Abstract
Spinal muscular atrophy (SMA), a recessive genetic disease, affects lower motoneurons leading to denervation, atrophy, paralysis and in severe cases death. Reduced levels of survival motor neuron (SMN) protein cause SMA. As a first step towards generating a genetic model of SMA in zebrafish, we identified three smn mutations. Two of these alleles, smnY262stop and smnL265stop, were stop mutations that resulted in exon 7 truncation, whereas the third, smnG264D, was a missense mutation corresponding to an amino acid altered in human SMA patients. Smn protein levels were low/undetectable in homozygous mutants consistent with unstable protein products. Homozygous mutants from all three alleles were smaller and survived on the basis of maternal Smn dying during the second week of larval development. Analysis of the neuromuscular system in these mutants revealed a decrease in the synaptic vesicle protein, SV2. However, two other synaptic vesicle proteins, synaptotagmin and synaptophysin were unaffected. To address whether the SV2 decrease was due specifically to Smn in motoneurons, we tested whether expressing human SMN protein exclusively in motoneurons in smn mutants could rescue the phenotype. For this, we generated a transgenic zebrafish line with human SMN driven by the motoneuron-specific zebrafish hb9 promoter and then generated smn mutant lines carrying this transgene. We found that introducing human SMN specifically into motoneurons rescued the SV2 decrease observed in smn mutants. Our analysis indicates the requirement for Smn in motoneurons to maintain SV2 in presynaptic terminals indicating that Smn, either directly or indirectly, plays a role in presynaptic integrity.
INTRODUCTION
Spinal muscular atrophy (SMA) is an autosomal recessive disease characterized by the degeneration of anterior horn spinal cord α-motoneurons. This degeneration subsequently leads to progressive muscle atrophy, paralysis, respiratory failure and in severe cases infant death. Humans have two copies of the SMN gene: SMN1 and SMN2, located on chromosome 5q13 (1,2). SMN1 is absent in ∼96% of SMA patients and those retaining the gene carry intragenic mutations (3). There are only a small number of nucleotide differences between SMN1 and SMN2 including a C/T transition at position +6 in exon 7 resulting in an exonic splicing silencer which removes exon 7 during splicing (4). Therefore, the vast majority of SMN2 transcripts give rise to an unstable truncated protein lacking the C-terminus (5). However, ∼10–15% of SMN2 product does give rise to full-length SMN protein. Therefore, deletion of, or mutations in, SMN1 leaves the patient to derive all full-length, functional SMN from the SMN2 gene. This also explains why patients with a greater number of SMN2 genes in their genome, in general, have less severe SMA since more full-length SMN is produced (6). Only humans have an SMN2 gene, whereas all other animals examined only have SMN1 (2).
Different animal models have been used to study the relationship between SMN and motoneuron development, function and survival. Transient knockdown of both maternal and zygotic Smn in zebrafish embryos to levels similar to those in SMA patients resulted in aberrant motor axon outgrowth including truncated and branched motor axons and decreased survival (7). While motor axons in SMA mice do not show such defects in vivo (8), cultured motoneurons from the SMA mice do exhibit short axons with smaller growth cones (9). This suggests that when removed from their normal environment, mouse motor axons are also compromised by low SMN levels. Mouse SMA models do display defects at the neuromuscular junction (NMJ) in vivo such as unoccupied synapses, morphologically immature synapses, some electrophysiological dysfunction and neurofilament accumulation (8,10–12). NMJs have also been examined in Drosophila Smn mutants and they are also compromised (13,14). Taken together, these data support that decreased SMN levels affect NMJ formation and function and that mice with less SMN have more severe defects. Where SMN function is needed for this process and why low levels of SMN lead to NMJ synaptic defects remains a central question in the field.
To analyze these questions in more detail, we have generated ENU-induced smn zebrafish mutants. The smn zebrafish mutants survive to larval stages due to maternally contributed Smn. In the homozygous mutant background, the maternal contribution is ultimately depleted during larval stages resulting in defects at the presynaptic terminal and decreased survival. To further understand the neuronal specificity of SMA, we have generated transgenic fish that expresses human SMN (hSMN) only in motoneurons and then generated mutant lines carrying this transgene. Interestingly, the motoneuron-specific hSMN corrected the NMJ defect. Thus, we conclude that SMN in motoneurons is necessary for presynaptic NMJ integrity.
RESULTS
Zebrafish smn mutations
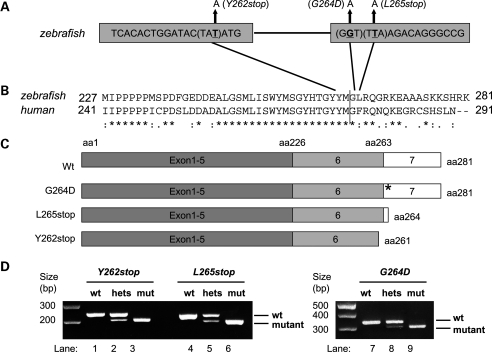
We identified eight putative smn mutations by screening over 8500 ENU mutated zebrafish genomes by TILLING (Targeted Induced Localized Lesions In Genomes) (15) (Supplementary Material, Fig. S1). Two of these, smnY262stop and smnL265stop, were stop mutations and both resulted in a C-terminal deletion of the protein (Fig. 1A–C). Because of their resemblance to the major, unstable SMN2 gene product in humans (4,5,16), we predicted they would give rise to an unstable Smn protein. A third mutation, smnG264D, occurred in exon 7 and corresponds to the human mutation SMNG279V, a point mutation that causes Type I SMA in humans (17). The other five mutations did not change the protein in a way suggestive of a change in gene function and thus were not investigated further. A dCAPS (derived cleavage amplified polymorphic sequence) strategy was developed to identify the three putative mutants at the DNA level (18) (Fig. 1D).
Figure 1.
Zebrafish smn mutations. (A) Schematic diagram showing the smn mutations smnY262stop in exon 6, smnG264D, and smnL265stop in exon 7. (B) Clustal W (1.82) alignment of zebrafish Smn exons 6 and 7 (black, vertical line demarcates the boundary) with the human homologue showing mutated amino acids. (C) Schematic diagram showing wild-type and mutant Smn proteins. (D) dCAPS of zebrafish smn mutations. The PCR product of wild-type (lane 1, 4, 7), heterozygous (lanes 2, 5, 8) and homozygous smn mutants (lanes 3, 6, 9). aa, amino acid; wt, wild-type. *= identical amino acid, : = conserved substitution, . = semi-conserved substitution.
SMNG279V replaces an unbranched glycine for a branched valine. In the zebrafish mutation SmnG264D, aspartic acid is also a branched substitution, thus we predicted that it would also affect function. To test whether the two stop mutations and the smnG264D mutation would indeed give rise to Smn with little or no function, we tested these mutations in our motor axon rescue assay. We have previously shown that knockdown of maternal and zygotic Smn in zebrafish using a translation blocking morpholino (MO) caused motor axon defects during embryonic and early larval development (7). Upon co-injection of smn MO and hSMN RNA, we found significant rescue of the motor axon defects. We then used this approach to determine whether human patient mutations could rescue the motor axon defects and found that they could not (19). One exception was the SMNA111G mutation that was shown to rescue only when expressed at high levels (20) indicating the sensitivity of this assay. Thus, to test whether the three putative zebrafish smn mutations lacked wild-type Smn function, we tested them in this rescue assay. For this, we knocked down the endogenous smn RNA using an smn MO that acted upstream of the start site in the 5′-UTR (5′-UTR MO) and added back the putative, in vitro transcribed, mutant smn RNA (i.e. smnY262stop, smnG264D and smnL265stop) that were generated using site-directed mutagenesis. Wild-type zebrafish smn RNA was used as a control. The 5′-UTR MO caused motor axon defects that were significantly rescued by co-injection of wild-type zebrafish smn RNA (7) (P < 0.001, Supplementary Material, Fig. S2). Upon co-injection with smn MO, none of the three mutant RNAs could rescue the motor axon defects. The two stop mutations were worse than the smnG264D mutation (smnY262stop P = 0.62 and smnL265stop P = 0.85 when compared with smn MO alone, Supplementary Material, Fig. S2). smnG264D (P = 0.05 compared with smn MO alone) weakly rescued but was also significantly different than wild-type smn RNA (P = 0.014). These data indicate that these three alleles are bona fide mutations and thus can be used to study Smn function. We proceeded, therefore, to establish lines of fish carrying these mutations.
smn zebrafish mutants survive to larval stages due to maternal Smn
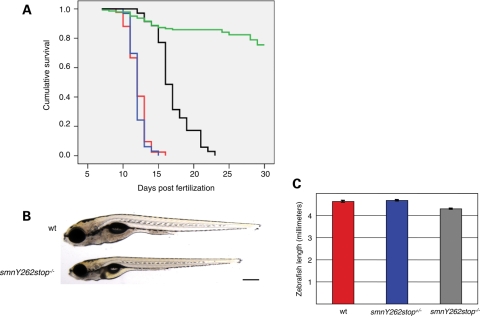
Heterozygous smn mutants (smn+/−) of all three alleles were fertile and had no phenotype. Homozygous mutants (smn−/−) obtained by heterozygous matings segregated following Mendelian ratios. To determine whether smn−/− embryos were lethal, embryos from smn+/− incrosses were grown in the same zebrafish nursery tank and any dead larvae were collected daily. At 20–30 days post-fertilization (dpf), the remaining zebrafish larvae were sacrificed and all larvae were then genotyped. Kaplan–Meier analysis revealed that both smnY262stop−/− and smnL265stop−/− mutants had an average survival of 12 dpf with a range of 9–16 and 11–15 dpf, respectively (Fig. 2A). Consistent with the results in the motor axon assay, the smnG264D−/− fish were less severe dying on average at 17 dpf with a range of 13–21 dpf. All three smn−/− lines were indistinguishable from wild-types and heterozygotes as embryos and early larvae. Before they died, however, smn−/− larvae were statistically smaller with an average reduction of 7–8% of body length compared with wild-types or smn+/− (Fig. 2B–C).
Figure 2.
smn mutants are larval lethal. (A) Progeny from incrosses were grown in the same nursery tank and monitored until 20 dpf (smnY262stop, and smnL265stop) or 30 dpf (smnG264D, wild-type). Fish were genotyped by dCAPS and survival of homozygous mutants was plotted (wild-type, green line, n = 143; smnY262stop−/−, red line, n = 42; smnG264D−/−, black line, n = 35; smnL265stop−/−, blue line, n = 33). The average survival for both smnY262stop−/− and smnL265stop−/− larvae was 12 dpf with ∼80% dying between 11 and 13 dpf. The average survival for smnG264D−/− larvae was 17 dpf with ∼80% dying between 15 and 21 dpf. Survival of all mutants was significantly different compared with wild-types (P < 0.0001). (B) Image of live 11 dpf wild-type and smnY262stop−/− larvae. (C) smnY262stop−/− (mutant, n=63) were smaller than wild-type (n = 55; P < 0.0001), and smnY262stop−/+ larvae (n = 114; P < 0.0001) at 11 dpf. Scale bar, 300 µm.
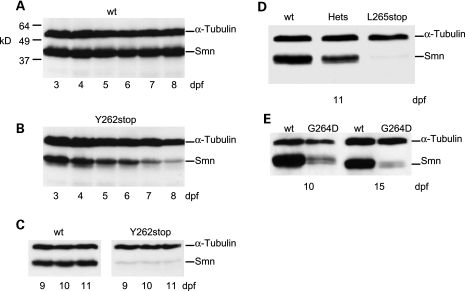
Since mouse Smn mutants die during early embryogenesis (21), we wondered whether maternal Smn was extending the survival of zebrafish smn−/− larvae. We also hypothesized that the three mutations would give rise to unstable proteins. Exon 7 is needed for SMN protein stability (16) and both the SmnY262stop and SmnL265stop resulted in proteins lacking this exon. In addition, SmnG264D corresponds to the human mutation SMNG279V, which has also been shown to produce an unstable protein (20). To examine both of these issues, we analyzed Smn protein expression in the mutant lines. Wild-type zebrafish had consistent Smn protein levels from day 3–11 dpf; however, smnY262stop−/− larvae had high levels of Smn on day 3–6 dpf, but these levels were substantially reduced by 7–8 dpf (Fig. 3A and B). By 9–11 dpf, Smn levels were depleted in smnY262stop−/− larvae and were only slightly above background (Fig. 3C). The SmnL265stop truncated protein showed an identical profile of expression as SmnY262stop and was also only present at very low levels at 11 dpf (Fig. 3D). We interpret these results to mean that since the zygotic mutant Smn is unstable and undetected on western blot, the protein observed at high levels between 3 and 6 dpf is from maternal smn transcripts or maternally deposited protein. Similarly, the SmnG264D protein was only present at low levels at 10 and 15 dpf suggesting that this mutation also results in an unstable Smn protein (Fig. 3E). This result is consistent with in vitro analysis of the corresponding human mutation, SMNG279V, showing that this mutation results in an unstable protein (22). The low level of SmnG264D protein at 12–15 dpf also explains the extended survival of smnG264D−/− larvae when compared with smnY262stop−/−, or smnL265stop−/− larvae (Fig. 2). Together, these data show that consistent with other species decreasing Smn to very low/undetectable levels in zebrafish is lethal.
Figure 3.
Smn protein levels in smn mutants. Smn protein levels from (A) wild-type (wt) and (B) smnY262stop−/− larvae at 3–8 dpf. (C) wt and smnY262stop−/− at day 9–11 dpf. (D) wt, heterozygous (hets) and smnL265stop−/− larvae at 11 dpf (E) wt and smnG264D−/− larvae at 10 and 15 dpf. Each lane was loaded with protein extracts from three larvae. Protein mass markers (kD) are present in A and α-tubulin served as loading control for all blots.
We also examined the protein expression of Gemin 2 and Profilin two SMN binding proteins (23,24). In smnY262stop−/− mutants, we found that Gemin 2 levels were decreased and the levels of Profilin were unchanged (Supplementary Material, Fig. S3). Both of these results are consistent with what is seen in a severe SMA mouse model (23,24) and indicate that low levels of Smn in zebrafish are affecting the organism in a matter similar to depletion in mammals.
Effect of smn mutations on zebrafish neuromuscular junctions
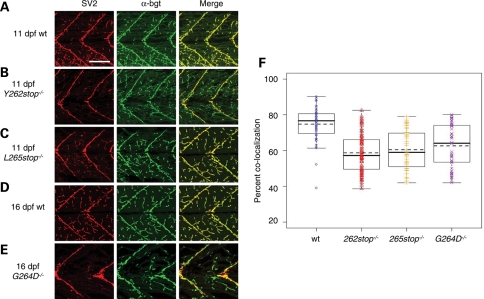
We have previously demonstrated that knocking down both maternal and zygotic Smn led to motor axon defects (7,19). Because the smn−/− embryos have high maternal Smn during the time of motor axon outgrowth (1–4 dpf), their motor axons developed normally (Supplementary Material, Fig. S4). Thus, we cannot examine the role of Smn in motor axon outgrowth in these mutants unless we deplete maternal Smn. To understand how low Smn protein levels affects the larval neuromuscular system, we investigated neuromuscular junctions (NMJs) in smn mutants. Normal NMJs have a tight juxtaposition of pre- and postsynaptic proteins. Decreases in presynaptic or postsynaptic proteins or a lack of co-localization is indicative of abnormal NMJs. Wild-type and smn−/− larvae were double immunolabeled with an antibody against synaptic vesicle protein 2 (SV2), a protein present in presynaptic vesicles and α-bungarotoxin (α-bgt) which binds to acetylcholine receptors found on the postsynaptic side of the NMJ. At 9 dpf, SV2 and α-bgt were co-localized and no NMJ defects were observed in smnY262stop−/− larvae (data not shown). However, on 11 dpf when smnY262stop−/− larvae had extremely reduced levels of Smn (Fig. 3), we found an overall reduction in the ratio of SV2/ α-bgt when compared with wild-types indicating a decrease in SV2 (Fig. 4, P < 0.0001). Similar to smnY262stop−/−, both smnL265stop−/− and smnG264D−/− mutants had reduced SV2/ α-bgt ratio when compared with the wild-type zebrafish larvae (Fig. 4, P < 0.0001). These data indicate that in zebrafish smn mutants, there is either a loss of presynaptic terminals at the NMJ or a decrease in NMJ SV2 protein. To address whether other presynaptic proteins were decreased, we examined synaptotagmin II and synaptophysin, which like SV2, are located on the presynaptic vesicle membrane (25). Immunohistochemistry revealed that both of these proteins were expressed normally at the NMJ in all three mutants (Fig. 5 and data not shown). These data indicate that presynaptic terminals are present in zebrafish Smn mutants, but there is a decrease in SV2 protein at the NMJ.
Figure 4.
SV2 is decreased at NMJs in smn mutants. (A) Wild-type (wt, n = 10), (B) smnY262stop−/− (n = 14), (C) smnL265stop−/− (n = 8) larvae at 11 dpf. (D) wt (n = 10) and (E) smnG264D−/− (n = 11) larvae at 16 dpf. Presynaptic regions were labeled with SV2 antibodies (red) and postsynaptic regions were labeled with α-bungarotoxin (α-bgt; green). Merge is an overlay (yellow). (F) The coefficients of co-localization were plotted. All mutants were significantly different compared with wild-types (P < 0.0001). Scale bar, 80 µm.
Figure 5.
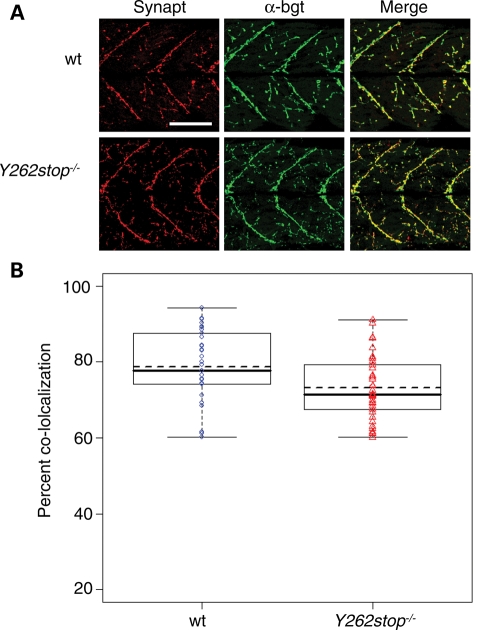
Synaptotagmin is not affected in smn mutants. (A) Wild-type (wt, n = 8) and smnY262stop−/− (n = 10) labeled with synaptotagmin II antibody (synapt, red), and α-bgt (green) at 11 dpf. (B) The coefficients of co-localization were plotted. Mutants were not significantly different from wild-types (P = 0.50). Scale bar, 80 µm.
Motoneuronal hSMN expression rescues the NMJ SV2 defect in smnY262stop mutants
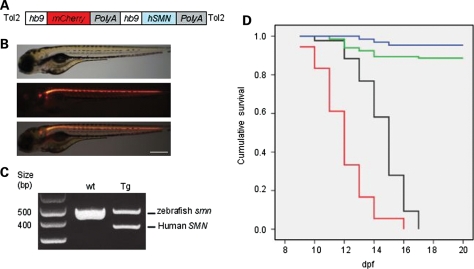
Since Smn is expressed in all tissues and cells, we next determined where Smn was needed to maintain SV2 NMJ expression. For these experiments, we generated a zebrafish transgenic line Tg(hb9:hSMN;hb9:mcherry) that expresses hSMN and mCherry using the zebrafish hb9 promoter (26) (Fig. 6A). This transgene will hereafter be referred to as Tg(hb9:hSMN). We verified that hSMN RNA was present in the transgenic fish and that mCherry fluorescence was expressed in motoneurons verifying that our construct was expressed in the appropriate cells (Fig. 6B and C). We then crossed Tg(hb9:hSMN) to the smnY262stop line to generate mutants expressing hSMN only in motoneurons. Quantitative real-time PCR revealed that the progeny from this cross had an average of six transgene copies (data not shown).
Figure 6.
Transgenic expression of hSMN in zebrafish motoneurons. (A) Schematic diagram showing the hb9:hSMN transgene construct with hb9:mCherry marker. (B) Lateral views of live Tg(hb9:hSMN;hb9:mCherry) (abbreviated Tg(hb9:hSMN)) zebrafish at 3 dpf. Top, brightfield; middle, fluorescence; bottom, merge. (C) Multiplex RT–PCR of wild-type (wt) and Tg(hb9:hSMN) (Tg) with human and zebrafish smn primers. (D) Kaplan–Meier survival curves of smnY262stop−/− larvae and smnY262stop−/−;Tg(hb9:hSMN). The average survival for smnY262stop−/− with the hb9:SMN transgene was significantly different from those without the transgene (P < 0.0001). Blue line, Tg(hb9:hSMN) (n = 132); Green line, Y262stop+/−; Tg(hb9:hSMN) (n = 64); black line, Y262stop−/−; Tg(hb9:hSMN) (n = 43); red line, Y262stop−/− (n = 42). Scale bar, 250 µm.
To determine whether driving hSMN exclusively in motoneurons affected survival, we repeated the survival assay on an incross of smnY262stop−/+;Tg(hb9:hSMN) fish. mCherry positive embryos were identified at 2 dpf and monitored over 20 days. Any larvae that died were kept and on day 20, all fish were genotyped to identify the smnY262stop−/−;Tg(hb9:hSMN) larvae. The hSMN protein that was expressed within the zebrafish motoneurons slightly extended the survival rate of smnY262stop−/− from an average of 12–14 dpf (P < 0.0001; Fig. 6D). It was not unexpected, however, that the fish still died since SMN is needed in all cells for survival (21).
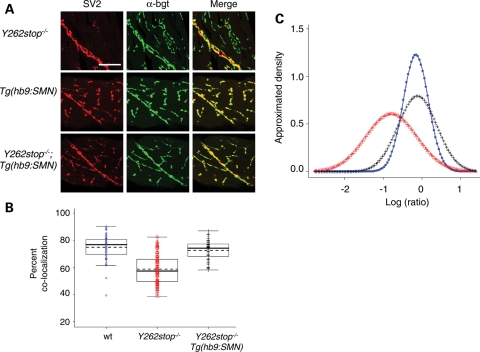
We next examined NMJs when hSMN was expressed only in motoneurons. smnY262stop−/−;Tg(hb9:hSMN) larvae were raised to 11 dpf and immunolabeled with SV2 and α-bgt. Interestingly, the introduction of hSMN specifically into motoneurons rescued the SV2 reduction seen in homozygous mutants (Fig. 7A). To quantify the NMJ defects, we plotted the co-localization efficiency of the pre- and postsynaptic regions as determined by NIH Image J (Fig. 7B). Wild-type zebrafish had an average coefficient of 77% compared to 60% in smnY262stop−/− zebrafish mutant. With the addition of hb9:hSMN transgene, the smnY262stop−/− larvae had an increased co-localization rate of 72%. This value is statistically different that smnY262stop−/− alone (P < 0.0001) but not different than wild-types (P = 0.482) indicating that the addition of hSMN to motoneurons rescued the SV2 defect. To look at this data in another way, we determined the density of areas that had only SV2 and only α-bgt and plotted this as a log ratio. In wild-type zebrafish, we found the same amount of pre- and postsynaptic only regions, therefore the log ratio was close to zero (Fig. 7C). However, in smnY262stop−/− larvae, the ratio of presynaptic versus postsynaptic labeling was decreased (P < 0.0001 compared with wild-type). The introduction of hSMN into motoneurons shifted this ratio closer to zero indicating that SV2 was rescued (P = 0.482 compared with wild-type). Since smnY262stop−/− larvae carrying the hb9:hSMN transgene lived slightly longer than mutants alone, we repeated the immunolabeling at 13 dpf. Even right before the fish died, we did not observe any SV2 defects (data not shown; n = 8) indicating that expressing hSMN in motoneurons rescued the presynaptic NMJ defects.
Figure 7.
hSMN expression in motoneurons rescues the NMJ defect in smn mutants (A) Immunolabeling of postsynaptic regions (α-bgt, green), and presynaptic regions (SV2, red) of 11 dpf smnY262stop−/− larvae (n = 14), Tg(hb9:SMN) (n = 10) and smnY262stop−/−;Tg(hb9:hSMN) larvae (n = 14). (B) The coefficients of co-localization were plotted and the means of each group were calculated. (C) The distribution of log ratio of presynaptic only regions versus postsynaptic only regions. Wild-type zebrafish had a ratio close to zero, indicating the same amount of pre- and postsynaptic regions. The smnY262stop−/− larvae (red) had a reduced ratio of co-localization when compared with wild-type zebrafish (blue), but increased with addition of the hb9:hSMN transgene (black). For co-localization, smnY262stop−/−;Tg(hb9:smn) versus wt, P = 0.1; smnY262stop−/−;Tg(hb9:smn) versus smnY262stop−/−, P < 0.0001. For pre/post log-ratios, smnY262stop−/−;Tg(hb9:smn) versus wt, P = 0.482; smnY262stop−/−;Tg(hb9:smn) versus smnY262stop−/−, P < 0.0001. For all the tests, the P-values for smnY262stop−/− versus wt <0.0001. Scale bar, 40 µm.
DISCUSSION
In this study, we report the isolation and characterization of mutant alleles in the zebrafish smn gene. The three zebrafish smn mutants described here are single base changes identified by TILLING. Two cause premature stop mutations resulting in the loss of the last 19 and 16 amino acids, respectively. As smn exon 7 encodes for the last 18 amino acids of the protein, these mutations lead to the loss of exon 7 producing a truncated, unstable protein. It has been shown in vitro that hSMN exon 7 is required for protein stability (16). Our data are consistent with this since both smnY262stop−/− and smnL265stop−/− had barely detectable Smn protein during the second postnatal week of development. In addition, Smn protein in smnG264D−/− was very low during the second postnatal week of development again suggesting it is an unstable protein. This is consistent with recent in vitro data showing that the corresponding mutation in humans, SMNG279V, is unstable (22). Thus, our in vivo data are consistent with in vitro data and indicate that some point mutations can lead to protein instability (22). During the first week of development, however, Smn levels were similar to wild-type levels suggesting that this Smn was derived from maternal RNA or protein. Maternal RNAs and proteins are deposited in the egg to drive early development with zygotic transcription starting at ∼3 hpf. How long a maternal protein is present depends on the amount deposited, how quickly the maternal RNA is degraded (27) and protein stability. These factors, along with the turnover of the mutant Smn protein, contribute to the Smn levels in these mutants. Due to the presence of Smn during early development, mutants live into the second postnatal week of development enabling examination of their neuromuscular system. Our analysis revealed that all three mutants had decreased SV2 NMJ expression. This decrease was rescued by driving hSMN in motoneurons revealing the importance of Smn in motoneurons for normal NMJ development. It is unclear at this time whether the function of Smn in this process is direct or indirect. It should also be noted that these mutants do not mimic the situation in human patients where there is constant low SMN levels. In the zebrafish mutants, levels of Smn are high early due to maternal RNAs/protein then the levels gradually decrease to very low/absent levels. These mutants do, however, allow us to look at the consequences of depleted Smn on the stability and/or maintenance of the neuromuscular system. Moreover, they serve as a first step toward generating a true genetic model of SMA in zebrafish, which will be accomplished by adding the hSMN2 to this mutant background.
Homozygous mutants were smaller, but otherwise formed normally. The fish did exhibit less activity before dying often lying at the bottom of their tank. The most striking neuromuscular defect in these fish was a decrease in the presynaptic protein SV2 in all three mutants. This indicates that NMJs are not maintained when Smn levels are decreased and that there is less presynaptic protein at the NMJs. We have previously shown that reducing both maternal and zygotic Smn using anti-sense morpholinos caused defects in motor axon outgrowth (7) and decreased longevity (19). Taken together, these data suggest that early depletion of Smn has more severe defects in zebrafish affecting the development of motor axons. Removing Smn after motor axon outgrowth and synapse formation, as is the situation in the smn mutants, however affects the maintenance of the presynaptic terminal. Thus, in both scenarios, motoneurons are affected but at difference stages, outgrowth and synaptic maintenance, respectively. Interestingly, this also suggests that depleting Smn after motor axon outgrowth and synapse formation still has an effect on the NMJs. Thus, the phenotypes exhibited strongly depend on the levels and timing of Smn protein expression.
The fact that we are seeing NMJ defects in zebrafish smn mutants is consistent with the analysis from SMA mouse models (8,10–12). In the most severe mice that have two copies of the SMN2 gene on the SMN null mouse background (SMN−/−; SMN2 2 copies) observed at embryonic day 18 and postnatal day 2, there is evidence of unoccupied synapses (∼50%) at NMJs in intercostal muscles, which are know to be affected in SMA patients (8). In mice considered still severe but less so (SMN−/−; SMN2 2 copies; SMNΔ7), there is only a low level of unoccupied NMJ synapses (<30%), but the presynaptic terminal is immature and structurally less complex (10–12). These mice also showed electrophysiological defects at the NMJ including decreased quantal content, decreased evoked endplate current and decreased facilitation when stimulated repetitively (11). Moreover, electrophysiological analysis in mild SMA mice (SMN−/−; SMN2 2 copies; SMNA2G) showed intermittent synaptic failures (10). Taken together, these data support that decreased SMN levels affect NMJ formation and function and that mice with less SMN have more severe NMJ defects. NMJs have also been examined in Drosophila Smn mutants. Like zebrafish, the NMJs in Drosophila are boutons and do not form elaborate structures thus analysis relies on quantifying bouton number/and or the density of pre- and postsynaptic protein juxtaposition. Drosophila Smn mutants exhibit a decrease in bouton number with the extent dependent on the mutant background line used (13,14). For example, a line with a moderate decrease in Smn levels had an ∼3.5-fold decrease in bouton number and a line with a mild decrease in Smn levels had an ∼1.6-fold decrease (14). Postsynaptic changes were also present in Drosophila with the loss of glutamate receptor subunit (GluRIIA) expression (13,14). Thus, in all animal models examined, low Smn levels compromise NMJs. It remains to be determined whether the defects at the NMJ caused by low Smn levels in any species can explain the severe neuromuscular defects seen in SMA. It is also possible that the presynaptic contacts onto motoneurons are also affected; however, these types of defects have yet to be reported.
Slightly different approaches have been used to address SMN cell autonomy in the different model systems. Driving expression of Smn only in motoneurons on an smn−/− background rescued the NMJ defects in zebrafish, but did not fully rescue survival (this report). In mice, the SMN−/− is on a background of low SMN levels due to introduction of the human SMN2 gene (28). This produces enough SMN for the mice to survive into early postnatal stages, but they die of SMA phenotypes. Using these mice and driving SMN expression to higher levels in all neurons using the prion promoter rescued survival. However, driving high expression in muscle on the same background of low SMN using the human skeletal actin promoter did not increase survival (29); synapses, however, were not examined in this study. In Drosophila, Smn knockdown using RNAi in either neurons or mesoderm, both decreased bouton number. Furthermore, in a hypomorphic Smn mutant Drosophila background, driving expression in mesoderm partially rescued the bouton loss as did driving expression in all neurons. The best rescue was achieved when the expression was driven in both neurons and mesoderm (14). Thus, in Drosophila, both neuronal and mesodermal Smn expression appears to be important for NMJs. In C. elegans, loss of the smn gene causes loss of motility, decreased pharyngeal pumping activity and premature death in larval stages again with maternal Smn keeping the animals alive until late larval stages (30). In this species, neuronal but not muscle expression partially rescues these phenotypes (30). The consistent result from these experiments is that neuronal Smn is essential and even on a null background can rescue motoneuron phenotypes. The fact that the presynaptic protein SV2 was decreased with no observable defect in the postsynaptic side in smn mutant zebrafish reflects a more profound perturbation on the presynaptic side of the NMJ. This is consistent with our rescue studies showing that driving Smn only in motoneurons rescues this phenotype; however, we cannot rule out a role for Smn in muscle as this has not been directly tested. The decrease in SV2 is not due to a decrease in motor axons as these are present and appear normal, however it may reflect a decrease in presynaptic vesicles, presynaptic docking or the ability to maintain the integrity of essential presynaptic components.
Our finding that SV2 and not other presynaptic proteins was decreased in Smn mutants suggests that SMN may have a unique role in SV2 production, stability or localization at the synapse. SV2 is a component of all vertebrate synapses (31) and is important for the efficacy of synaptic release (32). When synaptic vesicles fuse to the presynaptic membrane, SV2 can bind to laminin (α5, β1, γ), a component in the synaptic extracellular matrix (ECM), thus anchoring the nerve terminal to the synaptic ECM (33). Therefore, perturbations in SV2 could result in decreased nerve terminal stability. A defect in synapse vesicle localization has been described in mice (11) although a link to SV2 was not examined. Results from hippocampal neurons lacking the neuronal SV2A isoform suggest a role for SV2 in priming synaptic vesicles (34). These authors also showed that the decreased EPSC seen in these neurons is due to a decrease in the probability of vesicle release. Interestingly, it has been shown in mice that there is a decreased EPSC attributed to a decrease in vesicle release at the NMJ (11). SV2 decreases were not seen in these mice using immunofluorescense; however, it may be that the decrease in protein can only be visualized by immunohistochemistry on a null background. Because the SMN null mice die during early embryogenesis, this cannot be examined in mice. Our finding that SV2 is decreased when Smn levels are very low could provide a molecular link to the reported NMJ defects seen in SMA models.
There are a number of possibilities, but no direct evidence yet, for how SMN could affect NMJs. SMN is found associated with many different complexes (Reviewed in 35). SMN is known to function in snRNP assembly and its reduction causes reduced levels of snRNPs (36,37) which could affect the splicing of genes important for NMJs; however, this remains to be determined. Alternatively, SMN could have a function outside of splicing that affects that stability or maintenance of NMJ synapses. There is evidence supporting a connection between SMN and cytoskeletal elements such as β-actin (9). In support of this, over expression of Plastin 3, an actin bundling protein, rescues axon defects caused by decreased SMN in cultured cells and zebrafish (38). Such cytoskeletal elements are also important during synapse maintenance and function (39); however, whether Plastin 3 has an effect on NMJs remains to be determined.
A number of animal models of SMA now exist and can be exploited for their strengths. Drosophila and C. elegans models are being used for modifier screens to identify gene targets of smn (14). Mice continue to reveal aspects about the SMA phenotype and are also important for drug testing. A genetic model of SMA in zebrafish will be useful for performing genetic and drug screens, testing candidate modifier genes, and phenotypic analysis. The combined use of these model systems provides the best chance at understanding and finding therapies for this disease.
MATERIAL AND METHODS
Zebrafish maintenance
Adult zebrafish and embryos were maintained as previously described (40). All fish were maintained at temperatures between 27 and 29°C. Zebrafish used in this study were on the *AB/LF background.
TILLING
TILLING was performed as described in (18).
smn zebrafish genotyping and survival assay
smn mutants were genotyped by dCAPS. The dCAPS primer sequence were designed according to the point mutation nucleotides: smnY262stop (Forward primer 5′-GGGTTACATCACCCACCCAA-3′; reverse primer 5′-GGGGTACCTACAAGAAAAACAACTGTACAAT-3′), smnL265stop (Forward primer 5′-ACTTAATTATTTATTTTTCAAACAGGTTT-3′; reverse primer 5′-GCTCTTACAACAGGGATCGA-3′) and smnG264D (Forward primer 5′-GGGTTACATCACCCACCCAACA-3′; reverse primer 5′-ATGCAGCAGCCTCTTTACGGCCCTGTCTTGAA-3′). The dCAPS PCR products were digested with TSP509I restriction enzyme, New England Biolab (smnY262stop), Dra1 restriction enzyme, Invitrogen (smnL265stop), and TfiI restriction enzyme, New England Biolab (smnG264D) as described in the manufacturer's instruction. The digested PCR products were resolved on a 2% agarose gel.
For the survival assay, progeny from matings between heterozygous mutant adults were raised in the same nursery environment. Dead larvae were collected twice a day and frozen. On 20 dpf (smnY262stop and smnL265stop) or 30 dpf (smnG264D and wild-types), the remaining larvae were sacrificed and all larvae, including those that died earlier, were then genotyped. Survival of smn−/− larvae was evaluated using Kaplan–Meier survival curves. For each fish, time of death, survival status and classification (wild-type, heterozygote and mutant) were put into SPSS (SPSS version 15; SPSS, Chicago, IL, USA), then Kaplan–Meier survival tests were run to generate the survival curve and P-values were calculated by the log-rank test.
Transgenic line
To generate Tg(hb9:SMN), we cloned the zebrafish hb9 promoter (26) into the pminiTol2/MCS plasmid backbone (41,42). The human SMN cDNA with SV40 polyA sequence was then taken out from pCS2-hSMN (19) and placed under the regulation of the hb9 promoter. We then cloned another hb9 promoter driving mCherry-SV40-polyA gene (43) and placed it at the 5′-UTR of the previously cloned hb9:SMN (Fig. 6). The construct and transposase mRNA were then injected into embryos at the 1-cell stage as described (41). Injected zebrafish (F0s) with high levels of labeled mCherry motoneurons were selected, growth to adulthood, and then screened for transgenesis. Approximately 40% of injected F0s had successful transgene insertion into their germline as evidenced by transgene transmission to their progeny (F1s). All F1s were screened for mCherry fluorescence and the brightest ones kept and used to establish lines. One F1 was crossed to a smnY262+/−. This line was then grown to adulthood and incrossed to generate the line used in this analysis. Q-PCR revealed that this line had six copies of the transgene.
Total RNA isolation and RT–PCR
Total RNA from 24 to 36 hpf zebrafish embryos were isolated with Trizol reagent (Invitrogen) then treated with DNAse. RT–PCR was performed using Qiagen one-step RT–PCR kit following manufacturer's instructions. Both zebrafish and human SMN were amplified by gene specific primers: zebrafish smn (forward primer 5′-ACCCAGGGAAGAAGAGGAAA-3′; reverse primer 5′-ATCCAGGAAAGGAAGGACCA-3′), human SMN (forward primer 5′-CGAGCAGGAGATGGGAACC-3′; reverse primer 5′-CCCAAAGCATCAGCATCATC-3′).
Quantitative PCR
Quantitative PCR (qPCR) was performed essentially as described in (44). The primers FP2-HB9pQ (5′-TCATTTCAGAATTGGTCAGTGCATG-3′) and RP2-HB9pQ (5′-CGAGGGAAATCAACAAGTCACCG-3′) were used for amplification of part of the hb9 promoter, and primers FP-DrActQ (5′-TCATTTCAGAATTGGTCAGTGCATG-3′) and RP-DrActQ (5′-CGAGGGAAATCAACAAGTCACCG-3′) were used to amplify actin mRNA (45). The qPCRs were performed on the Mx3000P platform (Stratagene) using SYBR Green (Sigma). For data analysis, values were normalized to actin for input correction and the results of the wild-type samples analyzed in parallel set to 1.
Western blotting
Protein extracts from three zebrafish larvae were dissolved in blending buffer (10% SDS, 62.5 mm Tris pH6.8, 5 mm EDTA) and then resolved in a 12% bis-Tris gel, electro-blotted to a polyvinylidene difluoride membrane (Hybond-P; Amersham Bioscience). Membranes were probed with primary antibodies: anti-SMN (1:500, MANSMA7, (46)), anti-Gemin2 (1:10, 1G14, (47)), anti-Profilin (1:500, PF2A3, (48)), β-actin (1:3000, Santa Cruz, sc-47778) or α-Tubulin (1:1000, Santa Cruz Biotechnology, sc-5286). Signal was detected with horseradish peroxidase-conjugated goat anti-mouse antibodies (1:2000, Jackson ImmunoResearch Laboratories, Inc.) and ECL reagents (Amersham Bioscience).
Immunohistochemistry and NMJ analysis
Zebrafish larvae were anesthetized in tricaine (A-5040; Sigma) and then fixed in 4% paraformaldehyde overnight at 4°C. After fixation, larvae were washed with 1XPBS, and the skin along their trunk was peeled off. Postsynaptic regions were immunostained with Alexa Fluor 488 conjugated α-bungarotoxin (10 g/ml; Molecular Probes) for 30 min as described in (49). For presynaptic proteins, larvae were incubated with primary antibodies SV2 (1:100) (31), synaptotagmin II (znp1 1:100) (50) or synaptophysin (1:50, Invitrogen) followed by Alexa Fluor 594 conjugated secondary antibodies (1:200, Molecular Probes). In Tg(hb9:SMN;hb9:mCherry) zebrafish Alexa Fluor 633 conjugated secondary antibody was used instead of Alexa Fluor 594 so as to differentiate labeling from mCherry fluorescence. To examine motor axons in 11 dpf larvae, neurofilament antibodies (1:400, Abcam) followed by visualization with AlexaFluor 488-conjugated secondary antibodies (1:200, Molecular Probes) was used.
Coefficient of co-localization and log ratios were obtained by off-line analysis of confocal images (NIH Image J). For each hemisegment, we performed 0.3 µm scans for a distance of 10 µm on one side of the larvae. Each scan was repeated four times and then averaged. These averaged pictures for the red and green channels were projected into stacks, merged and used for the analysis. For all analysis, pixels exceeding the 50% threshold value for red and 60% threshold values for green were taken. The higher threshold value used for green pixels was to minimize the background effect of α-bgt labeling. For each fish, five hemisegments were analyzed and the co-localization coefficients for each data point were plotted. Each column represents a fish group (wild-type, Y262stop, L265stop and G264D). A box plot was added for each fish category to show the mean (solid line) and median (dash line). To show that all three mutant lines had reduced co-localization coefficients, we used one-tailed Mann–Whitney U test for comparison (R 2.6.0; GNU project).
Statistical analysis
smn mutant survival was plotted using Kaplan–Meier survival curves. Statistical significance was determined via the log-rank test (SPSS version 15; SPSS, Chicago, IL, USA). Fish length was evaluated using a student's t-test (Minitab version 15; Minitab Inc.). Decreases in the co-localization coefficients and log ratios of pre- and postsynaptic only regions were analyzed using a one-tailed Mann–Whitney U test (R 2.6.0; GNU project).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by postdoctoral fellowships from the Families of SMA to K.-L.B. and M.L.M. Additional support was provided by the National Institutes of Health (RO1 NS050414 and P30 NS045758 to C.E.B. and R01 HG002995 to C.B.M.), and the SMA Foundation (C.E.B.).
ACKNOWLEDGEMENTS
We thank the zebrafish facility staff at Ohio State for fish care, Dr Hao Le for help with the MO and RNA co-injections and Arthur Burghes for helpful comments. M.T.B. would like to thank Enrico Schleiff for support.
Conflict of Interest statement. The authors declare no conflict of interest.
REFERENCES
- 1.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 2.Rochette C.F., Gilbert N., Simard L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 3.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 6.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGovern V.L., Gavrilina T.O., Beattie C.E., Burghes A.H. Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marce M., Griffin J.W., Rich M.M., Sumner C.J. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 13.Chan Y.B., Miguel-Aliaga I., Franks C., Thomas N., Trülzsch B., Sattelle D.B., Davies K.E., van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 14.Chang H.C., Dimlich D.N., Yokokura T., Mukherjee A., Kankel M.W., Sen A., Sridhar V., Fulga T.A., Hart A.C., Van Vactor D., et al. Modeling spinal muscular atrophy in Drosophila. PLoS ONE. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCallum C.M., Comai L., Greene E.A., Henikoff S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 17.Talbot K., Ponting C.P., Theodosiou A.M., Rodrigues N.R., Surtees R., Mountford R., Davies K.E. Missense mutation clustering in the survival motor neuron gene: a role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum. Mol. Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 18.Draper B.W., McCallum C.M., Stout J.L., Slade A.J., Moens C.B. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- 19.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Workman E., Saieva L., Carrel T.L., Crawford T.O., Liu D., Lutz C., Beattie C.E., Pellizzoni L., Burghes A.H. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burnett B.G., Muñoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. Regulation of SMN protein stability. Mol. Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giesemann T., Rathke-Hartlieb S., Rothkegel M., Bartsch J.W., Buchmeier S., Jockusch B.M., Jockusch H. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J. Biol. Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 25.Sudhof T.C. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 26.Flanagan-Steet H., Fox M.A., Meyer D., Sanes J.R. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- 27.Schier A.F. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 28.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 29.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briese M., Esmaeili B., Fraboulet S., Burt E.C., Christodoulou S., Towers P.R., Davies K.E., Sattelle D.B. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckley K.M., Kelly R.B. Identification of a transmembrane glycoprotein specific for secretory vesicles of neurons and endocrine cells. J. Cell. Biol. 1985;100:1284–1294. doi: 10.1083/jcb.100.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang W.P., Sudhof T.C. SV2 renders primed synaptic vesicles competent for Ca2+-induced exocytosis. J. Neurosci. 2009;29:883–897. doi: 10.1523/JNEUROSCI.4521-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Son Y.J., Scranton T.W., Sunderland W.J., Baek S.J., Miner J.H., Sanes J.R., Carlson S.S. The synaptic vesicle protein SV2 is complexed with an alpha5-containing laminin on the nerve terminal surface. J. Biol. Chem. 2000;275:451–460. doi: 10.1074/jbc.275.1.451. [DOI] [PubMed] [Google Scholar]
- 34.Custer K.L., Austin N.S., Sullivan J.M., Bajjalieh S.M. Synaptic vesicle protein 2 enhances release probability at quiescent synapses. J. Neurosci. 2006;26:1303–1313. doi: 10.1523/JNEUROSCI.2699-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briese M., Esmaeili B., Sattelle D.B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;27:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- 36.Gabanella F., Butchbach M.E., Saieva L., Carissimi C., Burghes A.H., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oprea G.E., Krober S., McWhorter M.L., Rossoll W., Muller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cingolani L.A., Goda Y. Actin and action: interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- 40.Westerfield M. The Zebrafish Book. 3rd edn. Eugene: University of Oregon Press; 1995. [Google Scholar]
- 41.Balciunas D., Wangensteen K.J., Wilber A., Bell J., Geurts A., Sivasubbu S., Wang X., Hackett P.B., Largaespada D.A., McIvor R.S., et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawakami K. Transposon tools and methods in zebrafish. Dev. Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 43.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 44.Bohnsack M.T., Kos M., Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCurley A.T., Callard G.V. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young P.J., Man N.T., Lorson C.L., Le T.T., Androphy E.J., Burghes A.H., Morris G.E. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum. Mol. Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- 47.McWhorter M.L., Boon K.L., Horan E.S., Burghes A.H., Beattie C.E. The SMN binding protein Gemin2 is not involved in motor axon outgrowth. Dev. Neurobiol. 2008;68:182–194. doi: 10.1002/dneu.20582. [DOI] [PubMed] [Google Scholar]
- 48.Sharma A., Lambrechts A., Hao le T., Le T.T., Sewry C.A., Ampe C., Burghes A.H., Morris G.E. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp. Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 49.Ono F., Higashijima S., Shcherbatko A., Fetcho J.R., Brehm P. Paralytic zebrafish lacking acetylcholine receptors fail to localize rapsyn clusters to the synapse. J. Neurosci. 2001;21:5439–5448. doi: 10.1523/JNEUROSCI.21-15-05439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melancon E., Liu D.W., Westerfield M., Eisen J.S. Pathfinding by identified zebrafish motoneurons in the absence of muscle pioneers. J. Neurosci. 1997;17:7796–7804. doi: 10.1523/JNEUROSCI.17-20-07796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.