Abstract
Oxidative stress has been implicated in diverse disease states and aging. To date, induction of cellular responses to combat oxidative stress has been characterized largely at the transcriptional level, with emphasis on Nrf2-mediated activation of antioxidant response elements. In this study, we demonstrate that OLA1, a novel Obg-like ATPase, functions as a negative regulator of the cellular antioxidant response independent of transcriptional processes. Knockdown of OLA1 in human cells elicited an increased resistance to oxidizing agents including tert-butyl hydroperoxide (tBH) and diamide without affecting cell proliferation, baseline apoptosis, or sensitivity to other cytotoxic agents that target the mitochondria, cytoskeleton, or DNA. Conversely, overexpression of OLA1 increased cellular sensitivity to tBH and diamide. When challenged with oxidants, OLA1-knockdown cells had decreased production of intracellular reactive oxygen species and exhibited less depletion of reduced glutathione. Surprisingly, knockdown of OLA1 caused only minimal genomic response; no changes were found in the mRNA levels of genes encoding antioxidant enzymes, enzymes that produce antioxidants (including glutathione), or other genes known to respond to Nrf2. Moreover, when de novo protein synthesis was blocked by cycloheximide in OLA1-knockdown cells, they continued to demonstrate increased resistance to both tBH and diamide. These data demonstrate that OLA1 suppresses the antioxidant response through nontranscriptional mechanisms. The beneficial effects observed upon OLA1-knockdown suggest that this regulatory ATPase is a potential novel target for antioxidative therapy.
Keywords: drug target, oxidative stress, posttranslational regulation
Oxidative stress, caused by an imbalance between the production of reactive oxygen species (ROS) and the antioxidant defense, plays a key role in the pathogenesis of many chronic diseases including neurodegenerative disorders, atherosclerosis, cancer, aging, and acute tissue injuries due to ischemia-reperfusion, trauma, and radiation (1–3). Over the past few decades, a variety of antioxidants, such as natural antioxidant supplements (vitamins C and E) and synthetic free radical scavengers/trappers, have been tested for general or specific therapeutic benefits (4, 5). However, the results of these studies have been generally inconclusive and even contradictory, especially in recent clinical trials (5–8). These exogenously introduced antioxidants may fail to remove intracellular ROS efficiently or require higher than affordable doses to be effective.
An alternative strategy for increasing antioxidant potential is to augment the intrinsic antioxidant defense systems (9, 10). This approach is dependent on understanding the regulatory mechanisms of the cellular antioxidant response. Induction of antioxidant enzymes upon oxidative stress has been widely recognized as a genomic adaptive response, ultimately regulated via transcription (11–13). Although transcription factors including AP-1 and NF-κB have been implicated, the antioxidant response element (ARE)binding factor Nrf2 is thought to be a master regulator of antioxidant genes (12–14). A number of dietary chemopreventive compounds and synthetic compounds have been found to activate the Nrf2 pathway, in part by interrupting the interaction between Nrf2 and its cytosol inhibitor Keap1 (15–17). However, the specificity of these compounds is limited (18). Additionally, since Nrf2 also regulates the ARE-driven phase 2 detoxifying genes, inducers of Nrf2 have shown potential side effects, such as rendering cancer cells chemoresistant (19).
Although classical genomic responses are clearly indispensable during prolonged oxidative stress, it has long been hypothesized that posttranslational regulation must be important for survival responses during early phases of oxidant stress (20, 21), allowing cells to respond rapidly to an oxidative challenge without requiring de novo protein synthesis. Nonetheless, only a few examples of translational and posttranslational regulation of antioxidant enzymes are available (20–23), and studies on instant adaptation to oxidative stress or “express” antioxidant systems are limited. Accordingly, unidentified effectors of the oxidative response, acting via posttranslational mechanisms, may exist in the human proteome. The elucidation of these molecular regulators may reveal novel insights into the understanding of cellular defense systems as well as expose new therapeutic targets for antioxidant drugs.
Previously we selected a group of 32 mouse genes that were either up- or down-regulated during a progressive glutathione (GSH) deficiency generated in glutamate-cysteine ligase catalytic subunit (GCLC)-knockout cell lines incapable of synthesizing GSH (24, 25). Database analysis suggested that these genes fell within the functional categories of cell cycle, apoptosis, metabolism, and unknown function. In the present study, we have performed functional characterizations on one of the gene products, originally named GTPBP9 or PTD004, and recently defined as a new member of the Obg-like ATPase family [OLA1, (26)] in human cells. We now show that OLA1 is a negative regulator of the antioxidative response. Knockdown of OLA1 produces notable beneficial effect in defending cells against thiol-depleting agents and peroxide oxidants. We also demonstrate that OLA1 acts independently of gene transcription, which is distinctive from Nrf2-ARE and other known antioxidant mechanisms that are mediated by a genomic response. Our study suggests that this novel Obg-like ATPase could be a suitable target for enhancing redox protection.
Results
Identification of OLA1.
The mouse orthologue of OLA1 was initially identified in a microarray analysis as being up-regulated under the condition of progressive GSH depletion (25). The human OLA1 protein has a molecular weight of 45 kDa consisting of 396 aa and is expressed in the cytoplasm (27). We first confirmed the ubiquitous expression of OLA1 among 17 human cancer cell lines (supporting information (SI) Fig. S1A) including HeLa cells and one noncancerous cell line (BEAS-2B). In HeLa cells, OLA1 protein was up-regulated in response to moderate doses of H2O2 (Fig. S1B). This result is consistent with a previous report that identified the yeast homologue of OLA1, YBR025c, as an H2O2-inducible gene (28).
Knockdown of OLA1 Renders Cells More Resistant to Oxidative Agents.
To define possible biological functions of OLA1, we analyzed human cells in which OLA1 is specifically knocked down with the SMARTPool siRNAs. Reduced levels of OLA1 were confirmed by both RT-PCR and Western immunoblot analyses. At 24 to 48 h after the siRNA transfection, both HeLa and BEAS-2B cells showed a >75% decrease in OLA1 mRNA (Fig. S2). Moreover, at 48 to 72 h posttransfection, OLA1 protein levels decreased by more than 80% in these cells (Fig. 1). The OLA1-knockdown cells (both HeLa and BEAS-2B) had normal morphology and growth characteristics. The HeLa cells were exposed to a variety of cytotoxic compounds (Table 1), and cytotoxicity was recorded as both cell viability curves and ED50 values (concentrations yielding 50% viability). OLA1-knockdown cells were more resistant to the peroxides H2O2 and tBH, as well as the GSH-depleting agents diamide and dithiodipyridine, as compared to the control siRNA-transfected cells (Table 1). However, OLA1-knockdown cells showed no altered resistance to other stressors such as the ER stressors brefeldin A and tunicamycin, the lipoperoxidation inducer arachidonic acid, the cytoskeleton-damaging agent colchicine, and the DNA-targeting chemotherapeutic agents etoposide and doxorubicine. Interestingly, knockdown of OLA1 did not change the cytotoxicity caused by antimycin A and rotenone, which are mitochondrial electron transport chain targeting agents thought to generate endogenous ROS and initiate apoptosis. Resistance to H2O2 was seen only with serum-free medium. This may be due to the instability of H2O2 or due to the fact that H2O2 is a much weaker GSH-depleting agent than tBH and diamide (29, 30). Therefore, in subsequent experiments we used tBH and diamide as representative oxidative stressors. We found that resistance to tBH and diamide occurred as early as 5 h, as reflected by both increased viability (Fig. S3) and a better sustained morphology (Fig. 1A). After tBH or diamide treatment, most of the control cells exhibited rounded, swollen, or shrunken morphologies. In comparison, only a few OLA1-knockdown cells exhibited these features of cytotoxicity. Additionally, we tested cytotoxicity in the noncancerous BEAS-2B lung epithelial cell line (Fig. 1B). Correspondingly, OLA1-knockdown in BEAS-2B cells resulted in significantly increased resistance to tBH and diamide but not to antimycin A.
Fig. 1.
Knockdown of OLA1 renders cells more resistant to oxidative agents and overexpression of OLA1 sensitizes cells to them. (A) Morphological changes in OLA1-knockdown HeLa cells treated with tBH and diamide. Forty-eight hours after the siRNA transfection, cells were exposed to the indicated agents. Microscopic images were taken 5 h posttreatment. (Upper Inset) The effectiveness of the knockdown was evaluated by Western blot analysis; β-actin was used as the loading control. (B) Cytotoxicity assays in OLA1-knockdown BEAS-2B cells. The cells (48 h after the siRNA transfection) were treated with the indicated compounds for 5 h. ED50 values of each compound are presented (mean ± SD; n = 4). (Upper Inset) knockdown of OLA1 in BEAS-2B cells as evidenced by Western blot analysis. (C) Cytotoxicity assays in HeLa cells transfected with the OLA1-overexpression (Ola1) and the control plasmid. Twenty-four hours after the transfection, cells were exposed to different doses of tBH, diamide, and antimycin A for 5 h. ED50 values of each compound are presented (mean ± SD; n = 4). (Upper Inset) Western blot analysis at 24 h posttransfection showing overexpression of OLA1 in HeLa cells. *, P < 0.05; **, P < 0.01, as compared with control cells.
Table 1.
ED50 (μM) of cytotoxic compounds on the OLA1-knockdown cells (HeLa)
| Treatment* | Control siRNA | Ola1 siRNA |
|---|---|---|
| General oxidants (peroxides) | ||
| Tert-butylhydroperoxide | 225.66 ± 35.08 | 473.51 ± 42.72** |
| H2O2† | 935.97 ± 116.95 | 1505.23 ± 444.03** |
| Mitochondria-mediated ROS production | ||
| Antimycin A | 288.90 ± 4.85 | 298.36 ± 17.83 |
| Rotenone | 104.36 ± 4.54 | 111.47 ± 1.91 |
| Lipid peroxidation | ||
| Arachidonic acid | 325.50 ± 7.80 | 337.29 ± 16.99 |
| ER stress | ||
| Brefeldin A | 2.73 ± 0.35 | 2.46 ± 0.76 |
| Tunicamycin | 21.34 ± 2.75 | 23.26 ± 1.95 |
| Thiol depletion | ||
| Diamide | 367.20 ± 17.87 | 634.75 ± 7.47** |
| Dithiodipyridine | 93.98 ± 13.30 | 170.16 ± 3.43** |
| DNA–targeting chemotherapeutics | ||
| Doxorubincine | 2.14 ± 0.14 | 2.10 ± 0.08 |
| Etoposide | 111.10 ± 11.90 | 112.08 ± 6.84 |
| Cytoskeleton inhibition | ||
| Colchicine | 40.64 ± 1.19 | 40.15 ± 0.65 |
*Forty-eight hours after siRNA transfection, cells were treated with the listed compounds in complete medium for 24 h (except H2O2), and assayed for cell viability using the MTS assay. Data are shown as mean ± SD (n = 3 ∼ 6).
†Cells were exposed to H2O2 in serum-free medium for 5 h.
**, P < 0.01, as compared with control cells.
Overexpression of OLA1 Sensitizes Cells to Oxidant-Induced Cytotoxicity.
HeLa cells were transfected with the wild-type OLA1 sequence to induce transient overexpression. Western blot analysis confirmed an approximately 2-fold increase in OLA1 protein expression in these cells compared to cells transfected with the control plasmid (Fig. 1C). When the OLA1-overexpressing cells were treated with tBH or diamide, they showed increased cytotoxicity relative to the control cells. However, overexpression of OLA1 did not change the cell sensitivity to antimycin A. These results are consistent with the siRNA knockdown experiments, and indicate that our initial observations with siRNA were not attributable to off-target effects.
Cells Deficient in OLA1 Have Lower ROS.
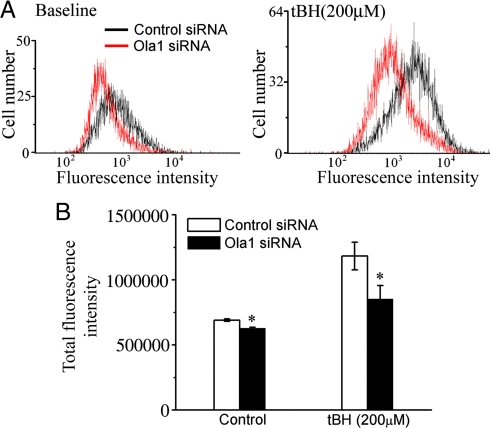
We next investigated whether resistance to oxidizing agents in OLA1-knockdown cells is associated with a decrease of intracellular ROS. The fluorescent probe DCFDA was used to monitor the production of ROS (Fig. 2). HeLa cells treated with OLA1 siRNA exhibited slightly lower fluorescence (−10.2%, P < 0.01), indicating a decreased baseline production of ROS (Fig. 2). Further, when cells were briefly challenged with tBH (200 μM, 1 h), ROS levels were found to rise in both the control cells and the OLA1-knockdown cells. However, the latter demonstrated a significantly smaller increase (36% vs. 71.4%), indicating that the OLA1-knockdown cells produced less ROS. This phenomenon of decreased ROS accumulation was also seen in OLA1-knockdown BEAS-2B cells after tBH challenge (Fig. S4).
Fig. 2.
Knockdown of OLA1 attenuated the production of ROS. Two days after the siRNA transfection, HeLa cells at baseline (without a challenge) and cells treated with 200 μM of tBH for 1 h were subjected to carboxy-H2DCFDA labeling and flow cytometry analysis. (A) Representative histograms of DCF fluorescence. (B) Quantitative analysis of the total DCF fluorescence intensity. The results are displayed as the mean of triplicate samples ± SD. *, P < 0.01, as compared with control cells.
Cells Deficient in OLA1 Are More Resistant to Thiol-Depletion.
GSH is the most abundant cellular nonprotein thiol, and plays a central role in maintaining cellular redox status and protecting cells from oxidative injury. Intracellular levels of reduced GSH in OLA1-knockdown cells were measured with a bioluminescence-based assay. At baseline, OLA1-knockdown and control cells had similar total GSH levels (Fig. 3). However, after treatment with 400 μM tBH for 1 h, a substantial reduction of cellular GSH was seen in the control cells, while the OLA1-knockdown cells showed no loss of GSH (Fig. 3A). Similarly, treatment with 400 μM diamide for 1 h resulted in a dramatic decrease in cellular GSH in the control cells, while only a moderate depletion of GSH was observed in the OLA1-knockdown cells (Fig. 3A). Flow cytometry was used to measure total thiol content (including GSH and protein thiol) labeled with monobromobimane (31). While the total thiol content was significantly decreased (−24%) in the control cells due to the treatment of 200 μM tBH, no such change was seen in OLA1-knockdown cells (Fig. 3B). Note that both GSH and thiol assays were performed at an early time point (1 h), at which they primarily reflect cellular susceptibility to the thiol depletion, rather than secondary effects (e.g., leakage) caused by cell death that may occur at later time points.
Fig. 3.
Knockdown of OLA1 protects cells against GSH depletion. (A) Total cellular GSH. Forty-eight hours after the siRNA transfection, HeLa cells were treated with tBH or diamide for 1 h, and intracellular GSH was measured by a bioluminescence-based assay. The GSH-induced signals in the microplate were imaged using a Xenogen IVIS200 optical imaging system. Representative images are shown with a color bar indicating the luminescence counts (Far Left). The experiment was repeated 2 more times with similar results. (B) Total intracellular thiol content. Transfected HeLa cells were treated with tBH and labeled with monobromobimane for flow cytometry analysis. Representative histograms are shown. The insets next to the histograms are the quantitative analysis of the mean fluorescence intensity (mean ± SD, n = 3). *, P < 0.05; **, P < 0.01, as compared with the “0” dose treatment; “n.s.,” statistically not significant.
Knockdown of OLA1 Caused No Changes in Cell Proliferation or Baseline Cell Death.
A simple cell counting method was used to monitor cell growth. There was no difference in cell growth between OLA1-knowndown and control cells up to 4 days after the siRNA transfection (Fig. 4A). Further, siRNA-transfected cells were grown for 48 h and then stained with propidium iodide (PI) for cell cycle analysis by flow cytometry. Knockdown of OLA1 did not produce any alteration in the cell cycle (Fig. 4B). We then tested whether knockdown of OLA1 affects baseline cell death. Cells transfected with OLA1 siRNA or control siRNA were stained with both Annexin V and PI and processed for flow cytometry. At 48 h posttransfection, OLA1-knockdown cells did not show any notable differences in cell death compared to control cells (Fig. 4C).
Fig. 4.
Knockdown of OLA1 has no effect on baseline cell growth and death in HeLa cells. (A) Cell growth rate as monitored by simple cell counting following the siRNA transfection. Data are represented by mean ± SD from 3 independent preparations. (B) Cell cycle analysis: 48 h after transfection with the control siRNA or the Ola1 siRNA, the cells were stained with PI and analyzed by flow cytometry. The percentages of cells in each cell cycle phase are shown in the inset above the histogram (mean ± SD; n = 3). Note the y axis is truncated at 1548 events. (C) Baseline cell death, as evaluated by the Annexin V/PI staining at 48 h after the siRNA transfection. Percentages of cells in each quadrant (Q1: viable, Q2: early apoptosis, Q3: late apoptosis/necrosis, and Q4, necrosis) are annotated.
Knockdown of OLA1 Does Not Induce Significant Alterations in Gene Expression.
To explore the underlying molecular basis for the OLA1 knockdown-mediated antioxidative protection, we compared gene expression profiles between OLA1-knockdown cells and control cells using the Agilent two-color microarray technology at 48 h after transfection. Two independent experiments with 4 dye-swap hybridizations were performed. A total of 13 differentially expressed genes was identified. Only one gene (CCL2) had increased expression in OLA1-knockdown cells as compared to the control cells with a fold change of 1.48. On the other hand, OLA1 (the siRNA target) and 11 other genes showed decreased transcription in OLA1-knockdown cells with fold changes ranging from 0.1 to 0.71 (see Table S1 for details). The up-regulated gene CCL2 encodes the monocyte chemoattractant protein (MCP)-1, a cytokine under the control of NF-κB. The most down-regulated gene (besides OLA1), arylacetamide deacetylase-like 1 (AADACL1), is an enzyme that regulates lipid signaling pathways by hydrolyzing 2-acetyl monoalkylglycerol (32). Based on function, these 2 genes are most likely not responsible for the observed phenotypes. Similarly, none of the other 10 down-regulated genes seem to account for the decreased cytotoxicity of OLA1-knockdown cells following oxidative challenge. The original data sets were also explored manually for genes that are known to encode antioxidant enzymes, Nrf2-responsive phase 2 enzymes, as well as enzymes that produce or regenerate antioxidants, including GSH, but none could be validated as up-regulated genes. RT-PCR was carried out on genes GCLC and glutamate-cysteine ligase modifier subunit (GCLM) in independent experiments. The results showed no notable mRNA level changes in both genes at 24, 36, 48, and 72 h after the OLA1 siRNA transfection (Fig. S5).
OLA1-Knockdown-Mediated Cellular Protection Is Independent of de Novo Protein Synthesis.
Since the gene expression profiling was performed in the absence of an oxidative challenge, it was conceivable that oxidative stress induced a differential genomic response that might contribute to the phenotype. We investigated whether cycloheximide (CHX), an inhibitor of protein synthesis, attenuated the increased oxidative resistance of OLA1-knockdown cells. HeLa cells were preincubated for 1 h with 20 μg/ml of CHX, a concentration known to inhibit protein translation by approximately 95% but have no notable cytotoxicity by itself (33, 34). The cells were then treated with tBH or diamide for another 5 h (in the presence of CHX) and cytotoxicity was measured. OLA1-knockdown cells exhibited a significantly increased resistance to both stressors, independent of the presence of CHX (Fig. 5A). To confirm that CHX treatment resulted in sufficient inhibition of protein synthesis, cells were collected at 4 h post-CHX treatment and protein levels of p53, a protein known to have short half-life (35), were analyzed by Western immunoblot. Indeed, levels of p53 were reduced dramatically following CHX treatment (Fig. 5B). These data demonstrate that de novo protein synthesis is neither required nor responsible for the resistant phenotype exhibited by the OLA1-knockdown cells.
Fig. 5.
Cytotoxicity analysis in OLA1-knockdown cells pretreated with protein synthesis inhibitor cycloheximide (CHX). (A) HeLa cells were pretreated with CHX (20 μg/ml, 1 h) and then the oxidants tBH or diamide were added (in the presence of CHX) for an additional 5 h; cells were then assayed for viability (MTS). Data represent ED50 values of tBH and diamide with or without the cotreatment of CHX (means ± SD, n = 3–5). **, P < 0.01, as compared with control cells. (B) Western blot analysis of OLA1 and p53 proteins in the cells treated with CHX (20 μg/ml, 4 h). The OLA bands are significantly decreased in the “Ola1 siRNA” transfected cells compared to the “Control siRNA” transfected cells, indicating a successful knockdown of the OLA1 protein (Top); suppression of the p53 bands in the CHX (+) group suggests that the CHX blocked protein synthesis successfully (Middle); β-actin was used as loading control (Bottom).
Discussion
OLA1 Is a Negative Regulator of the Antioxidant Response.
OLA1 belongs to the YchF subfamily of Obg-like GTPases containing a guanine nucleotide-binding domain (G domain) commonly found in p-loop GTPases. However, recent biochemical analyses (26, 36) have determined that YchF proteins bind and hydrolyze ATP more efficiently than GTP. In OLA1, the central G domain is flanked on either side by a coiled-coil domain and a ThrRS, GTPase, SpoT (TGS) domain of unknown function (26). Based on these structural features, OLA1 is predicted to be a regulatory protein that interacts with downstream effector protein(s) and exerts its downstream functions by the conformational switch between the ADP- and ATP-bound forms (26). To date, physiological functions of this subclass of ATPases are poorly understood. The yeast YBR025c/Ola1 has been identified as one of the proteasome-interacting proteins associated with the 26S proteasome (37, 38). However, a similar role for human OLA1 has not yet been established (39). The yeast YBR025c/Ola1 and human OLA1 are both inducible by H2O2 (28, Fig. S1B). More recently, the protozoan TcYchF/Ola1 has been shown to interact with the proteasome subunit RPN10, ribosomal subunits, and polysomes (36), suggesting a function in the regulation of protein translation and degradation.
Our results demonstrate that knockdown of OLA1 significantly attenuates the cytotoxicity induced by peroxide oxidants and thiol-depleting agents without affecting the cellular susceptibility to other forms of oxidative stressors and cytotoxic agents (Table 1 and Fig. 1). Knockdown of OLA1 results in decreased ROS production when cells are challenged with oxidants (Fig. 2) while concomitantly increasing cellular resistance to GSH depletion (Fig. 3). Thus, OLA1 is a negative regulator of the antioxidant response, yet one that appears to act upon only specific aspects of the response, likely the regulation of cellular thiols.
OLA1 Functions via Nontranscriptional Mechanisms.
Given the considerable cellular effects of OLA1 knockdown, we were surprised to find that OLA1 knockdown affected the expression of very few genes (Table S1); of these, none encoded known antioxidant or other cytoprotective enzymes, such as members of the Nrf2-ARE pathway. Perhaps more interesting was the fact that the inhibition of protein synthesis with CHX did not blunt the improved resistance of OLA1-knockdown cells to oxidative stress (Fig. 5). This strongly suggests that the improved resistance seen in OLA1-knockdown cells is due to posttranslational mechanisms, although we cannot completely rule out events at the level of protein translation that may have occurred following siRNA knockdown of OLA1 and before CHX treatment. We propose 2 mechanisms by which OLA1 knockdown may prepare cells for an oxidative challenge. Firstly, OLA1 may function through protein translation or posttranslational protein degradation pathways to suppress the levels of antioxidant enzymes. Knockdown of OLA1 would release this negative regulation, thus increasing the concentration of antioxidant enzymes and the cellular antioxidant potentials without showing changes at the mRNA level. Alternatively, OLA1 may be a suppressive cofactor or inhibitor of one or more antioxidant enzymes, likely those responsible for thiol redox balance. Consequently, inactivation of this inhibitor would result in the enhancement of the antioxidant response in the absence of gene transcription or protein synthesis.
In the majority of previous studies, the regulation of the antioxidant response has been attributable to transcriptional mechanisms. However, there are examples of nontranscriptional regulation. Ufer et al. (23) reported that expression of selenoprotein GPx4 is regulated at the translational level by the guanine-rich sequence-binding factor 1 that recognizes and recruits m-GPx4 mRNA to translationally active polysomes to enhance translation. Moreover, posttranslational regulation of enzymatic activity has been demonstrated in S. cerevisiae strains in response to oxidative challenge (20); when protein synthesis is blocked with CHX, SOD1 activity can be rapidly induced by activation of a preexisting pool of SOD1 protein via interaction with the copper chaperone CCS. Our present study on OLA1 provides new evidence that a regulatory factor of the mammalian antioxidant response acts independently of the genomic response pathways. This finding sheds light on a new therapeutic opportunity for certain clinical conditions, such as acute radiation injury and tissue injuries caused by ischemia and reperfusion. Under these conditions, up-regulation of the antioxidant response is immediately needed for survival, but induction of the adaptive response through transcriptional pathways could miss the therapeutic window.
The role of OLA1 as a novel negative regulator of the antioxidant response has potential therapeutic implications. Although positive antioxidant factors (e.g., antioxidant enzymes) have been proposed as therapeutic targets (40, 41), the development of compounds that directly stimulate enzymatic activity is challenging. Conversely, compounds that target OLA1 or other suppressive factors of the response may be more highly desirable; ultimately serving to boost the antioxidant response by “inhibiting the inhibitor.” In general, ATPases are attractive drug targets, and a number of ATPase inhibitors are already on the market (42). Consequently, OLA1, as a p-loop ATPase, would appear to be druggable. Additionally, the crystal structure of human OLA1, which was recently solved (26), may facilitate the structure-based design of small molecular inhibitors. Moreover, knockdown of OLA1 protects cells from specific types of oxidative stress, likely the thiol-oxidizing agents, but does not affect baseline cell growth and apoptosis (Fig. 4). These findings suggest that targeting of OLA1 may have a specific therapeutic effect on the antioxidant defense system, with a low probability of adverse side effects.
Materials and Methods
Chemicals and Cell Culture.
All chemicals used in this study were purchased from Sigma-Aldrich. Human cell lines HeLa and BEAS-2B (ATCC) were grown at 37 °C in DMEM supplemented with 10% FBS.
siRNA-Mediated Gene Knockdown.
Human OLA1 (NM_013341.3)-specific siRNAs (On-Target Plus SMARTpools, #L-015680–01) and the control siRNA (nontargeting siRNA, #D-001810–10) were acquired from Dharmacon. Cells seeded in 6-well plate were transiently transfected with 50 nM siRNA with the DharmaFECT1 siRNA Transfection Reagent (#T-2001–03) according to the manufacturer's instructions.
Transient Overexpression.
Cells in 6-well plates were transfected with pdEYFP-N1gen-OLA1 plasmid (4 μg/well) expressing the full-length OLA1 cDNA or the pdEYFP-N1gen plasmid expressing YFP using Lipofectamine 2000 (10 μl/well, Invitrogen). The parental plasmids were a kind gift from Stefan Wiemann of European Molecular Biology Laboratory, Heidelberg, Germany (27).
Cytotoxicity/Viability Assay.
Cells were seeded in a 96-well microplate at 1 × 104 cells/well; the following day cells were treated with chemical agents as indicated in Results. Cell viability was evaluated by the MTS-based CellTiter 96 Aqueous One Solution Reagent (Promega).
Intracellular ROS Measurement.
The measurement was obtained using the molecular probe 5-(and 6)-carboxy-2′, 7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCF-DA, Invitrogen). Cells were loaded with carboxy-H2DCFDA (10 μM in phenol-free medium) for 30 min and treated with 200 μM tBH for 1 h. Cells were then rinsed with PBS and evaluated for DCF fluorescence by flow cytometry (FACscan, Becton-Dickinson) using 492 nm for excitation and 520 nm for emission. A total of 10,000 events were collected for each histogram.
Intracellular GSH/Thiol Measurement.
GSH was assessed using a GSH-GLOTM kit (Promega). Luminescence was measured using a FLUOstar OPTIMA microplate reader (BMG LABTECH), and also imaged with a Xenogen IVIS200 optical imaging system with the Living Image 3.0 Software. To determine intracellular total thiol content (31), cells were suspended in PBS and labeled with the monobromobimane probe (Invitrogen) at a final concentration of 1 μM at 37 °C for 10 min, then immediately analyzed by flow cytometry (LSR II cytometer, BD) using 405 nm for excitation and 421 nm for emission. Data were collected from at least 10,000 cells.
Proliferation, Cell Cycle, and Apoptosis.
A manual trypan blue hemocytometer counting method was used to assess cell growth rate. Cell cycle was determined by flow cytometry analysis using PI staining (BD Pharmigen). For apoptosis analysis, cells were stained using the Annexin V-FITC Apoptosis Detection Kit (BD Pharmigen). In both experiments, cell fluorescence was analyzed using a BD LSR II cytometer.
Microarray-Based Gene Expression Analyses.
Total RNAs were extracted from siRNA-transfected HeLa cells using TRIzol reagent (Invitrogen) and further purified using an RNeasy Mini Kit (QIAGEN). Samples were sent to the Microarray Core Lab at Baylor College of Medicine for microarray assays using the Agilent 2-color hybridization platform (see SI Materials and Methods for procedure).
Quantitative RT-PCR.
The RT-PCR was carried out using the iScript one-step RT-PCR kit with SYBR Green reagent (Bio-Rad) in a Miniopticon thermal cycler (Bio-Rad), following the manufacturer's protocol. The mRNA level was analyzed by the relative quantification method using Ct values for the GAPDH as an internal reference. See SI Materials and Methods for primer sequences.
Western Blot Analysis.
Immunoblot analysis was performed according to standard procedures (SI Materials and Methods). Antibodies used in these studies were purchased from the following suppliers: OLA1 from Abcam Inc., p53 and β-actin from Sigma-Aldrich, and anti-rabbit IgG peroxidase linked whole antibody and anti-mouse Ig peroxidase linked whole antibody from GE Healthcare.
Supplementary Material
Acknowledgments.
This work is support by the Methodist Hospital Research Institute Faculty Seed Fund to Z.-Z.S. We thank Jennifer Jacobs and Sriram Bala for technical assistance, and Kathryn Stockbauer and Kevin Phillips for comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907213106/DCSupplemental.
References
- 1.Seis H. Oxidative Stress: Oxidants and Antioxidants. London: Academic; 1991. [Google Scholar]
- 2.Valko M, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Hensley K, Floyd RA. Reactive oxygen species and protein oxidation in aging: A look back, a look ahead. Arch Biochem Biophys. 2002;397:377–383. doi: 10.1006/abbi.2001.2630. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B. Drug antioxidant effects. A basis for drug selection? Drugs. 1991;42:569–605. doi: 10.2165/00003495-199142040-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slemmer JE, Shacka JJ, Sweeney MI, Weber JT. Antioxidants and free radical scavengers for the treatment of stroke, traumatic brain injury and aging. Curr Med Chem. 2008;15:404–414. doi: 10.2174/092986708783497337. [DOI] [PubMed] [Google Scholar]
- 6.Diener HC, et al. NXY-059 for the treatment of acute stroke: Pooled analysis of the SAINT I and II Trials. Stroke (Dallas) 2008;39:1751–1758. doi: 10.1161/STROKEAHA.107.503334. [DOI] [PubMed] [Google Scholar]
- 7.Bjelakovic G, Nikolova D, Gluud LL, Simonetti RG, Gluud C. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: Systematic review and meta-analysis. J Am Med Assoc. 2007;297:842–857. doi: 10.1001/jama.297.8.842. [DOI] [PubMed] [Google Scholar]
- 8.Sesso HD, et al. Vitamins E and C in the prevention of cardiovascular disease in men: The Physicians' Health Study II randomized controlled trial. J Am Med Assoc. 2008;300:2123–2133. doi: 10.1001/jama.2008.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent AM, Edwards JL, Sadidi M, Feldman EL. The antioxidant response as a drug target in diabetic neuropathy. Curr Drug Targets. 2008;9:94–100. doi: 10.2174/138945008783431754. [DOI] [PubMed] [Google Scholar]
- 10.Schreibelt G, et al. Therapeutic potential and biological role of endogenous antioxidant enzymes in multiple sclerosis pathology. Brain Res Rev. 2007;56:322–330. doi: 10.1016/j.brainresrev.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Crawford DR, Davies KJ. Adaptive response and oxidative stress. Environ Health Perspect. 1994;102(Suppl 10):25–28. doi: 10.1289/ehp.94102s1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osburn WO, Kensler TW. Nrf2 signaling: An adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 14.Lee JM, et al. Nrf2, a multi-organ protector? FASEB J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 15.Wakabayashi N, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 17.Liby K, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 18.Juge N, Mithen RF, Traka M. Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell Mol Life Sci. 2007;64:1105–1127. doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang XJ, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown NM, Torres AS, Doan PE, O'Halloran TV. Oxygen and the copper chaperone CCS regulate posttranslational activation of Cu,Zn superoxide dismutase. Proc Natl Acad Sci USA. 2004;101:5518–5523. doi: 10.1073/pnas.0401175101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusarov I, Nudler E. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci USA. 2005;102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuji Y, et al. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol Cell Biol. 2000;20:5818–5827. doi: 10.1128/mcb.20.16.5818-5827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ufer C, et al. Translational regulation of glutathione peroxidase 4 expression through guanine-rich sequence-binding factor 1 is essential for embryonic brain development. Genes Dev. 2008;22:1838–1850. doi: 10.1101/gad.466308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi ZZ, et al. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc Natl Acad Sci USA. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas E, et al. Cell survival and changes in gene expression in cells unable to synthesize glutathione. Biofactors. 2003;17:13–19. doi: 10.1002/biof.5520170102. [DOI] [PubMed] [Google Scholar]
- 26.Koller-Eichhorn R, et al. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J Biol Chem. 2007;282:19928–19937. doi: 10.1074/jbc.M700541200. [DOI] [PubMed] [Google Scholar]
- 27.Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, et al. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 29.Dierickx PJ, Nuffel GV, Alvarez I. Glutathione protection against hydrogen peroxide, tert-butyl hydroperoxide and diamide cytotoxicity in rat hepatoma-derived Fa32 cells. Hum Exp Toxicol. 1999;18:627–633. doi: 10.1191/096032799678839482. [DOI] [PubMed] [Google Scholar]
- 30.Alia M, Ramos S, Mateos R, Bravo L, Goya L. Response of the antioxidant defense system to tert-butyl hydroperoxide and hydrogen peroxide in a human hepatoma cell line (HepG2) J Biochem Mol Toxicol. 2005;19:119–128. doi: 10.1002/jbt.20061. [DOI] [PubMed] [Google Scholar]
- 31.Hedley DW, Chow S. Evaluation of methods for measuring cellular glutathione content using flow cytometry. Cytometry. 1994;15:349–358. doi: 10.1002/cyto.990150411. [DOI] [PubMed] [Google Scholar]
- 32.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Ursini MV, Parrella A, Rosa G, Salzano S, Martini G. Enhanced expression of glucose-6-phosphate dehydrogenase in human cells sustaining oxidative stress. Biochem J. 1997;323(Pt 3):801–806. doi: 10.1042/bj3230801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legros F, Lombès A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park K, et al. Homeobox Msx1 interacts with p53 tumor suppressor and inhibits tumor growth by inducing apoptosis. Cancer Res. 2005;65:749–757. [PubMed] [Google Scholar]
- 36.Gradia DF, et al. Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int J Parasitol. 2009;39:49–58. doi: 10.1016/j.ijpara.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci USA. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Stocker R, Perrella MA. Heme oxygenase-1: A novel drug target for atherosclerotic diseases? Circulation. 2006;114:2178–2189. doi: 10.1161/CIRCULATIONAHA.105.598698. [DOI] [PubMed] [Google Scholar]
- 41.Arrigo AP, et al. Hsp27 (HspB1) and alphaB-crystallin (HspB5) as therapeutic targets. FEBS Lett. 2007;581:3665–3674. doi: 10.1016/j.febslet.2007.04.033. [DOI] [PubMed] [Google Scholar]
- 42.Chene P. ATPases as drug targets: Learning from their structure. Nat Rev Drug Discov. 2002;1:665–673. doi: 10.1038/nrd894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.