Abstract
Many essential physiological processes are regulated by the modulation of calcium concentration in the cell. The EF-hand proteins represent a superfamily of calcium-binding proteins involved in calcium signaling and homeostasis. Secretagogin is a hexa-EF-hand protein that is highly expressed in pancreatic islet of Langerhans and neuroendocrine cells and may play a role in the trafficking of secretory granules. We present the X-ray structure of Danio rerio secretagogin, which is 73% identical to human secretagogin, in calcium-free form at 2.1-Å resolution. Secretagogin consists of the three globular domains each of which contains a pair of EF-hand motifs. The domains are arranged into a V-shaped molecule with a distinct groove formed at the interface of the domains. Comparison of the secretagogin structure with the solution structure of calcium-loaded calbindin D28K revealed a striking difference in the spatial arrangement of their domains, which involves approximately a 180-degree rotation of the first globular domain with respect to the module formed by the remaining domains.
Keywords: X-ray structure, calcium binding protein, EF-hand motif, structural genomics
INTRODUCTION
Danio rerio gene Zgc:100843 encodes a 272 amino acids long protein, which has been annotated as secretagogin based on its very high sequence identity (70–80%) to mammalian secretagogins. Secretagogins are hexa-EF-hand proteins that show sequence homology (40–45% sequence identity) to other known hexa-EF-hand proteins calbindins and calretinins. Human secretagogin has been identified in pancreatic beta cells of islets of Langerhans and in smaller amounts in neuroendocrine cells.1 Transfection of rat insulinoma cells (RIN-5FB23) with secretagogin resulted in an elevated Ca2+ flux and insulin secretion, suggesting a role of secretagogin in the trafficking of secretory granules.1 Secretagogin has been found expressed in high quantities in basket and stellate cells of the cerebellum and in the anterior part of the pituitary gland.2 Recent studies have established that secretagogin may be linked to the pathogenesis of neurological diseases such as Alzheimer’s.3 The protein has been detected in a human serum after ischemic strokes,2 and in the plasma of carcinoid patients suffering from distant metastases.4 Secretagogin could thus serve as a serum marker of neuronal damage and/or as a tumor biomarker. Indeed, secretagogin has been used as a biomarker to study carcinoid tumors of the lung and gastrointestinal tract.5
EF-hand calcium binding proteins can be divided into two functional groups, namely calcium buffers and calcium sensors/signal modulators.6 Calcium buffer proteins, like parvalbumin, have submicromolar affinities for Ca2+ ions and can sequester them efficiently while undergoing only minor structural alterations. On the other hand, calcium sensor proteins, like calmodulin or troponin C, have micromolar affinities for Ca2+ ions, undergo significant conformational changes upon calcium binding and play roles in signaling and allosteric molecular interactions. Secretagogin likely belongs to the calcium sensor/signal modulators family. Recent biophysical study of recombinant human secretagogin provided evidence for a significant conformational change of this protein upon Ca2+ binding. The change was accompanied by an increased exposure of hydrophobic residues.7 Isothermal titration calorimetry and chromophoric chelator titrations have suggested that human secretagogin binds four Ca2+ ions in one high affinity and three low affinity Ca2+-binding sites. The apparent affinity of these interactions (Kd = 25 μM) is relatively low compared to typical calcium sensor proteins but similar to Ca2+-dependent regulatory proteins such as synaptotagmin I (5–50 μM) and calpains. The search for secretagogin binding partners revealed a highly specific interaction of secretagogin with SNAP-25 (25 kDa synaptosome associated protein).7 SNAP-25 is involved in Ca2+-induced exocytosis in neurons and neuroendocrine cells. It binds secretagogin both in presence and absence of calcium with an apparent affinity of ∼0.12 μM and ∼1.5 μM, respectively.7 This finding further supports the putative role of secretagogin in the neuronal secretion and regulation of neuropeptide trafficking in endocrine cells.
Here we present the X-ray structure of the D. rerio secretagogin (UniProt ID SEGN_DANRE) in the uncomplexed Ca2+-free form at 2.1 Å resolution. The structure of D. rerio secretagogin, which had <30% overall sequence identity to any structure in the Protein Data Bank at the time of its selection and deposition, was chosen as a “sequence-to-structure” target under the National Institutes of Health Protein Structure Initiative. As highlighted above, this protein also has considerable medical relevance.
MATERIALS AND METHODS
Expression and purification of secretagogin
The gene Zgc:100843 encoding Danio rerio secretagogin was cloned and the selenomethionine (SeMet)-labeled protein was expressed and purified following the standard Center for Eukaryotic Structural Genomics pipeline protocols for cloning, protein expression, protein purification, and overall bioinformatics management.8-11 In short, the cDNA encoding secretagogin was for expression purposes cloned into a pVP16 plasmid, a custom vector derived from pQE80 (Qiagen, Valencia, CA). Protein was expressed in Escherichia coli B834 p(lacI+RARE) cells using 2 L of auto-inducing medium.12 Upon sonication of the harvested cells, the protein in the supernatant was purified via immobilized nickel affinity chromatography, and TEV protease was used to cleave the affinity/solubility tag consisting of His8-maltose binding protein. After tag capture by subtractive nickel affinity chromatography, and a final desalting step, the proteins were concentrated to 10 mg/mL and dialyzed against the protein buffer (50 mM NaCl, 3 mM NaN3, 0.3 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 5 mM bis[2-hydroxyethyl]amino-tris[hydroxymethyl]methane (BIS-TRIS) pH 8.0). Protein aliquots were then drop frozen in liquid nitrogen and stored at 193 K. Protein purifications resulted in 49.0 mg of protein with 89% SeMet incorporation.
Crystallization and structure solution
Crystals of secretagogin were grown at 277 K by the hanging drop method from a 10 mg ml-1 protein solution in the protein buffer mixed with an equal amount of well solution containing 26% (w/v) polyethylene glycol 2000 (PEG 2K), 100 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP) pH 9.0. Crystals were cryoprotected at 277 K by soaking in a solution containing 30% (w/v) PEG 2K, 5% ethylene glycol, 100 mM BTP pH 9.0 and were flash-frozen in a stream of cryogenic nitrogen gas at 100 K. X-ray diffraction data were collected near the selenium K absorption edge (12,663 eV and 12,860 eV) at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source at Argonne National Laboratory. The diffraction images were integrated and scaled using HKL2000.13 The selenium substructure of SeMet-labeled secretagogin crystals was determined using HySS and SHELXD.14,15 The programs identified 8 consensus anomalous sites. The structure was automatically phased and density-modified using autoSHARP16 with the help of auxiliary programs from the CCP4 suite.17 The initial model was built using ARP/wARP.18 The structure was completed with multiple cycles of iterative manual building in COOT19 and refinement in REFMAC5.20 All refinement steps were monitored using an Rfree value based on 5.1% independent reflections. TLS refinement was used in the final step with three groups defined that correspond to the three globular domains. The stereochemical quality of the final model was assessed using MOLPROBITY.21 The figures were prepared using PyMOL.22
RESULTS AND DISCUSSION
Structure quality
The structure of Danio rerio secretagogin was determined by multiple wavelength anomalous diffraction using two wavelengths and refined to a resolution of 2.1 Å. Data collection, phasing and refinement statistics are summarized in Table 1. A single molecule of secretagogin was present in the asymmetric unit. The final model contains residues 3–272 and 240 ordered water molecules. Residues 1 and 2 were not visible in the electron density map. Two regions spanning residues 177–192 and 227–243 showed increased disorder as judged by B-factors and real space density analysis in COOT. Residue Leu182 located in one of these regions could not be modeled satisfactorily and remained an outlier in the Ramachandran plot.
Table 1.
Crystal parameters, X-ray data collection, phasing and refinement statistics.
| Peak | HRem | |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 47.9 b = 52.7 c = 114.4 |
a = 47.7 b = 52.6 c = 114.1 |
| Data collection statistics | ||
| Wavelength (Å) | 0.97911 | 0.96411 |
| Energy (eV) | 12,663 | 12,860 |
| Resolution range (Å) | 44.17–2.10 (2.15–2.10) | 44.01–2.15 (2.20–2.15) |
| No. of reflections (measured/unique)a | 145306 / 17378 | 124618 / 16159 |
| Completeness (%) | 98.8 (98.3) | 98.1 (89.6) |
| Rmergeb | 0.093 (0.510) | 0.076 (0.539) |
| Redundancy | 8.4 (2.9) | 7.7 (5.6) |
| Mean I/sigma(I) | 11.0 (2.8) | 10.5 (2.0) |
| Phasing statistics c | ||
| Phasing power (isomorphous / anomalous) | 0.0 / 1.57 | 1.38 / 1.14 |
| Mean FOM (centric/acentric) | 0.53 / 0.39 | |
| Rcullis (isomorphous / anomalous) | 0.0 / 0.626 | 0.793 / 0.765 |
| Refinement and model statistics | ||
| Resolution range | 44.17–2.10 (2.15–2.10) | |
| No. of reflections (total / test) | 17336 / 885 | |
| Rcrystd | 0.175 (0.207) | |
| Rfreee | 0.253 (0.370) | |
| R.m.s.d. bonds (Å) | 0.019 | |
| R.m.s.d. angles (°) | 1.586 | |
| ESU from Rfree (Å) | 0.209 | |
| TLS groups (residue range) | 3–85, 86–181, 185–272 | |
| B factor - Wilson (Å2) | 36.7 | |
| Average B factor — protein / waters (Å2)f | 39.6 / 46.0 | |
| No. of protein molecules / all atoms | 1 / 2445 | |
| No. of waters | 240 | |
| Ramachandran Plot by MOLPROBITY (%) | ||
| Favored regions | 96.6 | |
| Additional allowed regions | 3.0 | |
| Outliers | 0.4 | |
| PBD code | 2be4 |
Values in parentheses are for the highest resolution shell.
Rmerge = ΣhΣi|Ii(h) - <I(h)>|/ ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
Phasing by SHARP in 38.77–2.1 Å resolution range.
Rcryst = Σh||Fobs|-|Fcalc||/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using ∼5.0% of the randomly selected unique reflections that were omitted from structure refinement.
B-factors computed from a model refined without use of TLS.
The X-ray structure of Danio rerio secretagogin
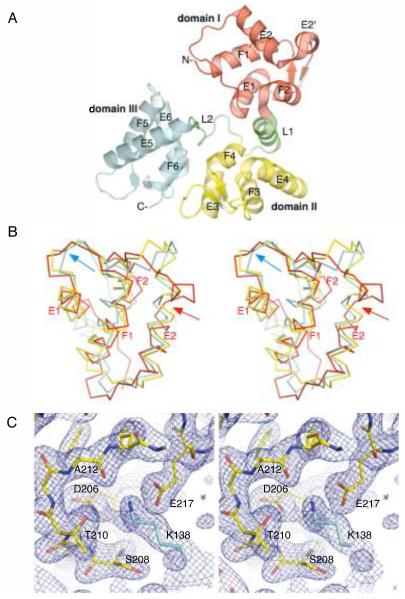
The structure of secretagogin revealed a modular protein with a radius of gyration of 20.6 Å that consists of three smaller globular domains, each with a radius of gyration of about 12 Å. The domains I, II, and III (red, yellow, and cyan, respectively in [Fig. 1(A)]) are arranged to form a bulky V-shaped molecule. The domains are joined together by two linker regions. The linker L1 (residues 85–98), which connects domains I and II, forms a well defined helix at the interface of these domains. The linker L2 (residues 176–185), which connects domains II and III, is extended and relatively disordered in our structure. Based on the PISA server,23 domains II and III associate through interdomain contacts with buried area of ∼690 Å2, while domain I seems to be less well attached to the core formed by domains II and III (buried area of ∼550 Å2). The TLS refinement confirms flexibility of the linker regions, with the N-terminal domain showing rotational variance along an axis perpendicular to the long axis of the molecule, and the C-terminal domain showing rotational variance along the long axis of the molecule. The central domain is relatively fixed. Each domain of secretagogin contains a pair of apposed EF-hand motifs; the whole protein contains six EF-hand motifs. In each domain, the metal binding loops of the EF-hand motifs form a solvent exposed two-stranded antiparallel β-sheet. A four-helix bundle formed from helices of the two EF-hand motifs in each domain gives rise to a domain core, which is stabilized by extensive hydrophobic interactions. The individual domains adopt a fold typical of other EF-hand proteins, such as calmodulin or troponin C.6 However, the overall modular arrangement of domains is quite unique. Based on the VAST server,24 the only other structurally characterized hexa-EF-hand protein is calbindin D28K.25
Figure 1.
The X-ray crystal structure of D.rerio secretagogin. (A) Secretagogin monomer consists of six EF-hand motifs arranged in pairs to form three globular domains (rendered consecutively in red, yellow and cyan). Linkers L1 and L2, highlighted in green, connect the individual domains. The apposed metal binding loops form antiparallel β-sheet on the outer surface of the V-shaped molecule. The helices of the individual EF-hand motifs are labeled for clarity. (B) A stereo view of Cα-trace of the superposed domains I, II and III (red, yellow and cyan, respectively) of secretagogin. The topologically equivalent motifs EF1, EF3 and EF5 overlap well, except for the Ca2+-binding loop which adopts the open, “Ca2+-ready” conformation in EF5-hand (cyan arrow) and closed, “Ca2+-free” conformation in EF1 and EF3 motifs. The EF2 hand differs from all the remaining EF-hands due to a break at Met63 in the helix E2 (red arrow). (C) A stereo image of the calcium-binding loop in EF5-hand motif of D. rerio secretagogin. A final 2mFo-DFc electron density map (blue mesh) is contoured at 1.2σ level. The refined protein model is shown in sticks. Residues of the calcium-binding motif at positions 1 (Asp206), 3 (Ser208), 5 (Thr210), 7 (Ala212) and 12 (Glu217) are labeled for clarity. A symmetry molecule of secretagogin from the crystalline lattice contributes Lys138 (cyan sticks), which mimics the positively charged calcium ion and stabilizes the loop in “Ca2+-ready”-like conformation.
Structural comparison of secretagogin domains
Despite the similar overall fold there are notable structural differences between secretagogin domains. These are illustrated best by their structural superposition [Fig. 1(B)]. Domains II and III are the most similar to each other (with an all-atom rmsd of 1.57 Å), while domain I is structurally less similar to both domain II (rmsd of 2.57 Å) and domain III (rmsd of 2.33 Å). The EF-hand motif EF1 of domain I aligns very well with topologically equivalent motifs EF3 and EF5 from domains II and III, respectively. However, the motif EF2 of domain I is distinct from all the remaining EF-hand motifs of secretagogin due to a break at Met63 in the helix E2 [Fig. 1(B), red arrow]. Interestingly, the two apposed EF-hands within each domain are not identical, but show variations in the loop conformations and interhelical angles.
Structure of secretagogin EF-hand motifs
The structure of secretagogin presented in this work represents a calcium-free form of the protein, and as such the structure provides the first ever glimpse at an apo-hexa-EF-hand protein. Comparison of the secretagogin structure with an archetypal Ca2+-binding protein calmodulin (PDB ID codes 1QX5 and 1CLL)26,27 revealed that the metal binding loops of motifs EF1, EF3, EF4, and EF6 of secretagogin adopt conformations similar to the closed (Ca2+-free) form of calmodulin. The divergent loop of the EF2 motif shows a poor overlap with either Ca2+-bound or Ca2+-free forms of calmodulin. Finally, the EF5 motif of domain III seems to be in the open, “Ca2+-ready” conformation despite the absence of metal in the binding site [Fig. 1(B), cyan arrow]. Detailed inspection of the structure revealed that conformation of EF5 loop is stabilized by electrostatic interaction with Lys138 of the symmetry molecule in the crystal lattice. Positively charged amino group thus partially mimics a Ca2+-ion, stabilizing an apparent “Ca2+-ready”—like conformation [Fig. 1(C)].
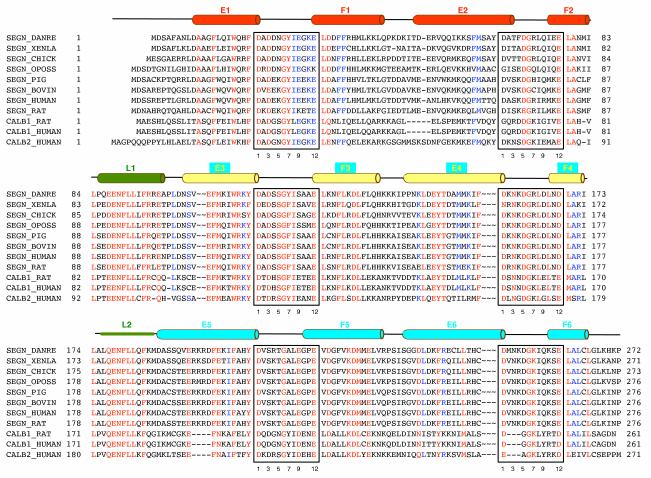
In a classical EF-hand motif, the calcium ion is coordinated in a pentagonal bipyramidal configuration by 6 residues in positions 1, 3, 5, 7, 9 and 12 of the loop.6 Residues in positions 1, 3, 5, 9, and 12 coordinate the calcium through their side-chains, while a residue in position 7 coordinates the calcium through the backbone carbonyl. The consensus sequence of the EF-hand calcium-binding loop is D—x—D/N—G—D/S/N—G—x—I/L/V—x—x—x—E/D, where the uppercase letters represent residues that occur with more than 50% frequency and ‘x’ stands in positions with less defined preference. The most conserved residues highlighted in bold letters are located in positions 1 (Asp, 100%), 6 (Gly, 96%), and 12 (Glu, 92%).6 The metal—binding residues in all six EF hand motifs of D. rerio secretagogin are consistent with the naturally occurring variations of the functional loop sequence [Fig. 2]. This suggests that D. rerio secretagogin could potentially bind six Ca2+ ions per molecule. However, multiple sequence alignment [Fig. 2] revealed that secretagogins of only some eukaryotes (including D. rerio, X. laevis, Monodelphis domestica, Gallus gallus, Ornithorhynchus anatinus (not shown)) could be fully competent in terms of Ca2+ binding. All other sequence homologs of D. rerio secretagogin, which include secretagogins from higher eukaryotes, calbindins, and calretinins, have at least one non-functional EF-hand motif due to the mutation(s) or deletions. In particular, asparagine in position 5 in the loop of EF1 motif is replaced by lysine in secretagogin of many mammalian species. The sequence of EF2 motif is the least conserved among mammalian secretagogins. The EF2 loop seems to be competent for Ca2+-binding in most mammalian secretagogins except for human and chimpanzee (not shown) orthologs. In these species arginine replaces a negative or polar residue in the consensus position 9. Similarly, loops of EF2 motifs of rat (not shown) and human calbindin D28k do not follow the consensus sequence and are probably non-functional in terms of Ca2+ binding. The EF-hand motifs EF3, EF4, EF5, and EF6 are highly conserved in all mammalian secretagogins and most likely competent for Ca2+-binding. This prediction is in accord with isothermal calorimetry experiments that revealed four Ca2+-binding sites in human secretagogin.7 Calbindin and calretinin subfamily of hexa-EF hand proteins contains a three-residue long deletion in the metal binding loop of EF6, which renders this domain inactive.
Figure 2.
Multiple sequence alignment of D. rerio secretagogin and selected hexa-EF-hand proteins. Amino acid sequences of proteins are labeled by their UniProt identifiers or pseudo identifiers (in italics): SEGN_DANRE, zebrafish secretagogin; SEGN_XENLA, African horned frog secretagogin; SEGN_CHICK, chicken secretagogin (Gene Bank gi:118086706); SEGN_OPOSS, opposum secretagogin (Gene Bank gi|126322175); SEGN_PIG, porcine secretagogin; SEGN_BOVIN, bovine secretagogin; SEGN_HUMAN, human secretagogin; SERGN_RAT, rat secretagogin; CALB1_HUMAN, human calbindin D28K; CALB2_HUMAN, human calretinin. Individual domains and linker regions of hexa-EF-hand proteins are aligned in three blocks to reveal interdomain conservation patterns. Positions of helices in D. rerio secretagogin are depicted above the alignment and secondary structure elements are labeled and color-coded as in Fig. 1(A). The calcium binding loops are boxed and position of the residues within the calcium-binding consensus motif is given for easy orientation.
Secretagogin and calbindin D28k show different spatial domain arrangement
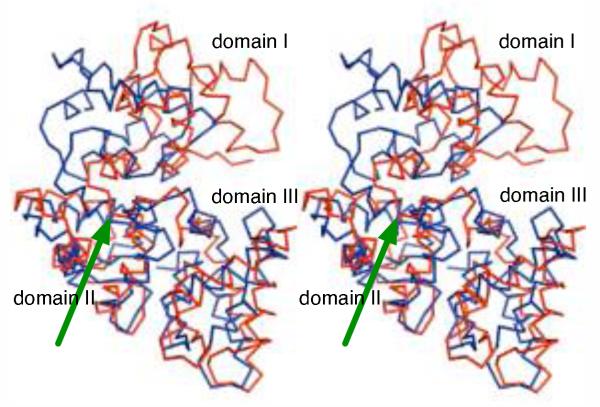
Pairing of two EF-hand motifs into a larger module is a common structural feature found in many Ca2+-binding proteins (calmodulin, troponin C, and members of S100 protein family),6 however, there have been only a few hexa-EF-hand proteins described so far. The Ca2+-free structure of D. rerio secretagogin represents the first X-ray structure of a hexa-EF-hand protein. Currently, the only other structurally characterized hexa-EF-hand protein is rat calbindin D28K. The solution NMR structure has been reported for the Ca2+-loaded form of this protein25 (PDB ID code 2f33). D. rerio secretagogin and rat calbindin D28K share 35% sequence identity. In addition, a successful crystallization of human calbindin D28K in the Ca2+-loaded form has been recently reported.28 Although secretagogin and calbindin D28K have similar overall topology they show significant structural differences. The most important change is a strikingly different spatial arrangement of the three domains in these structures. Specifically, domain I is rotated by almost 180° with respect to the core formed by domains II and III in both proteins [Fig. 3].
Figure 3.
D. rerio secretagogin and rat calbindin D28K have different quarternary domain arrangement. The domain II and III of D. rerio secretagogin (red) were superposed with the corresponding region of rat calbindin D28K (blue; PDB ID 2f33). In the resulting overlay, the domain I of secretagogin is rotated by almost 180 degrees with respect to the core formed by domains II and III in both proteins. The green arrow points to the point from which structures diverge.
Could a dramatic rearrangement involving domain I, as suggested by the above comparison, take place in secretagogin(s) upon calcium binding? Answering this question will require structural characterization of the calcium-loaded form of secretagogin. Our attempts to soak preexisting crystals of D. rerio secretagogin in calcium containing buffers lead to a complete loss of the diffraction, while no cracking of crystals was observed during the procedure. Our attempts to grow crystals of D. rerio secretagogin in the presence of calcium failed so far. Upon closer inspection of linker regions connecting the individual domains of secretagogin we noticed that despite their dramatically different structures (the L1 linker forms a three turns long helix, while the L2 linker is largely unstructured [Fig. 1(A)]) these regions show unexpectedly high sequence similarity [Fig. 2]. The linker region thus does not have clear propensity for a given secondary structure and can adopt significantly different context-dependent conformations. We envision that the helical linker between domains I and II may unwind and this could result in a spatial structure rearrangements in secretagogin and other hexa-EF-hand proteins. This conformational change could be triggered upon Ca2+-binding. Calcium-dependent conformational changes have been indeed documented in secretagogin by spectroscopic methods.7 Also, an independent behavior of domain I relative to the module consisting of domains II and III has been established experimentally in calretinin, a hexa-EF hand protein with 40% homology to D. rerio secretagogin.29
ACKNOWLEGMENTS
We acknowledge the financial support from NIH National Institute for General Medical Sciences grant P50 GM064598 and U54 GM074901 (John L. Markley, PI). Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No.W-31-109-ENG-38.
Special thanks go to all members of the CESG team involved in this work, especially Craig S. Newman, Zachary Eggers, John Kunert, Kory D. Seder, Frank C. Vojtik, Jason M. Ellefson, Grzegorz Sabat, David J. Aceti, Gary E. Wesenberg, Russell L. Wrobel.
Frequently used abbreviations
- VAST
Vector Alignment Search Tool
- SNAP-25
25 kDa synaptosome associated protein
Footnotes
ACCESSION NUMBERS: The atomic coordinates and structure factors for D. rerio secretagonin have been deposited in the Protein Data Bank under accession code 2be4.
REFERENCES
- 1.Wagner L, Oliyarnyk O, Gartner W, Nowotny P, Groeger M, Kaserer K, Waldhausl W, Pasternack MS. Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J Biol Chem. 2000;275(32):24740–24751. doi: 10.1074/jbc.M001974200. [DOI] [PubMed] [Google Scholar]
- 2.Gartner W, Lang W, Leutmetzer F, Domanovits H, Waldhausl W, Wagner L. Cerebral expression and serum detectability of secretagogin, a recently cloned EF-hand Ca(2+)-binding protein. Cereb Cortex. 2001;11(12):1161–1169. doi: 10.1093/cercor/11.12.1161. [DOI] [PubMed] [Google Scholar]
- 3.Nicolls MR, D’Antonio JM, Hutton JC, Gill RG, Czwornog JL, Duncan MW. Proteomics as a tool for discovery: proteins implicated in Alzheimer’s disease are highly expressed in normal pancreatic islets. J Proteome Res. 2003;2(2):199–205. doi: 10.1021/pr025576x. [DOI] [PubMed] [Google Scholar]
- 4.Birkenkamp-Demtroder K, Wagner L, Sorensen F Brandt, Astrup L Bording, Gartner W, Scherubl H, Heine B, Christiansen P, Orntoft TF. Secretagogin is a novel marker for neuroendocrine differentiation. Neuroendocrinology. 2005;82(2):121–138. doi: 10.1159/000091207. [DOI] [PubMed] [Google Scholar]
- 5.Adolf K, Wagner L, Bergh A, Stattin P, Ottosen P, Borre M, Birkenkamp-Demtroder K, Orntoft TF, Torring N. Secretagogin is a new neuroendocrine marker in the human prostate. Prostate. 2007;67(5):472–484. doi: 10.1002/pros.20523. [DOI] [PubMed] [Google Scholar]
- 6.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405(2):199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 7.Rogstam A, Linse S, Lindqvist A, James P, Wagner L, Berggard T. Binding of calcium ions and SNAP-25 to the hexa EF-hand protein secretagogin. Biochem J. 2007;401(1):353–363. doi: 10.1042/BJ20060918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon WB, Aceti DJ, Bingman CA, Vojtik FC, Olson AC, Ellefson JM, McCombs JE, Sreenath HK, Blommel PG, Seder KD, Burns BT, Geetha HV, Harms AC, Sabat G, Sussman MR, Fox BG, Phillips GN., Jr. High-throughput Purification and Quality Assurance of Arabidopsis thaliana Proteins for Eukaryotic Structural Genomics. J Struct Funct Genomics. 2005;6(2-3):143–147. doi: 10.1007/s10969-005-1908-7. [DOI] [PubMed] [Google Scholar]
- 9.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr., Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40(2):256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics. 2004;5(4):267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 11.Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr., Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;4(1):11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 12.Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnology progress. 2007;23(3):585–598. doi: 10.1021/bp070011x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 15.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 16.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Method Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 18.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6(5):458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 22.DeLano WL. The PYMOL Molecular Graphic System. DeLano Scientific LLC; San Carlos, CA, USA: 2002. [Google Scholar]
- 23.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23(3):356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 25.Kojetin DJ, Venters RA, Kordys DR, Thompson RJ, Kumar R, Cavanagh J. Structure, binding interface and hydrophobic transitions of Ca2+-loaded calbindin-D(28K) Nat Struct Mol Biol. 2006;13(7):641–647. doi: 10.1038/nsmb1112. [DOI] [PubMed] [Google Scholar]
- 26.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 Å resolution. J Mol Biol. 1992;228(4):1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 27.Schumacher MA, Crum M, Miller MC. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 2004;12(5):849–860. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Sun Y, Wang W, Zhang Y, Ma M, Lou Z. Crystallization and preliminary crystallographic analysis of human Ca2+-loaded calbindin-D28k. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 2):133–136. doi: 10.1107/S1744309108001905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palczewska M, Groves P, Batta G, Heise B, Kuznicki J. Calretinin and calbindin D28k have different domain organizations. Protein Sci. 2003;12(1):180–184. doi: 10.1110/ps.0215303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]