Abstract
Cellular DNA lesions are efficiently countered by DNA repair in conjunction with delays in cell cycle progression. Previous studies have demonstrated that Rad9, Hus1, and Rad1 can form a heterotrimeric complex (the 9-1-1 complex) that plays dual roles in cell cycle checkpoint activation and DNA repair in eukaryotic cells. Although the 9-1-1 complex has been proposed to form a toroidal structure similar to proliferating cell nuclear antigen (PCNA), which plays essential roles in DNA replication and repair, the structural basis by which it performs different functions has not been elucidated. Here we report the crystal structure of the human 9-1-1 complex at 3.2 Å resolution. The crystal structure, together with biochemical assays, reveals that the interdomain connecting loops (IDC loop) of hRad9, hHus1, and hRad1 are largely divergent, and further cocrystallization study indicates that a PCNA-interacting box (PIP box)-containing peptide derived from hFen1 binds tightly to the interdomain connecting loop of hRad1, providing the molecular basis for the damage repair-specific activity of the 9-1-1 complex in contrast to PCNA. Furthermore, structural comparison with PCNA reveals other unique structural features of the 9-1-1 complex that are proposed to contribute to DNA damage recognition.
Cellular DNA damage triggers the activation of the cell cycle checkpoint, leading to a delay or arrest in cell cycle progression to prevent replication and inducing DNA damage repair (1, 2). In response to DNA damage, the 9-1-13 complex can be loaded onto DNA lesion sites by Rad17-RFC2–5 (which consists of one large subunit, Rad17, and four small subunits, RFC2–5), where it triggers the activation of the cell cycle checkpoint (3, 4). Moreover, the 9-1-1 complex can also directly participate in DNA repair via physical association with many factors involved in base excision repair (BER), translesion synthesis, homologous recombination, and mismatch repair pathways (5–9).
Although both the 9-1-1 and the PCNA complexes perform critical functions in eukaryotic cells with predicted similar structures (10), their specific roles are distinct. First, the 9-1-1 complex is a DNA damage sensor in the cell cycle checkpoint but does not function as a scaffold for the major DNA replication factors; however, PCNA plays exactly the opposite role (1, 11). Second, although both the complexes function in DNA repair, their specific activities are different. Previous observations indicated that some BER enzymes, such as MYH (MutY glycosylate homolog) (12), TDG (thymine DNA glycosylate) (7), and NEIL (Nei-like glycosylate) (8), interact with the 9-1-1 complex via motifs that are located outside the conserved PCNA-interacting box (the PIP box), implying that the 9-1-1 complex functions as a damage repair-specific clamp, in contrast to PCNA. However, the structural basis for this hypothesis remains unclear. Another important unresolved issue concerns the damage-sensing mechanism of the 9-1-1 complex. During the DNA replication process, the PCNA·RFC clamp·clamp loader specifically recognizes the primer-template junction (13). However, the molecular basis by which the 9-1-1·Rad17-RFC2–5 clamp·clamp loader specifically recognizes the damaged DNA is little known. To address these questions, we performed structural and biochemical studies on the 9-1-1 complex.
EXPERIMENTAL PROCEDURES
Protein Preparation
The hRad9, hHus1, and hRad1 proteins were co-expressed in a Pichia pastoris expression system. To confirm that these three proteins interact with each other, only hHus1 was constructed with an N-terminal His6 tag, whereas the other two were constructed without the tag. The full length of His-tagged hHus1 sequence (residues 1–280) was cloned into a pPICZ C vector. A full-length hRad1 sequence (residues 1–282) and a truncated hRad9 sequence (residues 1–270) were separately cloned into PmeI-mutated pPICZ C vectors. Two expression cassettes separately containing the hRad1 gene and the hRad9 gene were then excised from these vectors using the BglII and BamHI sites. The cassettes were then sequentially ligated into the hHus1-pPICZ C vector, which was linearized with BamHI. The vector, containing hRad9, hHus1, and hRad1 genes, was then linearized with PmeI, transformed, and expressed in the GS115 strain of P. pastoris to produce the human 9-1-1 heterotrimeric complex. The expressed complex was purified to homogeneity via nickel affinity chromatography, Sepharose Q chromatography, and SuperdexTM200 HR 10/30 gel filtration, and concentrated to a final concentration of 7 mg/ml in 20 mm Tris-HCl, 150 mm NaCl, 1 mm dithiothreitol, and 1 mm EDTA, pH 8.0, for crystallization.
Crystal Growth
Crystals were grown at 16 °C by the hanging drop method, using equal volumes of protein and crystallization solution over a 1-ml reservoir, which was composed of 8% polyethylene glycol 2000 monomethyl ether, 100 mm Tris-HCl (pH 8.0), and 200 mm trimethylamine N-oxide dihydrate. The final plate-shaped single crystals were obtained by microseeding (14). Mercurated crystals were obtained by adding a final concentration of 1.5 mm Thimerosal (ethylmercuricthiosalicylic acid, sodium salt) in the hanging drop and soaking the crystals for about 30 h. The diffraction of the crystals was significantly improved to 3.2 Å by controlled crystal dehydration (15). The crystals were then flash-frozen in the liquid nitrogen before being taken to synchrotron radiation sources for data collection.
Data Collection and Structure Determination
3.2 Å native data were collected at Spring8 in Japan, on beamline BL41XU, and processed with HKL2000 (16). 3.2 Å mercury derivative data were collected at SLS (Swiss Light Source) in Switzerland, on beamline X06SA, and processed with MOSFLM (17). The crystal has space group P21 with one complex in the asymmetric unit. The phase is determined by single anomalous dispersion method. Electron density maps were calculated by PHINEX (18, 19). Density modification by SOLOMON (20) was applied at the final stage of phasing. Model building was performed with COOT (21), and refinement was carried out using REFMAC (22) and CNS (23). The final Rwork and Rfree values were 0.289 and 0.306, respectively. The structure of the 9-1-1 complex was analyzed with PROCHECK (24). Data collection and refinement statistics are presented in supplemental Table S1.
Proteins Prepared for Biochemistry Assays
The genes of truncated hRad17 (residues 78–337), hRad9 (residues 1–391), hHus1 (residues 1–280), and hRad1 (residues 1–282) are constructed in the pGEX-4T-1 vector separately. Each of the GST-tagged proteins and GST proteins are expressed in Escherichia coli BL21 cells and purified using glutathione-Sepharose 4 Fast Flow resin, respectively. His-tagged truncated hRad17 (residues 78–337), His-hRad9 (residues 1–262), His-hHus1 (residues 1–280), and His-hRad1 (residues 1–282) were expressed separately in E. coli BL21 cells using the pET24a(+) vector. His-tagged truncated hRad17 (residues 78–337) was purified by nickel affinity chromatography and SuperdexTM200 HR 10/30 gel filtration.
GST Pulldown Assays
The GST-tagged hRad17 (residues 78–337) and GST proteins were separately immobilized on glutathione-Sepharose 4 Fast Flow resin and then incubated with cell lysate separately containing His-hRad9 (residues 1–262), His-hHus1 (residues 1–280), and His-hRad1 (residues 1–282), respectively, for 2 h at 4 °C. The GST protein was used as a control. After washing the pellets five times with phosphate-buffered saline (pH 7.3), the bound proteins were eluted with 50 mm Tris-HCl, 10 mm reduced glutathione, pH 8.0. The proteins were subsequently analyzed by Western blotting with anti-His and anti-GST antibodies.
Biacore Analysis
Real-time binding and kinetic analyses by surface plasmon resonance were carried out on a Biacore 3000 instrument (GE Healthcare, Uppsala, Sweden). The eluent contained phosphate-buffered saline and 0.005% Tween 20. GST-hRad9 (residues 1–391), GST-hHus1 (residues 1–280), GST-hRad1 (residues 1–282), and GST (as control) were immobilized separately on CM5 chips using an amine coupling kit, and the remaining coupling sites were blocked with 1 m ethanolamine (pH 8.5). Three binding assays were carried out using 20 μm hFen1 peptide (residues 335–364), 20 μm p12 peptide (residues 4–11), and 3 μm His-tagged truncated hRad17 (residues 78–337), respectively, with 1 mm ATP at 25 °C. All data collected were analyzed with BIAevaluation software version 4.1, and the data were fitted to a 1:1 binding model to obtain equilibrium constants.
RESULTS AND DISCUSSION
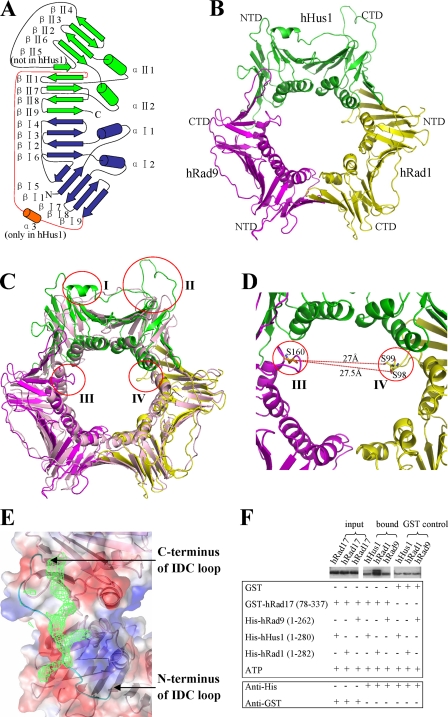
The crystal structure of the human 9-1-1 complex is a closed circular ring, with the C-terminal domain of one protein interacting with the N-terminal domain of the following protein in the order hRad9 → hHus1 → hRad1 (Fig. 1B). Each molecule in the 9-1-1 complex consists of 18 β sheets (βI1 to βI9 and βII1 to βII9) and four α helices (αI1, αI2, αII1, αII2) and is composed of two domains (the N- and C-terminal domains), which are connected by an interdomain connecting (IDC) loop (Fig. 1A). Among the intermolecular interfaces of the 9-1-1 complex, a zipper-like series of hydrogen bonds is formed between βII4 of the C-terminal domain of one molecule and βI9 of the N-terminal domain of the next molecule (supplemental Fig. S1). Additionally, intercomponent hydrophobic cores are formed by the hydrophobic residues in αII1 and βII4 in the C-terminal domain of one molecule and αI2 and βI9 in the N-terminal domain of the next molecule, resulting in a buried surface area of about 1,609 Å2 between hRad9 and hHus1, 1,475 Å2 between hHus1 and hRad1, and 1,500 Å2 between hRad1 and hRad9, which are comparable with that in PCNA (1,500 Å2).
FIGURE 1.
Structure and binding assay of the 9-1-1 complex. A, schematic diagram of the secondary structural elements in one molecule of the 9-1-1 complex. The N-terminal domain is colored blue, whereas the C-terminal domain is colored green. The IDC loop is colored red. The α3 helix inserted in the N terminus of the IDC loop in hHus1 is colored orange. B, three-dimensional diagram of the 9-1-1 complex. NTD and CTD indicate the N- and C-terminal domains, respectively. C, superposition of the 9-1-1 complex and PCNA. hRad9 is shown in magenta, hHus1 is in green, hRad1 is in yellow, and PCNA is in light pink. The four most notable differences are indicated by red circles. I indicates the α3 helix inserted in the N terminus of the IDC loop of hHus1; II indicates the βII4–βII6 loop connecting βII4 and βII6 in hHus1; and III and IV respectively indicate the αII1–βII2 loop of hRad9 and the αI2–βI7 loop of hRad1 near the channel in the 9-1-1 complex. D, the closest distance between the αII1–βII2 loop in hRad9 and the αI2–βI7 loop in hRad1. E, the electron density for the hFen1 peptide on the binding site of hRad1 in the 9-1-1 complex. The electron density is contoured at 0.7σ in the 2|Fo| − |Fc| map. F, GST pulldown assays to analyze the interactions of the truncated hRad17 with hHus1, hRad1, and hRad9. GST proteins are used as control. The lower bands in the samples labeled bound and GST control indicate nonspecific GST-interacting protein. The interaction of the truncated hRad17 with hRad1 is markedly stronger than its interaction with the other two proteins.
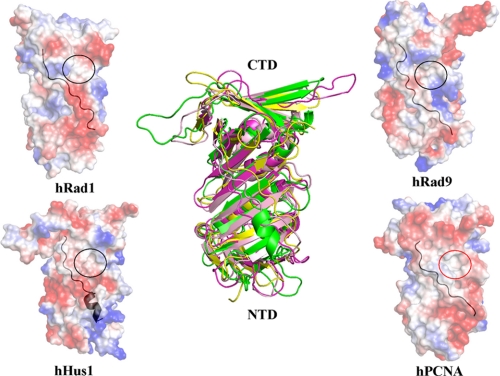
Unlike the almost three-fold symmetrical charge distribution on PCNA, the charge distribution on the 9-1-1 complex is extremely uneven (supplemental Fig. S2) with the inner surface highly positively charged and covered by multiple lysine and arginine residues (supplemental Fig. S3B), which are largely conserved from fission yeast to human (supplemental Fig. S4). Moreover, when compared with the near perfect three-fold symmetry of PCNA, the 9-1-1 complex shows a highly asymmetrical structure (Fig. 1C). Four major differences were observed between the 9-1-1 complex and PCNA. First, a short helix, α3, inserted in the N terminus of the IDC loop was observed exclusively in hHus1. Second, βII5 was replaced by a long loop connecting βII4 and βII6 (the βII4–βII6 loop) in hHus1 but not in hRad9, hRad1, or the PCNA monomer. It is highly probable that these two distinguishing structural features confer functional specificity to hHus1. In addition, loops extending near the core of the ring were present in hRad9 (αII1–βII2 loop) and hRad1 (αI2–βI7 loop), but not in hHus1 or the PCNA monomer, suggesting that these two loops have unique functional roles (this will be discussed later).
These differences provide structural evidence that the components of the 9-1-1 complex have diverged during evolution to possess unique specificities. It is known that a consensus binding motif (QXX(L/V/M)XXF(F/Y)) called the PIP box, which is present in most proteins that are essential for DNA replication or repair, interacts with a hydrophobic cavity (including some residues of the IDC loop) in PCNA (11). However, analysis of the corresponding regions in hRad9, hHus1, and hRad1 revealed that each of these regions has a distinct surface curvature and charge distribution (Fig. 2), suggesting that these regions show differences in interacting with the same proteins. To further understand this aspect, we synthesized a 30-amino acid PIP box-containing peptide (residues 335–364: STQGRLDDFFKVTGSLSSAKRKEPEPKGST) derived from the C terminus of hFen1 (flap endonuclease 1, an enzyme required for DNA replication and BER) and estimated its binding affinities with each component of the 9-1-1 complex using Biacore analysis. The results showed that the peptide interacted with all three proteins but with distinct affinities; hRad1 has the highest, hRad9 has the intermediate, and hHus1 has the lowest affinity (supplemental Fig. S5A). In contrast, the PCNA monomers possess equal affinity for binding to hFen1 (25). This observation suggests that the distinctive conformation and properties of the IDC loop in hHus1 largely disrupted its interaction with the PIP box and may be reserved for other DNA repair factor motifs. Next, we cocrystallized the 9-1-1 complex with the hFen1 peptide and solved the complex structure by molecular replacement at 3.4 Å resolution. In this complex structure, continuous residual density was observed around the IDC loops of hRad1 (Fig. 1E), disordered density around that of hRad9, and no residual electron density around the IDC loop or other regions of hHus1. This indicates that the hFen1 peptide interacts with the 9-1-1 complex mainly through the IDC loop of hRad1, which is consistent with our binding experiment results. The differences in the enzyme binding specificity of the three components are likely to confer the 9-1-1 complex with unique features in the DNA damage repair process, such as greater efficacy in simultaneously binding multiple DNA damage repair factors.
FIGURE 2.
The IDC loops of the 9-1-1 complex. The IDC loop on the surface is indicated in black. The PIP box-binding hydrophobic cavity of human PCNA is indicated by a red circle. Corresponding regions in hRad9, hHus1, and hRad1 are indicated by black circles. hRad9 is shown in magenta, hHus1 is in green, hRad1 is in yellow, and PCNA is in light pink. NTD and CTD indicate the N- and C-terminal domains, respectively.
Previous studies revealed that the 9-1-1 complex does not participate in DNA replication because it cannot bind to DNA polymerase δ, an essential factor in this process (26). Moreover, biochemical analyses indicated that two subunits (p66 and p12) of DNA polymerase δ interact with hydrophobic cavities in PCNA via their PIP boxes (27) and that the large subunit p125 interacts with PCNA via its putative variant PIP box (28). We proposed that the 9-1-1 complex has lost the ability to bind to the PIP box of one or more subunits of DNA polymerase δ because its IDC loops have diverged in the course of evolution. To confirm this hypothesis, we synthesized a peptide of the PIP box of p12 (residues 4–11: KRLITDSY) and evaluated its binding affinity with each component of the 9-1-1 complex using Biacore analysis. The results revealed that the peptide did not interact with any subunit of the 9-1-1 complex (data not shown), which supported our hypothesis. Moreover, when DNA is damaged, the normal replication process is inhibited by p21-mediated inactivation of PCNA via blocking the PIP box-binding hydrophobic cavity (29, 30), resulting in the inhibition of PCNA-mediated DNA repair because DNA repair factors become less likely to interact with PCNA. However, our structural data suggest that the three components of the 9-1-1 complex cannot be blocked by p21 simultaneously because of the divergent IDC loops, which is distinct from the case of PCNA. Taken together, our structural and biochemical data provided the further elucidation of the role of the 9-1-1 complex as a damage repair-specific clamp in the DNA repair pathway, in contrast to PCNA.
Moreover, how Rad17-RFC2–5 interacts with the 9-1-1 complex is still a manner of debate (3, 31). To illustrate this issue, we performed GST pulldown and Biacore assays to analyze the interaction between the truncated hRad17 (residues 78–337) and individual components of the 9-1-1 complex. The results indicated that hRad17 interacts with hRad1, but not hRad9 or hHus1 (Fig. 1F and supplemental Fig. S5B), which is in agreement with a previous study showing that mutation of residues 226–233 in hRad1 to polyalanine disrupts the interaction of hRad1 with hRad17 (31). In our structure, the αII2 helix in the C-terminal domain of hRad1 consisting of residues 226–233 is located in the inner ring of the 9-1-1 complex (supplemental Fig. S4). Because residues 228–231 are located inside the central channel and are likely to interact with double-stranded DNA, the residues of the N- or C-terminal ends of the αII2 helix present at the surface of the 9-1-1 ring are the probable binding sites for hRad17. Intriguingly, in the crystal structure of the Saccharomyces cerevisiae PCNA·RFC complex (13), a single subunit of PCNA interacts with RFC1 (the RFC large subunit homologous to hRad17) not only through the PIP box-binding hydrophobic cavity of PCNA but also through the N-terminal residues (210–211) of the PCNA αII2 helix (supplemental Fig. S6, A and B), which is present at the position corresponding to residues 226–227 in hRad1 (supplemental Fig. S6C). Taken together, these data suggest that hRad1 plays a dominant role in the interaction between the 9-1-1 complex and hRad17, most likely through the N-terminal residues (226–227) of its αII2 helix.
A prevailing view indicates that the 9-1-1 complex is a DNA damage sensor in the checkpoint signaling pathway (32). However, the mechanism by which this complex detects DNA lesions remains elusive. Although it has been suggested that the 9-1-1 complex may detect some types of DNA lesions via single-stranded DNA-binding replication protein A (33), it is unclear whether the 9-1-1 complex can be involved in DNA damage recognition in a more direct manner. Structural comparison reveals that the inner ring of the 9-1-1 complex shows significant differences from that of PCNA, including the αII1–βII2 loop in hRad9 and the αI2–βI7 loop in hRad1, both of which extend near the core of the ring. Additionally, our calculations using the CCP4 PISA (Protein Interfaces, Surfaces and Assemblies) server and the CCP4 SC program reveal that the hRad1-hRad9 interface has the lowest complementarity scores (supplemental Table S2), in conjugation with our experimental finding that hHus1 and hRad1 can form a stable 1:1 complex in solution (supplemental Fig. S7), suggesting that the hRad1-hRad9 interface could be the interface that opens to encircle DNA. Therefore, the distance variation between these two loops will be highly correlated to the opening and closing of the 9-1-1 ring via the hRad1-hRad9 interface, and the shortest distance is ∼27 Å from Ser-160 in the αII1–βII2 loop of hRad9 to Ser-98/Ser-99 in the αI2–βI7 loop of hRad1 (Fig. 1D), which is much shorter than the corresponding distance in PCNA (>30 Å). Such close loops can be expected to contact with some types of damaged DNA (such as bubbled and mismatched DNA) more tightly than undamaged DNA (supplemental Fig. S3A). This is in line with a recent study showing that the hRad9 S160A point mutation significantly reduced mismatch repair activity (6). In addition, the observations that both Rad9 and Rad1 possess 3′-5′-exonuclease activity (34, 35) also imply that they may possess an architecture specialized for recognizing nicked or gapped DNA.
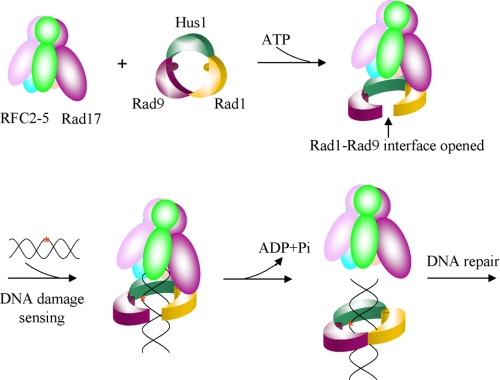
In summary, we propose a damage-sensing model. In this model, the clamp loader Rad17-RFC2–5 forms an ATP-dependent complex with the 9-1-1 sliding clamp in which Rad17 interacts with Rad1 and loads the 9-1-1 complex onto DNA via opening the Rad1-Rad9 interface. Sliding of the clamp·clamp loader along DNA will stop when certain types of DNA lesions are detected by some key residues inside the clamp, particularly Ser-160 of hRad9 and Ser-98/Ser-99 of hRad1. The detection of DNA lesions then leads to conformational changes that facilitate the ring closure and ATP hydrolysis, consequently stimulating the dissociation of Rad17-RFC2–5 clamp loader from the 9-1-1 clamp and ultimately triggering further DNA repair (Fig. 3). This proposed mechanism is distinct from the sensing mechanism of PCNA·RFC in the replication process, in which RFC is mainly responsible for recognizing the primer-template junction (13). However, further evidences are needed to confirm this hypothesis.
FIGURE 3.
DNA damage-sensing model. The clamp loader Rad17-RFC2–5 forms an ATP-dependent complex with the 9-1-1 sliding clamp, in which Rad17 interacts with Rad1, and loads the 9-1-1 complex onto DNA via opening the Rad1-Rad9 interface. The αII1–βII2 loop in hRad9 and the αI2–βI7 loop in hRad1 are proposed to detect some types of DNA lesions. The DNA damage recognition then leads to conformational changes that facilitate the ring closure and ATP hydrolysis, in turn stimulating the dissociation of Rad17-RFC2–5 clamp loader from the 9-1-1 clamp and ultimately triggering further DNA repair.
Supplementary Material
Acknowledgments
We thank the staff at Spring8 (beamline BL41XU) in Japan, at SLS (beamline X06SA) in Switzerland, and at BSRF (Beijing Synchrotron Radiation Facility) in China who helped with the data collection. We thank Y. Y. Chen for performing the Biacore analysis and X. D. Zhao for technical assistance.
This research was financially supported by the National Key Basic Research Program (Grant 2006CB910902), the National Natural Science Foundation of China (Grants 30770435, 30721003, and 30530180), the National High Technology Research and Development Program of China (Grant 2006AA02A319), and the National Natural Science Foundation of China (Grant 30530180).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and supplemental Tables S1 and S2.
The atomic coordinates and structure factors (code 3GGR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- 9-1-1
- Rad9-Hus1-Rad1
- PCNA
- proliferating cell nuclear antigen
- IDC loop
- interdomain connecting loop
- RFC
- replication factor C
- hFen1
- flap endonuclease 1
- PIP box
- PCNA-interacting box
- BER
- base excision repair
- GST
- glutathione S-transferase.
Addendum
While we were submitting the manuscript, an article on the 9-1-1 complex structure by Doré et al. (36) was published online. The authors proposed that a single repair enzyme-binding site on the 9-1-1 complex that can be blocked competitively by p21, and our cocrystallization data and binding assay suggest that Rad1 might be the most likely candidate of this binding site.
REFERENCES
- 1.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 2.Lukas J., Lukas C., Bartek J. (2004) DNA Repair 3, 997–1007 [DOI] [PubMed] [Google Scholar]
- 3.Bermudez V. P., Lindsey-Boltz L. A., Cesare A. J., Maniwa Y., Griffith J. D., Hurwitz J., Sancar A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L., Cortez D., Elledge S. J. (2002) Genes Dev. 16, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helt C. E., Wang W., Keng P. C., Bambara R. A. (2005) Cell Cycle 4, 529–532 [DOI] [PubMed] [Google Scholar]
- 6.He W., Zhao Y., Zhang C., An L., Hu Z., Liu Y., Han L., Bi L., Xie Z., Xue P., Yang F., Hang H. (2008) Nucleic Acids Res. 36, 6406–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan X., Madabushi A., Chang D. Y., Fitzgerald M. E., Shi G., Drohat A. C., Lu A. L. (2007) Nucleic Acids Res. 35, 6207–6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan X., Bai H., Shi G., Theriot C. A., Hazra T. K., Mitra S., Lu A. L. (2007) Nucleic Acids Res. 35, 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gembka A., Toueille M., Smirnova E., Poltz R., Ferrari E., Villani G., Hübscher U. (2007) Nucleic Acids Res. 35, 2596–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venclovas C., Thelen M. P. (2000) Nucleic Acids Res. 28, 2481–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldovan G. L., Pfander B., Jentsch S. (2007) Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 12.Shi G., Chang D. Y., Cheng C. C., Guan X., Venclovas C., Lu A. L. (2006) Biochem. J. 400, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowman G. D., O'Donnell M., Kuriyan J. (2004) Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]
- 14.Zhu D. Y., Zhu Y. Q., Xiang Y., Wang D. C. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 772–775 [DOI] [PubMed] [Google Scholar]
- 15.Heras B., Martin J. L. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17.Winn M. D. (2003) J. Synchrotron Radiat. 10, 23–25 [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger T. (2004) J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abrahams J. P., Leslie A. G. (1996) Acta Crystallogr. D. Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 24.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 25.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. (2005) EMBO J. 24, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toueille M., El-Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M. O., Hübscher U. (2004) Nucleic Acids Res. 32, 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Xie B., Zhou Y., Rahmeh A., Trusa S., Zhang S., Gao Y., Lee E. Y., Lee M. Y. (2006) J. Biol. Chem. 281, 14748–14755 [DOI] [PubMed] [Google Scholar]
- 28.Xu H., Zhang P., Liu L., Lee M. Y. (2001) Biochemistry 40, 4512–4520 [DOI] [PubMed] [Google Scholar]
- 29.Waga S., Hannon G. J., Beach D., Stillman B. (1994) Nature 369, 574–578 [DOI] [PubMed] [Google Scholar]
- 30.Gulbis J. M., Kelman Z., Hurwitz J., O'Donnell M., Kuriyan J. (1996) Cell 87, 297–306 [DOI] [PubMed] [Google Scholar]
- 31.Rauen M., Burtelow M. A., Dufault V. M., Karnitz L. M. (2000) J. Biol. Chem. 275, 29767–29771 [DOI] [PubMed] [Google Scholar]
- 32.Parrilla-Castellar E. R., Arlander S. J., Karnitz L. (2004) DNA Repair 3, 1009–1014 [DOI] [PubMed] [Google Scholar]
- 33.Xu X., Vaithiyalingam S., Glick G. G., Mordes D. A., Chazin W. J., Cortez D. (2008) Mol. Cell. Biol. 28, 7345–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bessho T., Sancar A. (2000) J. Biol. Chem. 275, 7451–7454 [DOI] [PubMed] [Google Scholar]
- 35.Parker A. E., Van de Weyer I., Laus M. C., Oostveen I., Yon J., Verhasselt P., Luyten W. H. (1998) J. Biol. Chem. 273, 18332–18339 [DOI] [PubMed] [Google Scholar]
- 36.Doré A. S., Kilkenny M. L., Rzechorzek N. J., Pearl L. H. (2009) Mol. Cell, in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.