Abstract
In genetic hybrids, the silencing of nucleolar rRNA genes inherited from one progenitor is the epigenetic phenomenon known as nucleolar dominance. An RNAi knockdown screen identified the Arabidopsis de novo cytosine methyltransferase, DRM2 and the methylcytosine binding domain proteins, MBD6 and MBD10 as activities required for nucleolar dominance. MBD10 localizes throughout the nucleus, but MBD6 preferentially associates with silenced rRNA genes, and does so in a DRM2-dependent manner. DRM2 methylation is thought to be guided by siRNAs whose biogenesis requires RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3). Consistent with this hypothesis, knockdown of DCL3 or RDR2 disrupts nucleolar dominance. In genetic hybrids, the silencing of nucleolar rRNA genes inherited from one progenitor is the epigenetic phenomenon known as nucleolar dominance. An RNAi knockdown screen identified the Arabidopsis de novo cytosine methyltransferase, DRM2 and the methylcytosine binding domain proteins, MBD6 and MBD10 as activities required for nucleolar dominance. MBD10 localizes throughout the nucleus, but MBD6 preferentially associates with silenced rRNA genes, and does so in a DRM2-dependent manner. DRM2 methylation is thought to be guided by siRNAs whose biogenesis requires RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3). Consistent with this hypothesis, knockdown of DCL3 or RDR2 disrupts nucleolar dominance. Collectively, these results indicate that in addition to directing the silencing of retrotransposons and noncoding repeats, siRNAs specify de novo cytosine methylation patterns that are recognized by MBD6 and MBD10 in the large-scale silencing of rRNA gene loci.
Introduction
In interspecific hybrids of plants, insects, mammals or invertebrates, it is often the case that the RNA Polymerase I-transcribed rRNA genes of only one progenitor are expressed, independent of maternal or paternal effects. This epigenetic phenomenon, known as nucleolar dominance (McStay, 2006; Preuss and Pikaard, 2007; Reeder, 1985), results from the preferential silencing of one parental set of rRNA genes (Chen and Pikaard, 1997a). The silenced rRNA genes are clustered at nucleolus organizer regions (NORs) in tandem arrays spanning millions of basepairs, making nucleolar dominance one of the most extensive gene silencing phenomena known, second in scope only to X chromosome inactivation in female eutherian mammals (Heard and Disteche, 2006; Huynh and Lee, 2005).
The mechanisms by which one parental set of rRNA genes in a hybrid is chosen for silencing is unclear. However, it is clear that a partnership between DNA methylation and repressive histone modifications carries out rRNA gene silencing. In Arabidopsis or Brassica allotetraploids (hybrids that possess diploid genomes of two progenitors), silenced rRNA genes can be derepressed by treatment with 5-aza-2’ deoxycytosine (aza-dC), a cytosine methyltransferase inhibitor, or by treatment with histone deacetylase inhibitors such as trichostatin A (TSA) (Chen and Pikaard, 1997a). Treatment with both aza-dC and TSA is no more effective than treatment with either chemical alone, indicating that DNA methylation and histone deacetylation act in the same repression pathway (Chen and Pikaard, 1997a). Moreover, loss of histone deacetylation causes decreased cytosine methylation at rRNA gene promoters; likewise, inhibiting cytosine methylation causes the loss of repressive histone modifications (Earley et al., 2006; Lawrence et al., 2004). Collectively, these observations support a model whereby cytosine methylation and repressive histone modifications specify one another in a self-reinforcing cycle that maintains rRNA gene silencing (Lawrence et al., 2004).
Reverse genetic approaches have begun to identify proteins involved in rRNA gene silencing in nucleolar dominance. A role for the histone deacetylase, HDA6 was revealed in a screen in which transgene-induced RNA interference (RNAi) was used to systematically knock down the activities of the sixteen predicted Arabidopsis histone deacetylases (Earley et al., 2006). Biochemical studies then showed that HDA6 is a broad-specificity, TSA-sensitive histone deacetylase capable of removing acetyl groups from multiple lysines of core histones (Earley et al., 2006). Therefore, it is likely that TSA derepresses silenced rRNA genes by inhibiting HDA6 activity. By contrast, the cytosine methylation machinery that can account for aza-dC’s ability to derepress silenced rRNA genes is unknown. Candidate activities include 11 predicted cytosine methyltransferases, 7 of which are expressed in A. thaliana, and 13 predicted methylcytosine binding domain (MBD) proteins (Scebba et al., 2003; Springer and Kaeppler, 2005; Zemach and Grafi, 2003; Zemach et al., 2005), ten of which are expressed. Of the eleven predicted cytosine methyltransferases, only three are known to function in DNA methylation: MET1, CMT3, and DRM2 (Bender, 2004; Chan et al., 2005). MET1 maintains CG methylation and also affects CHG methylation to some extent (where H is a nucleotide other than C). CMT3 is primarily responsible for CHG maintenance methylation. DRM2 is responsible for de novo methylation and can modify cytosines in any sequence context, including CG, CHG and CHH (Cao et al., 2003; Cao and Jacobsen, 2002).
Gene regulatory functions for the 13 predicted Arabidopsis methylcytosine binding domain (MBD) proteins have not yet been defined. However, mammalian MBD proteins interact with protein complexes that covalently modify chromatin. For example, mammalian MeCP2 interacts with histone deacetylase HDAC1 (Jones et al., 1998; Nan et al., 1998), DNA methyltransferase DNMT1 (Kimura and Shiota, 2003) and at least one histone H3 lysine 9 methyltransferase (Fuks et al., 2003), thereby mediating transcriptional repression. Plant MBD proteins are similar to MeCP2, but only within the MBD domain; no homology is apparent between the plant and animal proteins in other parts of the proteins (Springer and Kaeppler, 2005).
In the current study, we show that components of the siRNA-directed DNA methylation pathway are required for nucleolar dominance, including RDR2, DCL3 and the de novo cytosine methyltransferase, DRM2. Moreover, we show that the methylcytosine binding domain protein, MBD6 binds to rRNA gene loci in a DRM2-dependent manner. Collectively, our results suggest that siRNAs guide de novo cytosine methylation of rRNA genes by DRM2. We propose that MBD6 and MBD10 then recognize the DRM2-mediated methylation patterns, thereby helping establish a heterochromatic state that inactivates rRNA gene loci on a multi-megabase scale.
Results
The de novo cytosine methyltransferase DRM2 is required for nucleolar dominance
In Arabidopsis suecica, the allotetraploid hybrid of A. thaliana and A. arenosa, the A. thaliana-derived 45S rRNA genes that are transcribed by RNA polymerase I are inactivated during early post-embryonic development; as a result, only A. arenosa-derived rRNA genes are abundantly expressed in leaves of mature plants (Pontes et al., 2007). To identify cytosine methylation machinery required for nucleolar dominance, we transformed A. suecica with transgenes encoding transcripts capable of forming double-stranded RNA (dsRNA) hairpins (Fig. 1A) in order to bring about the RNAi-mediated knockdown of corresponding mRNAs. RNAi-inducing transgenes engineered using A. thaliana sequences knock down homologous mRNAs encoded by both A. suecica progenitors due to the ~90-95% nucleic acid sequence identity between A. thaliana and A. arenosa genes (Earley et al., 2006; Lawrence and Pikaard, 2003).
Figure 1. The cytosine methyltransferase DRM2 is required for rRNA gene silencing in nucleolar dominance.
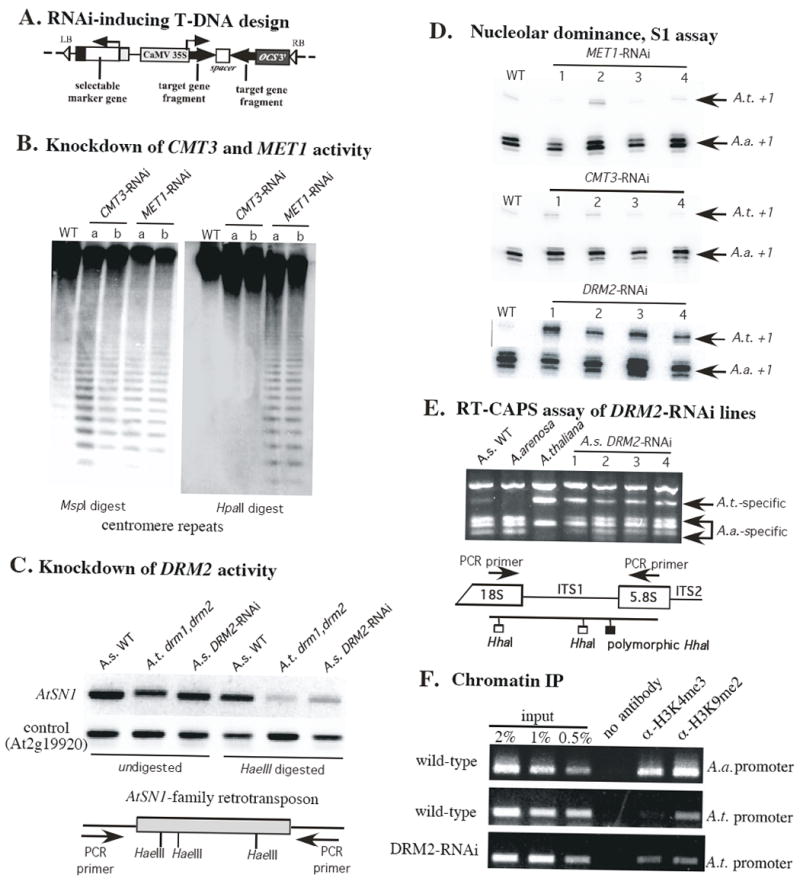
A. Organization of transgenes within the transferred DNA (T-DNA). LB and RB refer to left and right borders for T-DNA transfer. Target gene sequences of ~300-500 bp are cloned as inverted repeats separated by a spacer. Transcription by the strong Cauliflower Mosaic Virus (CaMV) 35S promoter generates double-stranded RNA hairpins that trigger RNAi. The selectable marker gene confers herbicide resistance.
B. RNAi-mediated knockdown of DNA methyltransferases CMT3 and MET1 causes loss of centromere repeat methylation. Genomic DNA of two CMT3-RNAi lines, two MET1-RNAi lines and a wild-type (WT) plant was digested with MspI or HpaII and subjected to Southern blot hybridization using a 180 bp centromere repeat probe.
C. Chop-PCR assay comparing AtSN1 retrotransposon methylation in an A. thaliana (A.t.) drm1 drm2 double mutant or in A. suecica (A.s.) wild-type (WT) or DRM2-RNAi lines. Genomic DNA was digested with HaeIII, or left undigested, prior to PCR using primers that flank the methylation-sensitive HaeIII sites (see diagram). Loss of HaeIII methylation results in cleavage of the template such that PCR fails.
D. S1 nuclease protection assays showing the loss of nucleolar dominance in four independent A. suecica DRM2-RNAi lines but no loss of dominance in four MET1-RNAi or CMT3-RNAi lines. RNA isolated from wild-type (WT) or RNAi lines was divided into equal aliquots and subjected to S1 nuclease protection assays using species-specific probes to detect transcripts initiated from the A. thaliana (A.t.) or A. arenos a (A.a)-derived rRNA gene promoters.
E. RT-CAPS assay confirming disruption of nucleolar dominance in A. suecica DRM2-RNAi lines. RNA isolated from A. suecica, A. arenosa or A. thaliana wild-type plants or four independent A. suecica DRM2-RNAi lines was subjected to reverse transcription (RT) followed by cleavage with HhaI to discriminate A. arenosa from A. thaliana gene transcripts (see diagram). An ethidium bromide-stained agarose gel image is shown.
F. Chromatin immunoprecipitation reveals a heterochromatic to euchromatic shift among A. thaliana-derived rRNA genes in DRM2-RNAi lines. Isolated chromatin from A. suecica wild-type or DRM2-RNAi plants was immunoprecipitated using antibodies specific for H3K4me3 or H3K9me2 and captured on protein A beads. Purified DNA was then subjected to PCR using primers specific for the A. arenosa (A.a.) or A. thaliana (A.t.)-derived rRNA gene promoter regions. Linear decreases in signal upon dilution of input chromatin show that the results are semi-quantitative.
There are eleven predicted DNA methyltransferases in A. thaliana, seven of which are expressed and were targeted for RNAi-mediated knockdown in A. suecica (Table S1). Methylation analyses confirmed that the three DNA methyltransferase genes of known function, MET1, CMT3 and DRM2 were knocked down (Fig. 1B, C). Centromeric 180 bp repeats are heavily methylated, making them resistant to the methylation-sensitive restriction endonucleases MspI and HpaII in wild-type plants (Fig. 1B, lanes labeled WT). Loss of CHG methylation in CMT3-RNAi and MET1-RNAi lines and loss of CG methylation in MET1-RNAi lines results in increased digestion by MspI and HpaII, respectively (Fig. 1B). The results indicate that CMT3 is required for CHG, but not CG methylation in A. suecica, as is also the case in A. thaliana. MET1 knockdown causes decreased CG and CHG methylation in A. suecica (Fig. 1B), as in A. thaliana (Bartee and Bender, 2001).
RNAi-mediated knockdown of DRM2 activity was verified by examining CHH methylation at AtSN1 family retrotransposons (Fig. 1C) that are methylated by DRM2 in a siRNA-directed fashion (Zilberman et al., 2003). Methylation of the internal C of GGCC motifs prevents digestion by the restriction endonuclease HaeIII, thereby allowing the uncut DNA template to be amplified by PCR using primers flanking the HaeIII sites (see diagram in Fig. 1C). Loss of methylation facilitates HaeIII digestion, resulting in a diminished PCR signal. In A. suecica DRM2- RNAi lines, HaeIII methylation is reduced to a degree comparable to the A. thaliana drm1drm2 double mutant (Fig. 1E, compare right-most two lanes). These data indicate that DRM2, and not DRM1, mediates AtSN1 HaeIII methylation, consistent with DRM2’s higher level of mRNA expression compared to DRM1 in A. thaliana (Chan et al., 2005).
To screen for the loss of nucleolar dominance in RNAi lines, multiple independent primary transformants (T1 generation) were tested using two assays capable of discriminating transcripts of A. thaliana or A. arenosa-derived rRNA genes: S1 nuclease protection (Fig. 1D) and RT-PCR-CAPS (Fig. 1E). The S1 nuclease protection assay makes use of A. thaliana or A. arenosa-specific probes to detect transcripts that are accurately initiated from the respective rRNA gene promoters. The RT-PCR-CAPS (reverse transcription-polymerase chain reaction-cleaved amplified polymorphic sequence) assay discriminates pre-rRNA transcripts of the two progenitors based on a single nucleotide polymorphism that creates an extra HhaI restriction site within internal transcribed spacer 1 (ITS1) of A. arenosa rRNA genes (Lewis and Pikaard, 2001). RNAi-mediated knockdown of the maintenance cytosine methyltransferases, MET1 and CMT3 had no appreciable effect on nucleolar dominance in the S1 nuclease protection assay (Fig. 1D) or in the RT-PCR-CAPS assay (data not shown). Likewise, nucleolar dominance was unaffected in RNAi lines targeting MET2, MET3, CMT1 or CMT2 (data not shown). Nucleolar dominance was also not disrupted by knocking down the SWI2/SNF2-related chromatin remodeller DDM1 (Fig. S1) which is known to affect rRNA gene methylation and approximately 70% of all cytosine methylation genome-wide (Vongs et al., 1993). However, in DRM2-RNAi lines, the S1 nuclease protection (Fig. 1D) and RT-PCR-CAPS assays (Fig. 1E) both revealed the co-expression of the A. thaliana and A. arenosa-derived rRNA genes, indicating a loss of nucleolar dominance, whereas in wild-type plants A. arenosa transcripts outnumber A. thaliana transcripts by more than ten-fold.
Derepression of the A. thaliana-derived rRNA genes in DRM2-RNAi lines is reflected by altered post-translational modifications of histones in the vicinity of their promoters (Fig. 1F). In wild-type A. suecica, the dominant A. arenosa rRNA genes are approximately equally associated with histone H3 that is trimethylated on lysine 4 (H3K4me3) or dimethylated on lysine 9 (H3K9me2), reflecting the fact that a subset of the genes is active (H3K4me3-associated) and the remainder are inactive (H3K9me2-associated) (Earley et al., 2006; Lawrence et al., 2004). The A. thaliana-derived rRNA genes in wild-type A. suecica are almost entirely associated with H3K9me2, reflecting their repressed state (Earley et al., 2006; Lawrence et al., 2004). However, in DRM2-RNAi plants, the A. thaliana-derived rRNA genes are nearly equally associated with H3K4me3 and H3K9me2, suggesting that approximately half of the genes becomes active when DRM2 is knocked down (Fig. 1F). In agreement with previous studies, the A. thaliana rRNA gene promoters that associate with H3K4me3 are cytosine-hypomethylated relative to promoters associated with H3K9me2 (Fig. S2).
The DCL3-dependent siRNA pathway regulates nucleolar dominance
Because DRM2 is responsible for siRNA-directed DNA methylation (Cao et al., 2003), we investigated the potential role of siRNAs in rRNA gene silencing. Oligonucleotides spanning 50 nt intervals of the rRNA gene promoter region were used to probe RNA blots, revealing abundant 24 nt siRNAs matching both strands of the rRNA gene promoter in A. thaliana (Fig. 2A). The siRNAs are most abundant between -50 and +1, which corresponds closely to the core promoter region (-55 to +6) defined by deletion analyses (Doelling and Pikaard, 1995; Saez-Vasquez and Pikaard, 1997). Analysis of A. thaliana mutants deficient for the four dicer endonucleases showed that promoter siRNAs are generated almost exclusively by DCL3 (Fig. 2B).
Figure 2. DCL3-dependent siRNAs match the rRNA gene intergenic spacer in A. thaliana.
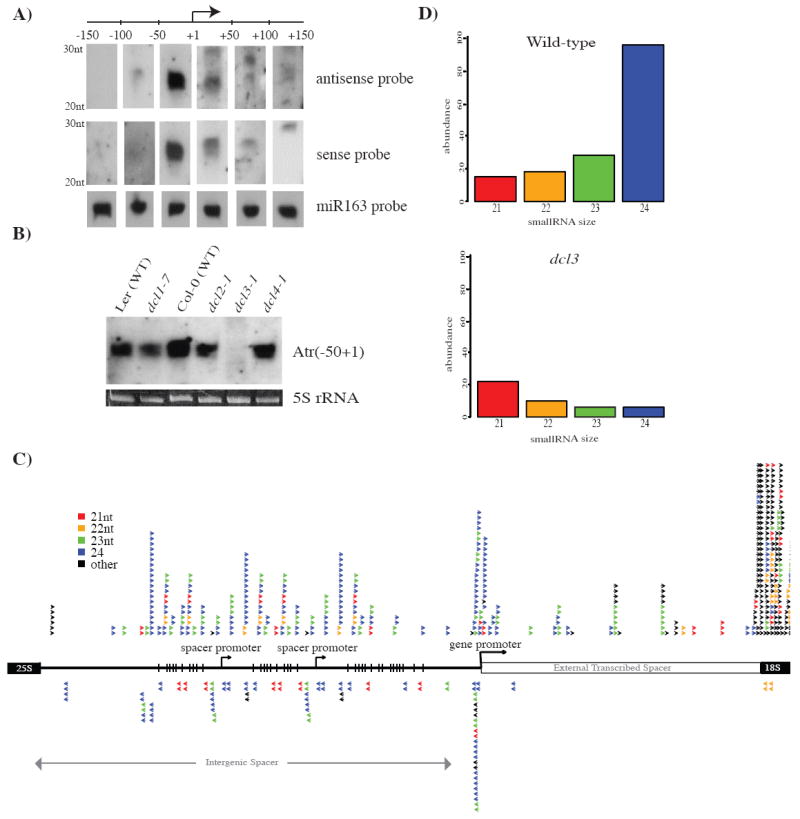
A. RNA blot of small RNAs detected using probes spanning 50 nt intervals on both strands of the rRNA gene promoter region, from -150 to +150 relative to the transcription start site (+1). The miRNA, miR163 serves as an RNA loading control.
B. siRNAs matching the rRNA gene promoter are generated by DCL3. An RNA blot using small RNAs isolated from wild-type plants (ecotypes Ler or Col-0) or from plants mutant for dcl1, dcl2, dcl3 or dcl4 was hybridized to the -50 to +1 antisense probe. The ethidium bromide stained 5S rRNA band on the agarose gel prior to blotting serves as a loading control.
C. Deep sequencing reveals abundant siRNAs matching both strands of the intergenic spacer, including the gene promoter. siRNAs are color-coded by size. Downstream of the gene promoter, within the external transcribed spacer and coding regions for 18S rRNA, small RNAs almost exclusively match the rRNA strand. Long arrows indicate duplicated spacer promoters and the gene promoter.
D. siRNAs throughout the ~3kb intergenic spacer are generated primarily by DCL3. Histograms show the number of sequenced siRNAs matching the intergenic spacer in wild-type and dcl3 mutant plants. Color-coding for siRNAs of specific sizes is the same as for panel C.
Analysis of small RNA libraries subjected to deep sequencing (Mosher et al., 2008) (Kasschau et al., 2007) confirmed that the rRNA gene promoter is a hotspot for siRNAs that are predominantly 23 and 24 nt in size (Fig. 2C). However, siRNAs are not restricted to the promoter region but correspond to both DNA strands throughout the ~3 kb intergenic spacer. Downstream of the gene promoter, within the coding sequences (only a small part of the 18S coding region is shown), small RNAs that are heterogeneous in size derive almost exclusively from the rRNA strand. Their coding strand-specificity and size heterogeneity suggests that they are mostly rRNA degradation products rather than siRNAs (Fig. 2C; see Fig. S3 for additional details). By contrast, intergenic spacer small RNAs are primarily 23 and 24 nt in size and are depleted upon mutation of DCL3 (Fig. 2D; Fig. S3), consistent with the RNA blot analyses of promoter siRNAs (Fig. 2B).
To test the role of DCL3 in nucleolar dominance, we targeted DCL3 mRNAs for RNAi-mediated knockdown in A. suecica (Fig. 3). DCL3 mRNA levels were substantially reduced relative to non-transformed wild-type plants in multiple independent transgenic DCL3-RNAi lines (Fig. 3A), with line #1 showing the greatest degree of knockdown and line #4 showing the least knockdown (~ 25% of wild-type). In DCL3-RNAi plants, nucleolar dominance was disrupted, as shown using both the S1 nuclease protection and RT-PCR-CAPS assays (Figs. 3B, C). The extent of A. thaliana rRNA gene derepression correlates with the degree of DCL3 mRNA knockdown (compare panels A, B and C). Nucleolar dominance remained disrupted in T2 generation siblings (Fig. 3D) in which 24 nt siRNAs corresponding to both the A. thaliana and A. arenosa-derived rRNA gene promoters were substantially reduced, as were siRNAs corresponding to 5S RNA genes (Fig. 3E).
Figure 3. DCL3 and RDR2 are required for rRNA gene silencing in nucleolar dominance.
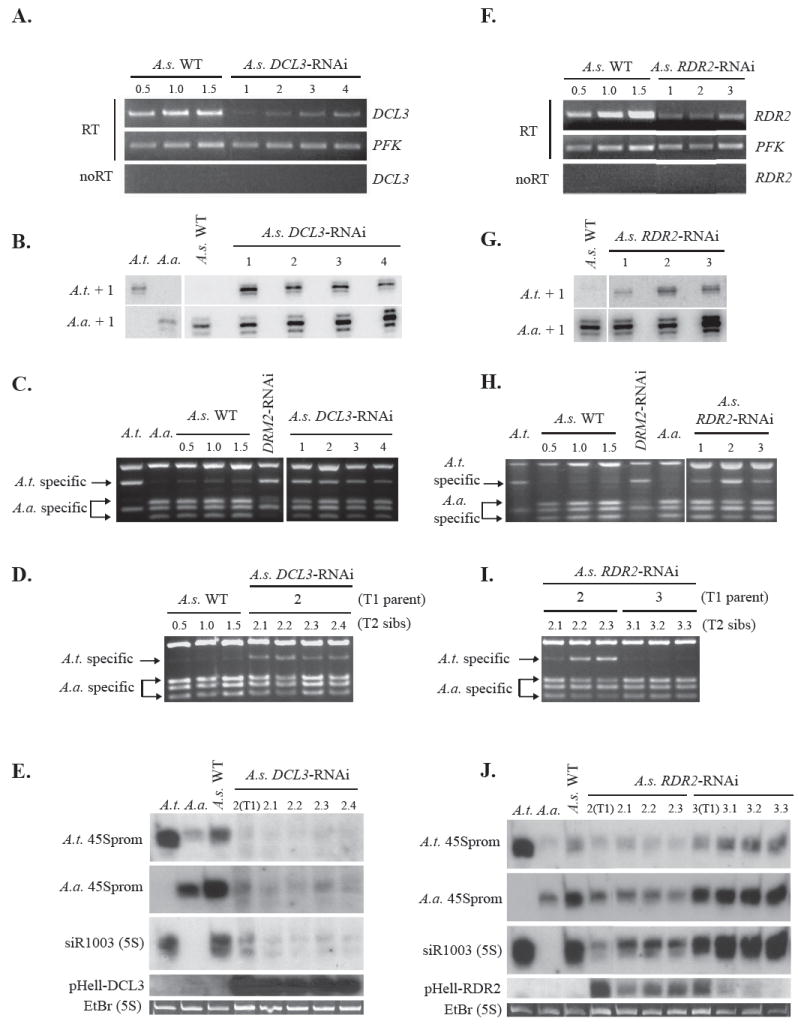
A. RNAi-mediated knockdown of DCL3 mRNA in four A. suecica (A.s.) T1 transgenic plants. For the wild-type controls, RT-PCR reactions were conducted using 0.5X, 1.0X or 1.5X of the RNA quantity tested for each transgenic plant. Phosphofructokinase (PFK) was amplified as a loading control. Primer sequences are provided in Table S5. Reactions lacking reverse transcriptase (noRT) control for potential DNA contamination.
B. Loss of nucleolar dominance in DCL3-RNAi lines detected by S1 nuclease protection. A. thaliana, A. arenosa and A. suecica RNAs (from wild-type or four independent DCL3-RNAi T1 plants) were subjected to S1 nuclease protection using species-specific probes.
C. Loss of nucleolar dominance in DCL3-RNAi lines detected by RT-PCR-CAPS. A. thaliana, A. arenosa and A. suecica RNAs (from wild-type, DRM2-RNAi or DCL3-RNAi plants) were subjected to RT-PCR followed by HhaI digestion.
D. Loss of nucleolar dominance persists in T2 progeny of DCL3-RNAi primary transformant (T1 generation) #2 . RT-PCR-CAPS analysis was conducted using 0.5X, 1.0X or 1.5X quantities of wild-type A. suecica RNA or 1.0X quantities of RNA from four T2 siblings (2.1-2.4).
E. Reduced 24 nt siRNA levels in DCL3-RNAi lines. RNA of A. thaliana, A. arenosa, A. suecica wild-type, primary transformant (T1) plant #2 or its T2 siblings (2.1-2.4) was blotted and probed for siRNAs matching the A. thaliana or A. arenosa 45S rRNA gene promoters or an siRNA matching the A. thaliana 5S rRNA gene intergenic spacer (siR1003). The second row from the bottom shows 21 nt siRNAs derived from the RNAi-inducing transgene (pHellsgate-DCL3 inverted repeat). Ethidium bromide-stained 5S rRNA serves as a loading control (bottom row).
F. RNAi-mediated knockdown of RDR2 mRNA in A. suecica (A.s.) T1 transgenic plants. For the wild-type controls, RT-PCR reactions were conducted using 0.5X, 1.0X or 1.5X of the amount of RNA tested for each transgenic plant. PFK was amplified as a loading control. RDR2 reactions lacking reverse transcriptase (noRT) control for DNA contamination.
G. Derepression of A. thaliana-derived rRNA genes in A. suecica RDR2-RNAi lines. RNA from wild-type or three independent T1 generation RDR2-RNAi transgenic plants was subjected to S1 nuclease protection using probes that specifically detect A. thalian a or A. arenosa-derived rRNA gene transcripts.
H. Disruption of nucleolar dominance in RDR2-RNAi T1 plants detected using the RT-PCR-CAPS assay. A. thaliana, A. arenosa and A. suecica RNAs (from wild-type, DRM2-RNAi or RDR2-RNAi plants) were subjected to RT-PCR followed by HhaI digestion and agarose gel electrophoresis. 0.5X, 1.0X or 1.5X quantities of wild-type A. suecica RNA were amplified; all other reactions used 1.0X amounts.
I. Disruption of nucleolar dominance persists in T2 progeny of RDR2-RNAi primary transformant #2 (lines 2.2-2.3) but not in T2 progeny of transformant #3 (lines 3.1-1.3), as shown by using the RT-PCR-CAPS assay.
J. Reduced 24 nt siRNA accumulation persists in T2 progeny (lines 2.2-2.3) of RDR2-RNAi T1 plant #2 [labeled 2(T1)] but not in T2 progeny (lines 3.1-1.3) of primary transformant #3 [labeled 3(T1)]. RNA of A. thaliana, A. arenosa or A. suecica plants was blotted and probed for A. thaliana or A. arenosa 45S rRNA gene promoter siRNAs. In the second row from the bottom, siRNAs derived from the RNAi-inducing transgene (pHellsgate-RDR2 inverted repeat) are detected. Ethidium bromide stained 5S rRNA serves as a loading control (bottom row).
In the RNA-directed DNA methylation pathway, the double-stranded RNA precursors that are cleaved into 24 nt siRNAs by DCL3 are thought to be generated by RNA-DEPENDENT RNA POLYMERASE 2 (RDR2). Consistent with this expectation, RNAi-mediated knockdown of RDR2 (Fig. 3F) disrupted nucleolar dominance in A. suecica (Fig. 3G, H). In T2 sibling progeny of RDR2-RNAi line 2, in which nucleolar dominance remained disrupted, siRNA levels were several fold lower than in wild-type plants (Fig. 3I, J), but not diminished to the same extent as in DCL3- RNAi lines. A fortuitous observation was that in RDR2-RNAi line 3, in which nucleolar dominance was disrupted in the T1 generation, nucleolar dominance became reestablished (Fig. 3I) and siRNAs returned to wild-type levels (Fig. 3J) in T2 progeny. Reestablishment of nucleolar dominance in line 3 T2 progeny correlated with the loss of siRNAs derived from the RNAi-inducing transgene (Fig. 3J, 2nd row from the bottom), providing additional evidence for RDR2 involvement in nucleolar dominance.
Evidence for DRM2, DCL3 and RDR2-dependent methylation of rRNA genes
Collectively, the data of Figures 1-3 indicate that DRM2, DCL3 and RDR2 are involved in the silencing of Pol I-transcribed 45S rRNA genes subjected to nucleolar dominance, thereby implicating the siRNA-directed DNA methylation pathway. Although this pathway is known to silence retrotransposons and foreign transgenes and to methylate the Pol III-transcribed 5S RNA genes (Chan et al., 2005; Huettel et al., 2007; Matzke et al., 2006), siRNA-directed de novo methylation of 45S rRNA genes has not been demonstrated. To search for sites of DRM2-dependent methylation within potential regulatory sequences of 45S rRNA genes, we examined publicly accessible methylation profiling datasets for wild-type and drm1 drm2 double mutant A. thaliana plants. Zhang et al. performed ChIP using an anti-methylcytosine antibody followed by DNA microarray analyses on a whole-genome tiling array (Zhang et al., 2006). Our analysis of their raw data did not reveal significant changes in methylation in drm1 drm2 mutants relative to wild-type A. thaliana in the vicinity of the rRNA gene promoter. However, drm-dependent methylation was detected near the duplicated spacer promoters that share >80% identity with the gene promoter and program weak Pol I transcription (Doelling et al., 1993). Bisulfite-mediated DNA sequencing (Frommer et al., 1992) in this region revealed an ~50% loss of cytosine methylation on both DNA strands in DRM2-RNAi lines (Fig. 4), in both CG and non-CG sequence contexts. Methylation is also reduced in RDR2-RNAi and DCL3-RNAi lines, thereby implicating the 24 nt siRNA-directed DNA methylation pathway (Fig. 4).
Figure 4.
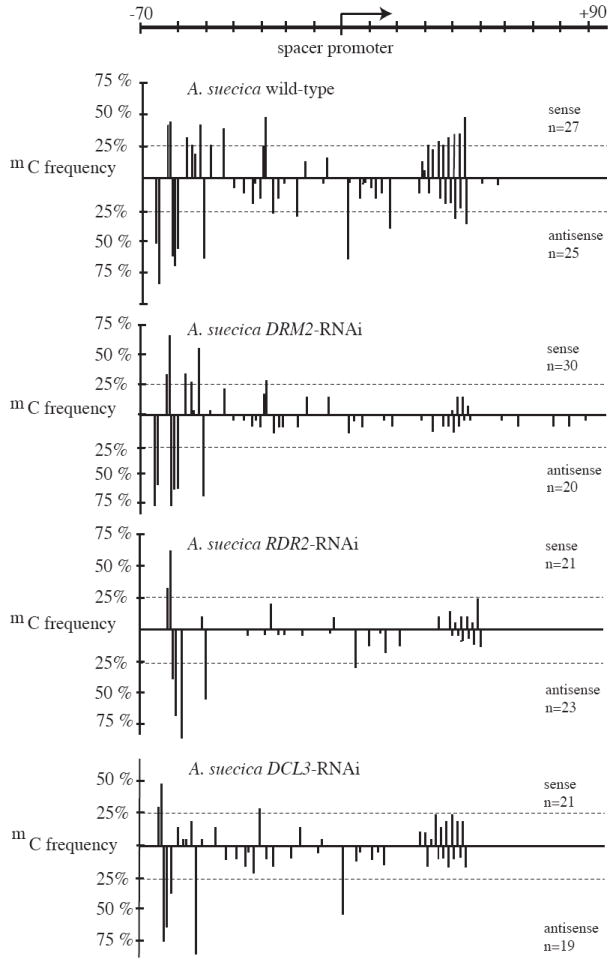
RNAi-mediated knockdown of DRM2, RDR2 or DCL3 reduces cytosine methylation in the rRNA gene intergenic spacer in the vicinity of the spacer promoters. Positions of cytosine methylation were determined by bisulfite-mediated sequencing and are denoted as vertical bars; the height of each bar reflects the frequency at which that cytosine is methylated. Sense (rRNA) strand cytosines are denoted above the horizontal line and antisense cytosines below. The number of independent clones sequenced (n) is shown for each strand.
Methylcytosine binding domain proteins recognize RNA-directed DNA methylation
The A. thaliana genome includes thirteen predicted MBD proteins (Springer and Kaeppler, 2005), ten of which are expressed (Table S2). RNAi-mediated knockdown of MBD1, MBD2, MBD4, MBD5, MBD7, MBD8, MBD9 and MBD11 had no significant effect on nucleolar dominance (data not shown). However, in multiple independent RNAi lines targeting MBD6 and MBD10, nucleolar dominance was disrupted (Figs. 5A, B). Coincident with their derepression, A. thaliana-derived rRNA gene promoters shifted from exclusive association with H3K9me2 to a partial association (~30%) with H3K4me3 (Fig. 5D), as in DRM2-RNAi lines (Figs. 5D and 1F).
Figure 5. MBD6 and MBD10 are required for rRNA gene silencing in nucleolar dominance.
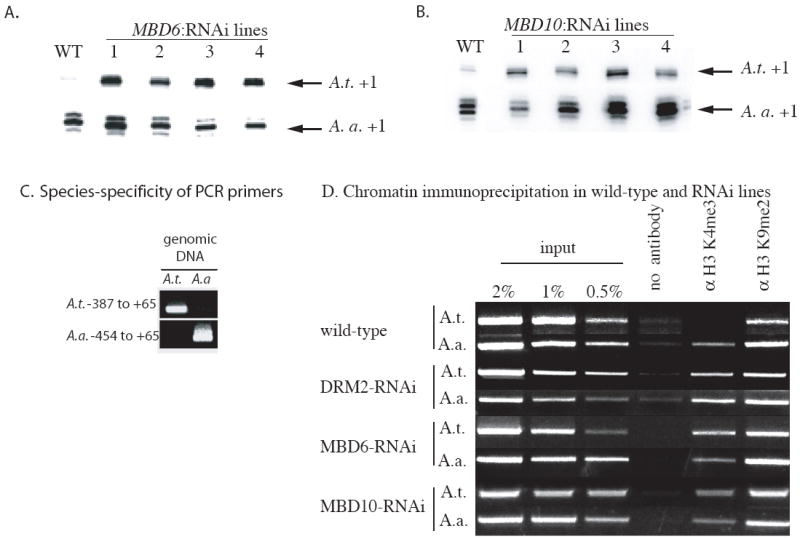
A and B. S1 nuclease protection assays comparing expression of A. thaliana and A. arenosa-derived rRNA genes in wild-type A. suecica (WT) or four independent RNAi lines targeting MBD6 or MBD10.
C. Specificity of PCR primer pairs for promoter regions of A. thaliana or A. arenosa-derived rRNA genes in A. suecica. These primer pairs are used in panel D.
D. Activation of A. thaliana-derived rRNA genes in MBD6- and MBD10-RNAi lines is associated with their increased association with H3K4me3. ChIP was performed as in Figure 1F.
In nuclei stained with the fluorescent DNA-binding dye DAPI (blue signal), the nucleolus, where rRNA gene transcription takes place, appears as a black hole due to the paucity of nucleolar DNA (Figs. 6A, S4). At the outer periphery of the nucleolus, the portions of the NORs that are composed of inactive, excess rRNA genes are condensed into heterochromatin and yield distinct DNA-FISH signals (red signals in Figs. 6A, S4). Detection of MBD6 using an antibody raised against the native protein (Fig. 6A), or detection of MBD6 fused to YFP (Yellow Fluorescent Protein; Fig. S4) revealed that the strongest sites of MBD6 localization correspond to the condensed portions of the NORs, in agreement with a prior study (Zemach et al., 2005). MBD6 also localizes to all chromocenters, the bright DAPI-positive foci where centromere repeats and other heterochromatic repeats coalesce. By contrast, MBD10-YFP is broadly distributed throughout the nucleus (data not shown).
Figure 6. MBD6 colocalization with A. thaliana NORs is DRM-dependent.
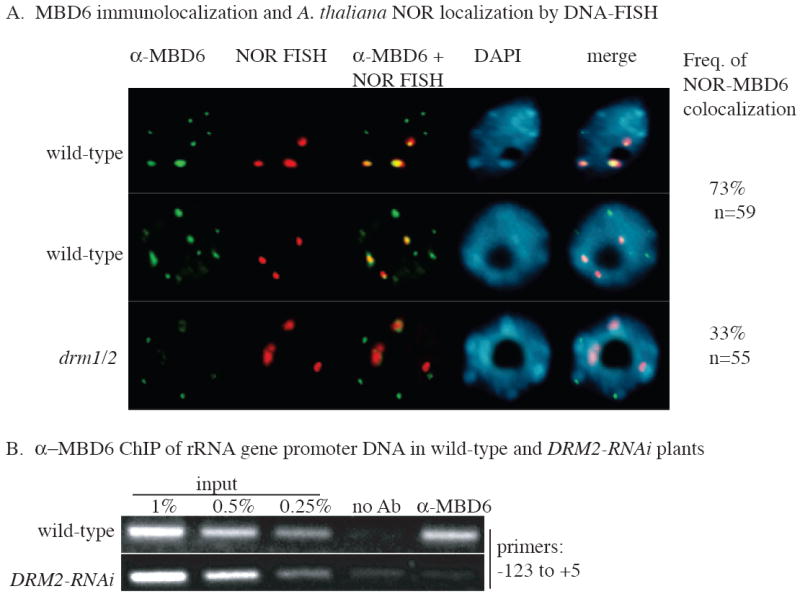
A. MBD6 was immunolocalized using an anti-MBD6 antibody (green signals) and NORs were detected by DNA-FISH (red signals) in isolated nuclei of A. thaliana wild-type or drm1/2 mutant plants. DNA was counterstained with DAPI (blue signals). Yellow signals result from colocalization of MBD6 with the DAPI-intensive chromocenters that include the NORs.
B. Physical association of MBD6 with rRNA genes is DRM2-dependent. Chromatin of wild-type or DRM2-RNAi plants was immunoprecipitated using anti-MBD6 antibody and rRNA gene promoter sequences from -123 to +5 were amplified by PCR. No antibody controls (no Ab) reveal background signals. Input chromatin dilutions show that results are semi-quantitative.
Loss of DRM activity has consequences on NOR condensation and association of MBD6 with NORs (Fig. 6A). There are four NORs in diploid A. thaliana but they tend to coalesce such that one typically (69% frequency, n=93) observes three NOR FISH signals in wild-type nuclei; four signals are observed only 13% of the time. However, in the drm mutant, four NOR FISH signals were observed in 48% of the nuclei examined (n=113) and these FISH signals are decondensed relative to wild-type nuclei. Moreover, strong MBD6 signals are no longer detected at NORs in drm2 mutant nuclei, although MBD6 signal strength at others chromocenters is unaffected in drm2 mutants.
To test whether MBD6 physically interacts with A. thaliana-derived rRNA genes in A. suecica in a DRM2-dependent manner, we performed ChIP using anti-MBD6 antibodies. A. thaliana rRNA gene promoter sequences were readily detected in association with MBD6 in chromatin of wild-type plants (Fig. 6B, top row) but were not detected above background levels in DRM2-RNAi plants t (Fig. 6B, bottom row). Collectively, the immunolocalization and ChIP data suggest that MBD6 is recruited to rRNA genes in a DRM2-dependent manner.
Discussion
DRM2, the de novo cytosine methyltransferase, is required for rRNA gene silencing in nucleolar dominance, unlike the maintenance methyltransferases MET1 and CMT3. These findings suggest that nucleolar dominance is regulated by dynamic changes in DNA methylation, consistent with its developmental regulation (Chen and Pikaard, 1997b; Neves et al., 1995; Pontes et al., 2007). The data also fit with evidence that nucleolar dominance is a manifestation of an rRNA gene dosage control system that operates in non-hybrids to regulate rRNA gene activity in response to physiological needs (Lawrence et al., 2004). We had hypothesized that switching rRNA genes from the “on” state to the “off” state when fewer ribosomes are needed might involve de novo cytosine methylation (Lawrence et al., 2004). DRM2’s involvement in nucleolar dominance fits this prediction.
RNA-directed DNA methylation is carried out by DRM2 [for reviews see (Chan et al., 2005; Huettel et al., 2007; Matzke et al., 2006)], with histone deacetylase HDA6 also participating in the silencing of affected loci (Aufsatz et al., 2002; Aufsatz et al., 2007). Both DRM2 (this study) and HDA6 (Earley et al., 2006) are required for nucleolar dominance. Moreover, the RNA-directed DNA methylation pathway involves the plant-specific RNA polymerases Pol IV and Pol V (previously known as Pol IVa and Pol IVb), which co-localize with NORs (Pontes et al., 2006). Pol IV is thought to work in partnership with RDR2, generating dsRNA substrates that are then diced into siRNA duplexes by DCL3. As we have shown, abundant DCL3-dependent siRNAs match virtually all of the rRNA gene intergenic spacer and knocking down DCL3 or RDR2 disrupts nucleolar dominance coincident with the loss of DRM2-dependent DNA methylation (see Fig. 4). The simplest explanation for these observations is that 45S rRNA gene silencing involves siRNA-directed DNA methylation, with DRM2-dependent DNA methylation patterns then recognized by MBD6 (Fig. 7). We speculate that MBD6 helps form the highly condensed heterochromatin typical of chromocenters, which includes the silenced portions of the NORs (Fig. 7). MBD10 is also involved in rRNA gene silencing but localizes throughout the nucleoplasm, and not specifically at NORs or chromocenters, suggesting that MBD10 may play a more general role in the nucleus. Interestingly, siRNA levels increase slightly in A. suecica MBD6 knockdown lines but are decreased in DRM2-RNAi lines (Fig. S5). Current hypotheses suggest that DRM-dependent cytosine methylation positively influences the production of aberrant RNAs or Pol IV transcripts that serve as siRNA precursors (e.g. see Pontes et al, 2006). If so, MBD6 may limit the levels of siRNA precursor transcripts originating from methylated DNA such that siRNA levels increase when MBD6 is knocked down.
Figure 7.
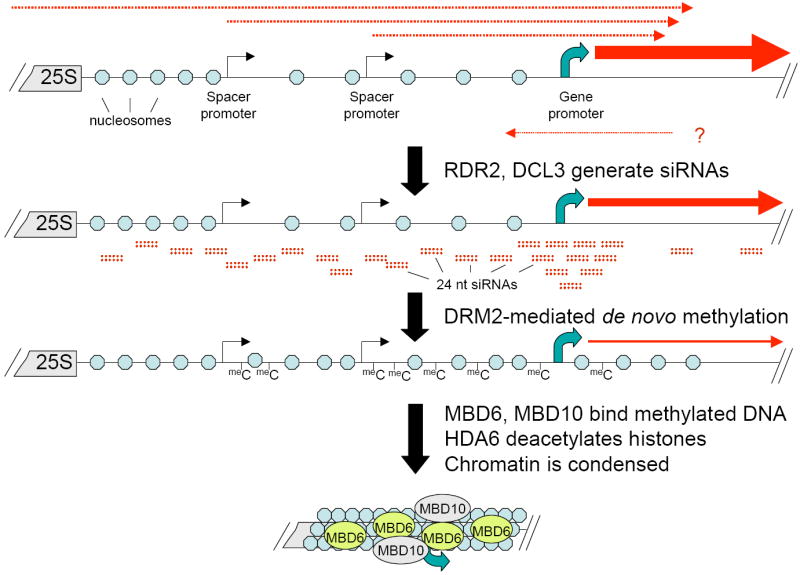
Model for RNA-directed cytosine methylation and MBD-mediated heterochromatin condensation. Noncoding spacer transcription (red dashed arrows) initiates 24 nt siRNA production by RDR2 and DCL3. Bidirectional transcription in the vicinity of the promoter may account for the siRNA hotspot. siRNAs direct de novo methylation of spacer sequences by DRM2. Binding of MBD6 and MBD10 to-methylated DNA in conjunction with histone deacetylation by HDA6 reduces transcription and nucleosome displacement, facilitating heterochromatin compaction.
rRNA gene silencing in plants and mammals has intriguing parallels as well as important differences. In mouse, a key activity for rRNA gene silencing is NoRC (Nucleolar Remodelling Complex), a complex of TIP5 (TTF-I-interaction protein #5) and SNF2h, an ATP-dependent chromatin remodeller (Strohner et al., 2001). TIP5 over-expression inhibits transcription in an aza-dC and TSA-reversible manner (Santoro et al., 2002), implicating DNA methylation and histone deacetylation in rRNA gene silencing in mouse, as in plants. NoRC physically interacts with histone deacetylase HDAC1 (Zhou et al., 2002), which is in the same gene family as Arabidopsis HDA6 (Earley et al., 2006). NoRC also interacts with the DNA methyltransferases DNMT1 and DNMT3 (Santoro and Grummt, 2005; Santoro et al., 2002), the latter being a de novo DNA methyltransferase that is a homolog of Arabidopsis DRM2 (Cao and Jacobsen, 2002). Chromatin modifications marking active and silenced rRNA genes are also similar in mouse and plants. In both cases, active gene promoters associate with histone H3 that is trimethylated on lysine 4 (H3K4me3) and with hyperacetylated histones H3/H4. Likewise, silenced rRNA genes associate with methylated H3K9, deacetylated H3/H4, and have cytosine-hypermethylated promoters. Although there is no obvious ortholog of TIP5 in Arabidopsis, the overall similarities of rRNA gene regulation in plants and mammals is striking (Grummt and Pikaard, 2003; McStay, 2006).
Recently, highly structured RNAs of 200-300 nucleotides, initiating within the intergenic spacer and reading through the promoter, were reported to bind to TIP5 and control its nucleolar localization and the repressive activity of NoRC (Mayer et al., 2008; Mayer et al., 2006). These repressive RNAs are presumed to initiate from upstream spacer promoters, but this has not been demonstrated definitively. Unlike our study, siRNAs corresponding to the mouse rRNA gene promoter have not been reported, making it unclear whether RNA-mediated regulation of rRNA gene transcription is accomplished differently in plants and mammals.
In Arabidopsis, the source of the spacer transcripts that serve as the precursors for RDR2 polymerase activity and siRNA biogenesis is unknown. Possibilities include Pol I transcripts initiated from the spacer promoters, however siRNAs are also produced upstream of the distal spacer promoter (see Fig. 7). Therefore, Pol I transcripts reading through the spacer from the preceding rRNA gene, or transcripts of other polymerases may be sources of siRNA precursors. It is conceivable that these spacer transcripts are the true regulatory molecules, with siRNA production being merely a by-product of their degradation. Although this seems unlikely, a simple relationship between promoter siRNA levels in mature plants and the degree of nucleolar dominance is not apparent in all RNAi knockdown lines. It may be that siRNAs produced at a specific time in early development are critical or that siRNAs matching regulatory elements other than the promoter are key. Potential regulatory sequences include repetitive elements located between the gene promoter and proximal spacer promoter and between the duplicated spacer promoters. In Xenopus and mouse, repetitive elements located in analogous positions act as enhancers of the gene promoter (Labhart and Reeder, 1984; Pikaard et al., 1990). However, full enhancer activity, including enhancer involvement in a nucleolar dominance-like competition between minigenes that are co-injected into Xenopus oocytes, requires one of more spacer promoters upstream of the enhancers (Caudy and Pikaard, 2002; DeWinter and Moss, 1987). Although enhancer activity has not been demonstrated for repetitive spacer elements in plants, Arabidopsis intergenic spacer repeats enhance Xenopus rRNA minigene transcription in injected Xenopus oocytes (Doelling et al., 1993). Because siRNAs and their presumptive precursor transcripts are generated throughout the intergenic spacer, including potential regulatory sequences that differ between the two parental types of rRNA genes in A. suecica hybrids, pinpointing sequences subjected to RNA-mediated control may reveal the basis for selectively silencing one parental set of rRNA genes in nucleolar dominance.
Experimental Procedures
Plant growth conditions
Arabidopsis suecica plants were grown and induced to flower as described (Lawrence, 2003). For chromatin immunoprecipitation (ChIP), plants were grown on semi-solid, half-strength Murashige-Skoog medium, pH 5.2 (Sigma-Aldrich Company) in deep petri dishes for two weeks at 22°C under continuous light.
Generation of RNAi lines
Inverted repeat constructs targeting DNA methyltransferases and MBDs were cloned in the pFGC5941 plasmid vector and are available at http://chromdb.org. DCL3 and RDR2 RNAi constructs were generated using the pHELLSGATE8 vector and contain RDR2 sequences amplified using primers 5’-CACCCTCAATGCGCTTGTTCATGC-3’ and 5’-AAATCCGAGACATGCTCTGC-3’ or DCL3 sequences amplified using 5’-CACCGCCACCTTTCAGGCTTAT-3’ and 5’- CGGATGAGGTATTGCACTGA-3’. Agrobacterium-mediated transformation of A. suecica was as described (Lawrence and Pikaard, 2003).
Nucleic acid isolation, S1 nuclease protection, and RNA blot assays
Purification of genomic DNA and total RNA from 30 day old plants and S1 nuclease protection were as described (Chen et al., 1998). S1 nuclease assay details are provided in the supplement. Small RNAs were extracted from inflorescence (A. thaliana) or leaves (A. suecica) using mirVana miRNA isolation kits (Ambion). Probes for RNA blot hybridization, as described in Onodera et al., 2005, were labeled using mirVana miRNA probe construction kits. Tables S3 and S4 provide probe sequences.
Small RNA libraries
Construction and sequencing of small RNA libraries used to generate Figures 2C and 2D is described in Mosher et al., 2008 and Kasschau et al, 2007, respectively. Additional details are provided in the supplement.
Chromatin immunoprecipitation assays
ChIP was performed according to published protocols (Gendrel et al., 2002; Lawrence et al., 2004) with minor modifications. Briefly, two-week-old seedlings were vacuum-infiltrated with 1% formaldehyde. Following homogenization, isolated nuclei were sonicated four times using a Branson sonifier (output setting 2, 40% duty cycle). Chromatin was incubated with anti-Histone H3K4me3 (Abcam ab8580), anti-Histone H3K9me2 (Abcam ab7312) or anti-MBD6 antibodies raised in rabbits against peptide CTSRNPSKVSA and affinity purified. Antibody-chromatin complexes captured on protein-A agarose beads (Upstate 16-157) were washed four times. Protein-DNA crosslinks were reversed by boiling for 10 min in 100 μl of 10% (w/v) Chelex-100 chelating resin (Biorad 142-1253) slurry (in water) and 200 μg/ml proteinase K (Nelson et al., 2006). DNA was eluted in 100 μL TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). 1% of the eluate was used in PCR reactions (Lawrence et al., 2004) using rRNA gene-specific primers and 26 cycles of amplification. A. thaliana-specific primers, numbered relative to the transcription start site (+1), with F for forward and R for reverse, were: -397F: ACCGGGTCCGAGGATT, -123F: CCTTATGATGCATGCCAAAAAGAATT, +5R: CCCCCTATATAGCTTAATAGCCCTTTT, +81R: ATCCCTCGATCGCTACCCA. A. arenosa-specific promoter sequences were PCR amplified using primer -454F: ATGCCTCAATGAAGAGTAACGTT with primer +81R.
Cytosine methylation analysis
ChIP-Chop PCR assays (Lawrence et al., 2004) were performed by treating 2% of the ChIP-isolated DNA with 10 units of McrBC (New England Biolabs) for 3 hours at 37°C. McrBC-digested DNA was then subjected to PCR amplification for 28 cycles. Bisulfite sequencing was performed using the EpiTect bisulfite conversion kit (Qiagen) and 2 μg of genomic DNA. Bisulfite-treated DNA was PCR-amplified using -397F and +81R primers (see above) and cloned into the pCR4-Topo vector (Invitrogen). Fifty-one clones from A. suecica wild-type plants and forty-seven clones from drm2- RNAi plants were sequenced. DNA methylation assays using methylation-sensitive restriction endonucleases were as described (Onodera et al., 2005).
Immunolocalization and DNA-FISH
Nuclei of leaves of 28 day-old plants were isolated as described (Onodera et al., 2005). Following fixation in 4% paraformaldehyde/PBS (phosphate buffered saline), four washes in PBS, and blocking at 37°C, slides were exposed to primary antiserum in PBS and 0.5% blocking reagent (Roche) overnight, using 1:200 dilutions of anti-MBD6 antisera or 1:500 mouse anti-GFP/YFP (BD Biosciences, CA, USA). Following four washes in PBS, slides were incubated at 37°C with FITC-conjugated anti-mouse or anti-rabbit secondary antibodies (Sigma-Aldrich) diluted 1:100. Nuclei were counterstained with 1μg/mL DAPI (Sigma-Aldrich) in Vectashield (Vector Laboratories). DNA-FISH using biotin-dUTP labeled 45S rRNA gene probes was as described (Pontes et al., 2003). Biotin labeled probes were detected using goat anti-biotin conjugated with avidin (1:200, Vector Laboratories) followed by streptavidin-Alexa 543 (Molecular Probes). For dual protein/nucleic acid localization, immunolocalization was followed by post-fixation in 4% formaldehyde/PBS and DNA-FISH.
Supplementary Material
Acknowledgments
Pikaard lab research was supported by United States NIH grant GM60380. RNAi vector development was supported by United States National Science Foundation grant 9975930. P.C.N. was supported by fellowships SFRH/BD/6520/2001 and SFRH/BPD/30386/2006 from the Fundação para a Ciência e Tecnologia, Portugal. The content of this paper is solely the responsibility of the authors and does not necessarily reflect the views of our sponsors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
S.P. conducted DNA methyltransferase and MBD knockdown screens and S1 nuclease protection, RT-PCR, DNA methylation and ChIP analyses of these RNAi lines. P.C.N. performed siRNA blots, DCL3 and RDR2 knockdown screens, and related S1 nuclease protection and RT-PCR assays of these lines. O.P. performed all microscopy. S.T. performed bioinformatic analyses identifying the hypermethylated intergenic spacer site and generated the figures showing siRNA positions and abundance. R.M, D. B., K. K. and J. C. provided siRNA deep sequencing data. W.V. and C.S.P. co-mentored P.C.N. C.S.P. wrote the manuscript.
References
- Aufsatz W, Mette MF, Van Der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. Embo J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Stoiber T, Rakic B, Naumann K. Arabidopsis histone deacetylase 6: a green link to RNA silencing. Oncogene. 2007;26:5477–5488. doi: 10.1038/sj.onc.1210615. [DOI] [PubMed] [Google Scholar]
- Bartee L, Bender J. Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 2001;29:2127–2134. doi: 10.1093/nar/29.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Pikaard CS. Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J Biol Chem. 2002;277:31577–31584. doi: 10.1074/jbc.M202737200. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental rRNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci USA. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997a;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA. 1997b;94:3442–3447. doi: 10.1073/pnas.94.7.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWinter R, Moss T. A complex array of sequences enhances ribosomal transcription in Xenopus laevis. J Mol Biol. 1987;196:813–827. doi: 10.1016/0022-2836(87)90407-4. [DOI] [PubMed] [Google Scholar]
- Doelling JH, Gaudino RJ, Pikaard CS. Functional analysis of Arabidopsis thaliana rRNA gene and spacer promoters in vivo and by transient expression. Proc Natl Acad Sci U S A. 1993;90:7528–7532. doi: 10.1073/pnas.90.16.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Pikaard CS. The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J. 1995;8:683–692. doi: 10.1046/j.1365-313x.1995.08050683.x. [DOI] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. Epigenetic mechanisms controlling RNA polymerase I transcription. Nature Rev Mol Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat Rev Genet. 2005;6:410–418. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase. Dnmt1 J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- Labhart P, Reeder RH. Enhancer-like Properties of the 60/81 bp Elements in the Ribosomal Gene Spacer of Xenopus laevis. Cell. 1984;37:285–289. doi: 10.1016/0092-8674(84)90324-6. [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Pikaard CS. Transgene-induced RNA interference: a strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant J. 2003;36:114–121. doi: 10.1046/j.1365-313x.2003.01857.x. [DOI] [PubMed] [Google Scholar]
- Lewis MS, Pikaard CS. Restricted chromosomal silencing in nucleolar dominance. Proc Natl Acad Sci USA. 2001;98:14536–14540. doi: 10.1073/pnas.251424098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harb Symp Quant Biol. 2006;71:449–459. doi: 10.1101/sqb.2006.71.028. [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008 doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Neves N, Heslop-Harrison JS, Viegas W. rRNA gene activity and control of expression mediated by methylation and imprinting during embryo development in wheat x rye hybrids. Theor Appl Genet. 1995;91:529–533. doi: 10.1007/BF00222984. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Pape LK, Henderson SL, Ryan K, Paalman MH, Lopata MA, Reeder RH, Sollner-Webb B. Enhancers for RNA polymerase I in mouse ribosomal DNA. Mol Cell Biol. 1990;10:4816–4825. doi: 10.1128/mcb.10.9.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Neves N, Silva M, Lee JH, Chen ZJ, Viegas W, Pikaard CS. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:11418–11423. doi: 10.1073/pnas.1932522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Silva M, Preuss S, Costa-Nunes P, Earley K, Neves N, Viegas W, Pikaard CS. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS ONE. 2007;2:e1157. doi: 10.1371/journal.pone.0001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Preuss S, Pikaard CS. rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim Biophys Acta. 2007;1769:383–392. doi: 10.1016/j.bbaexp.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH. Mechanisms of nucleolar dominance in animals and plants. J Cell Biol. 1985;101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J, Pikaard CS. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc Natl Acad Sci USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- Scebba F, Bernacchia G, De Bastiani M, Evangelista M, Cantoni RM, Cella R, Locci MT, Pitto L. Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Mol Biol. 2003;53:715–731. doi: 10.1023/B:PLAN.0000019118.56822.a9. [DOI] [PubMed] [Google Scholar]
- Springer NM, Kaeppler SM. Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiol. 2005;138:92–104. doi: 10.1104/pp.105.060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. Embo J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Zemach A, Grafi G. Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 2003;34:565–572. doi: 10.1046/j.1365-313x.2003.01756.x. [DOI] [PubMed] [Google Scholar]
- Zemach A, Li Y, Wayburn B, Ben-Meir H, Kiss V, Avivi Y, Kalchenko V, Jacobsen SE, Grafi G. DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant Cell. 2005;17:1549–1558. doi: 10.1105/tpc.105.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. Embo J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


