mut, HFPmn, and HFPtr in antiparallel β sheet structure. The β strand conformation was supported by the 13CO peak chemical shifts for the labeled residues (Table S1) and the antiparallel β sheet structures for HFPmn and HFPtr were based on previous experiments (14, 20). The depths of insertion follow the trend that HFPmn_mut < HFPmn < HFPtr. As described in SI Text, quantitative analysis of the REDOR data were done by first removing the natural abundance 13CO contributions and then fitting the remaining (ΔS/S0)lab which represent the signals of only the labeled 13COs. The fitting model was two populations of spin pairs (e.g., two 13CO-31P or two 13CO-19F pairs) with one pair having fractional population f and magnitude of dipolar coupling d > 0 and the other pair having fractional population 1 − f and d = 0. For a single spin pair, d is quantitatively related to the internuclear distance r by a r−3 dependence. The existence of the 1 − f, d = 0 population is ascribed to 13CO nuclei in the β sheet interior that are far from any region of the membrane and for the 13CO-19F data, the dilute 19F spin density because of the small mol fraction of fluorinated lipid. Because each dataset only contained 4 or 5 points, it was not reasonable to fit the data to more sophisticated structural models, e.g., multiple 31P nuclei. The two spin pair model was at least reasonable as evidenced by typical best-fit χ2min < 5.
The SI includes tables that summarize the spin pair populations and best-fit internuclear distances for the f fractional populations. A summary of the quantitative data analysis includes: (i) For all HFPmn_mut samples, the best-fit 13CO-31P distances are in the 5.0–6.3-Å range. These data and reasonable values of van der Waals radii are consistent with close contact of the β sheet region of HFPmn_mut with the phosphate headgroups. (ii) For HFPmn and HFPtr samples labeled at Ala-1 or Ala-14, the best-fit 13CO-31P distances are in the 4.8 – 5.9-Å range with best-fit f > 0.7. For more interior β sheet residues, the (ΔS/S0)lab are small, e.g., Ala-6 or Leu-9, and could not be reliably fitted or the fitted distances are in the 8- to 10-Å range, e.g., Ile-4 or Leu-12. These data suggest membrane insertion of the Ile-4 to Leu-12 region of HFPmn and HFPtr with the termini of the β sheet, e.g., Ala-1 and Ala-14, in contact with the lipid headgroups (Fig. 3 B and C). For the HFPmn sample labeled at Ala-21, the best-fit 13CO-31P distance is 6.9 Å with f = 0.98 which suggests that the C-terminal region of HFPmn also contacts the headgroups. (iii) For all HFPmn_mut samples and most HFPmn and HFPtr samples, the 13CO-19F (ΔS/S0)lab are small and could not be reliably fitted. The exceptions are the samples containing 5-19F-DPPC lipid and HFPmn-A6 or HFPmn-L9 and the samples containing 16-19F-DPPC and HFPtr-A6 or HFPtr-L9. The best-fit 13CO-19F distances in these samples are in the 6.9 – 8.1-Å range and the best-fit f are in the 0.34 – 0.39 range. These analyses are consistent with partial membrane insertion of the interior β sheet residues of HFPmn and deeper insertion of HFPtr.
Discussion
Insertion Models of β Sheet HFP Constructs.
The membrane location of the HFP provides useful information to understand the perturbation of membranes and the catalysis of membrane fusion. The present study provides residue-specific membrane locations based on solid-state NMR experiments for three HFP constructs with very different fusogenicities. Insertion models for β strand HFPs will be discussed in the context of previous and present work. Earlier solid-state NMR studies on HFPmn showed that relative to the Gly-5 to Gly-13 13COs, the Ala-1 to Gly-3 and Ala-14 to Gly-16 13COs were closer to the lipid 31Ps (15, 19). Two insertion models were proposed with either insertion into a single leaflet or membrane traversal of both leaflets. Another study focused on the secondary and tertiary structure of HFPmn in membranes with physiologically relevant cholesterol content and supported the formation of small oligomers in an antiparallel β sheet structure with adjacent strand crossing near Phe-8 and Leu-9 (14). Therefore, Ala-1 to Gly-3 and Ala-14 to Gly-16 were close to one another in adjacent strands of the sheet and were at the termini of the sheet. It was therefore reasonable that both regions could be close to the phosphate groups. All of these results are consistent with the results of the present study and with the HFPmn insertion model present in Fig. 3B. Furthermore, the proximity of interior β sheet residues to 5-19F but not 16-19F lipid nuclei supports membrane insertion into a single leaflet rather than membrane traversal by HFPmn. This partial insertion model is also consistent with earlier fluorescence studies showing proximity of residue 8 of HFPmn to the middle region of a single leaflet (9, 10). In virus-cell fusion, partial fusion peptide insertion into the outer leaflet of the target cell membrane would likely perturb this leaflet and lead to increased lipid mixing with the viral membrane. Such lipid mixing is thought to be a prerequisite for formation of a fusion pore (1).
In Fig. 3, HFPmn_mut and HFPtr are also represented by antiparallel β sheet structure. Evidence for this structure includes: (i) earlier 13CO-15N REDOR measurements on HFPtr; (ii) peak 13CO chemical shifts in HFPmn_mut and HFPtr which are typically within 0.5 ppm of the corresponding shift of HFPmn (Table S1); and (iii) the large (ΔS/S0)exp for all constructs labeled at Ala-1 or Ala-14 as would be expected for the antiparallel β sheet structure with the strand termini near the phosphate groups (20). Predominant β sheet secondary structure was also observed in the infrared spectra of peptides with sequences close to that of HFPmn or HFPmn_mut and bound to membranes with large fractions of choline lipids and cholesterol (11). Unlike HFPmn, HFPmn_mut is primarily located on the membrane surface as evidenced by Figs. 2, 4, and 5, and by the quantitative distance analysis presented in SI Text. It is very interesting that the charged Glu-2 residue near the terminus of the β sheet induces a significant change in HFP membrane location. The HFPtr antiparallel β sheet is most reasonably described with a minimal unit of two HFPtr molecules “A” and “B” and adjacent antiparallel β strands arranged in an ABABAB structure. Because of the close contact of HFPtr Ala-6 and Leu-9 13COs with the 16-19F lipid nuclei, it is not possible to discount membrane traversal by HFPtr, i.e., molecules A and B on opposite sides of the membrane. However, the displayed model in Fig. 3C is more consistent with viral fusion in which multiple gp41 trimers would initially bind to the same outer leaflet of the target cell membrane. The Fig. 3C model also correlates with the membrane locations of HFPmn and HFPmn_mut. It is definitive that relative to HFPmn, HFPtr is more deeply inserted in the membrane. This may be related to formation of larger and more hydrophobic oligomers by HFPtr relative to HFPmn.
The present study focuses on membranes with biologically relevant cholesterol content in which all of the constructs have predominant β strand conformation. For HFPmn and HFPtr associated with membranes that do not contain cholesterol, there are significant populations of molecules with helical conformation (15). Helical conformation is also observed for HFPmn in detergent micelles (4, 12). There is a reasonable correlation between the location of β sheet HFPs in membranes and the current data on the locations of helical HFPs in membranes and micelles. One point of agreement between all of the data are that in either helical or β strand conformation, the Ala-14 and Ala-15 residues are near the membrane or micelle surface. These residues are on the border between the apolar and polar regions of the HFP sequence which approximately matches the polarity change at the membrane or micelle surface. The location of these residues appears to be an intrinsic property of the HFP sequence that is independent of conformation. For one model of helical HFPmn in a micelle, the Gly-5 to Ala-15 region traverses the micelle interior (12). This correlates with the similar membrane location of this region in β sheet HFPmn in the present study (Fig. 3B). Molecular dynamics simulations of a single helical molecule of HFPmn_mut or HFPmn in a membrane show a surface location or shallow insertion, respectively, which correlates with the β sheet HFPs of the present study (Fig. 3 A and B) (16).
Biological Relevance of the Membrane Location Studies.
Two requirements of virus-cell fusion are assembly of multiple fusion peptides and destabilization of the target cell membrane (1–3). The present study provides insight into these requirements including a possible link between them. There is a clear positive correlation between the depth of HFP membrane insertion and fusogenicity. The correlation can be understood by a second correlation between depth of membrane insertion and membrane destabilization. For insertion into a single leaflet, there will be perturbation in the packing of lipids near the HFPs which will likely destabilize this region of the leaflet and reduce the activation energy needed to form membrane fusion intermediate states such as stalks and fusion pores (1). It is reasonable that deeper insertion into a single leaflet will cause greater destabilization and therefore faster fusion rate which correlates with experimental observations for HFPmn_mut, HFPmn, and HFPtr.
For these three constructs, there may also be a positive correlation between number of molecules in the β sheet assembly and depth of membrane insertion. A larger assembly would likely be more hydrophobic and therefore more stable in the membrane interior. Evidence to support this hypothesis includes: (i) HFPtr has the deepest insertion and is preorganized into trimers; and (ii) HFPmn_mut has the shallowest insertion and relative to HFPmn and HFPtr, HFPmn_mut has the greatest helical population which is likely helical monomers (Fig. 1). Inhibition of HFP oligomeric assembly in HFPmn_mut may be due to charge repulsion between Glu-2 sidechains on different molecules. The HFP assembly/fusion correlation is also supported by virus-cell and cell-cell fusion studies with the gp41 V2E mutant. Viruses or cells expressing 91% wild-type gp41 and 9% gp41 V2E mutant had only 40% of the fusion activity of the corresponding system expressing 100% wild-type gp41 (2). This “transdominant” effect suggests that assembly of multiple wild-type HFPs is required for efficient fusion. Consider that “n” wild-type HFPs are needed in the oligomer so that (0.91)n = 0.4. The resulting n ≈ 10 is consistent with multiple HFPs and trimers at the fusion site as shown in Fig. 3.
In summary, the membrane locations have been determined for three different HFP constructs in membranes with biologically relevant cholesterol content. All three constructs adopt predominant β strand conformation for the N-terminal region and are less structured in the C-terminal region. HFPmn_mut is the least fusogenic construct and is located on the membrane surface. HFPmn has intermediate fusion rate and its Ile-4 to Leu-12 region is inserted into one leaflet of the bilayer. HFPtr is preassembled in the putative trimeric state of gp41 and is the most fusogenic construct with the deepest membrane insertion that extends to the bilayer center. This study therefore correlates membrane insertion depth in a single leaflet and fusion rate and this correlation may be a general structure-function model for enveloped virus fusion peptides. The correlation is reasonably understood in terms of destabilization of the lipid packing. In addition, the present work including use of 5-19F-DPPC lipid describes a general approach to study the membrane locations of specifically labeled peptides and proteins, and may also be applicable to more uniformly labeled systems with appropriately modified REDOR pulse sequences.
Materials and Methods
5-fluoropalmitic acid, was synthesized by using literature methods and details are provided in the SI. The 5-19F-DPPC lipid was synthesized by Avanti Polar Lipids using the 5-fluoropalmitic acid. The general protocols for peptide synthesis and NMR sample preparation have been described and a brief summary is provided in SI Text (15, 19). Solid-state NMR experiments were conducted on a 9.4 T spectrometer. Most of the experimental setup has been described previously and the new parts of the setup are detailed in SI Text (15, 19). Spectra were processed with 200- Hz Gaussian line broadening and polynomial baseline correction.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant AI47153.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907360106/DCSupplemental.
References
- 1.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed EO, Delwart EL, Buchschacher GL, Jr, Panganiban AT. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 4.Li YL, Tamm LK. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieva JL, Nir S, Muga A, Goni FM, Wilschut J. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: Different structural requirements for fusion and leakage. Biochemistry. 1994;33:3201–3209. doi: 10.1021/bi00177a009. [DOI] [PubMed] [Google Scholar]
- 6.Reichert J, et al. A critical evaluation of the conformational requirements of fusogenic peptides in membranes. Eur Biophys J. 2007;36:405–413. doi: 10.1007/s00249-006-0106-2. [DOI] [PubMed] [Google Scholar]
- 7.Yang R, Prorok M, Castellino FJ, Weliky DP. A trimeric HIV-1 fusion peptide construct which does not self-associate in aqueous solution and which has 15-fold higher membrane fusion rate. J Am Chem Soc. 2004;126:14722–14723. doi: 10.1021/ja045612o. [DOI] [PubMed] [Google Scholar]
- 8.Gordon LM, et al. The amino-terminal peptide of HIV-1 glycoprotein 41 interacts with human erythrocyte membranes: Peptide conformation, orientation and aggregation. Biochim Biophys Acta. 1992;1139:257–274. doi: 10.1016/0925-4439(92)90099-9. [DOI] [PubMed] [Google Scholar]
- 9.Agirre A, et al. Interactions of the HIV-1 fusion peptide with large unilamellar vesicles and monolayers. A cryo-TEM and spectroscopic study. Biochim Biophys Acta. 2000;1467:153–164. doi: 10.1016/s0005-2736(00)00214-5. [DOI] [PubMed] [Google Scholar]
- 10.Haque ME, Koppaka V, Axelsen PH, Lentz BR. Properties and structures of the influenza and HIV fusion peptides on lipid membranes: Implications for a role in fusion. Biophys J. 2005;89:3183–3194. doi: 10.1529/biophysj.105.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira FB, Goni FM, Muga A, Nieva JL. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: Dose and sequence effects. Biophys J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaroniec CP, et al. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 13.Brugger B, et al. The HIV lipidome: A raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiang W, Bodner ML, Weliky DP. Solid-state NMR spectroscopy of human immunodeficiency virus fusion peptides associated with host-cell-like membranes: 2D correlation spectra and distance measurements support a fully extended conformation and models for specific antiparallel strand registries. J Am Chem Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiang W, Weliky DP. HIV fusion peptide and its cross-linked oligomers: Efficient syntheses, significance of the trimer in fusion activity, correlation of β strand conformation with membrane cholesterol, and proximity to lipid headgroups. Biochemistry. 2009;48:289–301. doi: 10.1021/bi8015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamath S, Wong TC. Membrane structure of the human immunodeficiency virus gp41 fusion domain by molecular dynamics simulation. Biophys J. 2002;83:135–143. doi: 10.1016/S0006-3495(02)75155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddox MW, Longo ML. Conformational partitioning of the fusion peptide of HIV-1 gp41 and its structural analogs in bilayer membranes. Biophys J. 2002;83:3088–3096. doi: 10.1016/S0006-3495(02)75313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toke O, Maloy WL, Kim SJ, Blazyk J, Schaefer J. Secondary structure and lipid contact of a peptide antibiotic in phospholipid bilayers by REDOR. Biophys J. 2004;87:662–674. doi: 10.1529/biophysj.103.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiang W, Yang J, Weliky DP. Solid-state nuclear magnetic resonance measurements of HIV fusion peptide to lipid distances reveal the intimate contact of beta strand peptide with membranes and the proximity of the Ala-14-Gly-16 region with lipid headgroups. Biochemistry. 2007;46:4997–5008. doi: 10.1021/bi6024808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Z, Yang R, Bodner ML, Weliky DP. Conformational flexibility and strand arrangements of the membrane-associated HIV fusion peptide trimer probed by solid-state NMR spectroscopy. Biochemistry. 2006;45:12960–12975. doi: 10.1021/bi0615902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venable RM, Brooks BR, Pastor RW. Molecular dynamics simulations of gel (LβI) phase lipid bilayers in constant pressure and constant surface area ensembles. J Chem Phys. 2000;112:4822–4832. [Google Scholar]
- 22.Bloom M, Evans E, Mouritsen OG. Physical properties of the fluid lipid-bilayer component of cell membranes: A perspective. Q Rev Biophys. 1991;24:293–397. doi: 10.1017/s0033583500003735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.