Abstract
Shwachman-Diamond syndrome (SDS; OMIM 260400) results from loss-of-function mutations in the Shwachman-Bodian Diamond syndrome (SBDS) gene. It is a multi-system disorder with clinical features of exocrine pancreatic dysfunction, skeletal abnormalities, bone marrow failure and predisposition to leukemic transformation. Although the cellular functions of SBDS are still unclear, its yeast ortholog has been implicated in ribosome biogenesis. Using affinity capture and mass spectrometry, we have developed an SBDS-interactome and report SBDS binding partners with diverse molecular functions, notably components of the large ribosomal subunit and proteins involved in DNA metabolism. Reciprocal co-immunoprecipitation confirmed the interaction of SBDS with the large ribosomal subunit protein RPL4 and with DNA-PK and RPA70, two proteins with critical roles in DNA repair. Function for SBDS in response to cellular stresses was implicated by demonstrating that SBDS-depleted HEK293 cells are hypersensitive to multiple types of DNA damage as well as chemically induced endoplasmic reticulum stress. Furthermore, using multiple routes to impair translation and mimic the effect of SBDS-depletion, we show that SBDS-dependent hypersensitivity of HEK293 cells to UV irradiation can be distinguished from a role of SBDS in translation. These results indicate functions of SBDS beyond ribosome biogenesis and may provide insight into the poorly understood cancer predisposition of SDS patients.
INTRODUCTION
Shwachman-Diamond syndrome (SDS; OMIM 260400) is an autosomal recessive cancer predisposition disorder characterized by exocrine pancreatic dysfunction, skeletal abnormalities and bone marrow failure with neutropenia and poor growth (1–3). SDS is caused by mutations in the highly conserved gene, Shwachman-Bodian Diamond syndrome (SBDS) (4). Targeted deletion of Sbds leads to embryonic lethality in mice by E6.5 (5). Despite a common occurrence in patients, homozygosity for null alleles has not been reported, suggesting that SBDS is also essential in humans. The greatest concerns for SDS patients are risks of severe infections, bone marrow failure and leukemic transformation, most commonly leading to acute myelogenous leukemia (reviewed in 2). SDS patients exhibit acquired chromosomal changes in cells or subgroups of hematopoetic cells, implying that loss of SBDS predisposes patients to leukemic transformation (6–8). Consistent with a chronic state of stress or susceptibility to genetic instability, increased p53 protein expression has also been reported in the bone marrow cells of SDS patients (9,10).
To date, the characterized function of the yeast ortholog of SBDS, Sdo1, is in ribosome biogenesis. Loss leads to perturbed ribosome profiles, and Sdo1 was found to be required for the release and recycling of the ribosome ancillary factor Tif6 (11). A number of Sdo1 interacting proteins have been identified, but the functional relevance of these interactions remains to be determined (12,13). SBDS involvement in ribosome biogenesis has also been implicated in mammalian cells, but reduction of SBDS protein expression using siRNA methods leads to conflicting results with regard to cell growth consequences (14–17) and has been reported to lead to defective spindle formation with an increased incidence of aneuploidy (18). Additionally, it has been reported that SBDS-depleted HeLa cells demonstrate increased Fas-ligand induced apoptosis (19). Given the diversity of these findings, the function or functions of SBDS remain unclear.
In order to better understand the cellular functions of SBDS, we used affinity capture and mass spectrometry to identify SBDS interacting proteins. By protein network inference and the PANTHER classification system (20), we have developed an SBDS interactome that supports diverse biological functions for SBDS. Proteins with an array of biological functions interact with SBDS, with a notable enrichment of ribosomal proteins and proteins involved in DNA metabolism. Depletion of SBDS from human cells leads to defects in protein translation and sensitizes cells to both DNA damage and endoplasmic reticulum (ER) stress, implying functions for SBDS in these processes. Furthermore, using multiple methods to impair global translation, we demonstrate that SBDS-dependent hypersensitivity of HEK293 cells to UV irradiation is distinct from a role of SBDS in translation. These findings support that SBDS is a multi-functional protein and provide a foundation for obtaining insight into the molecular mechanisms underlying the leukemia-predisposition observed in SDS patients.
RESULTS
Identification of novel SBDS interacting proteins
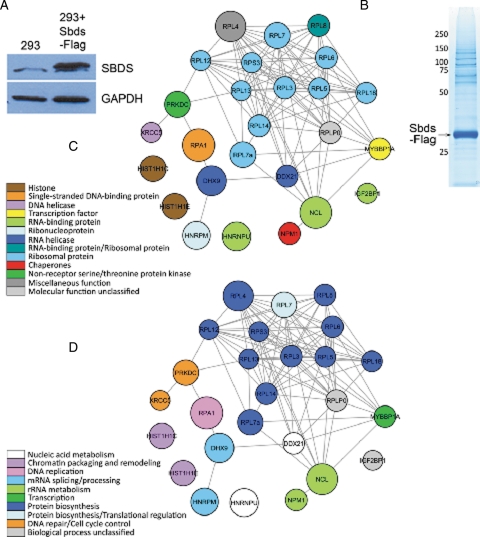
In order to elucidate functions of SBDS, we sought to identify interacting proteins using a broad, unbiased approach. We first generated an HEK293 cell line that stably expressed Sbds with a Flag-epitope at the C terminus. Immunoblot analysis of HEK293 lysates and Sbds-Flag HEK293 lysates demonstrated that the engineered cells express Sbds-Flag at steady-state levels that are modestly higher than endogenous SBDS (Fig. 1A). Sbds-Flag and co-associated proteins were captured from the Sbds-Flag HEK293 cells by affinity capture with Flag antibody immobilized on agarose beads. A control capture was conducted with HEK293 extract to recognize and eliminate proteins that bound directly to the resin and/or immobilized Flag antibody. Immunocomplexes were disassociated by native elution with Flag peptide and eluted proteins were separated by SDS–PAGE. Protein-rich slices were extracted for analysis by capillary liquid chromatography/electrospray ionization/mass spectrometry/mass spectrometry (LC–MS–MS) using reverse-phase HPLC coupled directly to a linear ion trap mass spectrometer (Fig. 1B). Peptides from both the Sbds-Flag and control capture were identified, and statistical analysis of spectral count for each protein was used to generate a list of proteins enriched in the Sbds-Flag purification relative to the control purification (Table 1). Using the PANTHER classification system, Sbds interacting proteins with low false discovery rates (FDR) were classified by molecular function and biological process and SBDS-interactomes were generated (Fig. 1C and D) (20). Consistent with a proposed function for SBDS in ribosome biogenesis, many of the proteins identified are components of the ribosome with a notable enrichment of proteins of the large ribosomal subunit such as RPL4, RPL7A, RPL7 and others (Table 1). Additionally, proteins with diverse roles in chromatin packaging, transcription and RNA processing were identified (Fig. 1C). In yeast, Sdo1 was reported to interact with proteins with diverse functions including many ribosomal proteins but also Histone H2A, Serine/Threonine kinase Ksp1 and a component of the mTOR signaling pathway, Ssd1 (12). The nucleolar protein nucleophosmin (NPM) was previously reported to interact with SBDS and was identified in our analysis, indicating consistency with our proteomic data set (21). The previously reported SBDS-interacting protein Nip7 was not found in our data set, however, evidence for this interaction remains limited to heterologous expression and in vitro analysis methods (16). When Sbds-interacting proteins were classified by biological process, the majority of proteins fall into one of two groups: ribosomal/protein biosynthesis or DNA metabolism (Fig. 1D). DNA metabolism proteins replication protein A (RPA70) and DNA-dependent protein kinase catalytic subunit (DNA-PK) ranked high in our data set with low false discovery scores. These interactomes suggested an additional role for SBDS beyond the implicated function in ribosome biogenesis.
Figure 1.
SBDS interacts with functionally diverse proteins. (A) Expression of Sbds-Flag compared with endogenous SBDS in HEK293 cells. (B) Native elution and separation of Sbds-Flag and co-precipitating proteins. (C) Color-coded Sbds interactome based on the molecular function of interacting proteins. (D) Color-coded Sbds interactome based on the biological process attributed to interacting proteins. Both molecular function and biological processes were assigned using Protein Analysis Through Evolutionary Relationships (PANTHER) classification (20). Node size is proportional to spectral counts for each protein. All networks were generated in Cytoscape (40).
Table 1.
Sbds interacting proteins identified by mass spectrometry
| Ranka | Protein FDRb | Protein listc | Gene symbol | Descriptiond |
|---|---|---|---|---|
| 1 | 1.61E−144 | IPI00427330 | SBDS | Shwachman-Bodian-Diamond syndrome protein [Acc: Q9Y3A5] |
| 2 | 4.75E−38 | IPI00020127 | RPA1 | Replication protein A 70 kDa DNA-binding subunit (RP-A) (RF-A) (Replication factor-A protein 1) (Single-stranded DNA-binding protein) (p70) [Acc: P27694] |
| 3 | 9.24E−38 | IPI00604620 | NCL | Nucleolin (Protein C23) [Acc: P19338] |
| 4 | 2.69E−33 | IPI00003918 | RPL4 | 60S ribosomal protein L4 (L1) [Acc: P36578] |
| 5 | 1.76E−29 | IPI00844578 | DHX9 | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) [Acc: Q08211] |
| 6 | 2.55E−28 | IPI00479217 | HNRPU | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120) [Acc: Q00839] |
| 7 | 7.55E−27 | IPI00171903 | HNRPM | Heterogeneous nuclear ribonucleoprotein M (hnRNP M) [Acc: P52272] |
| 8 | 3.70E−24 | IPI00299573 | RPL7A | 60S ribosomal protein L7a (Surfeit locus protein 3) (PLA-X polypeptide) [Acc: P62424] |
| 9 | 5.57E−23 | IPI00296337 | PRKDC | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460) [Acc: P78527] |
| 10 | 1.04E−22 | IPI00217467 | HIST1H1E | Histone H1.4 (Histone H1b) [Acc: P10412] |
| 11 | 1.58E−21 | IPI00030179 | RPL7 | 60S ribosomal protein L7 [Acc: P18124] |
| 12 | 2.97E−21 | IPI00217465 | HIST1H1C | Histone H1.2 (Histone H1d) [Acc: P16403] |
| 13 | 1.53E−18 | IPI00549248 | NPM1 | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38) [Acc: P06748] |
| 14 | 9.15E−17 | IPI00550021 | RPL3 | 60S ribosomal protein L3 (HIV-1 TAR RNA-binding protein B) (TARBP-B) [Acc: P39023] |
| 15 | 1.75E−16 | IPI00329389 | RPL6 | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107) (Neoplasm-related protein C140) [Acc: Q02878] |
| 16 | 3.36E−16 | IPI00012772 | RPL8 | 60S ribosomal protein L8 [Acc: P62917] |
| 17 | 1.24E−15 | IPI00005024 | MYBBP1A | Myb-binding protein 1A [Acc: Q9BQG0] |
| 18 | 2.40E−15 | IPI00015953 | DDX21 | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21) [Acc: Q9NR30] |
| 19 | 4.65E−15 | IPI00555744 | RPL14 | 60S ribosomal protein L14 (CAG-ISL 7) [Acc: P50914] |
| 20 | 9.02E−15 | IPI00008530 | RPLP0 | 60S acidic ribosomal protein P0 (L10E) [Acc: P05388] |
| 21 | 3.54E−14 | IPI00215719 | RPL18 | 60S ribosomal protein L18 [Acc: Q07020] |
| 22 | 4.82E−13 | IPI00008557 | IGF2BP1 | Insulin-like growth factor 2 mRNA binding protein 1 [Acc: NP_006537] |
| 23 | 4.82E−13 | IPI00024933 | RPL12 | 60S ribosomal protein L12 [Acc: P30050] |
| 24 | 9.46E−13 | IPI00000494 | RPL5 | 60S ribosomal protein L5 [Acc: P46777] |
| 25 | 3.56E−12 | IPI00220834 | XRCC5 | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Lupus Ku autoantigen protein p86) (Ku86) (Ku80) (86 kDa subunit of Ku antigen) (Thyroid-lupus autoantigen) (TLAA) (CTC box-binding factor 85 kDa subunit) [Acc: P13010] |
| 26 | 3.56E−12 | IPI00011253 | RPS3 | 40S ribosomal protein S3 [Acc: P23396] |
| 27 | 7.01E−12 | IPI00465361 | RPL13 | 60S ribosomal protein L13 (breast basic conserved protein 1) [Acc: P26373] |
aPutative Sbds interacting proteins were ranked by descending protein false discovery rate (FDR).
bFDR corresponds to an adjusted P-value for each prediction in the data set.
cProtein list numbers represent international protein index (IPI) database references.
dDescription includes gene accession numbers and names of identified interacting proteins.
SBDS binds functionally diverse proteins
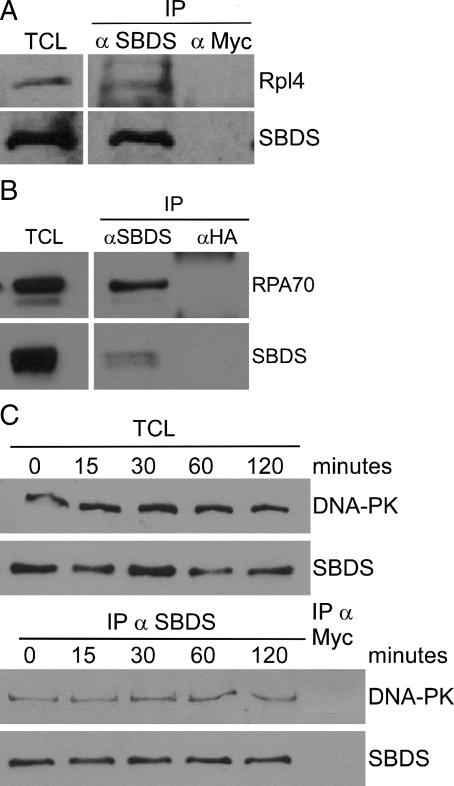
To validate our data set, we investigated the specificity of the interaction of endogenous SBDS with proteins of distinct biological processes. Given the previously established connections between SBDS and translation, we first sought to confirm the interaction of SBDS with at least one large ribosomal subunit by validating the specificity of the interaction between SBDS and ribosomal protein L4 (RPL4), the highest ranking ribosomal protein from our data set. SBDS was immunoprecipitated from HEK293 cell lysates using an anti-SBDS polyclonal antibody (α-N-SBDS). Immunocomplexes were separated by SDS–PAGE and subjected to immunoblotting using antibodies to endogenous SBDS (α-M-SBDS) or RPL4. Endogenous SBDS and RPL4 were both detected in the immunoprecipitation (IP) eluate obtained with the SBDS antibody, but not with a control antibody (Rabbit α–Myc), supporting that endogenous SBDS and RPL4 interact in human cells (Fig. 2A). Given the size and complexity of the ribosome, it is challenging to examine the functions of individual protein–protein interaction(s), or to determine which surface of the ribosome may have affinity, but these results confirm that SBDS interacts directly or indirectly with RPL4, a component of the large ribosomal subunit.
Figure 2.
Interaction of SBDS with RPL4, RPA70 and DNA-PK. (A) Co-IP of RPL4 with SBDS from HEK293 cells. (B) Co-IP of RPA70 with SBDS from HEK293 cells. (C) SBDS interacts with DNA-PK, and binding does not change following UV irradiation.
To investigate the connection between SBDS and DNA metabolism, we sought to confirm the interactions of SBDS with RPA70 and DNA-PK in HEK293 cell lysates using co-IP with anti-SBDS (α-N-SBDS) or control (Rabbit α-HA) antibodies. Immunocomplexes were eluted from beads, separated by SDS–PAGE and subjected to immunoblotting with antibodies to endogenous SBDS and RPA70 (Fig. 2B). RPA70 was readily and specifically detected in the α-SBDS eluate but not in the control (α-HA) eluate (Fig. 2B). Similarly, DNA-PK also co-precipitated with SBDS using anti-SBDS (α-N-SBDS) but not with a control (Rabbit α–Myc) antibody (Fig. 2C), confirming that SBDS binds proteins with well-characterized functions in DNA metabolism and repair. Complementary co-IPs were obtained from lysates of U2OS cells that had been engineered to stably express Sbds-HA. Co-association of Sbds-HA could be detected in anti-DNA-PK eluates using anti-HA antibody (Supplementary Material, Fig. S1A). DNA-PK antibody was then used for co-IPs with lysates of HEK293 cells following transient expression of a series of Sbds-HA variant vectors. Epitope-tagged wild-type Sbds as well as variant proteins possessing SDS-associated missense mutations (K33E, R126T, K148T and R169C) could be detected in α-DNA-PK complexes with anti-HA antibody, further supporting an interaction between SBDS and DNA-PK (Supplementary Material, Fig. S1B). Addition of ethidium bromide to IPs from HEK293 cell lysates did not alter the binding of SBDS to either DNA-PK or RPA70, suggesting that these interactions were not mediated by DNA (data not shown).
We hypothesized that interactions between SBDS and RPA70 or DNA-PK may be regulated by DNA damage to facilitate coordination of ribosomal and translational functions with cellular DNA metabolism. Therefore, we sought to examine whether the interaction between SBDS and DNA-PK was altered following DNA damage. HEK293 cells were treated with 50 J/m2 of ultraviolet (UV) radiation and harvested at 0, 15, 30, 60 and 120 min post-treatment. Co-IP reactions were performed using anti-SBDS (α-N-SBDS) antibody and eluted proteins were analyzed by immunoblotting using antibodies to endogenous DNA-PK or SBDS. DNA-PK co-immunoprecipitated with SBDS, however, no change in the interaction was detectable following UV treatment with the time points examined (Fig. 2C). Consistently, no significant change in the binding of SBDS to DNA-PK or RPA70 was detected following treatment of cells with the DNA-damaging agents etoposide or bleomycin (data not shown). To better understand the interaction of DNA-PK and SBDS, we sought to assess whether both proteins were found in the same cellular component using cellular fractionation or microscopy. Untreated and UV-irradiated HEK293 cells were separated into cytoplasmic, insoluble nuclear and soluble nuclear fractions, proteins were separated by SDS–PAGE, and DNA-PK and SBDS were detected by immunoblotting. DNA-PK was found primarily in the insoluble nuclear fraction, with moderate amounts in the cytoplasmic fraction (Supplementary Material, Fig. S2A). The majority of SBDS was found in the cytoplasm, however, a small portion of SBDS localized to the soluble and insoluble nuclear fractions. No significant relocalization of either protein was detected following UV irradiation (Supplementary Material, Fig. S2A). Immunoflourescence using anti-DNA-PK and anti-HA antibodies in U2OS cells expressing Sbds-HA demonstrated that, consistent with previous reports, DNA-PK was located diffusely throughout the nucleus (22). Prominent cytoplasmic and modest nuclear staining was evident for SBDS and we failed to detect relocalization following UV irradiation (Supplementary Material, Fig. S2B). Although the diffuse nuclear localization of SBDS and DNA-PK prohibited a precise co-localization, we can conclude that a portion of both SBDS and DNA-PK is located in the nucleus. Together, we conclude that SBDS and DNA-PK do interact in human cells, however, the function and regulation of this interaction remain to be elucidated.
Transient SBDS depletion causes SDS-associated defects in global protein synthesis
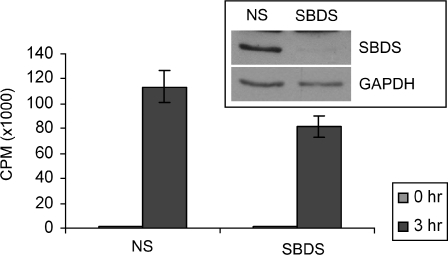
Preliminary evidence from our laboratory has demonstrated that mouse embryonic fibroblasts (MEFs) from SDS-model mice exhibit abnormal ribosomal profiles with low levels of the 60S ribosomal subunit components and reduced global protein synthesis capacity (Zhang et al., unpublished data). These MEFs grow slowly in culture, so to further examine the connection between SBDS and DNA metabolism, we considered an acute cell-culture system that mimicked SDS using transient depletion of SBDS from HEK293 cells. SBDS protein was depleted using siRNA and total cell extracts were analyzed by SDS–PAGE and immunoblot (Fig. 3, inset). After an extended incubation with human SBDS-targeting siRNA oligonucleotides to compensate for the long half-life of SBDS (23), SBDS levels were routinely reduced by 80–90% compared with cells treated with non-specific (NS) siRNA oligonucleotides (Fig. 3, inset). No alterations in the population doubling times of HEK293 cells treated with NS or SBDS-specific siRNA were detected during the time frame of our experiments (data not shown). To determine if transient knockdown with these conditions was sufficient to recapitulate the cellular phenotypes associated with SDS, we monitored total protein synthesis using a radiotracer for a 3 h period. SBDS-depleted HEK293 cells displayed reduction in the amount of global protein synthesis compared with NS-siRNA treated cells (Fig. 3), mimicking the defects observed in MEFs derived from SDS-model mice.
Figure 3.
Transient depletion of SBDS recapitulates SDS-associated translation deficiency. Incorporation of 35S-methionine in HEK293 cells treated with SBDS-specific (SBDS) siRNA is reduced compared with cells treated with non-specific (NS) siRNA oligonucleotides. The inset immunoblot indicates the extent of SBDS protein depletion.
SBDS-deficient cells are sensitive to diverse cellular stressors
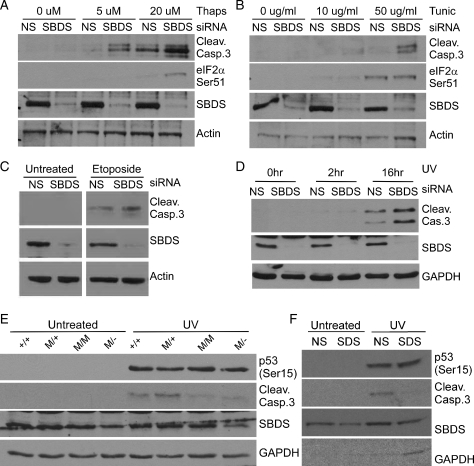
The identification of SBDS interacting proteins with diverse roles in the cell implicated SBDS in many functional pathways. To gain further insight, we sought to determine the impact of the loss of SBDS upon cellular responses to different stressors. First, we examined the effect of reduced SBDS protein levels in cells treated with thapsigargin, which activates ER stress by depleting Ca+ stores in the ER, and tunicamycin, which activates ER stress by blocking N-glycosylation of proteins leading to the accumulation of unfolded proteins in the ER (24). HEK293 cells were treated with siRNA to transiently deplete SBDS, and then treated with thapsigargin (5 or 20 μm) or tunicamycin (10 or 50 μg/ml) for 16 h. We examined the cleavage of Caspase 3 as an indicator of apoptosis and the phosphorylation of eIF2α on Serine 51 as an indicator of ER-stress signaling by immunoblot. Increased Caspase 3 cleavage was observed in SBDS-depleted cells in response to both thapsigargin and tunicamycin compared with cells treated with NS-siRNA (Fig. 4A and B). In response to tunicamycin, eIF2α (Ser51) phosphorylation demonstrated a dose-dependent induction of ER-stress signaling in SBDS-depleted cells even as late as 16 h after treatment (Fig. 4B). These results suggest that SBDS plays a role in cellular responses to ER stress.
Figure 4.
SBDS depletion disturbs cellular responses to cell stressors. HEK293 cells treated with SBDS-specific siRNA display hyper-activation of apoptosis (Caspase 3 cleavage) and ER-stress signaling (eIF2αSer51) following (A) thapsigargin and (B) tunicamycin treatment compared with control (NS-siRNA treated) cells. HEK293 cells treated with SBDS-specific siRNA display hyper-activation of apoptosis following (C) etoposide treatment or (D) UV irradiation compared with control (NS-siRNA treated) cells. (E) Fibroblasts (MEFs) from littermate Sbds+/+; +/+ and SbdsR126T/+; M/+ or SDS embryos SbdsR126T/R126T; M/M and SbdsR126T/−; M/− were UV irradiated or left untreated. MEFs harvested from SDS embryos had reduced Caspase 3 cleavage following UV irradiation compared with WT MEFs. (F) Wild-type MEF cells (Sbds+/+) treated with SBDS-specific siRNA display decreased activation of apoptosis (Caspase 3 cleavage) following UV irradiation compared with control (NS-siRNA treated) cells. Reduction of Sbds by siRNA silencing was typically less effective in MEFs (mouse) compared with HEK293 (human) cells.
To consider stressors quite distinct from ER stress and given the binding of SBDS to multiple proteins with functions in DNA metabolism, we hypothesized that SBDS may play a role in response to DNA damage by analyzing survival of SBDS-depleted cells following exposure to damaging agents. SBDS was transiently depleted from HEK293 cells using siRNA, as described above, followed by treatment with etoposide, which inhibits DNA topoisomerase II and leads to DNA strand breaks (25,26). Cells were harvested after 16 h of treatment, and proteins were separated by SDS–PAGE and analyzed by immunoblotting. SBDS-depleted HEK293 cells displayed an increase in Caspase 3 cleavage after treatment with etoposide compared with cells treated with NS-siRNA (Fig. 4C), suggesting that SBDS depletion sensitizes cells to at least some types of DNA damage. To assess if this SBDS-dependent hypersensitivity to DNA damage is demonstrated in response to UV irradiation, SBDS was transiently depleted from HEK293 cells using siRNA, followed by treatment with 50 J/m2 of UV light. Cells were harvested 2 or 16 h after irradiation, proteins were separated by SDS–PAGE and analyzed by immunoblotting (Fig. 4D). SBDS-depleted cells displayed an increase in Caspase 3 cleavage 16 h after UV damage compared with cells treated with NS-siRNA (Fig. 4D). These findings indicate that SBDS-deficiency leads to hyper-activation of DNA-damage stress responses in HEK293 cells and support a functional role for SBDS in DNA metabolism.
We then sought to determine whether SBDS depletion also influences DNA damage in other cell types. Analysis of UV responses in fibroblast cell lines derived from E15 embryos homozygous for the SDS-associated missense allele R126T (R126T/R126T), compound heterozygotes for mutant alleles (R126T/−) versus their littermate controls (+/+, R126T/+) were investigated. In contrast to siRNA-treated HEK293 cells, SDS MEFs displayed decreased cleavage of Caspase 3 following UV treatment, despite evidence of DNA-damage induced phosphorylation of p53 on Serine 15. This residue is known to be phosphorylated by DNA damage response kinases such as ATR, ATM and DNA-PK (Fig. 4E; 27). To ensure that the discrepancy between the sensitivity of HEK293 and MEF cells was not due to adaptation of MEFs in culture to down-regulate DNA-damage responses and permit cell survival in the absence of Sbds, we also monitored UV sensitivity of wild-type MEFs treated with siRNA to transiently deplete SBDS. MEF cells treated with mouse SBDS-specific siRNA oligonucleotides displayed decreased Caspase 3 cleavage following UV irradiation compared with cells treated with NS-siRNA (Fig. 4F), consistent with the findings of the constitutive mutant MEF cell lines. The hypersensitivity of HEK293 versus the dampened sensitivity of MEF cell lines to UV irradiation is interesting and could reflect different roles of SBDS in these cell types. Alternatively, there may be different kinetics of UV-induced apoptosis in these cells. It is clear that SBDS depletion disturbs cellular responses to DNA damage, supporting a functional role of SBDS in DNA metabolism. Taken together, our findings reveal that depletion sensitizes cells to DNA damage as well as ER stress indicating a role or roles for SBDS in general cellular stress responses.
Reduced protein synthesis does not sensitize cells to UV irradiation
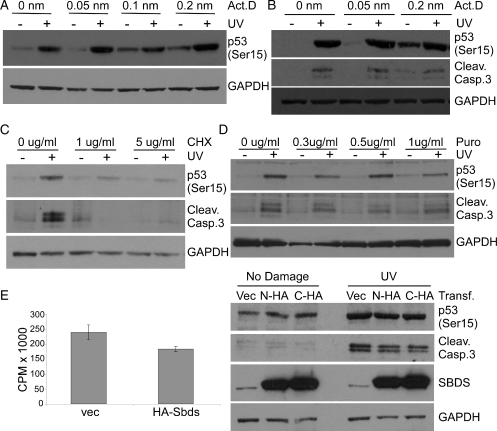
Protein synthesis is tightly coordinated with diverse response pathways to allow cells to conserve energy and alter transcriptional and translational profiles in response to stress (28). There is evidence for at least two roles of SBDS, one in protein translation and one in cellular stress responses. We considered that the sensitivity of SBDS-depleted cells to ER and DNA stress may result directly from the reduced translation capacity of SBDS-deficient cells (Fig. 3). To test this, we sought to separate translation and stress response roles of SBDS. SBDS is proposed to function in ribosome biogenesis (11), therefore, to mimic the effect of SBDS depletion upon translation, we treated HEK293 cells with Actinomycin D at concentrations known to inhibit RNA Polymerase I and cause nucleolar stress (29). Cells were incubated with various concentrations of Actinomycin D for 1 h before UV irradiation and harvested 16 h post-treatment. Actinomycin D treatment at low levels (0.05–0.2 nm) did not sensitize HEK293 cells to UV irradiation, as measured by phosphorylation of p53 on Serine 15 (Fig. 5A and B). At the highest concentration of Actinomycin D (0.2 nm), Caspase 3 clevage was detected in the absence of UV irradiation, indicating that inhibition of RNA Polymerase I alone could lead to apoptosis (Fig. 5B). However, even at this level of Actinomycin D treatment (0.2 nm), HEK293 cells do not demonstrate increased Caspase 3 cleavage or phosphorylation of p53 following UV irradiation compared with UV-irradiated cells that were treated with no Actinomycin D (Fig. 5B), suggesting that inhibition of ribosome biogenesis alone does not cause UV hypersensitivity in HEK293 cells. To confirm this finding, we also used multiple translation inhibitors including cycloheximide and puromycin, both inhibitors of translation elongation, and emitine, an inhibitor of translation initiation. HEK293 cells were incubated with low concentrations of translation inhibitors for 1 h before UV irradiation. Cells were harvested 16 h after treatment, and induction of apoptosis (Caspase 3 cleavage) and DNA-damage signaling (p53Ser15) were monitored by immunoblotting. Treatment of cells with 1 μg/ml cycloheximide before UV irradiation resulted in reduced p53 phosphorylation (p53 Ser15) and Caspase 3 cleavage following DNA damage relative to cells that were not treated with cycloheximide (Fig. 5C). Treatment of cells with various concentrations of puromycin (Fig. 5D) and emitine (data not shown) also led to dampened p53 phosphorylation and Caspase 3 cleavage following UV irradiation. These observations are in contrast to the hyper-sensitivity of SBDS-depleted HEK293 cells responding to UV irradiation (Fig. 4D). Together, these findings indicate that the nucleolar stress and/or translation deficiency generated by SBDS depletion in HEK293 cells is not the cause of UV hypersensitivity in these cells.
Figure 5.
SBDS has distinct functions in translation and cellular responses to stress. Inhibition of RNA Polymerase I using Actinomycin D had no effect on (A) phosphorylation of p53 or (B) cleavage of Caspase 3 following UV irradiation of HEK293 cells. Inhibition of translation using (C) cycloheximide or (D) puromycin reduces cleavage of Caspase 3 and phosphorylation of p53 following UV irradiation in HEK293 cells. (E) Overexpression of HA-Sbds decreases global translation (left panel) but does not influence Caspase 3 cleavage or p53 phosphorylation following UV irradiation in HEK293 cells (right panel).
To address concerns that chemical methods to disrupt translation (actinomycin D, cycloheximide, puromycin and emitine) may have off-target effects, we also devised a genetic strategy to impair protein translation. Overexpression of translation initiation factors such as eIF6 are known to decrease global translation by reducing the formation of active ribosomes (30). During the course of our studies, we observed that overexpression of SBDS by transient transfection reduces the global translation capacity of cells, similar to the effect of SBDS depletion (Fig. 3). We took advantage of this finding to address more directly if SBDS-dependent translation defects led to UV hypersensitivity. HEK293 cells were transfected with vectors expressing N- or C-terminally tagged Sbds, or vector only as a control. Thirty-six hours after transfection, cells were irradiated with UV and then harvested 16 h post-treatment. Aliquoted portions of the cells were incubated with 35S-methionine for 3 h to monitor global protein synthesis and the remaining cells were used to generate protein lysates for SDS–PAGE and immunoblot analysis. Transfection of cells with vectors encoding N- or C-terminally tagged Sbds led to a dramatic overexpression of SBDS compared with endogenous protein levels (Fig. 5E, right panel) and a concomitant decrease in global protein synthesis over the 3 h labeling period (Fig. 5E, left panel). Despite the impaired translation that was measured, phosphorylation of p53 (Ser15) and induction of apoptosis (Caspase 3 cleavage) were not altered relative to cells transfected with vector alone (Fig. 5E, right panel). This is in contrast to the increased Caspase 3 cleavage evident after SBDS depletion in HEK293 cells (Fig. 4D), further supporting that the UV hypersensitivity phenotype seen following SBDS depletion is independent of the translation deficiency phenotype. In summary, both overexpression and underexpression of SBDS lead to decreased global protein synthesis in HEK293 cells, but hypersensitivity to UV irradiation occurs only in the absence of SBDS. These data indicate that the role of SBDS in translation and/or ribosome biogenesis is distinct from its function in cellular responses to DNA damage.
DISCUSSION
SBDS interacting proteins have diverse molecular functions
Using affinity capture and mass spectrometry, we have developed an SBDS-interactome highlighting connections between SBDS and proteins with diverse biological functions in the cell. Consistent with a role of SBDS in translation, the interactome revealed a notable number of ribosomal proteins, particularly components of the large ribosomal subunit. We have confirmed that SBDS binds to at least one protein component of the large ribosomal subunit (RPL4). This is consistent with reports that SBDS co-fractionates with the 60S ribosomal subunit and may bind ribosomal RNA (21). Although SBDS has been reported to be enriched in the nucleolus using immunoflourescence (31), fractionation of cellular lysates demonstrates that the majority of SBDS is in the cytoplasm (Supplementary Material, Fig. S2A). It is possible that, like other proteins involved in ribosome biogenesis, SBDS shuttles between the nucleolus and cytoplasm. It is likely that the binding of SBDS to the large ribosomal subunit is dynamic and specific to different stages of ribosome biogenesis; however, detailed characterization of SBDS interactions with ribosomal proteins is required to understand the molecular functions of SBDS in the formation of active ribosomes. Identification of SBDS interacting proteins with ribosomal functions provides an excellent starting platform from which these experiments can be conducted.
In addition to interactions between SBDS and ribosomal proteins, we uncovered novel SBDS interacting proteins with other functions. Of particular interest, given the cancer-predisposition observed in SDS patients, was the enrichment of proteins involved in DNA replication and repair, including DNA-PK and RPA70. We failed to uncover regulation of these interactions following DNA damage, however, it is possible that these interactions are regulated under alternate conditions or that our detection methods were not sufficiently sensitive. Many nucleolar proteins such as NPM and dyskerin are known to relocalize from the nucleolus to the nucleoplasm following DNA damage (32,33). It is possible that some portion of SBDS follows a similar pattern of relocalization to alter its interactions with DNA-associated proteins. Indeed, although the majority of SBDS is located in the cytoplasm, a portion of SBDS is located diffusely in the nucleus and, following UV irradiation, nucleoli are known to be disrupted to release nucleolar proteins (NPM and possibly SBDS) (Supplementary Material, Fig.S2) (33). Although we have not seen evidence by gel migration, mass spectrometry analysis of SBDS immunoprecipitated from human cells identified a peptide with a mass corresponding to phosphorylation on a threonine residue followed by a glutamine residue (TQ) (H.B. and J.M., unpublished data). This is a well-characterized consensus sequence (S/TQ) for PIKK kinases, including DNA-PK, ATM and ATR (34). The interaction of SBDS with DNA-PK may be transient and difficult to capture, as is typical of many kinase–substrate interactions. Further studies are needed to understand the function of SBDS binding to DNA-PK and RPA70.
SBDS-deficiency sensitizes cells to a variety of stress conditions
In addition to decreasing global translation, we found that depletion of SBDS sensitized cells to a variety of cellular conditions including ER stress (tunicamycin and thapsigargin) and DNA damage (UV and etoposide). In response to exogenous stress, SBDS-depleted cells demonstrated increased stress signaling (eIF2α or p53 phosphorylation) and apoptosis (Caspase 3 cleavage), indicating that SBDS is required for cells to deal efficiently with adverse conditions. The sensitivity of SBDS-depleted cells to ER stress is interesting, and tight connections between the ER and protein translation regulation are known (35).
We chose to characterize the hypersensitivity of SBDS-depleted HEK293 cells to UV irradiation based on the cancer predisposition of SDS patients and the identification of multiple SBDS-interacting proteins with functions in DNA metabolism. Treatment of SBDS-depleted HEK293 cells with DNA damage leads to increased cell death compared with non-depleted cells. Using distinct methods, including different types of chemical treatments and SBDS overexpression, to impair translation and mimic the SDS cellular phenotype, we determined that SBDS-dependent UV hypersensitivity is generally independent of decreased translation in SBDS-deficient HEK293 cells. These findings demonstrate that in the presence of reduced translation and/or nucleolar stress, cells with endogenous SBDS protein levels are able to respond to exogenous DNA damage.
We suspect that the addition of exogenous DNA damage exacerbates the underlying defects associated with loss of SBDS. SBDS-depleted HEK293 cells demonstrate a mild accumulation in G1 phase of the cell cycle as well as elevated p21 protein levels, indicating that cell growth requires SBDS (data not shown). Taken together with reports that bone marrow cells from SDS patients exhibit an increased incidence of abnormal mitoses (18), we conclude that the genomic instability demonstrated by SDS patients need not be explained by a role of SBDS in protein biosynthesis alone, and SBDS may also have a more specific function or functions in responding to cellular stresses. It remains unclear what extra-ribosomal functions SBDS may possess and our findings do not preclude that protein biosynthesis deficiency of SDS cells may alter the pool of translating messenger RNAs, leading to indirect aberrations in chronic stress response situations.
Leukemia in SDS patients is associated with clonal cell changes, indicating that the mutation of SBDS may be a first event in a multi-step tumor-formation process, particularly in the bone marrow (6,7). There may be a strong selective pressure with loss of SBDS for mutations that can compensate to allow cells to escape growth arrest or apoptosis, predisposing them to genomic instability. It is clear that cells lacking SBDS show perturbed responses to DNA damage and chemical stressors. The development of an SBDS-interactome provides a platform from which we can recognize the diverse functions of SBDS in the cell and come to understand the direct and indirect consequences of their loss. These findings support that SBDS is a multi-functional protein and provides a framework to obtain insight into the molecular mechanisms underlying the leukemia-predisposition observed in SDS patients.
MATERIALS AND METHODS
Proteomics analysis
HEK293 cells expressing Sbds-Flag (source clone MGC:6426) were generated by transfection with pcDNA3.1 expression vector using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. Cells were split 24 h after transfection to avoid confluency. Cells were selected with 600 μg/ml of geneticin for 10 days, beginning 48 h after transfection. HEK293 and HEK293 + Sbds-Flag cells were harvested, washed with PBS and lysed in NETN (50 mm Tris pH 7.0, 125 mm NaCl, 0.5% NP-40). Anti-Flag M2 affinity gel (Sigma) was used to purify Sbds-Flag and co-associated proteins from expressing cell lysates, or background associated proteins from HEK293 cell lysates. Bound proteins were released from affinity gel using native elution with Flag peptide (Sigma) and eluted proteins were fractionated by SDS–PAGE. Protein-containing bands were excised and proteins were reduced, alkylated and digested in gel with trypsin. The resulting peptides were extracted and analyzed by capillary LC–MS–MS using reverse-phase HPLC chromatographic separation coupled directly to a Thermo LTQ linear ion trap mass spectrometer. Peptides were matched to spectra using the TurboSEQUEST v.27 (reviewed in 12) algorithm (36), and the human international protein index database (version 3.33) concatenated with reverse sequences for false discovery calculations. Control cells and Sbds-Flag purifications were compared by spectral counts and protein false discovery to reveal Sbds-Flag specific interacting proteins.
Statistical analysis
The Flag and Sbds-Flag purifications were compared by spectral counts using the Poisson regression model (37) as a Poisson distribution and have been demonstrated for differential proteome data sets (38). The P-values generated by the model were further adjusted for multiple comparisons using the Benjamini and Hochberg correction (39) to generate adjusted P-values or FDR.
Network analysis
A global human protein interaction network with 10 454 proteins and 72 428 interactions was generated by integrating data collected from HPRD, MINT, IntAct, REACTOME and DIP databases. Only manually curated interactions were considered to assure the reliability of the network. SBDS interacting proteins identified in this study were superimposed upon the global network. A sub-network centered around SBDS was extracted and visualized in Cytoscape (17).
Cell culture, transfections, chemicals and antibodies
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM)-10% fetal bovine serum (FBS). MEFs were harvested from E14.5 embryos of different genotypes (Sbds+/+, SbdsR126T/+, SbdsR126T/R126T and SbdsR126T/−) as described previously (5) and maintained in DMEM + 10% FBS for up to three passages. Plasmid transfections were performed with Lipofectamine (Invitrogen). HA-Sbds, Sbds-HA and Sbds-Flag vectors were generated using standard PCR and subcloning procedures with Xho1 and Kpn1 restriction sites and the pcDNA3.1 expression vector (Invitrogen). Small-interfering RNA (siRNA) transfections were performed using Dharmafect 4 and human or mouse SBDS-specific or non-targeting siGENOME SMARTpool oligonucleotides (Dharmacon). Etoposide, thapsigargin and tunicamycin were purchased from Sigma. Antibodies against Phospho-p53 Ser15 (Cell Signaling), Cleaved Caspase 3 Asp175 (Cell Signaling), phospho-eIF2-α Ser51 (Cell Signaling), Nucleolin C23 (Santa Cruz Biotechnology) and GAPDH (Abcam) were used for immunoblotting. Antibodies used for both IP and immunoblotting included Rabbit polyclonal SBDS (α-N-terminal-SBDS, raised against peptide CYKNKVVGWRSGVEKD) and Chicken polyclonal SBDS (α-Middle-SBDS, raised against peptide KGEVQVSDKERHTQLE), Rabbit polyclonal α-HA (Abcam), Rabbit polyclonal α-Myc (Santa Cruz Biotechnology), Mouse monoclonal α-DNA-PK (Neomarkers), Rabbit polyclonal α-RPA70 (Santa Cruz Biotechnology) and mouse monoclonal α-RPL4 (Abnova Corporation).
Global protein synthesis assay
Cells were cultured to ∼80% confluency for radio-tracing in DMEM containing 150 µCi/ml 35S-methionine for 0 or 3 h time points (in triplicate). Following incubation, cells were immediately harvested by trypsin digestion, washed once with phosphate buffered saline (PBS) and lysed in RIPA buffer (50 mm Tris pH 7.0, 150 mm NaCl, 1% Triton, 0.5% sodium deoxycholate, 0.1% SDS). Protein concentration was quantitated by Lowry assay (BioRad); 25 μg aliquots (in triplicate) of total protein were precipitated by bringing each sample volume to 500 μl using RIPA and then adding 500 μl of 20% trichloroacetic acid (TCA). Samples were incubated on ice and the precipitates were applied to 37.5 mm glass fiber membranes (Whatman) using vacuum filtration, washing once with 500 μl of cold 10% TCA, then twice with 500 μl of 100% ethanol. Membranes were dried overnight in scintillation vials, 2 ml of Ready Safe scintillation fluid (Beckman) was added and incorporation of radioactivity was quantitated using a Beckman LS6500 scintillation counter.
Inhibition of translation
HEK293 cells were incubated with various concentrations of cycloheximide, emitine (0.1–0.5 μg/ml), actinomycin D or puromycin (0.1–1.0 μg/ml) (Sigma Aldrich) in DMEM + 10% FBS for 1 h before irradiation at 50 J/m2. Cells were harvested 16 h post-irradiation, lysed in RIPA buffer, separated by SDS–PAGE and immunoblotted using standard procedures.
Immunoprecipitations
HEK293 cells were lysed in NET buffer (50 mm Tris pH 7.0, 150 mm NaCl) + 0.75% CHAPS, and 600 μg aliquots of total protein were diluted to 0.375% CHAPS with NET buffer. Rabbit polyclonal SBDS antibody (α-N-SBDS), rabbit polyclonal HA, rabbit polyclonal Myc or mouse monoclonal DNA-PK were added and incubated for 1.5 h at 4°C. Protein A/G beads were added and incubated for an additional 2 h to overnight at 4°C. Beads were washed three times with NET + 0.375% CHAPS. Proteins were released by resuspension in Laemmli buffer, boiled for 5 min and separated by SDS–PAGE. Proteins were transferred onto supported nitrocellulose membranes (Hybond-C, Amersham) and immunoblotted with antibodies to SBDS, HA-epitope, DNA-PK, RPA70 or RPL4 and visualized using Western Lightning Enhanced chemiluminescence reagent (Perkin Elmer) and Amersham Hyperfilm (GE Healthcare).
ER stress or DNA damage treatment of SBDS-depleted cells
HEK293 cells at 95% confluency were transfected with siGENOME SMART pool SBDS-specific or non-targeting (NS) siRNA (Dharmacon). Cells were incubated for 72 h, split into 35 mm dishes and treated UV at 50 J/m2 using a Stratalinker (Stratagene), 20 μm etoposide, 5 or 20 μm thapsigargin, 10 or 50 μg/ml tunicamycin or left untreated at 80 h post-transfection. Cells were then incubated 16 h, harvested using trypsin and washed one time with PBS. Cell pellets were lysed in RIPA buffer (50 mm Tris pH 7.0, 150 mm NaCl, 1% Triton, 0.5% sodium deoxycholate, 0.1% SDS). Proteins were quantitated by Lowry assay (BioRad) and separated by SDS–PAGE.
Generation of U2OS stable cell lines
pcDNA3.1 vector (Invitrogen) or pcDNA3.1 with Sbds-HA were linearized and transfected into U2OS cells using Lipofectamine (Invitrogen). Cells were plated sparsely and successful transformation was achieved by selection with Neomycin for 7 days. Single colonies were picked and propagated from each transfection and screened by western blot for expression of Sbds-HA. Whole cell protein extracts were prepared and screened by immunoblot for expression of Sbds-HA with antibodies to Sbds and the HA-epitope.
Construction of vectors with SDS-associated missense mutations
Wild-type and, subsequently, variant expression vectors were generated by PCR mutagenesis using established methods. Forward and reverse oligonucleotide primers included: K33E-For 5′CGA AAT CGC CTG CTA TGA AAA CAA GGT CGT C, K33E-Rev 5′GTC GAC CTT GTT TTC ATA GCA GGC GAT TTC G, R126T-For 5′CCA GAA ACA AAG ACA CCT TAC ACC GTT ATC C, R126T-Rev 5′GGA TAA CGG TGT AAG GTG TCT TTG TTT CTG G, K148T-For 5′CGT GAA ACC CAA CAC GAG CAC AAA GCA ACA G, K148T-Rev 5′CTG TTG CTT TGT GCT CGT GTT GGG TTT CAC G, R169C-For 5′GAA GAT AGA GTG GGC CCA CAT GCG ATT GC, R169C-Rev 5′GCA ATC GCA TGT GGG CCC ACT CTA TCT TC, SBDS-For 5′GGG GTA CCG CCA CCA TGT CGA TCT TCA CCC CCA CCA A and SBDS-Rev 5′CTA CTC GAG TCA AGC GTA ATC TGG AAC ATC GTA TGG GTA TTC AAA CTT CTC ATC GCC TTC CT. Separate rounds of PCR for each mutation were carried out using pCDNA3.1 with Sbds-HA as template with SBDS-For and mutant Rev primers and with SBDS-Rev and mutant For primers. Corresponding DNA fragments were purified, combined and used in an additional PCR reaction using only the SBDS-For and SBDS-Rev primers. These products were then digested with Xho1 and Kpn1 restriction enzymes and subcloned into the pcDNA3.1 vector. All Sbds gene and mutation fragments were verified by sequencing.
Localization by immunoflourescence
U2OS cell lines stably expressing Sbds-HA were grown on four-chamber culture slides and left untreated or UV irradiated with 50 J/m2. One hour after UV irradiation, cells were washed twice with PBS, then fixed for 20 min at room temperature in 4% paraformaldehyde. Cells were washed three times with PBS, then permeabilized for 5 min in 0.1% Triton/PBS at room temperature. Cells were washed three times with PBS, then blocked with 5% BSA in PBS. Slides were then incubated with Mouse α-DNA-PK (1:100) and Rabbit α-HA antibodies in 1% BSA/PBS for 1 h at room temperature. Cells were washed three times with PBS then incubated with FITC-conjugated Goat α-Rabbit (1:250) and Rhodamine-conjugated Rabbit α-Mouse (1:250) in 1% BSA/PBS for 1 h at room temperature. Cells were washed three times in PBS then mounted with Prolong anti-fade gold with DAPI (Invitrogen) overnight at room temperature. Indirect immunofluorescence was measured with a Nikon E1000 fluorescence microscope. Images were captured using a Photometrics CCD camera and Simple PCI6 software.
Cellular fractionation
HEK293 cells were grown to 75% and left untreated, or treated with UV at 50 J/m2, incubated for an additional 2 or 4 h and harvested using trypsin digestion. Cells were washed one time in PBS, then resuspended in buffer A (10 mm HEPES pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 0.1 mm phenylmethylsulfonyl fluoride). Triton X-100 (0.1%) was added and the cells were incubated for 5 min on ice. Nuclei were pelleted by low speed centrifugation (4 min, 1300g) and the supernatant (cytoplasmic fraction) was retained. The pellet fractions (nuclei) were washed once in buffer A, then lysed in buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 0.1 mm phenylmethylsulfonyl fluoride). The insoluble nuclear fraction was again collected by centrifugation (4 min, 1700g), and the supernatant was retained as the soluble nuclear fraction. Insoluble nuclear fractions were resuspended in Laemmli buffer and sheared using a syringe and 20 guage needles. Equivalents (cell numbers) of each fraction were separated by SDS–PAGE and analyzed by immunoblot. GAPDH was used as a cytoplasmic control protein and Nucleolin was used as an insoluble chromatin control protein.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by National Institutes of Health grant number HL079573. H.L.B. is a recipient of the Markel Training Fellowship Award of the Hospital for Sick Children.
ACKNOWLEDGEMENTS
We thank Daniel C. Liebler for support in completing this project. We thank R. Tsai for preparation of Sbds-mutant expression vectors and M. Tourlakis for help with the RPL4 immunoprecipitations. This work has been conducted in part with resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN, USA.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Makitie O., Ellis L., Durie P.R., Morrison J.A., Sochett E.B., Rommens J.M., Cole W.G. Skeletal phenotype in patients with Shwachman-Diamond syndrome and mutations in SBDS. Clin. Genet. 2004;65:101–112. doi: 10.1111/j.0009-9163.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- 2.Dror Y. Shwachman-Diamond syndrome. Pediatr. Blood Cancer. 2005;45:892–901. doi: 10.1002/pbc.20478. [DOI] [PubMed] [Google Scholar]
- 3.Ginzberg H., Shin J., Ellis L., Morrison J., Ip W., Dror Y., Freedman M., Heitlinger L.A., Belt M.A., Corey M., et al. Shwachman syndrome: phenotypic manifestations of sibling sets and isolated cases in a large patient cohort are similar. J. Pediatr. 1999;135:81–88. doi: 10.1016/s0022-3476(99)70332-x. [DOI] [PubMed] [Google Scholar]
- 4.Boocock G.R., Morrison J.A., Popovic M., Richards N., Ellis L., Durie P.R., Rommens J.M. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S., Shi M., Hui C.C., Rommens J.M. Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol. Cell. Biol. 2006;26:6656–6663. doi: 10.1128/MCB.00091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dror Y., Durie P., Ginzberg H., Herman R., Banerjee A., Champagne M., Shannon K., Malkin D., Freedman M.H. Clonal evolution in marrows of patients with Shwachman-Diamond syndrome: a prospective 5-year follow-up study. Exp. Hematol. 2002;30:659–669. doi: 10.1016/s0301-472x(02)00815-9. [DOI] [PubMed] [Google Scholar]
- 7.Maserati E., Minelli A., Pressato B., Valli R., Crescenzi B., Stefanelli M., Menna G., Sainati L., Poli F., Panarello C., et al. Shwachman syndrome as mutator phenotype responsible for myeloid dysplasia/neoplasia through karyotype instability and chromosomes 7 and 20 anomalies. Genes Chromosomes Cancer. 2006;45:375–382. doi: 10.1002/gcc.20301. [DOI] [PubMed] [Google Scholar]
- 8.Maserati E., Pressato B., Valli R., Minelli A., Sainati L., Patitucci F., Marletta C., Mastronuzzi A., Poli F., Lo Curto F., et al. The route to development of myelodysplastic syndrome/acute myeloid leukaemia in Shwachman-Diamond syndrome: the role of ageing, karyotype instability, and acquired chromosome anomalies. Br. J. Haematol. 2009;145:190–197. doi: 10.1111/j.1365-2141.2009.07611.x. [DOI] [PubMed] [Google Scholar]
- 9.Dror Y. P53 protein overexpression in Shwachman-Diamond syndrome. Arch. Pathol. Lab Med. 2002;126:1157–1158. doi: 10.5858/2002-126-1157b-PPOISS. [DOI] [PubMed] [Google Scholar]
- 10.Elghetany M.T., Alter B.P. p53 protein overexpression in bone marrow biopsies of patients with Shwachman-Diamond syndrome has a prevalence similar to that of patients with refractory anemia. Arch. Pathol. Lab. Med. 2002;126:452–455. doi: 10.5858/2002-126-0452-PPOIBM. [DOI] [PubMed] [Google Scholar]
- 11.Menne T.F., Goyenechea B., Sanchez-Puig N., Wong C.C., Tonkin L.M., Ancliff P.J., Brost R.L., Costanzo M., Boone C., Warren A.J. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 12.Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 13.Savchenko A., Krogan N., Cort J.R., Evdokimova E., Lew J.M., Yee A.A., Sanchez-Pulido L., Andrade M.A., Bochkarev A., Watson J.D., et al. The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism. J. Biol. Chem. 2005;280:19213–19220. doi: 10.1074/jbc.M414421200. [DOI] [PubMed] [Google Scholar]
- 14.Nihrane A., Sezgin G., Dsilva S., Dellorusso P., Yamamoto K., Ellis S.R., Liu J.M. Depletion of the Shwachman-Diamond syndrome gene product, SBDS, leads to growth inhibition and increased expression of OPG and VEGF-A. Blood Cells Mol. Dis. 2009;42:85–91. doi: 10.1016/j.bcmd.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Rawls A.S., Gregory A.D., Woloszynek J.R., Liu F., Link D.C. Lentiviral-mediated RNAi inhibition of Sbds in murine hematopoietic progenitors impairs their hematopoietic potential. Blood. 2007;110:2414–2422. doi: 10.1182/blood-2006-03-007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hesling C., Oliveira C.C., Castilho B.A., Zanchin N.I. The Shwachman-Bodian-Diamond syndrome associated protein interacts with HsNip7 and its down-regulation affects gene expression at the transcriptional and translational levels. Exp. Cell Res. 2007;313:4180–4195. doi: 10.1016/j.yexcr.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi M., Fujimura K., Toga H., Khwaja A., Okamura N., Chopra R. Shwachman-Diamond syndrome is not necessary for the terminal maturation of neutrophils but is important for maintaining viability of granulocyte precursors. Exp. Hematol. 2007;35:579–586. doi: 10.1016/j.exphem.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Austin K.M., Gupta M.L., Coats S.A., Tulpule A., Mostoslavsky G., Balazs A.B., Mulligan R.C., Daley G., Pellman D., Shimamura A. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J. Clin. Invest. 2008;118:1511–1518. doi: 10.1172/JCI33764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rujkijyanont P., Watanabe K., Ambekar C., Wang H., Schimmer A., Beyene J., Dror Y. SBDS-deficient cells undergo accelerated apoptosis through the Fas-pathway. Haematologica. 2008;93:363–371. doi: 10.3324/haematol.11579. [DOI] [PubMed] [Google Scholar]
- 20.Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganapathi K.A., Austin K.M., Lee C.S., Dias A., Malsch M.M., Reed R., Shimamura A. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110:1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson A., Sirzen F., Lewensohn R., Wang N., Skog S. Cell cycle-dependent regulation of the DNA-dependent protein kinase. Cell Prolif. 1999;32:239–248. doi: 10.1046/j.1365-2184.1999.3240239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erdos M., Alapi K., Balogh I., Oroszlan G., Rakoczi E., Sumegi J., Marodi L. Severe Shwachman-Diamond syndrome phenotype caused by compound heterozygous missense mutations in the SBDS gene. Exp. Hematol. 2006;34:1517–1521. doi: 10.1016/j.exphem.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Chen L.H., Jiang C.C., Watts R., Thorne R.F., Kiejda K.A., Zhang X.D., Hersey P. Inhibition of endoplasmic reticulum stress-induced apoptosis of melanoma cells by the ARC protein. Cancer Res. 2008;68:834–842. doi: 10.1158/0008-5472.CAN-07-5056. [DOI] [PubMed] [Google Scholar]
- 25.van Maanen J.M., Retel J., de Vries J., Pinedo H.M. Mechanism of action of antitumor drug etoposide: a review. J. Natl Cancer Inst. 1988;80:1526–1533. doi: 10.1093/jnci/80.19.1526. [DOI] [PubMed] [Google Scholar]
- 26.Yarbro J.W. Mechanism of action of hydroxyurea. Semin. Oncol. 1992;19:1–10. [PubMed] [Google Scholar]
- 27.Abraham J., Spaner D., Benchimol S. Phosphorylation of p53 protein in response to ionizing radiation occurs at multiple sites in both normal and DNA-PK deficient cells. Oncogene. 1999;18:1521–1527. doi: 10.1038/sj.onc.1202454. [DOI] [PubMed] [Google Scholar]
- 28.Brostrom C.O., Brostrom M.A. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- 29.Iapalucci-Espinoza S., Franze-Fernandez M.T. Effect of protein synthesis inhibitors and low concentrations of actinomycin D on ribosomal RNA synthesis. FEBS Lett. 1979;107:281–284. doi: 10.1016/0014-5793(79)80390-7. [DOI] [PubMed] [Google Scholar]
- 30.Ceci M., Gaviraghi C., Gorrini C., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 31.Austin K.M., Leary R.J., Shimamura A. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood. 2005;106:1253–1258. doi: 10.1182/blood-2005-02-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruggero D., Pandolfi P.P. Does the ribosome translate cancer? Nat. Rev. Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 33.Rubbi C.P., Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abraham R.T. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 36.Yates J.R., III, Eng J.K., McCormack A.L., Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 37.Dobson A.J. An Introduction to Generalized Linear Models. 2nd edn. London: Chapman and Hall/CRC; 2001. [Google Scholar]
- 38.Chourey K., Thompson M.R., Shah M., Zhang B., Verberkmoes N.C., Thompson D.K., Hettich R.L. Comparative temporal proteomics of a response regulator (SO2426)-deficient strain and wild-type Shewanella oneidensis MR-1 during chromate transformation. J. Proteome Res. 2009;8:59–71. doi: 10.1021/pr800776d. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 40.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.