Abstract
Genome-wide levels of DNA methylation vary between tissues, and compared with other tissues, the placenta has been reported to demonstrate a global decrease in methylation as well as decreased methylation of X-linked promoters. Methylation is one of many features that differentiate the active and inactive X, and it is well established that CpG island promoters on the inactive X are hypermethylated. We now report a detailed analysis of methylation at different regions across the X in male and female placenta and blood. A significant (P < 0.001) placental hypomethylation of LINE1 elements was observed in both males and females. Relative to blood placental promoter hypomethylation was only observed for X-linked, not autosomal promoters, and was significant for females (P < 0.0001) not males (P = 0.9266). In blood, X-linked CpG island promoters were shown to have moderate female methylation (66% across 70 assays) and low (23%) methylation in males. A similar methylation pattern in blood was observed for ∼20% of non-island promoters as well as 50% of the intergenic or intragenic CpG islands, the latter is likely due to the presence of unannotated promoters. Both intragenic and intergenic regions showed similarly high methylation levels in male and female blood (68 and 66%) while placental methylation of these regions was lower, particularly in females. Thus placental hypomethylation relative to blood is observed globally at repetitive elements as well as across the X. The decrease in X-linked placental methylation is consistently greater in females than males and implicates an inactive X specific loss of methylation in the placenta.
INTRODUCTION
The human placenta is a unique organ in that while it is critical for in utero healthy development, it is no longer needed after birth. The placenta has been shown by HPLC to have ∼20% fewer 5-methylcytosines compared with the vast majority of other tissues (1–3). Regions previously shown to have reduced placental methylation include repeat elements (Alus, LINEs, and satellites) and several X-linked genes with CpG island associated promoters (2,4–7). Methylation occurs at CpG dinucleotides in mammals, and the human genome contains over 27 million CpGs, ∼70% of which are typically methylated (1,8). CpGs are not evenly distributed across the genome, with regions of high CpG density known as CpG islands often found at promoters. The majority of CpG islands at promoters are unmethylated and methylation of these islands is typically linked with gene silencing (9). The distinction between CpG islands and non-islands is based on sequence composition and the criteria traditionally used to define a CpG island are GC content and observed CpG to expected CpG ratio (10–12).
Of the approximately 1.2 million CpGs on the human X chromosome only 5% are located within CpG islands [University of California Santa Cruz (UCSC) Genome Browser], and ∼50% of these islands are associated with known promoters, while the remaining islands are equally distributed within gene bodies and between genes across the X chromosome (12). With the exception of genes which escape X inactivation, CpG island-containing promoters show limited methylation in males and partial methylation in females (10,13). We term this classic methylation pattern of active X (Xa) hypomethylation and inactive (Xi) hypermethylation as MeXiP (Methylation of Xi Promoters).
There is some conflicting data as to the methylation status of the X chromosome as a whole. Using methylation-sensitive restriction enzyme analysis, the Xi appears to be hypomethylated compared with the Xa, conversely, in situ nick translation data demonstrated the Xa to be hypomethylated compared with the Xi and antibody staining demonstrated no difference between the Xs (14–16). Southern analysis of a limited number of X-linked single copy loci and two tandem repeats have suggested that the Xa is hypermethylated relative to the Xi, although non-promoter methylation levels were high and variable (17–20). Jones discussed such patterns as a ‘methylation paradox’ and proposed that hypermethylation of gene bodies might be caused by gene transcription (21). This hypothesis is supported by a recent microarray study that found overall hypermethylation of the X in males relative to females (22). Furthermore, using single-nucleotide polymorphisms in clonal lymphoblast cell lines, Hellman and Chess (23) were able to specifically demonstrate that the Xa has over two times more methylation than the Xi, particularly within gene bodies.
Our comparison of methylation between placenta and blood has revealed global LINE1 placental hypomethylation, as well as placental hypomethylation of X-linked promoters but not autosomal promoters. Using a combination of Illumina GoldenGate methylation analysis and pyrosequencing, three different types of X-linked regions (promoter, intragenic and intergenic) were shown to have less methylation in placenta compared with blood. This hypomethylation was greater in female than male placenta suggesting the difference is predominantly due to the Xi. These findings suggest a different system for the establishment or maintenance of methylation on the active and inactive X chromosomes.
RESULTS
Placental hypomethylation is specific to X-linked promoters and repetitive elements
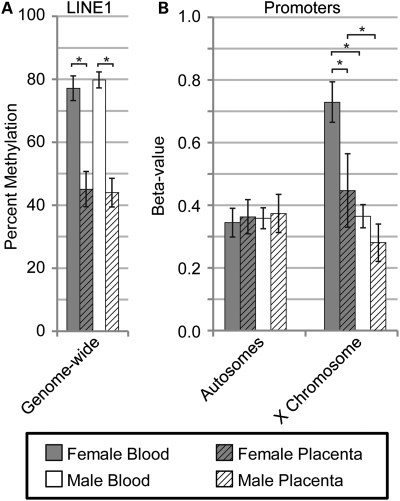
Average LINE1 methylation across the genome, as determined by pyrosequencing, displayed significant (P = 0.0009) placental hypomethylation compared with blood in both females (32% lower methylation) and males (36% lower methylation) (Fig. 1A). To determine if the placental hypomethylation previously reported at X-linked promoters also extended to autosomal promoters, the Illumina GoldenGate panel was used to assess the level of promoter methylation at 1421 promoter sites on the autosomes and 84 sites on the X chromosome in both blood and placenta (Fig. 1B). Average autosomal methylation levels showed no significant difference (P = 0.05) between male and female blood and placenta, whereas dramatic sex and tissues differences were observed for the X chromosome (P < 0.001). The X chromosomes were examined in more detail to determine the extent of the X-linked placental hypomethylation.
Figure 1.
Reduced placental methylation found at LINE1 repetitive elements and promoters on the X chromosome. Average level of methylation for female blood (grey), female placenta (grey hatched), male blood (white) and male placenta (white hatched) are shown with error bars (one standard deviation) based on the average sample deviation at a single site. Significance calculated using Mann–Whitney test with P < 0.001 (*). (A) LINE1 percent methylation as determined by pyrosequencing at LINE1 repetitive elements across the genome. (B) Illumina Golden Gate Promoter methylation array data averaged separately for 1421 sites on the autosomes and 84 X-linked sites. Beta-values represent average percent methylation.
CpG density influences the methylation patterns of X-linked promoters
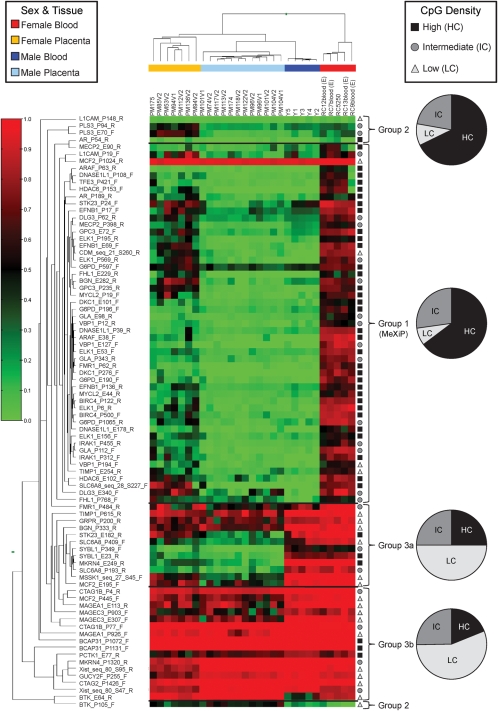
The Manhattan Hierarchical Cluster Metric (Illumina BeadStudio) separated the 29 samples analyzed into four clusters that corresponded to male blood, female blood, male placenta and female placenta (Fig. 2). To assess the impact of CpG density, we utilized the expanded nomenclature of Weber et al. (10) to include an intermediate CpG density category (IC) in addition to the high density (HC) and low density (LC) categories (see Materials and Methods). In blood, 51 of 84 X-linked promoter assays examined demonstrated moderate methylation in females (average beta-value of 0.67) and negligible methylation in males (average beta-value of 0.08), the anticipated pattern for X-linked CpG island promoters of genes subject to inactivation, MeXiP (labeled group 1 on Fig. 2). MeXiP assays tended to have less methylation in the placenta. One-third of all assays demonstrated high methylation in both male and female blood (group 3 on Fig. 2) and could be subdivided into those with generally less placental methylation (group 3a) and those that were also highly methylated in placenta (group 3b). A small number of assays demonstrated extremely low methylation in both males and females (group 2 on Fig. 2). As shown in the pie charts on Figure 2, LC assays were found in each group. However, the majority of assays in LC regions were located within group 3; while the promoters displaying MeXiP were generally in HC and IC regions. A complete list of the HC and IC genes that do not display MeXiP can be found in Supplementary Material, Table S1. The majority of these exceptions can be explained by expression pattern or proximity to repetitive elements, however, there are some CpG island promoter assays with no obvious reason for deviation from the MeXiP pattern.
Figure 2.
Heatmap of methylation levels at 84 sites across the X chromosome demonstrates that the majority of X-linked promoter assays demonstrate MeXiP and are of high and intermediate CpG density while low CpG density assays tend to be highly methylated. Methylation levels determined by Illumina Golden Gate promoter methylation array are represented as a gradient from red (high methylation) to green (low methylation). BeadStudio software used the Manhattan Hierarchical Cluster Metric to group samples that were separated by tissue and sex (coding of samples as follows: yellow = female placenta, blue = male placenta, dark blue = male blood, orange = female blood). Assays were visually divided into four groups based on methylation trends. Group 1 shows high female methylation and low male in blood (MeXiP), group 2 shows low methylation in all samples and group 3 shows high methylation in both male and female blood. Group 3a had variable placenta methylation while group 3b had high methylation in the placenta. The CpG density of each assay, high (HC) (black square), intermediate (IC) (dark grey circle) or low (LC) (light grey triangle), is shown to the right and the assay names to the left of the heatmap. The percent of assays within each group based on CpG density is shown as a pie chart to the far right of the heatmap.
For regions that display MeXiP the methylation provides a means to examine the distance over which promoter methylation is correlated with inactivation status. Plotting DNA methylation levels against the distance from the transcription start site (TSS) for each promoter CpG density class, sex and tissue (Supplementary Material, Fig. S1) demonstrated that a majority of HC assays outside the −700 to +200 bp promoter window had nearly complete methylation in both tissues (10). The HC and IC regions examined were not significantly different from each other in either sex or tissue and thus, both will subsequently be referred to as CpG islands. Non-island (LC) assays showed no relationship between methylation and the distance from the TSS.
DNA methylation is female-specific at X-linked island promoters but is consistently high at intragenic and intergenic regions
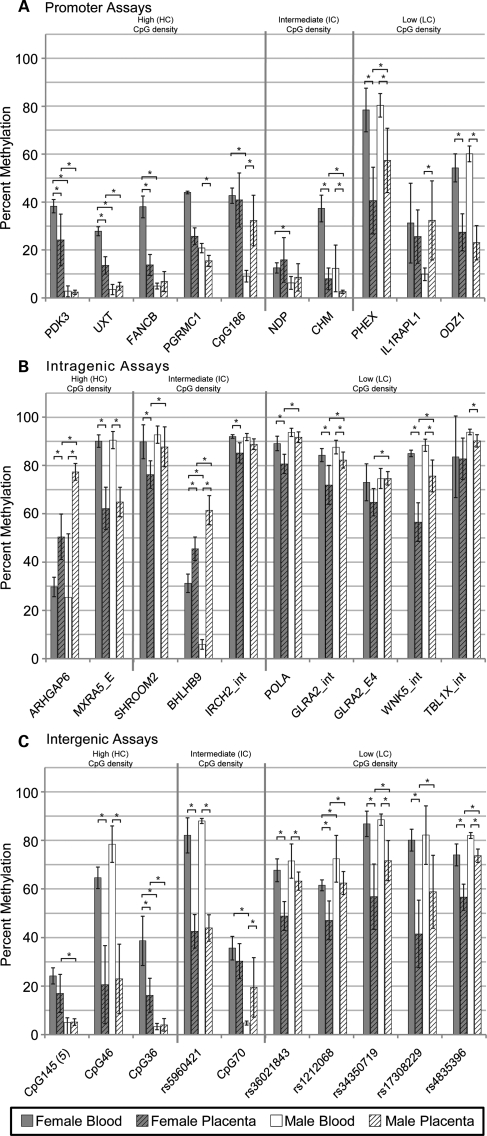
The Illumina GoldenGate panel only provided data on CpGs in promoters, so pyrosequencing was used to confirm the level of methylation at promoters of all three CpG densities, as well as to determine methylation at intragenic and intergenic regions (10 assays each). Another advantage to pyrosequencing is a larger number of CpGs within a small region can be examined. The complete data set of 30 assays for six blood samples and six placenta samples (two sites sampled per placenta) for a total of 5076 data points is shown in Supplementary Material, Figure S2. There was more variation in methylation levels between placental samples and between sites of the same placenta than for blood samples across all males and females. An individual CpG in a region was on average only 7% different from the average of all CpGs assayed from that region, with only one assay (rs1212068) showing an average difference of more than 12% due to a single outlier CpG. Therefore, the average percent methylation for each assay was compared in Figure 3 for the six male and six female blood as well as all placenta samples. Assays were subdivided into panels for location in promoters, intragenic or intergenic regions and ordered according to the CpG density (HC/IC/LC) of the region. Consistent with the Illumina GoldenGate promoter methylation data, female placenta showed an average of 16% less methylation compared with female blood, whereas male placenta showed an average of only 4% less methylation compared with male blood.
Figure 3.
X-linked CpG island promoters show female-specific methylation, whereas methylation is high in both X-linked intragenic and intergenic regions in females and males. Average percent methylation from 30 pyrosequencing assays for six female blood (grey), six female placenta (grey hatched), six male blood (white) and six male placenta (white hatched). Each placenta was sampled from two sites within a single placenta for a total of 12 placental samples. Assays are separated into CpG density, high (HC), intermediate (IC) and low (LC), from the left to the right, by vertical lines. (A) Promoter assays; (B) intragenic assays; (C) intergenic assays. The region assayed is listed below each set of averages. Significance calculated using Mann–Whitney test with P < 0.01 (*). Error bars are one standard deviation.
Ten promoter regions of varying CpG density were examined by pyrosequencing to determine if they followed MeXiP (Fig. 3A). One assay, NDP, stood out as an outlier with low methylation (below 20%) in all samples despite carriers manifesting Norrie's disease with X chromosome rearrangements suggesting that NDP undergoes X inactivation (24). For other assays the methylation patterns of the island promoter assays were very similar, with the highest methylation being detected in female blood which averaged 38%, followed by female placenta which averaged 21%. Male blood and placenta generally showed low methylation averaging 10%. One of the three non-island promoters examined (ILRAPL1) showed methylation levels comparable to island promoters in blood, while the others showed higher methylation levels than the island promoter assays in all samples. Therefore, in both the Illumina GoldenGate panel and the pyrosequencing data the HC and IC promoter assays examined showed MeXiP as did a subset of LC promoters.
Intragenic regions analyzed included both exons and introns which showed similar patterns of methylation. For LC intragenic and intergenic regions there was an average of 80 and 70% methylation, respectively, across all assays with less difference observed between sexes for blood than for placenta. While five of the intragenic and intergenic island assays showed a similar high methylation, others (ARHGAP6, BHLHB9, CpG145, CpG36 and CpG70) were much more reminiscent of the MeXiP pattern of methylation of the promoter assays. Examination of the histone modifications present in the regions of CpG36 and BHLHB9 using the histone modification tracks of UCSC Genome Browser showed that they possess histone modification patterns reminiscent of a promoter (12,25,26). Thus the MeXiP pattern may provide an additional approach to determine the location of unannotated promoters on the X chromosome.
Two assays, ARHGAP6 and BHLHB9, showed hypermethylation of the placenta compared with blood in males and females and are located within 2 kb of an alternative promoter for an isoform of their respective genes and thus may reflect tissue-specific methylation. One male blood sample showed a methylation level very different from the others for ARHGAP6 at all five CpG sites in the assay, despite sex-normal methylation at all other loci examined. This could potentially reflect allele-specific methylation in this individual (27). Overall, with the exception of likely unannotated promoters, intragenic and intergenic assays were heavily methylated independent of CpG density. Less methylation was still observed in the placenta, particularly in female samples, suggesting that the Xi shows more placental hypomethylation than the Xa.
The inactive X has less methylation than the active X in placenta compared with blood
To distinguish how the Xa and Xi differed in methylation the assumption was made that males and females would have equivalent methylation on their respective Xa's and, therefore, the methylation of the male X was used as the value for the Xa in both males and females. The Xi methylation level was then calculated as described in Materials and Methods. The difference in methylation levels between blood and placenta for each pyrosequencing assay is shown for the Xa and the Xi in Supplementary Material, Figure S3 and summarized according to location and CpG density in Table 1. The assays located in promoters on average had ∼3% less methylation on the Xa in placenta compared with blood and a significant (P < 0.05) decrease in methylation on the Xi of 35%. The island-containing promoters had limited Xa methylation and thus would not be anticipated to differ in placenta. The non-island promoters, as well as the intragenic and intergenic regions, however, showed equivalent Xa and Xi methylation in blood, whereas in placenta the Xi showed 2-fold less methylation relative to the Xa. In placenta, the Xa did appear to show a slight decrease in methylation compared with blood, however, this was not significant, whereas the decrease in methylation from the Xi was significant in all regions (P < 0.01). The methylation difference was also greatest from females overall for X-linked assays on the Illumina GoldenGate panel (Supplementary Material, Fig. S4), further supporting that the majority of the methylation decrease observed in placenta is from differences in methylation of the Xi.
Table 1.
Average percent methylation as determined by pyrosequencing at promoter, intragenic and intergenic regions across the X chromosome in blood and placenta for high (HC), intermediate (IC) and low (LC) CpG density
| Location and CpG density | Number of assaysa | Male blood (Xa)b, % | Female blood (XaXi), % | BloodXic, % | BloodXa:Xi ratiod | Male placenta (Xa)b, % | Female placenta (XaXi) | PlacentaXic, % | PlacentaXa:Xi ratiod | Blood–Placenta |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔXae, % | ΔXie, % | ||||||||||
| Promoter | 9 | 23 | 44 | 64 | 0.35* | 20 | 24 | 29 | 0.68 | −3 | −35** |
| HC | 5 | 8 | 38 | 68 | 0.12 | 12 | 24 | 35 | 0.35 | 4 | −33 |
| ICf | 1 | 12 | 37 | 62 | 0.20 | 3 | 8 | 13 | 0.19 | −10 | −49 |
| LC | 3 | 50 | 55 | 59 | 0.85 | 38 | 31 | 25 | 1.52 | −13 | −34 |
| Intragenic | 8 | 89 | 86 | 83 | 1.08* | 82 | 73 | 63 | 1.30** | −7 | −19** |
| HCg | 1 | 91 | 90 | 90 | 1.01 | 65 | 62 | 60 | 1.09 | −26 | −30 |
| ICg | 2 | 92 | 91 | 90 | 1.03 | 88 | 81 | 73 | 1.21 | −4 | −16 |
| LC | 5 | 88 | 83 | 78 | 1.12 | 83 | 71 | 60 | 1.39 | −5 | −19 |
| Intergenic | 10 | 58 | 62 | 65 | 0.88 | 43 | 38 | 33 | 1.29* | −15 | −32*** |
| HC | 3 | 29 | 42 | 56 | 0.52 | 11 | 18 | 25 | 0.43 | −18 | −31 |
| IC | 2 | 46 | 59 | 71 | 0.65 | 32 | 36 | 41 | 0.77 | −15 | −30 |
| LC | 5 | 79 | 74 | 69 | 1.15 | 66 | 50 | 34 | 1.92 | −13 | −34 |
aNumber of independent regions assessed as shown on Figure 3, excluding the genes noted below (NDP, ARHGAP6 and BHLHB9) and discussed in the text.
bMethylation in males was used as Xa methylation level.
cXi methylation calculated assuming that Xa in female is equivalent to Xa in males.
dXa and Xi were compared to determine if they differed in blood and placenta. Significance calculated using Mann–Whitney test with significance shown as P = 0.01 to 0.05 (*) and P = 0.001 to 0.01 (**).
eBlood and placenta were compared to determine if the Xa and Xi differed in their tissue specific methylation levels. Significance calculated using Mann–Whitney test with significance shown as P = 0.001 to 0.01 (**) and P < 0.001 (***).
fNDP removed from the average as it showed low methylation in all samples.
gARHGAP6 and BHLHB9 removed from the average as both appeared to be an alternative promoter for an isoform of their respective genes.
DISCUSSION
Previous studies have shown a global reduction in DNA methylation of placenta compared with the vast majority of other tissues and a specific placental hypomethylation of repetitive elements and X-linked promoters (1–3,28). In this study, a comparison of autosomal and X-linked promoters showed that methylation of placental promoters was strikingly reduced only on the X chromosome, particularly in females, with the placenta showing an average of 27% less promoter methylation than blood in females and 8% less in males. The greater placental hypomethylation observed in females implicates an Xi-specific methylation decrease. To calculate Xi methylation we make the assumption that the male and female Xa are equivalently methylated, a common assumption in the study of X inactivation. For methylation this assumption has been supported by studies of the Xi isolated in mouse/human somatic cell hybrids, or distinguished by SNPs in clonal female population of cells; however, differences at individual loci could arise due to hormonal differences, or the different gene content of the sex chromosomes (23,29).
In general, IC regions are not classified as CpG islands, however, in this study HC and IC regions were not significantly different in their methylation levels suggesting that functionally on the X chromosome IC regions behave as CpG islands. For promoters demonstrating MeXiP, we propose that the small degree of methylation seen in males (averaging 8%) is not biologically relevant in preventing expression from the Xa, whereas the higher level observed in females reflects the additional methylation on the Xi which is associated with gene silencing. Methylation levels varied between promoters and were also dependent on the assay technology. Nonetheless, consistent with a recent study, it is clear that for individual gene promoters silencing on the Xi can be maintained with substantially less than 100% methylation (30).
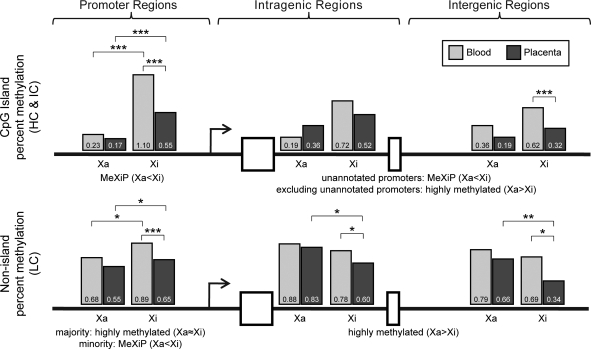
Figure 4 summarizes the changes in placental X-linked methylation and illustrates that not only is placenta less methylated than blood but the majority of this difference is due to the Xi. The Xi appeared to show the greatest decrease in methylation at promoters, however, the limited number of assays and the combination of two methylation detection techniques precludes a definitive conclusion as to the degree of Xi placental hypomethylation between regions. MeXiP is the clear pattern for X-linked CpG island promoters, as well as a subset of non-island promoters. This trend is maintained for the intragenic and intergenic regions for CpG islands, likely due to the presence of unannotated gene promoters. The exclusion of possible unannotated promoters (regions demonstrating MeXiP) resulted in higher intragenic and intergenic methylation on the Xa than the Xi for both blood and placenta regardless of CpG density.
Figure 4.
Summary of methylation analyses showing placental reduction in methylation predominately on the Xi. Data from both Illumina and pyrosequencing are combined and shown separately for CpG island (HC & IC) assays and non-island (LC) assays in promoter regions, intragenic regions (includes both introns and exons) and intergenic regions. In order to combine Illumina GoldenGate data (which is only for promoter regions), we converted beta-values to percent methylation. These values were consistent with pyrosequencing data at the low range, but generally higher than pyrosequencing in the midrange, accounting for the Xi value over 1 for promoters. Percent methylation is the average of all CpGs in the indicated region. Percent methylation is divided into Xa and Xi with grey bars representing methylation in blood and black bars for placenta with the average percent methylation value written in each bar. The summary of the methylation trends for the different regions is described below each bar graph. Significance calculated using Mann–Whitney test with significance shown as P = 0.01 to 0.05 (*), P = 0.01 to 0.001 (**) and P < 0.001 (***).
The observation of MeXiP at CpG island promoters is consistent with previous array-based studies that have shown an inverse correlation between DNA methylation and expression at island promoters (10). The regulatory nature of the promoter methylation has been demonstrated by the removal of methylation through 5-azacytidine treatment resulting in the reactivation of genes on the X chromosome (31). In contrast, it has been suggested that a consequence of transcription may be subsequent gene-body methylation, a finding supported by higher intragenic Xa methylation detected by array-based technologies (21,23,32). If gene body methylation were reflecting transcription then the methylation patterns of intragenic and intergenic regions should be very different. We observe, however, a similar reduction in methylation in intergenic regions on the Xi compared with the Xa, consistent with the observation that 5 of the 17 most consistently Xa methylated SNPs examined by Hellman and Chess (23) were located outside of the gene bodies. Therefore, the relative Xa:Xi hypermethylation cannot be solely attributed to the transcription of currently annotated genes. Hansen (33) has previously proposed that the Xa and Xi are methylated by different de novo methyltransferases based on the hypomethylation of LINE1 elements on the Xi but not the Xa or autosomes in ICF syndrome cells (33,34). While ICF syndrome cells, which have a mutation in DMNT3B, also show hypomethylation of X-linked promoters and several classes of satellite elements no reduction in DNA methyltransferases has been observed in the placenta (35–37).
The lower methylation in placenta could impact the stability of silencing. Indeed, some normally silent repetitive elements, imprinted genes and individual X-linked genes have been shown to become expressed in placenta (38–41). Expression of both alleles of the X-linked gene G6PD were observed in a subset of placental cells, and when chorionic villi cells were used to create somatic cell hybrids some hybrids showed global X chromosome reactivation, a process which normally occurs only during oogenesis (38,42,43). There does not, however, appear to be a general over-expression of placental X-linked genes compared with other somatic tissues, thus the synergistic silencing action of chromatin changes and the non-coding XIST RNA are apparently sufficient to maintain the majority of placental X inactivation (40). Nonetheless, the decreased placental methylation should be considered when using assays based on methylation, such as those to evaluate skewing of X inactivation (38).
CpG islands are generally unmethylated, but genome-wide studies have demonstrated tissue-specific hypermethylation of some island promoter regions as well as hypermethylation in cancer (44–46). While we did not observe a significant methylation difference in autosomal promoter methylation between blood and placenta, a recent study comparing blood and placenta reported many regions showing tissue-specific differences on chromosomes 13, 18 and 21, with hypomethylation being more common than hypermethylation (47). In a minority of our X-linked assays (4 pyrosequencing assays and 14 Illumina GoldenGate assays out of a total of 114) the level of methylation was significantly (P < 0.05) higher in placenta than in blood for at least one sex, perhaps reflecting sex and tissue-specific gene silencing on the X. Little is known about the methylation at tissue-specific island promoters on the X except for AR which has been shown to maintain MeXiP in a variety of tissues forming the basis of a commonly used methylation based X inactivation skewing assay (48).
Here we have shown that reduced DNA methylation in the human placenta is not consistent across the genome, rather it occurs in repetitive elements and across the X chromosome. The Xa consistently showed less decrease in placental methylation than the Xi, even when the Xa was equivalently or more methylated than the Xi. This suggests that the facultative heterochromatin of the Xi behaves similarly to repetitive elements in the placenta. As previously reported, we observe that X-linked island promoters are methylated in females not males, a pattern which we have termed MeXiP. Intriguingly, MeXiP is also seen for 20% of non-island promoters as well as 50% of non-promoter islands, which we attribute to unannotated promoters. Outside of promoters, the Xa is slightly more methylated than the Xi in both intragenic and intergenic regions, with intragenic regions tending to be more methylated than intergenic regions. Further study is required to determine the contribution of transcription or other processes to the establishment and/or maintenance of such methylation patterns.
MATERIALS AND METHODS
Sample collection
Placentas were collected with consent from pregnancies delivered at British Columbia's Women's Hospital. Placentas from females (n = 11) were an average of 37.8 months gestational age and had an average maternal age of 34.6 years. Placentas from males (n = 15) were an average of 38.9 months gestational age and had an average maternal age of 35.2 years. Blood samples were from anonymous males (n = 6) and females (n = 6) ranging in age from 2 to 49 years for males and 22 to 44 years for females. Ethics approval was obtained from the University of British Columbia clinical research ethics board.
DNA extraction and bisulfite conversion
DNA was extracted from fresh whole blood samples following a standard salting out DNA extraction (49). After removal of amniotic and chorionic membranes DNA was extracted from two separate samples of chorionic villi from the fetal side of the placenta as outlined in Penaherrera et al. (50). Five hundred nanograms of DNA were then used for a bisulfite conversion following the instructions in the EZ DNA Methylation Gold Kit (Zymo Research Corporation). Complete conversion was confirmed using the internal bisulfite conversion controls in each pyrosequencing assay and the bisulfite conversion control summary graph for the Illumina GoldenGate panel.
Illumina goldengate panel
Samples were applied to an Illumina GoldenGate bead array that assays 1505 CpG sites located in promoters across the genome. Eighty-four of these sites were located on the X chromosome and associated with 39 X-linked genes. The location of autosomal CpGs assayed on this panel ranged from 1499 bp upstream of the transcription start site (TSS) to 497 bp downstream. The X chromosome assays were located up to 1681 bp upstream of the TSS and 472 bp downstream. Data underwent average normalization using the Methylation Module (version 3.2.0) in BeadStudio (version 3.1.3.0 Illumina, Inc.) to ensure that the background intensities of each array were comparable. The heatmap and dendrogram in Figure 2 were generated using the Manhattan Hierarchical Cluster Metric in BeadStudio.
Pyrosequencing
Pyrosequencing was performed using a Pyromark ID machine and the PyroGoldSQA reagent kit (Biotage). Each 25 µL pyrosequencing PCR contained the following final concentration of reagents: 1× PCR Buffer (Qiagen), 0.2 mm dNTPs, 0.025U HotStart Taq DNA Polymerase (Qiagen), 0.25 mm forward primer, 0.25 mm reverse primer and ∼25 ng bisulfite converted DNA. Cycling conditions for each assay were the same for all primers except for the annealing temperature which is listed for each assay individually in Supplementary Material, Table S2. Cycling conditions were: 95°C for 15 min, 50 cycles of 94°C for 30 s, annealing temperature (listed in Supplementary Material, Table S2) for 30 s, 72°C for 60 s, followed by a final step of 72°C for 10 min. Template preparation and pyrosequencing was then done according to Tost and Gut with sequencing primers listed in Supplementary Material, Table S2 (51). Global methylation of LINE1 elements was performed using the LINE1 assay from Biotage with PCR and cycling conditions as specified by the supplier. The UCSC Genome Browser was used to compare non-promoter regions against four histone modifications (H2AK9ac, H2BK5ac, H3K18ac, H3K36ac) that are concentrated around the TSS and eight histone modifications (H2BK12ac, H3K4ac, H4K5ac, H4K8ac, H4K12ac, H2BK20ac, H2BK120ac, H4K16ac) that are elevated in promoters and the transcribed regions of active genes (12,25). Four histone methylation modifications (H3K4me1, H3K4me2, H3K4me3, H2A.Z) previously found to be in promoter regions were also examined (26).
Statistical analysis
Statistical analysis of the Illumina GoldenGate panel was performed using the Mann–Whitney test as calculated by GraphPad prism. For the comparison of Illumina Golden Gate results the data were analyzed separately for CpG islands and non-islands based on information from the manufacturer. Male methylation levels were used as the methylation level of the Xa. Since the methylation level obtained from females is the average of both Xs this was multiplied by two and then the Xa methylation was subtracted resulting in the calculated amount of methylation on the Xi.
CpG density definitions used
The definition of a CpG island used by Illumina and UCSC to define a CpG island (GC content >50%, a ratio of greater than 0.6 for ObservedCpG/ExpectedCpG and a length >200 bp) is the same definition first proposed by Gardiner-Garden and Frommer in 1987 (11). In a recent genome-wide analysis of promoter methylation Weber et al. (10) introduced three CpG density classes, the strong CpG islands (high CpG density or HC) and the non-CpG islands (low or LC) as well as an intermediate class (IC). To ensure that all potential regions of CpG islands were recognized in this study we have also classified CpG islands based on these three density classes. Each class was defined as follows, HC, >55% GC content, >0.75 ObservedCpG/ExpectedCpG and a length >500 bp; LC had a ObservedCpG/ExpectedCpG ratio <0.48 and were shorter than 500 bp. Regions which were neither HC or LC were classified as IC. Using this system most regions classified as CpG islands by UCSC are defined as HCs, whereas only approximately one-quarter of ICs would be classified by UCSC as islands (10).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by Canadian Institutes of Health Research [GMH-79041]; and the Interdisciplinary Women's Reproductive Health Research Training Program.
ACKNOWLEDGEMENTS
We thank M. Kobor for collaborations with Illumina data collection, A.M. Devlin for use of the pyrosequencing machine, L. Currie and J. Sloan for the recruitment of placenta donors and R. Jiang for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ehrlich M., Gama-Sosa M.A., Huang L.-H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gama-Sosa M.A., Midgett R.M., Slagel V.A., Githens S., Kuo K.C., Gehrke C.W., Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta. 1983;740:212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- 3.Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 4.Hellmann-Blumberg U., Hintz M.F., Gatewood J.M., Schmid C.W. Developmental differences in methylation of human Alu repeats. Mol. Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen H.M., Nakamura A., Sugimoto J., Sakumoto N., Oda T., Jinno Y., Okazaki Y. Tissue specificity of methylation and expression of human genes coding for neuropeptides and their receptors, and of a human endogenous retrovirus K family. J. Hum. Genet. 2006;51:440–450. doi: 10.1007/s10038-006-0382-9. [DOI] [PubMed] [Google Scholar]
- 6.Driscoll D., Migeon B. Sex differences in methylation of single-copy genes in human meiotic germ cells: implications for X chromosome inactivation, parental imprinting, and origin of CpG mutations. Somat. Cell Mol. Genet. 1990;16:267–282. doi: 10.1007/BF01233363. [DOI] [PubMed] [Google Scholar]
- 7.Luo S., Robinson J.C., Reiss A.L., Migeon B.R. DNA methylation of the fragile X locus in somatic and germ cells during fetal development: relevance to the fragile X syndrome and X inactivation. Somat. Cell Mol. Genet. 1993;19:393–404. doi: 10.1007/BF01232750. [DOI] [PubMed] [Google Scholar]
- 8.Saxonov S., Berg P., Brutlag D. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 10.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 11.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 12.Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamieson R.V., Tam P.P., Gardiner-Garden M. X-chromosome activity: impact of imprinting and chromatin structure. Int. J. Dev. Biol. 1996;40:1065–1080. [PubMed] [Google Scholar]
- 14.Viegas-Pequignot E., Dutrillaux B., Thomas G. Inactive X chromosome has the highest concentration of unmethylated Hha I sites. Proc. Natl Acad. Sci. USA. 1988;85:7657–7660. doi: 10.1073/pnas.85.20.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller D.A., Okamoto E., Erlanger B.F., Miller O.J. Is DNA methylation responsible for mammalian X chromosome inactivation? Cytogenet. Cell Genet. 1982;33:345–349. doi: 10.1159/000131782. [DOI] [PubMed] [Google Scholar]
- 16.Prantera G., Ferraro M. Analysis of methylation and distribution of CpG sequences on human active and inactive X chromosomes by in situ nick translation. Chromosoma. 1990;99:18–23. doi: 10.1007/BF01737285. [DOI] [PubMed] [Google Scholar]
- 17.Yen P.H., Patel P., Chinault A.C., Mohandas T., Shapiro L.J. Differential methylation of hypoxanthine phosphoribosyltransferase genes on active and inactive human X chromosomes. Proc. Natl Acad. Sci. USA. 1984;81:1759–1763. doi: 10.1073/pnas.81.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen C.R., Hubberman P., Kaslow D.C., Migeon B.R. Comparison of factor IX methylation on human active and inactive X chromosomes: implications for X inactivation and transcription of tissue-specific genes. EMBO J. 1986;5:2223–2229. doi: 10.1002/j.1460-2075.1986.tb04488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd Y., Fraser N.J. Methylation patterns at the hypervariable X chromosome locus DXS255 (M27B); correlation with X inactivation status. Genomics. 1990;7:182–187. doi: 10.1016/0888-7543(90)90539-7. [DOI] [PubMed] [Google Scholar]
- 20.Giacalone J., Friedes J., Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat. Genet. 1992;1:137–143. doi: 10.1038/ng0592-137. [DOI] [PubMed] [Google Scholar]
- 21.Jones P.A. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 22.Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 23.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 24.Ohba N., Yamashita T. Primary vitreoretinal dysplasia resembling Norrie's disease in a female: association with X autosome chromosomal translocation. Br. J. Ophthal. 1986;70:64–71. doi: 10.1136/bjo.70.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M.Q., et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V.V., Schupf N., Vilain E., et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 28.Estecio M.R., Gharibyan V., Shen L., Ibrahim A.E., Doshi K., He R., Jelinek J., Yang A.S., Yan P.S., Huang T.H., et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen R.S., Canfield T.K., Stanek A.M., Keitges E.A., Gartler S.M. Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proc. Natl Acad. Sci. USA. 1998;95:5133–5138. doi: 10.1073/pnas.95.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeschnigk M., Martin M., Betzl G., Kalbe A., Sirsch C., Buiting K., Gross S., Fritzilas E., Frey B., Rahmann S., et al. Massive parallel bisulfite sequencing of CG-rich DNA fragments reveals that methylation of many X-chromosomal CpG islands in female blood DNA is incomplete. Hum. Mol. Genet. 2009;18:1439–1448. doi: 10.1093/hmg/ddp054. [DOI] [PubMed] [Google Scholar]
- 31.Mohandas T., Sparkes R.S., Shapiro L.J. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 32.Ball M.P., Li J.B., Gao Y., Lee J.H., LeProust E.M., Park I.H., Xie B., Daley G.Q., Church G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen R.S. X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum. Mol. Genet. 2003;12:2559–2567. doi: 10.1093/hmg/ddg268. [DOI] [PubMed] [Google Scholar]
- 34.Lyon M.F. X-chromosome inactivation: a repeat hypothesis. Cytogenet. Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 35.Jeanpierre M., Turleau C., Aurias A., Prieur M., Ledeist F., Fischer A., Viegas-Pequignot E. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 36.Hansen R.S., Stoger R., Wijmenga C., Stanek A.M., Canfield T.K., Luo P., Matarazzo M.R., D'Esposito M., Feil R., Gimelli G., et al. Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum. Mol. Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- 37.Tsien F., Fiala E.S., Youn B., Long T.I., Laird P.W., Weissbecker K., Ehrlich M. Prolonged culture of normal chorionic villus cells yields ICF syndrome-like chromatin decondensation and rearrangements. Cytogenet. Genome Res. 2002;98:13–21. doi: 10.1159/000068543. [DOI] [PubMed] [Google Scholar]
- 38.Migeon B.R., Axelman J., Jeppesen P. Differential X reactivation in human placental cells: implications for reversal of X inactivation. Am. J. Hum. Genet. 2005;77:355–364. doi: 10.1086/432815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiss D., Zhang Y., Mager D.L. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res. 2007;35:4743–4754. doi: 10.1093/nar/gkm455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen D.K., Disteche C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 41.Lambertini L., Diplas A.I., Lee M.J., Sperling R., Chen J., Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics. 2008;3:251–269. doi: 10.4161/epi.3.5.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Migeon B.R., Wolf S.F., Axelman J., Kaslow D.C., Schmidt M. Incomplete X chromosome dosage compensation in chorionic villi of human placenta. Proc. Natl Acad. Sci. USA. 1985;82:3390–3394. doi: 10.1073/pnas.82.10.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer-Sam J., Goldstein L., Dai A., Gartler S.M., Riggs A.D. A potentially critical Hpa II site of the X chromosome-linked PGK1 gene is unmethylated prior to the onset of meiosis of human oogenic cells. Proc. Natl Acad. Sci. USA. 1992;89:1413–1417. doi: 10.1073/pnas.89.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakyan V.K., Down T.A., Thorne N.P., Flicek P., Kulesha E., Gräf S., Tomazou E.M., Bäckdahl L., Johnson N., Herberth M., et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 46.Straussman R., Nejman D., Roberts D., Steinfeld I., Blum B., Benvenisty N., Simon I., Yakhini Z., Cedar H. Developmental programming of CpG island methylation profiles in the human genome. Nat. Genet. 2009;41:178–186. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 47.Papageorgiou E.A., Fiegler H., Rakyan V., Beck S., Hulten M., Lamnissou K., Carter N.P., Patsalis P.C. Sites of differential DNA methylation between placenta and peripheral blood: molecular markers for noninvasive prenatal diagnosis of aneuploidies. Am. J. Pathol. 2009;174:1609–1618. doi: 10.2353/ajpath.2009.081038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubota T., Nonoyama S., Tonoki H., Masuno M., Imaizumi K., Kojima M., Wakui K., Shimadzu M., Fukushima Y. A new assay for the analysis of X-chromosome inactivation based on methylation-specific PCR. Hum. Genet. 1999;104:49–55. doi: 10.1007/s004390050909. [DOI] [PubMed] [Google Scholar]
- 49.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penaherrera M.S., Ma S., Ho Yuen B., Brown C.J., Robinson W.P. X-chromosome inactivation (XCI) patterns in placental tissues of a paternally derived bal t(X;20) case. Am. J. Med. Genet. 2003;118A:29–34. doi: 10.1002/ajmg.a.10041. [DOI] [PubMed] [Google Scholar]
- 51.Tost J., Gut I.G. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.