Abstract
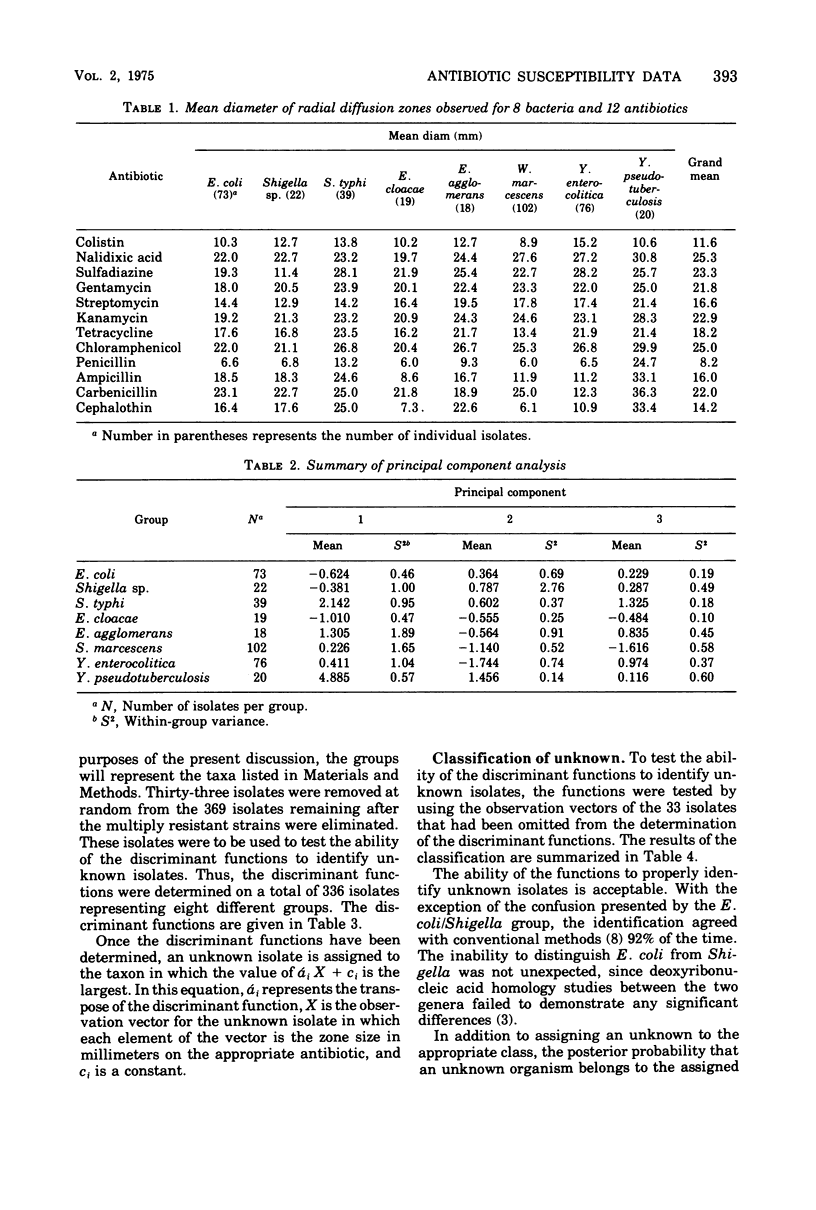
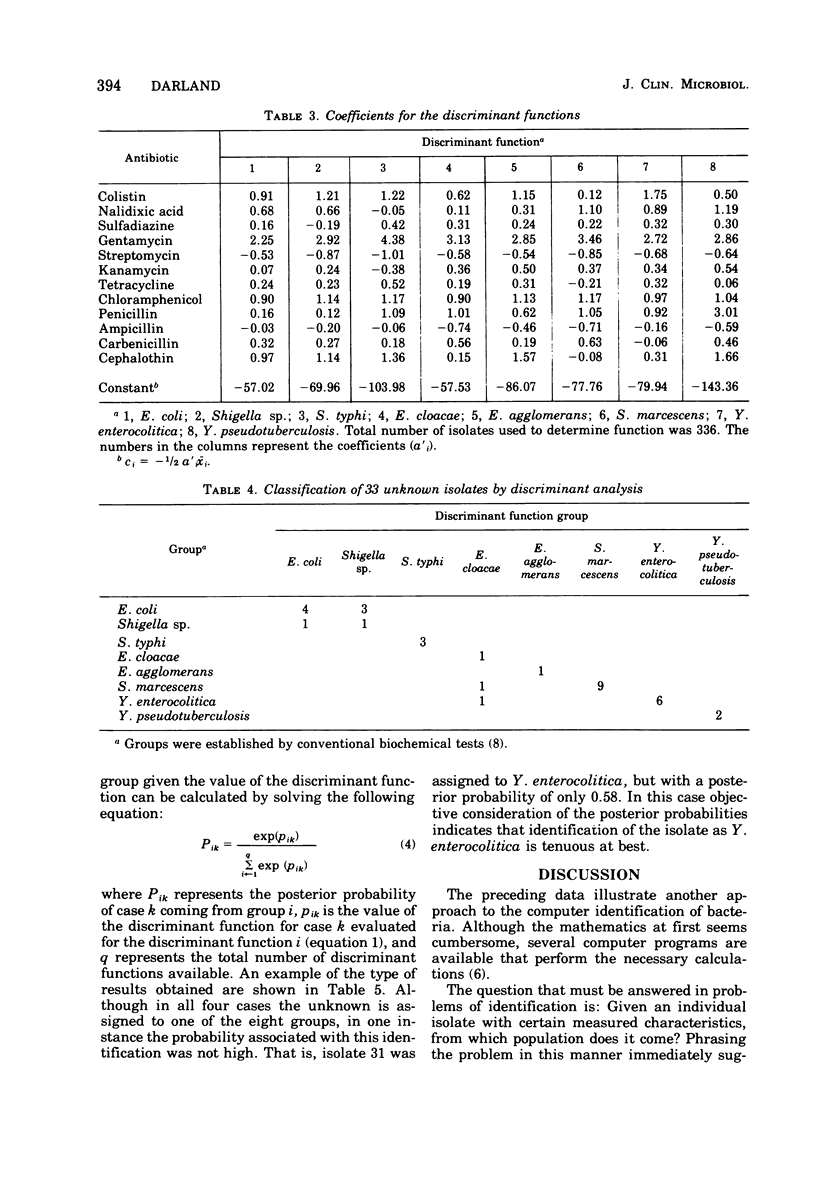
This study shows that antibiotic susceptibility data can be used effectively in the presumptive identification of bacteria. Using 12 antibiotics and determining the zone sizes for each, 82% of the isolates considered were correctly identified without any other information. If the inability to distinguish between Escherichia coli and Shigella is disregarded, the percentage of correct identification is 92%. The method involves determining a set of discriminant functions and defining each taxon by a unique function. An unknown isolate is identified by evaluating each discriminant function and assigning the isolate to the taxon whose discriminant function has the largest value. A total of 468 isolates were examined. After eliminating the multiply resistant isolates, the remaining 369 isolates were used to determine the discriminant functions for the eight taxa considered.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Skerman F. J., Falkow S. Polynucleotide sequence divergence among strains of Escherichia coli and closely related organisms. J Bacteriol. 1972 Mar;109(3):953–965. doi: 10.1128/jb.109.3.953-965.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darland G. Principal component analysis of infraspecific variation in bacteria. Appl Microbiol. 1975 Aug;30(2):282–289. doi: 10.1128/am.30.2.282-289.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybowski W., Franklin D. A. Conditional probability and the identification of bacteria: a pilot study. J Gen Microbiol. 1968 Dec;54(2):215–229. doi: 10.1099/00221287-54-2-215. [DOI] [PubMed] [Google Scholar]
- Friedman R. B., Bruce D., MacLowry J., Brenner V. Computer-assisted identification of bacteria. Am J Clin Pathol. 1973 Sep;60(3):395–403. doi: 10.1093/ajcp/60.3.395. [DOI] [PubMed] [Google Scholar]
- Friedman R., MacLowry J. Computer identification of bacteria on the basis of their antibiotic susceptibility patterns. Appl Microbiol. 1973 Sep;26(3):314–317. doi: 10.1128/am.26.3.314-317.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllenberg H. G. A model for computer identification of micro-organisms. J Gen Microbiol. 1965 Jun;39(3):401–405. doi: 10.1099/00221287-39-3-401. [DOI] [PubMed] [Google Scholar]


