Abstract
Neuroglobin (Ngb) is a hexacoordinate globin expressed in the brain of vertebrates. Ferrous Ngb binds dioxygen with high affinity and the O2 adduct is able to scavenge NO. Convincing in vitro and in vivo data indicate that Ngb is involved in neuroprotection during hypoxia and ischemia. The 3D structure of Ngb reveals the presence of a wide internal cavity connecting its heme active site with the bulk. To explore the role of this “tunnel” in the control of ligand binding, we determined the structure of metNgb and NgbCO equilibrated with Xe or Kr. We show four docking sites for Xe (only two for Kr); two of the four Xe sites are within the large cavity. They are only partially conserved in globins, since the two proximal Xe sites identified in myoglobin (Xe1 and Xe2) are absent in Ngb, as well as in cytoglobin. The Xe docking sites in Ngb map a pathway within the protein matrix, leading to the heme, which becomes more accessible in the ligand-bound species. This may be of significance in connection with the redox chemistry that may be the primary function of this hexacoordinate globin.
Introduction
During the last decade, several new members of the globin superfamily have been discovered, including neuroglobin (Ngb), cytoglobin (Cygb), globin E (GbE, eye-specific in chickens), and globin X (GbX, in fish and amphibians) (1–5). Since both Cygb and Ngb are intracellular and expressed at low concentrations, there was considerable interest in understanding their physiological role.
In the case of Ngb, which is expressed in the brain, in vivo studies have shown that it counteracts the damage due to hypoxic states, such as stroke and ischemia, since 1), its expression in neurons is upregulated under hypoxia; and 2), its overexpression reduces the extent of tissue damage after experimental stroke in rats (6–8). The mechanism underlying Ngb-mediated neuroprotection is still under debate, however, as is its function under normoxic conditions. It has been proposed that Ngb promotes O2 supply to mitochondria (9). Ngb indeed binds O2 reversibly with high affinity (P1/2 ≈ 2 torr (1,10)), and therefore, in normoxic cells, Ngb should be present in the oxygen-bound form (oxyNgb), although its in vivo concentration in the micromole range seems hardly compatible with O2 transport alone. During hypoxia, the population of deoxy ferrous Ngb, which was shown in vitro to reduce cytochrome c (Cyt-c) (11), increases. Since the release of oxidized Cyt-c into the cytosol is a proapoptotic signal, reduction of Cyt-c may prevent apoptosis (12). Moreover, hypoxia is associated with an increase in NO and related radicals; in vitro, oxyNgb and nitrosyl-Ngb have been shown to scavenge these radicals, yielding harmless species and metNgb (13,14).
The transition to the unbound state makes (ferrous and ferric) Ngb competent for binding the α-subunit of heterotrimeric G-proteins (15,16), possibly through the structural transition observed upon ligation in the CO bound structure (17,18). In summary, given the scavenging activity and the reduction of Cyt-c, the primary role of Ngb may be as an NO/O2 sensor that regulates apoptotic pathways and, by interacting with G proteins, induces signaling of hypoxia and activation of defense mechanisms in neurons and retinal cells (12,19).
Ngb and Cygb share little amino acid sequence similarity with vertebrate myoglobins and hemoglobins (∼25%) (1–3); nevertheless, they adopt the typical globin fold (20,21). Three remarkable peculiarities of Ngb can be pointed out: 1), the ferric and ferrous heme iron are both hexacoordinate, with the distal His(E7) and the proximal His(F8) directly bound to the metal; 2), a wide, branched cavity (∼300 Å3) connecting the heme pocket with the solvent is present in the core structure; and 3), binding of CO to ferrous Ngb is associated with a conformational change involving a sliding motion of the heme and a moderate shift in the position of helix F and loops CD and EF (17). Extensive kinetic data (22–26) have clarified the relevance of the endogenous hexacoordination for Ngb reactions. This feature requires that binding of external ligands (e.g., O2) to the heme iron can only occur upon rupture of the bond with His(E7). It implies equilibrium between hexa- and pentacoordinate states, which limits the ligand-binding rate. Moreover, bis-histidyl coordination enhances the rate of electron transfer to or from the heme iron (27), consistent with a role of Ngb in redox reactions such as in vitro NO scavenging (13) and Cyt-c reduction (11).
On the other hand, the functional role of the huge internal cavity has not been investigated in an equally exhaustive manner. This tunnel extends around the heme with two branches, connecting the distal and proximal heme pockets with the bulk. It has been shown that structural rearrangements coupled with CO binding affect the topology of this cavity. The distal branch is enlarged and the proximal branch drastically reduced, suggesting that the wide cavity may be necessary to accommodate the heme during the ligand-linked sliding motion (17,18). Molecular dynamics (MD) simulations also highlight some similarities between this large cavity and the four Xe binding sites of sperm whale myoglobin (swMb) (28).
Crystallographic studies of photodissociated intermediates of wild-type (wt) and mutant Mb (29–31) have shown that these Xe binding cavities play a role as transient docking sites for heme ligands, providing a structural interpretation for geminate rebinding kinetics (32). These results convey the general picture that internal cavities of Mb are involved in regulating the diffusion dynamics of small gaseous ligands through the protein matrix (32–34).
Four Xe binding sites also have been recently observed in Cygb (35), a vertebrate globin that shares with Ngb two structural peculiarities: endogenous hexacoordination and a large internal cavity. It seems unlikely that packing defects, which imply a relevant offset in terms of thermodynamic stability, would be conserved during evolution without fulfilling a functional requirement. Therefore, the large cavities found in Ngb and Cygb could play a similar role with respect to Mb Xe sites, possibly sharing a common origin.
We have investigated the large branched cavity of Ngb to better pin down its similarities with, as well as how it differs from, those characterized in Mb and Cygb. We determined the structure of metNgb after derivatization with Xe and Kr to identify internal hydrophobic niches compatible with the size of gaseous heme ligands (NO, O2, and CO). The crystal structure of oxidized murine Ngb(FeIII) (metNgb) and reduced carbonmonoxy Ngb(FeII) (NgbCO) showed four Xe docking sites that only partially correlate with the Xe sites of Mb and Cygb (28,35). Comparison of the sequences and the Xe-bound structures of Ngb, Cygb, and Mb allowed us to elucidate how far these hydrophobic niches are conserved in these monomeric globins. We believe that the tunnel may be a pathway for gaseous ligands to enter into the protein matrix, providing an alternative to the classical “histidine gate” (32,36,37) and possibly having significance with reference to the hexacoordinate configuration of the heme in Ngb and Cygb.
Materials and Methods
Ngb purification, crystallization, and derivatization
Expression, purification, and crystallization of recombinant metNgb were carried out as described in Arcovito et al. (38). To allow Xe diffusion and docking within the protein matrix, both metNgb and NgbCO crystals in their own cryoprotectant solutions were incubated in a pressure chamber (Xcell, Oxford Cryosystems, Oxford, United Kingdom) at 20 bar Xe for 20 min. Kr derivatization was carried out in the BESSY-MX sample preparation lab, where metNgb crystals were exposed to 40 bar Kr for 52 min in a Hampton Research (Aliso Viejo, CA) xenon chamber. After incubation, all the crystals were frozen in liquid N2. To prepare the NgbCO derivative, metNgb crystals were transferred into a glass slide filled with mother liquor, which contained 25% glycerol as cryoprotectant and 4 mM sodium dithionite that had been previously purged with N2, to achieve reduction of the heme iron. The reduced crystals were then soaked in the same solution, which contained 1 mM CO to allow binding. As previously observed (17), CO binding to Ngb determines deterioration, and eventually loss, of crystal order, which could be due to the rearrangements observed in the CO structure. Nevertheless, the crystals were good enough to describe and analyze Xe atom positions. Crystallization of Ngb in the CO state (which has been unsuccessful) will make it possible to clarify whether the transition upon ligand binding exceeds that observed to date; nevertheless, we know, from MD simulations in solution and in crystals, that the only conformational change involved in crystal disorder may be thus related to the motion of the CD loop on the surface (18).
However, NgbCO crystals are stable for 4–6 h, thus allowing incubation in the pressure chamber and freezing with N2(l). This procedure, involving three soakings in sequence (in dithionite, CO, and Xe), leads to some loss of crystal order, as inferred from the increased mosaicity and a reduced diffraction limit, with respect to untreated metNgb crystals (see Table S1 in the Supporting Material). Furthermore, it was impossible to fully occupy the CO binding site of Ngb after Xe derivatization. The best result we obtained was a CO occupancy of 83% (slightly lower than previously observed, i.e., 90% (17)). We attribute this result to the fragility of NgbCO crystals, although we cannot exclude the possibility that exposure to high Xe pressure may cause CO dissociation and promote iron oxidation.
Data collection and structure determination
X-ray diffraction data sets were all collected at 100 K, at three different synchrotron sources: metNgb-Xe at the European Synchrotron Radiation Facility, beamline ID14.2 (Grenoble, France; detector ADSC Quantum 4, x-ray wavelength 0. 933 Å); NgbCO-Xe at Elettra, beamline XRD1 (Trieste, Italy; detector MAR165, x-ray wavelength 1.20025 Å); and metNgb-Kr at the BESSY, BL14.1 (Berlin, Germany; detector MX225, x-ray wavelength 0.86437 Å, corresponding to the Kr k-edge).
All crystals were isomorphous with those previously measured (17,21). Data for metNgb-Xe and NgbCO-Xe were indexed, integrated, and scaled using the programs DENZO and SCALEPACK (39) or MOSFLM and SCALA (40). The CCP4 program suite (41) was used for subsequent data evaluation and structure refinement (Table S1).
The processing of metNgb-Kr data was carried out using XDS and XSCALE programs (42). After anomalous scaling, preliminary analysis of the anomalous difference Fourier map indicates the presence of two Kr atoms per Ngb molecule. Model refinement was carried out on data without anomalous contributions, taking into account the presence of two Kr atoms per molecule.
Initial phases were derived from the deposited structures of Ngb and NgbCO (17,21). The models were subjected to iterative rounds of refinement and model building. Refinement was carried out with Refmac5 in CCP4 (43), followed by model adjustment and water addition with COOT (44). Final cycles of refinement were carried out using TLS refinement as implemented in Refmac5 (45).
Noble gas atoms were added to the models after refinement of the protein model, in positions corresponding to positive electron density peaks. Kr atoms were assigned based on the anomalous difference Fourier map. To assign an electron density peak to a Xe atom, before modeling it, we inspected the Fo-Fc difference map using a cutoff of 6σ. Xe and Kr occupancies were refined, starting from a value of 100% and decreasing the occupancy until there were no residuals in the difference maps and their B-factors were in the same range as those of the protein atoms. The quality of the final model was analyzed by using PROCHECK (46).
The final models do not contain amino acids 1, 2, and 151. NgbCO•Xe structure lacks also residues Asp149 and Gly150; in this model, Arg3 is refined as Ala, since the corresponding electron density shows no side chain. The N- and C-terminal regions, as well as the CD and EF corners, are more mobile than the rest of the structure, showing higher values of B factors, as already reported (38). Superposition and structure alignment of main chains between mNgb•Xe, hCygb•Xe, swMb•Xe, and Hb from antarctic fishes were carried out by means of an incremental combinatorial extension (CE) method (47).
To facilitate comparison with previously published data on swMb and human Cygb (28,35), we chose the same numbering for sites located in the same topological positions and labeled with roman numerals the Xe and Kr atoms bound to Ngb.
The protein cavity represented in Fig. 1 A was created using the program CAVER (48). All the figures, with the exception of one (see Fig.3 A) were drawn using PyMol (49).
Figure 1.
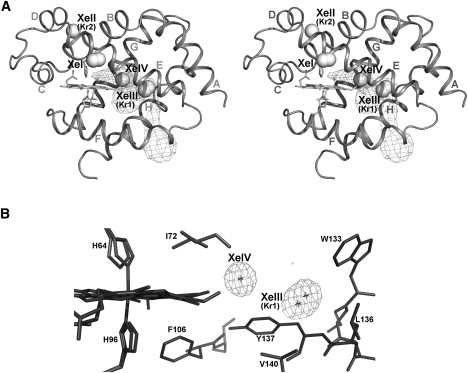
Xenon and krypton binding to Ngb. (A) Stereo view of Ngb, showing the location of the four Xe atoms and the two Kr atoms represented as spheres. α-Helices are labeled according to the globin fold nomenclature. The shape and position of the main tunnel, connected with the bulk, and its distal branch were calculated using CAVER (46). (B) The XeIII and XeIV atoms in the main cavity, limited by the heme and neighboring side chains, are shown. The electron density for XeIII and XeIV atoms is depicted (2Fo-Fc map, contoured at 1.8σ).
Figure 3.
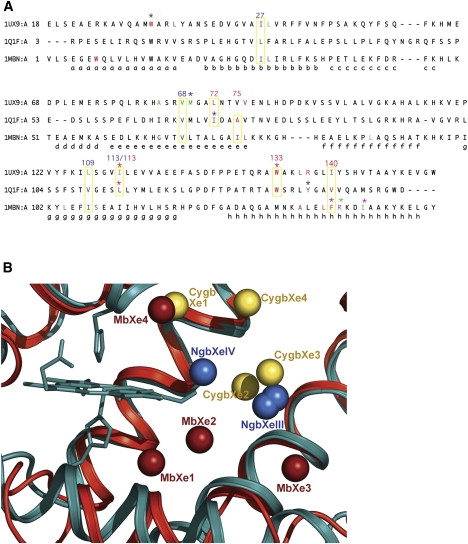
Structural superposition of Ngb•Xe with swMb•Xe and hCygb•Xe. (A) Structure-based sequence alignments were performed using CE (46). Sequences are reported with their relative PDB code (1UX9 for Cygb•Xe; 1Q1F for Ngb; and 1MBN for swMb), and the α-helices are labeled according to the globin fold topology rules. Amino acids that form a xenon site are color coded green (Xe1), magenta (Xe2), red (Xe3 and XeIII), and blue (Xe4 and XeIV). Amino acids that participate at two sites are treated with the same color code but have an asterisk above them. The alignments show that some residues belonging to the NgbXeIII site (red; I72, A75, L113, W133, and V140) or the NgbXeIV site (blue; L27, V68, V109, and L113) are conserved or have the same chemical nature with respect to the amino acids that form the Xe sites of Cygb and/or Mb. We enclose these residues in yellow boxes, also indicating their position in Ngb. (B) Comparison of the Xe atom positions in Ngb (blue), Cygb (yellow), and Mb (red) after superposition of the protein main chain only. The Xe atoms are represented as spheres. Cygb and the heme group of Mb are not shown. Distances between Xe atoms are reported in Table 2. Xe proximal sites are present only in Mb.
Coordinates were deposited at the Protein Data Bank (www.rcsb.org) with accession numbers 3GK9, 3GKT, and 3GLN for metNgb-Xe, metNgb-Kr, and NgbCO-Xe, respectively.
Results
Xenon and krypton binding to metNgb
We determined the crystal structure of metNgb derivatized with either Xe or Kr at 1.8 Å and 1.9 Å resolution, respectively (Ngb-Xe and Ngb-Kr; see Table S1). Since binding of Xe to internal packing defects may be limited by its radius of 2.16 Å, we decided to use Kr as an additional probe, given its smaller van der Waals radius of 2.02 Å. Electron density maps show the presence of four Xe atoms in metNgb, which we numbered XeI–XeIV (Fig. 1 A). On the other hand, we detected only two Kr binding sites, called KrII and KrI (equivalent to XeII and XeIII in Fig. 1 A). Since at low occupancy Kr atoms can hardly be distinguished from H2O, we used the Kr anomalous signal for its unambiguous identification. The data show that Kr derivatization identifies no alternative hydrophobic pockets in addition to those mapped with Xe. However, we cannot exclude that mobility of Kr in cavities may prevent its detection by x-ray diffraction.
The XeI atom is on the external Ngb surface at the interface between two symmetry-related molecules; this site hosts two alternative positions for XeI, each with partial occupancy.
XeII and KrII are both placed near the interface with a symmetry-related molecule in a small breach accessible to the solvent (Fig. 1 A); the distance between XeII and the heme iron is 13.3 Å. Moreover, XeII is in contact with the conserved Cys55 that in human Ngb is involved in a functionally relevant disulphide bond (24).
Xe atoms in positions III and IV are hosted in two adjacent hydrophobic niches located inside the large internal cavity (Fig. 1, A and B). The XeIII docking site is an apolar pocket close to the EF corner, at the entrance to the tunnel that connects the heme iron with the bulk (21). The distance between XeIII and the heme vinyl of pyrrole C is 6.38 Å. This pocket displays the highest occupancy for both Xe and Kr, hosting KrI with 47% occupancy and two XeIII atoms in alternative positions (XeIII a/b), which together account for an occupancy of 80% (Table 1).
Table 1.
Distances between noble gas atoms and protein atoms
| Xe atom | Occupancy (%) | B-factor (Å2) | Neighbor amino acid | Distance (Å) |
|---|---|---|---|---|
| Ngb•Xe distances | ||||
| XeI a/b | 40/15 | 20/19 | D63-Cβ | 4.84 |
| K67-Cɛ | 3.91 | |||
| XeII | 48 | 14.8 | T25-CO | 4.78 |
| F28-Cβ | 4.26 | |||
| F32-Cδ2 | 3.69 | |||
| P52-Cβ | 4.67 | |||
| S56-Oγ | 3.33 | |||
| L56-Cδ2 | 4.37 | |||
| F61-Cɛ2 | 4.21 | |||
| XeIII a/b | 45/35 | 16.2/13.2 | I72-Cγ1 | 4.40 |
| A75-Cβ | 4.54 | |||
| L113-Cδ2 | 4.35 | |||
| W133-Cζ3 | 4.55 | |||
| Y137-Cδ1 | 3.97 | |||
| V140-Cγ2 | 3.88 | |||
| XeIV | 38 | 18.7 | L27-Cδ1 | 4.41 |
| V68-Cγ1 | 4.22 | |||
| I72-Cδ | 3.56 | |||
| V109-Cg1 | 3.99 | |||
| L113-Cδ2 | 4.36 | |||
| Y137-Cɛ1 | 3.81 | |||
| Ngb•Kr distances | ||||
| Kr1 | 47 | 43.2 | L36- Cδ | 3.95 |
| I72−Cγ2 | 3.87 | |||
| W133-Cζ | 4.03 | |||
| Y137-Cδ1 | 4.21 | |||
| V140-Cγ2 | 4.08 | |||
| Kr2 | 32 | 41.6 | A29-Cα | 3.72 |
| F32-Cδ2 | 3.53 | |||
| L56-Cδ2 | 3.31 | |||
| F61-Cɛ2 | 4.06 | |||
| NgbCO•Xe distances | ||||
| XeI a/b | 40/40 | 61.1 | K67-Cγ | 4.39 |
| L72-Cδ2 | 4.19 | |||
| V71-Cγ2 | 3.88 | |||
| Y88-OH | 3.56 | |||
| XeII | 40 | 40.8 | F28-Cβ | 4.27 |
| A29-Cβ | 3.48 | |||
| F32-Cδ1 | 3.69 | |||
| S55-Oγ | 3.30 | |||
| L56-Cδ2 | 3.12 | |||
| F61-Cɛ2 | 4.15 | |||
| XeIIIa/b | 53/25 | 29.4/20.8 | I72-Cγ2 | 4.53 |
| A75-Cβ | 4.71 | |||
| L113-Cδ2 | 4.25 | |||
| W133-Cζ3 | 4.60 | |||
| L136-Cδ2 | 4.37 | |||
| Y137-Cδ1 | 4.01 | |||
| V148-Cγ2 | 3.77 | |||
| XeIV | 40 | 26.8 | L27-Cδ1 | 4.36 |
| V68-Cγ1 | 4.16 | |||
| I72-Cδ1 | 4.36 | |||
| V109-Cγ1 | 3.72 | |||
| L113-Cδ2 | 4.65 | |||
| Y137-Cɛ1 | 4.14 | |||
| hemeA, vynil pyrrole B | 4.06 | |||
Distances between noble gas atoms and protein atoms are reported for the metNgb-Xe model, metNgb-Kr model, and NgbCO-Xe model. We list only distances <5.0 Å.
XeIV is the closest to the iron and is located in the distal branch of the large cavity, between XeIII and the heme. After refinement, the XeIV occupancy was only 38%, possibly due to proximity to the heme, which is known to occur in two conformers called A and B (ratio 70:30) (21,50). Indeed, in the B conformer, the porphyrin vinyl group of pyrrole B introduces some steric hindrance that may interfere with Xe docking given their short distance (2.56 Å).
As shown in Fig. 1 A, XeIII and XeIV seem to define a path inside the Ngb large cavity, which connects the protein surface to the heme distal site.
There is no evidence for binding of Xe or Kr on the proximal heme side of metNgb, similar to the case of Cygb (35).
Xenon binding in NgbCO
Ligand binding to ferrous Ngb promotes heme sliding in a preformed pocket, which in metNgb is occupied by F106 and is part of the large tunnel; as a result, the heme sliding obliterates the tunnel proximal branch and enlarges the distal branch, inducing a shift in the F helix and a rearrangement in the CD corner, E helix, and EF loop (17,18).
To investigate whether this transition affects Xe docking, we exposed NgbCO crystals to Xe. In the dataset of higher quality, Ngb was not fully saturated with CO (83%), possibly due to the fact that the procedures to obtain NgbCO•Xe crystals required three soakings in sequence. To date, there is no other way to produce a CO-bound Ngb crystal, and we had to compromise. The presence of some ligand-free hexacoordinate Ngb introduces a significant heterogeneity in the structure, mainly in the position of the heme and nearby residues. Nevertheless electron density maps clearly show the presence of four Xe atoms that occupy the same binding pockets observed for metNgb, with the exception of XeI, which is still at the interface between two symmetry-related molecules but in a different position. The occupancy of the four Xe sites is slightly decreased, with the exception of XeIV, which is marginally increased (from 38% to 40%; Table 1). It is, however, significant that after CO binding, the distance between the Fe and XeIV decreases from 9.06 Å (metNgb) to 6.97 Å.
Unlike the metNgb•Xe structure, in NgbCO the XeII atom displays a B factor higher than that of the other sites (see Table 1), without significant differences in the orientation of the side chains forming this niche.
We noticed (Fig. 2) some electron density between CO and the XeIV atom, which we could not satisfactorily refine as a water molecule. It may be accounted for by a small fraction of photodissociated CO, which we know to occur due to x-ray radiation breakage of the Fe-CO bond, as recently seen (38) by x-ray absorption near-edge spectroscopy.
Figure 2.
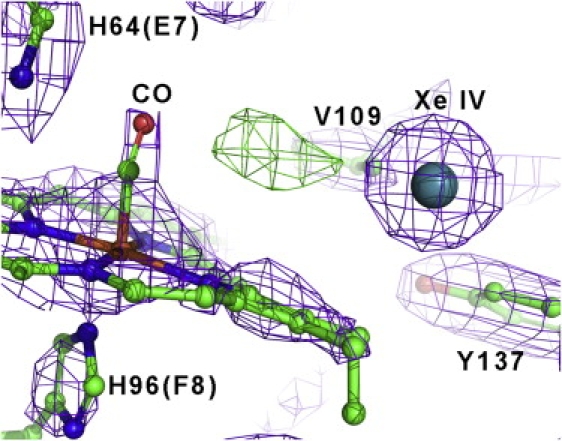
Xenon binding in the distal heme pocket of CO-Ngb electron density map in the XeIV niche, for the carbonmonoxy-Ngb•Xe structure (2Fo-Fc map, contoured at 1.7σ). XeIV is depicted as a sphere. The Fo-Fc map at 3.3σ is also displayed in green, showing an unmodeled positive electron density, which may be due to a small fraction of photodissociated CO (see Results).
In summary, from the experiment on NgbCO, we conclude that Xe docking is not affected by the structural rearrangements coupled to ligand binding. Remarkably, after heme sliding, the CO bound to the heme is located in front of XeIV at a distance of 6.97 Å. This location of XeIV with respect to the bound ligand could be functionally relevant; we speculate that in oxyNgb, a second ligand (e.g., NO (13)), either already docked in the XeIV site or incoming through the distal branch of the tunnel, would be positioned favorably for reacting with the O2 bound to the iron.
Structural superposition and sequence alignments with Mb and Cygb
The structure of metMb from sperm whale was shown to contain four Xe atoms that occupy preexisting cavities (28). Some of these are closer to the heme pocket: Xe1 and Xe2 on the proximal heme side, and Xe4 on the distal side. By using the same approach, de Sanctis et al. (35) recently analyzed the human Cygb structure, identifying four Xe atoms per molecule. It was surprising to find that in Cygb there are no proximal Xe binding sites like those found in Mb (Xe1 and Xe2), similar to our observations for Ngb.
To elucidate similarities and differences among Ngb, Cygb, and Mb, we used CE (47) for superposition of the structures in complex with Xe, and for calculation of a structure-based multiple sequence alignment. Sequence alignment (Fig. 3 A) shows that all these pockets are coated by residues belonging to the same secondary structure elements, analogous in their polarity and structurally aligned or fully conserved in some positions (see boxes 72, 113, 133, and 137 in Fig. 3 A). Indeed, we notice a remarkable conservation for the amino acids coating pockets MbXe4, NgbXeIV (blue) and CygbXe1 (and, partially, CygbXe4; see boxes 27, 68, 109, and 113 in Fig. 3).
The root mean-squared deviation (RMSD) between Cα positions was, respectively, 1.7 Å and 2.0 Å for Ngb versus Cygb and Ngb versus Mb. The number of structurally equivalent residues was 141 for Ngb versus Cygb and 136 for Ngb versus Mb. The optimized structural superposition of Ngb and Cygb shows that NgbXeIII is close to CygbXe3 and CygbXe2 (see Table 2), and their positions appear by and large equivalent (see Fig. 3 B). The Xe atoms in Cygb (Xe1, Xe2, and Xe3) and Ngb (XeIII and XeIV) all map in the large tunnel, but overlap only partially. Moreover, the comparison with Mb shows that NgbXeIII and XeIV have roughly similar but not identical positions compared to MbXe3 and MbXe4, respectively (see Fig.2 B). The NgbXeIV site is located between two different Mb sites (see Fig. 3 B and Table 2), i.e., MbXe2 and MbXe4 (at distances of 5.01 Å and 5.89 Å, respectively). On the other hand, it has been reported that the MbXe4 site is coincident with CygbXe1 (35).
Table 2.
Distances among xenon sites after superposition
| Ngb Xe and swMb Xe atoms | ||
|---|---|---|
| Ngb•Xe atom | swMb•Xe atom | Distance (Å) |
| XeIII | Xe2 | 5.51 |
| Xe3 | 4.95 | |
| XeIV | Xe2 | 5.01 |
| Xe4 | 5.89 | |
| Ngb Xe and hCygb Xe atoms | ||
| Ngb•Xe atom | hCygb•Xe atom | Distance (Å) |
| XeIII | Xe2 | 4.03 |
| Xe3 | 2.03 | |
| XeIV | Xe1 | 5.45 |
| Xe2 | 4.66 | |
| Xe3 | 4.75 | |
| Xe4 | 5.84 | |
Distances of Ngb XeIII and XeIV atoms with respect to Xe positions in Mb and Cygb. The superposition of protein structures was carried out using CE (46).
Using CE, we compared the structures of Hb β-chains from the antarctic fishes T. newnesii and T. bernacchii (HbTn-β and HbTb-β, respectively (51,52)) with both Ngb•Xe and Mb•Xe, to clarify whether the peculiar cavity pattern of Ngb is correlated with hexacoordination. The RMSD for each alignment was in the same range as for the structural alignment of Ngb versus Mb, and the resulting sequence identity was low (RMSDs of 1.8 Å2 for Ngb versus HbTn-β, 2.1 Å2 for Ngb versus HbTb-β, 1.6 Å2 for HbTn-β versus Mb, and 1.7 Å2 HbTb-β versus Mb; sequence identity of 20.1% for Ngb versus HbTn-β, 18.7% for Ngb versus HbTb-β, 21.1% for Mb versus HbTn-β, and 22.1% for Mb versus HbTb-β).
In both HbTn-β and HbTb-β, we observed that Xe atoms docked internally to Ngb (NgbXeIII and XeIV) cannot be positioned in these fish Hb β-chains without collision with protein atoms. On the other hand, when we superposed Mb in complex with xenon onto HbTn-β and HbTb-β, the atoms MbXe2, MbXe3, and MbXe4 could be accommodated without hindrance, whereas there was no space for the proximal MbXe1 because of amino acid substitutions that cancel this pocket in Mb.
In summary, this comparison indicates that 1), in analogy with Cygb, the large inner cavity in Ngb, coated by hydrophobic residues, is able to host noble gases; 2), NgbXeIII displays a topology fairly similar with respect to the Xe3 sites in Cygb and Mb; 3), the NgbXeIV site shares similar residues with MbXe4 and CygbXe1, but its topological location is peculiar; 4), the MbXe1 and MbXe2 on the proximal side of the heme are absent in Ngb, as well as in Cygb (35).
Discussion
Mb and Cygb form a common branch within the tree of vertebrate globins. Ngb instead belongs to a different lineage that diverged 700 million years ago, before the split of Protostomia and Deuterostomia, and includes annelid nerve Mb and Chordata hemoglobins (Hbs) (5). Nevertheless, despite low sequence homology (1–3) Mb, Cygb, and Ngb share the typical globin fold.
Within the framework of the classical globin fold, Ngb and Cygb share two peculiar structural features, i.e., an endogenous hexacoordination with the distal His(E7), which regulates ligand binding (22,23) and promotes electron transfer rates (27); and a wide, branched cavity (∼300 Å), connecting the heme proximal and distal pockets with the exterior (20,21).
In Mb, the four Xe binding cavities, with volumes ranging from 50 to 80 Å3 (28), were reported to regulate the internal dynamics of small gaseous ligands (i.e., O2, CO, and NO). These cavities are supposed to facilitate protein motions and side-chain conformational changes involved in the gated migration of ligands through the protein matrix (32,33). Ligand docking within, and hopping between, Mb cavities is one of the factors controlling the rate of geminate rebinding, which involves rapid intramolecular diffusion to the heme after photodissociation (34). These findings suggest that the Mb cavities have been conserved, despite their thermodynamic cost, because they fulfill a role in the protein dynamics and in the control of ligand binding (33).
Why do Ngb and Cygb maintain a large internal cavity that exceeds the sum of the packing defects in Mb, and does the cavity have a functional role?
In the case of Ngb, MD simulations and crystallography (17,18) suggest that the huge cavity provides space for the rapid (∼90-ns) heme sliding motion coupled to CO binding, thus accounting for the similar thermodynamic stability of the met and CO-bound states. This transition is associated with the shift of helix F, which, together with H64 rotation (18), determines the repositioning of CD and EF loops upon ligand binding. Since the CD-D region of Ngb, especially residues Glu51 and Glu60, is responsible for Ngb binding to the regulatory Gα subunit of heterotrimeric Gαβγ proteins, the heme-linked structural rearrangement may regulate this interaction (15,19). We suggest that the presence of a wide cavity facilitates the signaling activity of Ngb by fast ligand-linked conformational changes.
Our results show that the wide Ngb tunnel contains two hydrophobic pockets that bind noble gases, called XeIII (KrI) and XeIV. The positions of these pockets suggest that they may play a role as ligand docking sites, in analogy with the function of Xe sites in Mb. Based on structure superposition and sequence alignment, we note the presence of residue conservation and a similar topology between MbXe4, CygbXe1, and NgbXeIV. Indeed, they are all on the distal heme side, a location favorable for ligand docking on its way onto and off the iron. Therefore, a photodissociated ligand might temporarily occupy a niche on the distal side (NgbXeIV, MbXe4, and CygbXe1), and eventually diffuse either back toward the heme iron or farther away. The pathway, however, is clearly different for the three proteins. In Mb the ligand diffuses from the distal primary docking site into MbXe4, populating the proximal cavities MbXe2 and MbXe3 and finally ending in the proximal side MbXe1 (see Fig. 3 B). The description of this complex route is due mainly to time-resolved crystallography experiments and confirmed by MD simulations (53–56). However, because the proximal cavities are absent in Ngb and Cygb, the most likely pathway for ligand diffusion is from the distal NgbXeIV niche toward the solvent via the NgbXeIII pocket, which exhibits an affinity for nonionic gases, and through the tunnel into the solvent.
As a consequence of the heme sliding associated with binding of CO to ferrous Ngb, the NgbXeIV niche is closer to the CO-bound iron, and it directly faces the bound Xe (Fig. 2). We speculate that this configuration may be a component of the high reactivity of oxyNgb with NO (13), if the latter was momentarily trapped in the matrix and possibly docked in the XeIV site.
Finally, Fourier transform infrared spectroscopy, temperature derivative spectroscopy, and kinetic analyses of photolyzed NgbCO showed the presence of multiple ligand docking sites in the distal heme pocket and a very fast rate for geminate rebinding (57). In our opinion, the XeIII and XeIV pockets, which are located inside the tunnel and are directly connected to the heme by a wide empty space, may be compatible with the docking and kinetics shown in these experiments.
In line with a popular hypothesis, we assume that these sites are part of a pathway for ligands through the protein matrix connecting the heme iron to the exterior via the tunnel gate, which may be either complementary or alternative to the classical “histidine gate” (32,36,37). A similar viewpoint has been proposed for the apolar channels typical of group I truncated globins, namely M. tuberculosis HbN (58) and others (59). Moreover, in the mini-Hb from C. lacteus, the apolar tunnel is the major pathway for ligand entry and exit; kinetic, structural, and mutational analysis showed that this pathway accounts for the ligand association and dissociation rates, and for geminate kinetics (60).
One might speculate that the cavity found in Ngb could allow the entry of bulky ligands, in analogy to the behavior of Vitreoscilla Hb, which displays binding of cyclopropanated phospholipids and hydroperoxide reductase activity (61). Nevertheless, to our knowledge, no lipid binding by Ngb has yet been shown.
Concerning the conservation of cavities among globins, previous crystallographic data suggested that the Ngb tunnel partially overlaps the MbXe sites (21). Moreover, MD simulations exploring fluctuations of the Ngb internal space pointed out some correspondence between the MbXe sites and the Ngb packing defects due to the presence of the wide tunnel (18).
The idea that the Ngb tunnel is evolutionarily related to a subset of Mb cavities is supported by our comparative analysis, which shows that some of the Xe docking sites are common to Mb, Cygb, and Ngb. These are lined by partially or totally conserved residues belonging to the same secondary structure elements in all three proteins (see Fig. 3 A) and are mostly characterized by apolar side chains. On the other hand, especially for NgbXeIV, they do not coincide exactly after superposition (distances reported in Table 2); more significantly, both Ngb and Cygb lack the proximal docking sites detected in Mb (Xe1 and Xe2; see Fig. 3 B), as well as HbA (62).
We compared the hexacoordinated β-chains of antarctic fish Hbs with Ngb-Xe and Mb-Xe and concluded that in this case, the pattern of internal cavities is very different with respect to Ngb, despite the common trait of heme iron hexacoordination. The possible docking sites in β-chains of antarctic fishes seem more compatible with the sites described for Mb and HbA β chains (62).
In conclusion, we believe that, starting from a common ancestral precursor, packing defects evolved into a discrete cavity network in Mb, or in a unique huge cavity endowed with distal and proximal branches in Ngb and Cygb. Indeed, Cygb and Ngb share with Mb the presence of hydrophobic pockets that are partially similar in topology. At variance with Mb, however, these pockets are internal to the tunnel, and none is observed in the proximal heme site.
If the tunnel indeed provides a passageway from the solvent to the distal heme site, such a different configuration may be correlated with endogenous hexacoordination with His(E7), which enhances redox reactivity by reducing the reorganization energy term at the heme level (27,38), but also represents a barrier to ligand binding. In the presence of endogenous hexacoordination, the Ngb and Cygb tunnel would facilitate ligand access to the sixth coordination position and also provide a reaction chamber for catalysis and radical scavenging, which are suggested to be early functions of globins with respect to the more recent adaptation for O2 storage and delivery.
Acknowledgments
We thank Caterina Rondinelli, Stefano Franceschini, Georg Zocher, and Stefano Lucidi for their skillful assistance, and Veronica Morea for useful discussions about sequence alignment. Data were collected on beamlines BL14.2 of BESSY (Helmholtz-Zentrum Berlin and BESSY II), ID.14.2 of the European Synchrotron Radiation Facility, Grenoble, France, and BL5.2R (XRD1) of Elettra (Basovizza, Trieste, Italy).
This project was partially supported by MiUR of Italy (FIRB RBLA03B3KC_004 and PRIN 2007 to M.B.).
Supporting Material
References
- 1.Burmester T., Weich B., Reinhardt S., Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 2.Burmester T., Ebner B., Weich B., Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol. Biol. Evol. 2002;19:416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 3.Trent J.T., III, Hargrove M.S. A ubiquitously expressed human hexacoordinate hemoglobin. J. Biol. Chem. 2002;277:19538–19545. doi: 10.1074/jbc.M201934200. [DOI] [PubMed] [Google Scholar]
- 4.Kugelstadt D., Haberkamp M., Hankeln T., Burmester T. Neuroglobin, cytoglobin, and a novel, eye-specific globin from chicken. Biochem. Biophys. Res. Commun. 2004;325:719–725. doi: 10.1016/j.bbrc.2004.10.080. [DOI] [PubMed] [Google Scholar]
- 5.Roesner A., Fuchs C., Hankeln T., Burmester T. A globin gene of ancient evolutionary origin in lower vertebrates: evidence for two distinct globin families in animals. Mol. Biol. Evol. 2005;22:12–20. doi: 10.1093/molbev/msh258. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y., Jin K., Mao X.O., Zhu Y., Greenberg D.A. Neuroglobin is up-regulated by and protects neurons from hypoxic-ischemic injury. Proc. Natl. Acad. Sci. USA. 2001;98:15306–15311. doi: 10.1073/pnas.251466698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y., Jin K., Peel A., Mao X.O., Xie L. Neuroglobin protects the brain from experimental stroke in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:3497–3500. doi: 10.1073/pnas.0637726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan A.A., Wang Y., Sun Y., Mao X.O., Xie L. Neuroglobin-overexpressing transgenic mice are resistant to cerebral and myocardial ischemia. Proc. Natl. Acad. Sci. USA. 2006;103:17944–17948. doi: 10.1073/pnas.0607497103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hankeln T., Ebner B., Fuchs C., Gerlach F., Haberkamp M. Neuroglobin and cytoglobin in seach of their role in the vertebrate globin family. J. Inorg. Biochem. 2005;99:110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Giuffrè A., Moschetti T., Vallone B., Brunori M. Neuroglobin: enzymatic reduction and oxygen affinity. Biochem. Biophys. Res. Commun. 2008;367:893–898. doi: 10.1016/j.bbrc.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Fago A., Mathews A.J., Moens L., Dewilde S., Brittain T. The reaction of neuroglobin with potential redox protein partners cytochrome b5 and cytochrome c. FEBS Lett. 2006;580:4884–4888. doi: 10.1016/j.febslet.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Fago A., Mathews A.J., Brittain T. A role for neuroglobin: resetting the trigger level for apoptosis in neuronal and retinal cells. IUBMB Life. 2008;60:398–401. doi: 10.1002/iub.35. [DOI] [PubMed] [Google Scholar]
- 13.Brunori M., Giuffrè A., Nienhaus K., Nienhaus G.U., Scandurra F.M. Neuroglobin, nitric oxide, and oxygen: functional pathways and conformational changes. Proc. Natl. Acad. Sci. USA. 2005;102:8483–8488. doi: 10.1073/pnas.0408766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold S., Fago A., Weber R.E., Dewilde S., Moens L. Reactivity studies of the Fe(III) and Fe(II)NO forms of human neuroglobin reveal a potential role against oxidative stress. J. Biol. Chem. 2003;279:22841–22847. doi: 10.1074/jbc.M313732200. [DOI] [PubMed] [Google Scholar]
- 15.Wakasugi K., Nakano T., Morishima I. Oxidized human neuroglobin acts as a heterotrimeric Gα protein guanine nucleotide dissociation inhibitor. J. Biol. Chem. 2003;278:36505–36512. doi: 10.1074/jbc.M305519200. [DOI] [PubMed] [Google Scholar]
- 16.Kitatsuji C., Kurogochi M., Nishimura S., Ishimori K., Wakasugi K. Molecular basis of guanine nucleotide dissociation inhibitor activity of human neuroglobin by chemical cross-linking and mass spectrometry. J. Mol. Biol. 2007;368:150–160. doi: 10.1016/j.jmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Vallone B., Nienhaus K., Matthes A., Brunori M., Nienhaus G.U. The structure of carbonmonoxy neuroglobin reveals a heme-sliding mechanism for control of ligand affinity. Proc. Natl. Acad. Sci. USA. 2004;101:17351–17356. doi: 10.1073/pnas.0407633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anselmi M., Brunori M., Vallone B., Di Nola A. Molecular dynamics simulation of deoxy and carboxy murine neuroglobin in water. Biophys. J. 2007;93:434–441. doi: 10.1529/biophysj.106.099648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giuffrè A., Moschetti T., Vallone B., Brunori M. Is neuroglobin a signal transducer? IUBMB Life. 2008;60:410–413. doi: 10.1002/iub.88. [DOI] [PubMed] [Google Scholar]
- 20.Pesce A., Dewilde S., Nardini M., Moens L., Ascenzi P. Human brain neuroglobin structure reveals a distinct mode of controlling oxygen affinity. Structure. 2003;11:1087–1095. doi: 10.1016/s0969-2126(03)00166-7. [DOI] [PubMed] [Google Scholar]
- 21.Vallone B., Nienhaus K., Brunori M., Nienhaus G.U. The structure of murine neuroglobin: novel pathways for ligand migration and binding. Proteins. 2004;56:85–92. doi: 10.1002/prot.20113. [DOI] [PubMed] [Google Scholar]
- 22.Dewilde S., Kiger L., Burmester T., Hankeln T., Baudin-Creuza V. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J. Biol. Chem. 2001;276:38949–38955. doi: 10.1074/jbc.M106438200. [DOI] [PubMed] [Google Scholar]
- 23.Trent J.T., 3rd, Watts R.A., Hargrove M.S. Human neuroglobin, a hexacoordinate hemoglobin that reversibly binds oxygen. J. Biol. Chem. 2001;276:30106–30110. doi: 10.1074/jbc.C100300200. [DOI] [PubMed] [Google Scholar]
- 24.Hamdane D., Kiger L., Dewilde S., Green B.N., Pesce A. The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J. Biol. Chem. 2003;278:51713–51721. doi: 10.1074/jbc.M309396200. [DOI] [PubMed] [Google Scholar]
- 25.Van Doorslaer S., Dewilde S., Kiger L., Nistor S.V., Goovaerts E. Nitric oxide binding properties of neuroglobin. A characterization by EPR and flash photolysis. J. Biol. Chem. 2003;278:4919–4925. doi: 10.1074/jbc.M210617200. [DOI] [PubMed] [Google Scholar]
- 26.Nienhaus K., Kriegl J.M., Nienhaus G.U. Structural dynamics in the active site of murine neuroglobin and its effects on ligand binding. J. Biol. Chem. 2000;279:22944–22952. doi: 10.1074/jbc.M401561200. [DOI] [PubMed] [Google Scholar]
- 27.Weiland T.R., Kundu S., Trent J.T., 3rd, Hoy J.A., Hargrove M.S. Bis-histidyl hexacoordination in hemoglobins facilitates heme reduction kinetics. J. Am. Chem. Soc. 2004;126:11930–11935. doi: 10.1021/ja046990w. [DOI] [PubMed] [Google Scholar]
- 28.Tilton R.F., Jr., Kuntz I.D., Jr., Petsko G.A. Cavities in proteins: structure of a metmyoglobin-xenon complex solved to 1.9 Å. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 29.Chu K., Vojtchovský J., McMahon B.H., Sweet R.M., Berendzen J. Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature. 2000;403:921–923. doi: 10.1038/35002641. [DOI] [PubMed] [Google Scholar]
- 30.Ostermann A., Waschipky R., Parak F.G., Nienhaus G.U. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 31.Brunori M., Vallone B., Cutruzzola F., Travaglini-Allocatelli C., Berendzen J. The role of cavities in protein dynamics: crystal structure of a photolytic intermediate of a mutant myoglobin. Proc. Natl. Acad. Sci. USA. 2000;97:2058–2063. doi: 10.1073/pnas.040459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott E.E., Gibson Q.H. Ligand migration in sperm whale myoglobin. Biochemistry. 1997;36:11909–11917. doi: 10.1021/bi970719s. [DOI] [PubMed] [Google Scholar]
- 33.Brunori M., Gibson Q.H. Cavities and packing defects in the structural dynamics of myoglobin. EMBO Rep. 2001;2:674–679. doi: 10.1093/embo-reports/kve159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson J.S., Soman J., Phillips G.N., Jr. Ligand pathways in myoglobin: a review of Trp cavity mutations. IUBMB Life. 2007;59:552–562. doi: 10.1080/15216540701230495. [DOI] [PubMed] [Google Scholar]
- 35.de Sanctis D., Dewilde S., Pesce A., Moens L., Ascenzi P. Mapping protein matrix cavities in human cytoglobin through Xe atom binding. Biochem. Biophys. Res. Commun. 2004;316:1217–1221. doi: 10.1016/j.bbrc.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Perutz M.F. Myoglobin and haemoglobin: role of distal residues in reactions with haem ligands. Trends Biochem. Sci. 1989;14:42–44. doi: 10.1016/0968-0004(89)90039-x. [DOI] [PubMed] [Google Scholar]
- 37.Olson J.S., Phillips G.N., Jr. Kinetic pathway and barriers for ligand binding to myoglobin. J. Biol. Chem. 1996;271:17593–17596. [PubMed] [Google Scholar]
- 38.Arcovito A., Moschetti T., D'Angelo P., Mancini G., Vallone B. An x-ray diffraction and x-ray absorption spectroscopy joint study of neuroglobin. Arch. Biochem. Biophys. 2008;475:7–13. doi: 10.1016/j.abb.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z., Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Leslie A.G.W. The integration of macrolecular diffraction data. Acta Crystallogr D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project No. 4 “The CCP4 Suite”: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 43.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Winn M.D., Isupov M.N., Murshudov G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 47.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 48.Damborsky J., Petrek M., Banas P., Otyepka M. Identification of tunnels in proteins, nucleic acids, inorganic materials and molecular ensembles. Biotechnol. J. 2007;2:62–67. doi: 10.1002/biot.200600208. [DOI] [PubMed] [Google Scholar]
- 49.DeLano W. DeLano, Scientific; San Carlos, CA: 2002. The PyMOL User's Manual. [Google Scholar]
- 50.Du W., Syvitski R., Dewilde S., Moens L., La Mar G.N. Solution 1h NMR characterization of equilibrium heme orientational disorder with functional consequences in mouse neuroglobin. J. Am. Chem. Soc. 2003;125:8080–8081. doi: 10.1021/ja034584r. [DOI] [PubMed] [Google Scholar]
- 51.Riccio A., Vitagliano L., Di Prisco G., Zagari A., Mazzarella L. The crystal structure of a tetrameric hemoglobin in a partial hemichrome state. Proc. Natl. Acad. Sci. USA. 2002;99:9801–9806. doi: 10.1073/pnas.132182099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vergara A., Franzese M., Merlino A., Vitagliano L., Verde C. Structural characterization of ferric hemoglobins from three Antarctic fish species of the suborder Notothenoidei. Biophys. J. 2007;93:2822–2829. doi: 10.1529/biophysj.107.105700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srajer V., Ren Z., Teng T.Y., Schmidt M., Ursby T. Protein conformational relaxation and ligand migration in myoglobin: a nanosecond to millisecond molecular movie from time-resolved Laue x-ray diffraction. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 54.Bourgeois D., Vallone B., Schotte F., Arcovito A., Miele A.E. Complex landascape of protein structural dynamics unveiled by nanosecond Laue crystallography. Proc. Natl. Acad. Sci. USA. 2003;100:8704–8709. doi: 10.1073/pnas.1430900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hummer G., Schotte F., Anfinrud P.A. Unveiling functional protein motions with piosecond x-ray crystallography and molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2004;101:15330–15334. doi: 10.1073/pnas.0405295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anselmi M., Di Nola A., Amadei A. The kinetics of ligand migration in crystallized myoglobin as revealed by molecular dynamics simulations. Biophys. J. 2008;94:4277–4281. doi: 10.1529/biophysj.107.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kriegl J.M., Bhattacharyya A.J., Nienhaus K., Deng P., Minkow O. Ligand binding and protein dynamics in neuroglobin. Proc. Natl. Acad. Sci. USA. 2002;99:7992–7997. doi: 10.1073/pnas.082244399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milani M., Pesce A., Ouellet Y., Ascenzi P., Guertin M. Mycobacterium tuberculosis hemoglobin N displays a protein tunnel suited for O2 diffusion to the heme. EMBO J. 2001;20:3902–3909. doi: 10.1093/emboj/20.15.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milani M., Pesce A., Ouellet Y., Dewilde S., Friedman J. Heme-ligand tunneling in group I truncated hemoglobins. J. Biol. Chem. 2004;279:21520–21525. doi: 10.1074/jbc.M401320200. [DOI] [PubMed] [Google Scholar]
- 60.Salter M.D., Nienhaus K., Nienhaus G.U., Dewilde S., Moens L. The apolar channel in Cerebratulus lacteus in hemoglobin is the route for O2 entry and exit. J. Biol. Chem. 2008;283:35689–35702. doi: 10.1074/jbc.M805727200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rinaldi A.C., Bonamore A., Macone A., Boffi A., Bozzi A. Interaction of Vitreoscilla hemoglobin with membrane lipids. Biochemistry. 2006;45:4069–4076. doi: 10.1021/bi052277n. [DOI] [PubMed] [Google Scholar]
- 62.Savino C., Miele A.E., Draghi F., Johnson K.A., Sciara G. Pattern of cavities in globins: the case of human hemoglobin. Biopolymers. 2009 doi: 10.1002/bip.21201. In press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


