Abstract
The human DLX homeobox genes, which are related to Dll (Drosophila distal-less gene), encode transcription factors that are expressed primarily in embryonic development. Recently, DLX5 was reported to act as an oncogene in lymphomas and lung cancers, although the mechanism is not known. The identification of target genes of DLX5 can facilitate our understanding of oncogenic mechanisms driven by overexpression of DLX5. The MYC oncogene is aberrantly expressed in many human cancers and regulates transcription of numerous target genes involved in tumorigenesis. Here we demonstrate by luciferase assay that the MYC promoter is specifically activated by overexpression of DLX5 and that two DLX5 binding sites in the MYC promoter are important for transcriptional activation of MYC. We also show that DLX5 binds to the MYC promoter both in vitro and in vivo and that transfection of a DLX5 expression plasmid promotes the expression of MYC in a dose-dependent manner in mammalian cells. Furthermore, overexpression of DLX5 results in increased cell proliferation by up-regulating MYC. Knockdown of DLX5 in lung cancer cells overexpressing DLX5 resulted in decreased expression of MYC and reduced cell proliferation, which was rescued by overexpression of MYC. Because DLX5 has a restricted pattern of expression in adult tissues, it may serve as a potential therapeutic target for the treatment of cancers that overexpress DLX5.
The MYC protooncogene encodes a DNA-binding factor that can activate or repress transcription. MYC regulates expression of numerous target genes that control key cellular functions, including cell proliferation (1, 2). Deregulation of MYC has been observed in most human cancers (3–7). MYC activation can result from gene amplification or various chromosomal rearrangements to promote oncogenesis (6–11). The MYC promoter can also be regulated by various transcription factors (12–14). Studies in mouse models of lymphoma, osteogenic sarcoma, lung cancer, and pancreatic tumors have shown that Myc is a potentially important therapeutic target (15–19). For example, brief inactivation of Myc in osteogenic sarcoma cells resulted in sustained regression of tumors (19). However, the application of a small molecule inhibitor of MYC in human cancer cells has limited effectiveness (20, 21).
Homeobox genes are characterized by a 180-base pair DNA segment encoding a highly conserved 60-amino acid DNA-binding domain. The homeobox domain interacts with DNA elements containing a TAAT core motif. It is estimated that the human genome includes more than 200 homeobox genes (22). These genes play important roles in various developmental processes (23, 24). A growing number of homeobox genes have been shown to be deregulated in various human cancers and have the potential to serve as therapeutic targets (25–27). Among the homeobox gene superfamily, DLX (distal-less homeobox) genes, related to the distal-less gene (Dll) of Drosophila, are expressed primarily in the developing forebrain and craniofacial structures. In humans, there are six distal-less homeobox (DLX) genes, represented by three bigene clusters, DLX1/DLX2, DLX5/DLX6, and DLX3/DLX7.
The DLX5/DLX6 locus was implicated in split hand/split foot malformation (28, 29). Mice lacking Dlx5 and Dlx6 die shortly after birth and are characterized by complex defects in craniofacial, vestibular, and bone structures (24, 30). Overexpression of Dlx5 in mouse osteoblastic cells can induce osteoblast differentiation resembling the phenotype of cells incubated with the bone morphogenetic protein BMP4 (31, 32). Various Dlx5 target genes involved in osteoblast differentiation have been identified (33–35). For example, Dlx5 can up-regulate the expression of the ALP (alkaline phosphatase) gene either directly by binding to the ALP promoter or indirectly by stimulating Runx2 expression (34).
Recently, Dlx5 was reported to be overexpressed in a subset of lymphomas and lung cancers. We reported that expression of Dlx5 could be up-regulated as a result of a novel recurrent chromosome 6 inversion, inv(6), seen in spontaneous thymic lymphomas from Lck-Myr-Akt22 transgenic mice, in which the Lck promoter is used to drive expression of a myristoylated, constitutively active form of Akt2 in the early stages of thymocyte development (36). Moreover, knockdown of Dlx5 was shown to result in decreased cell proliferation in lymphoma cells harboring an inv(6). Another group reported overexpression of DLX5 in most human lung cancers (37). DLX5 protein was present mainly in the nucleus, suggesting that DLX5 could play an important role in transcriptional regulation and thereby directly or indirectly transactivate various downstream genes in lung cancer cells. Intriguingly, inhibition of DLX7 in erythroleukemia cells leads to a reduction in MYC expression (38). Here we report that DLX5 directly binds the MYC promoter and can promote tumor cell proliferation by up-regulating MYC.
EXPERIMENTAL PROCEDURES
Cell Line and Reagents
Jurkat cells and NCI-H322M cells were cultured in RPMI containing 10% fetal bovine serum. HeLa cells and HEK293 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. DLX5 and ODC antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). MYC antibody was from Cell Signaling Technology. Antibody against β-actin was from Sigma. Validated Stealth RNAiTM siRNA against MYC, DLX5 Stealth Select RNAiTM, and non-targeting siRNA were from Invitrogen.
Plasmids and Site-directed Mutations
Luciferase reporter plasmids driven by a MYC promoter (i.e. pGL2-XNM (dubbed XNM-Luc herein) and pGL2-SNM (SNM-Luc)) were gifts from Dr. Mark D. Minden (12). pcDNA3.1-DLX5 was generated by cloning the cDNA of human DLX5 into EcoRI and BamHI of pcDNA3.1. Primers used to make DLX5 binding site mutations are indicated in the text. A site-directed mutation kit from Stratagene was employed to make mutated MYC reporter constructs.
RNA Extraction and cDNA Synthesis
RNAs for mouse samples were isolated by TRIzol (Invitrogen) from either wild-type lymphocytes or fresh T-cell lymphoma samples prepared from Lck-Myr-Akt2 transgenic mice (36). All cDNAs were synthesized by using SuperScript II reverse transcriptase (Invitrogen) with oligo(dT).
Real Time RT-PCR
Total RNA samples were treated with TURBO DNA-free (Ambion) to remove possible contaminating DNA. RNA was quantified using a Nanodrop UV spectrophotometer and reverse-transcribed using the Moloney murine leukemia virus reverse transcriptase (Ambion) with a mixture of anchored oligo(dT) and random decamers. Aliquots of cDNAs were used for PCR. Real time Taqman PCR assays were performed using an ABI 7900 HT instrument. The primers and dual labeled 6FAM and BHQ1 (Black Hole Quencher) probes for human DLX5, mouse Dlx5, and actin were designed using Primer ExpressTM version 1.5 software from Applied Biosystems. Assays-on-Demand from Applied Biosystems were used for human ACTIN (assay ID number Hs99999903_m1), human MYC (assay ID number Hs00153408_m1), and mouse Myc (assay ID number, Mm00487803_m1). PCR master mix from Applied Biosystems was used for PCR. Cycling conditions were 95 °C for 15 min followed by 40 (two-step) cycles (95 °C for 15 s, 60 °C for 60 s). A 4-fold dilution standard curve established with a calibrator sample was used to convert the Ct values into quantities. For each sample, the values were averages from two PCRs performed with two different amounts of total RNA in the RT reaction. Primer sequences for human DLX5 were as follows: forward primer, 5′-CAACTTTGCCCGAGTCTTCA-3′; reverse primer, 5′-GTTGAGAGCTTTGCCATAGGAA-3′; probe, 5′-6FAM-CTACCGATTCTGACTACTACAGCCCTACGGG-BHQ1-3′. Primer sequences for mouse Dlx5 were as follows: forward primer, 5′-CCGCTTTACAGAGAAGGTTTCA-3′; reverse primer, 5′-TCTTCTTGATCTTGGATCTTTTGTT-3′; probe, 5′-6FAM-ACTCAGTACCTCGCCCTGCCAGAAC-BHQ1-3′. Primer sequences for mouse Actin were as follows: forward primer, 5′-CCAGCAGATGTGGATCAGCA-3′; reverse primer, 5′-CTTGCGGTGCACGATGG-3′; probe, 5′-6FAM-CAGGAGTACGATGAGTCCGGCCCC-BHQ1-3′.
Transient Transfection
An Amaxa Nucleofector kit V was used to transfect the DLX5 plasmid into Jurkat cells, using the manufacturer's protocol. Briefly, 5 × 106 cells were centrifuged in 15-ml conical tubes. Cell pellets were suspended in Nucleofector solution V with DLX5 plasmids (or empty vector) in the indicated amounts. The mixture was then transferred to a cuvette and processed using program X-001 of the Amaxa Nucleofector II system. Cells were transferred immediately into prewarmed media using pipettes provided by the manufacturer and then placed in culture. Cells were harvested at the indicated time point(s) with either radioimmune precipitation assay lysis buffer for protein analysis or the RNeasy Plus mini kit (Qiagen) for RNA extraction.
Luciferase Assay
HeLa cells or HEK293 cells at 90% confluence were washed with PBS once and trypsinized. Then 2 × 106 cells were centrifuged at 1,200 rpm for 5 min. Cell pellets were suspended in 100 μl of Nucleofector solution R (Amaxa, Nucleofector kit R) with plasmids pCMV-Renilla luciferase (0.5 μg; Promega), pGL2-XNM-Luc, or pGL2 SNM-Luc (4 μg each) and pcDNA3.1-DLX5 or pcDNA3.1 (4 μg each). All samples were processed using program I-13 and immediately transferred into 6 ml of warm culture media. Each 6-ml sample was evenly plated into three wells of a 6-well plate (2 ml/well). 12 h following transfection, the media were replaced with normal media. Cell extracts were collected in 200 μl of 1× Passive Lysis Buffer 48 h after transfection. Luciferase activity was evaluated using a dual luciferase reporter assay system (Promega).
ChIP Assay
ChIP assays were performed using the chromatin immunoprecipitation assay kit from Upstate Biotechnology, Inc. (Lake Placid, NY). Briefly, HeLa cells were transfected with pcDNA3.1-DLX5. After 48 h, these cells (or untransfected NCI-H322M cells, which overexpress DLX5) were used for cross-linking of histones to DNA by the addition of formaldehyde. Cells were then washed and centrifuged, and then the cell pellet was suspended in SDS lysis buffer. The lysates were sonicated to shear DNA into fragments of 200–1000 base pairs. The samples were diluted 10-fold in ChIP Dilution Buffer, and the sheared chromatin was precleared with salmon sperm DNA/protein A-agarose beads and subjected to immunoprecipitation with 2 μg of either anti-DLX5 (C-20) antibody or goat IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) overnight at 4 °C. Then 60 μl of protein A-agarose/salmon sperm DNA (50% slurry) was added; after rotation for 1 h at 4 °C, the antibody-histone complex was collected. The immunoprecipitated chromatin complexes were washed and eluted with freshly prepared elution buffer. Following the reversal of cross-links, the DNA was extracted, resuspended in water, and used as template for subsequent PCRs. The primer sequences for endogenous human MYC promoter were as follows: forward primer, 5′-TCTCCGCCCACCGGCCCTTTATAATGCGA-3′; reverse primer, 5′-CCGTTCTTTTTCCCGCCAAGC-3′. The primer sequences for upstream negative control were as follows: forward primer, 5′-AACTCCTCTTTCTTCGGACCT-3′; reverse primer, 5′-ACCAATCGCTATGCTGGATT-3′. The primer sequences for downstream negative control were as follows: forward primer, 5′-TTCACTAAGTGCGTCTCCGA-3′, reverse primer, 5′-TCCTGTTGGTGAAGCTAACG-3′. PCR products were checked on agarose gel and confirmed by sequencing.
Gel Shift Assay
Gel shift assays were performed by using a kit from Promega. Recombinant protein DLX5 (Prosci) was incubated in a reaction containing 1× gel shift binding buffer in a volume of 9 μl for 10 min, followed by the addition of 1 μl of 32P-labeled duplex oligonucleotide probes corresponding to the MYC promoter region (as shown under “Results”). The reactions were incubated at room temperature for 20 min, and then 2 μl of 5× TBE loading buffer was added to each reaction. The samples were loaded onto a 6% DNA retardation gel (Invitrogen) and run at 300 V for 15 min. The gel was then dried and exposed to x-ray film.
Immunoblot Analysis
Whole cell extract proteins (20 μg) were separated electrophoretically on 4–12% Novex Tris-glycine polyacrylamide gels (Invitrogen). Proteins were transferred to Immobilon-P membrane (Millipore), blocked in 5% nonfat milk with 0.1% Tween 20 in phosphate-buffered saline. Membranes were probed with different antibodies, as indicated. Immunoreactive bands were visualized using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences) and exposed to x-ray film or captured by the FluorchemTM SP imaging system (Alpha Innotech). All immunoblot experiments were performed at least three times.
RESULTS
Expression of Dlx5 Correlates with Myc Expression in Lck-Myr-Akt2 Lymphomas
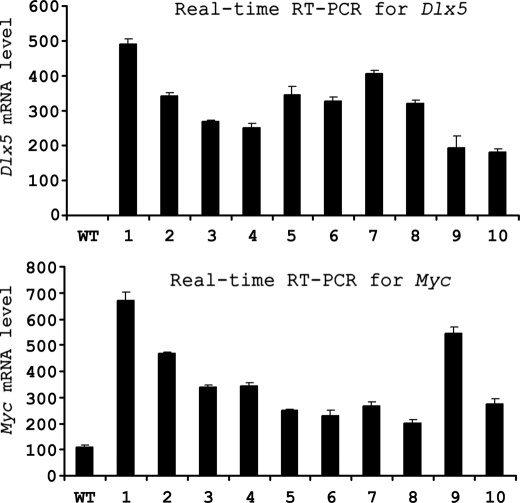
In previous studies of thymic lymphomas from Lck-Myr-Akt2 mice, we reported a novel recurrent inv(6) that leads to robust expression of Dlx5 (36). Interestingly, Myc expression was consistently up-regulated in tumors from mice harboring the inv(6) (Fig. 1). Based on this observation, we hypothesized that Myc expression may be regulated, at least in part, by Dlx5. To test this hypothesis, we performed the following series of experiments.
FIGURE 1.
Expression of Dlx5 and Myc in inv(6)-positive T-cell lymphomas from Lck-Myr-Akt2 transgenic mice. Real time RT-PCR analysis was performed with total RNA samples extracted from wild-type T cells (WT) or T-cell lymphomas from 10 different mice represented by numbers 1–10. Values were normalized against expression of Actin.
The MYC Promoter Is Directly Activated by Overexpression of DLX5
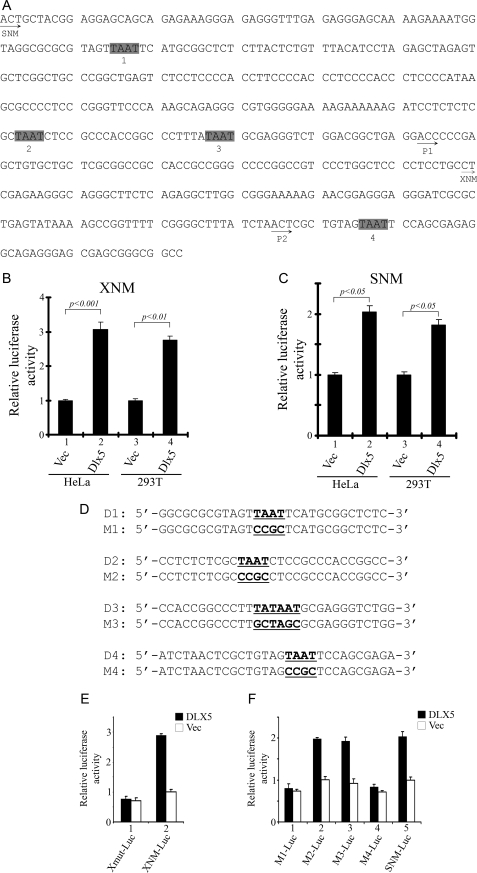
We initially analyzed the conventional MYC promoter (12), and we identified four putative homeobox binding sites having the core binding sequence TAAT. In this report, these sites are designated D1, D2, D3, and D4 (Fig. 2, A and D), and the mutated binding sites are referred to as M1, M2, M3, and M4, respectively (Fig. 2D). The luciferase constructs (dubbed SNM-Luc and XNM-Luc) used in this investigation for the MYC promoter activity assay have been reported elsewhere (12). SNM-Luc contains the entire conventional MYC promoter region, including the major transcription start sites P1 and P2 (39), whereas XNM-Luc contains only the P2 site. In HeLa cells, transient transfection of DLX5 together with XNM-Luc or SNM-Luc resulted in a 3- or 2-fold increase, respectively, in MYC promoter activity (Fig. 2B). In HEK293T cells, co-transfection of DLX5 with XNM-Luc or SNM-Luc resulted in a 2.7- or 1.8-fold increase, respectively, in MYC promoter activity (Fig. 2, B and C). These data suggest that DLX5 can directly regulate MYC transcription.
FIGURE 2.
DLX5 regulates MYC transcription in vitro. A, annotation of the MYC promoter. SNM and XNM, sequences inserted in SNM-Luc and XNM-Luc reporter vectors, respectively. The MYC promoter contains four putative DLX5 binding sites with the core motif of TAAT. P1 and P2 sites are major transcription start sites. B and C, luciferase activity in cells transfected with XNM-Luc or SNM-Luc, respectively, normalized by Renilla luciferase. pcDNA3.1 (Vec) in samples 1 and 3 or pcDNA3.1-DLX5 (DLX5) in samples 2 and 4 was co-transfected with luciferase reporter constructs and Renilla luciferase plasmid. Statistical analysis was performed by a two-tailed t test. D, MYC promoter reporter constructs bearing four different DLX5 binding sites were subjected to site-directed mutagenesis to substitute putative DLX5 binding sequences with the designated sequences (underlined). E, promoter activity measured using XNM-Luc or XNM-M4-Luc (Xmut-Luc), which contains a mutation in the D4 binding site of DLX5 in HeLa cells. Luciferase activity was normalized by Renilla luciferase. F, promoter activity of four different SNM-Luc mutant vectors (M1-Luc, M2-Luc, M3-Luc, and M4-Luc) containing different mutations in putative DLX5 binding sites in SNM-Luc in HeLa cells. Luciferase activity was normalized by Renilla luciferase.
Our site-directed mutagenesis studies demonstrated that mutations in D1 and D4 binding sites can inhibit MYC transcriptional activation (Fig. 2, E and F). MYC promoter activity was assayed by transient transfection of DLX5 together with XNM-Luc or SNM-Luc. Interestingly, when the D4 binding site in either XNM-Luc or SNM-Luc was mutated, the MYC promoter did not respond to overexpression of DLX5. Likewise, when the MYC promoter was mutated from D1 to M1 in the SNM-Luc construct, impaired transcription was observed, whereas M2 and M3 mutations did not inhibit MYC promoter activity induced by DLX5. Collectively, these data suggest that the binding sites D1 and D4, but not D2 and D3, are important for the maintenance of MYC promoter activation in response to DLX5.
DLX5 Binds to the MYC Promoter in Vitro and in Vivo
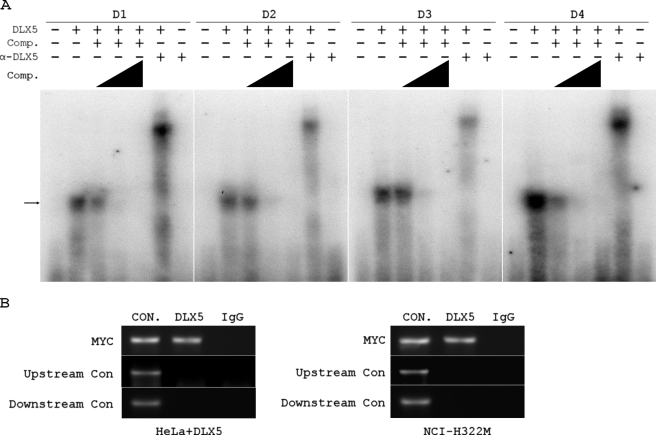
To investigate whether DLX5 can directly bind to the MYC promoter, we employed a gel shift assay and a ChIP assay. Individual DNA probes corresponding to the four DLX5 binding sites within the MYC promoter region (Fig. 2D) were used to perform the gel shift assay. These probes were labeled by ATP (α-32P) and incubated with recombinant human DLX5 protein. Each probe was found to bind to recombinant DLX5 protein, and the binding was competed by an excess molar concentration of unlabeled probe. When an anti-DLX5 antibody together with recombinant human DLX5 protein were added into the reaction, we detected a supershift of the DNA probe. No band was detected by adding anti-DLX5 antibody alone into the reaction, indicating that the supershift is DLX5-specific. These results imply that DLX5 binds to the MYC promoter in vitro (Fig. 3A). In addition, ChIP assays were carried out in HeLa cells transiently transfected with DLX5 or in NCI-H322M cells, which express high endogenous levels of DLX5. Following ChIP with a DLX5 antibody, PCR analysis using specific primers against the MYC promoter revealed a single PCR amplification product. Sequence analysis confirmed that the PCR products represented a portion of the MYC promoter. No PCR band was detected when goat IgG was used in the ChIP assay. Importantly, PCR analysis using specific primers against upstream or downstream sequences of the MYC promoter did not show bands in ChIP samples (Fig. 3B). Thus, these findings suggest that DLX5 can directly bind to the MYC promoter in vivo.
FIGURE 3.
DLX5 binds to MYC promoter in vitro and in vivo. A, gel shift assays were performed using recombinant human DLX5 protein and radiolabeled oligonucleotides. DLX5 binding could be competed by excess molar concentrations (25-, 50-, or 250-fold excess) of unlabeled probes, as indicated by the triangles. Supershift was observed by adding a DLX5-specific antibody (α-DLX5) into each reaction. The arrow indicates oligonucleotides binding to DLX5. B, ChIP assay carried out using ChIP-grade antibody against DLX5 (or normal goat IgG as a negative control) with HeLa cells transiently transfected with DLX5 or NCI-H322M cells. PCR products represent a portion of the MYC promoter (MYC). The upstream control (Upstream Con) represents a portion of the upstream MYC promoter containing nucleotides −2305 to −2015. The second control (Downstream Con) represents a downstream sequence containing nucleotides 1801–2062. Nucleotide numbers are relative to the P2 transcription start site (+1) in the MYC promoter. CON, positive control using total genomic DNA as template; DLX5, PCR products amplified from the ChIP samples mediated by anti-DLX5 antibody; IgG, absence of PCR product corresponding to MYC promoter in sample mediated by IgG.
Overexpression of DLX5 Up-regulates MYC Expression in a Dose-dependent Manner
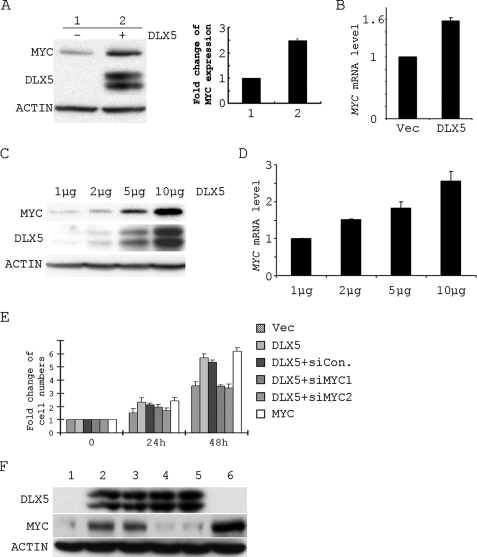
To test whether DLX5 can regulate MYC expression, we performed a series of transient transfection experiments. In Jurkat cells, overexpression of DLX5 resulted in a 2.5-fold increase in the expression of MYC protein (Fig. 4A) as well as a 1.6-fold increase in MYC mRNA. These findings suggest that expression of DLX5 can up-regulate MYC expression. To further confirm that DLX5 can regulate the expression of MYC, we tested MYC expression in Jurkat cells transfected with increasing amounts of DLX5 expression plasmids. Parallel increases in MYC protein levels (Fig. 4C) and MYC mRNA levels (Fig. 4D) were observed. These data indicate that up-regulation of MYC correlates with expression of DLX5.
FIGURE 4.
DLX5 promotes Jurkat cell proliferation by regulating the expression of MYC. A, expression of MYC protein in Jurkat cells transfected by vector (1) or DLX5 (2). The histogram in A represents the density of the MYC bands in immunoblot. B, real time RT-PCR analysis of MYC mRNA levels in Jurkat cells transfected by vector and DLX5. C, expression of MYC in Jurkat cells transfected with increasing amounts of DLX5 plasmid. D, real time RT-PCR analysis of MYC mRNA level in Jurkat cells transfected with increasing amounts of DLX5 plasmid. E, cell numbers counted at 0, 24, and 48 h in Jurkat cells transfected with vector (Vec), DLX5, DLX5 + non-targeting siRNA (siCon), DLX5 + siRNA1 against MYC (siMYC1), DLX5 + siRNA2 against MYC (siMYC2), and MYC. F, expression of MYC, DLX5, and β-ACTIN in Jurkat cells transfected with Vec (1), DLX5 (2), DLX5 + non-targeting siRNA (3); DLX5 + siRNA1 against MYC (4), DLX5 + siRNA2 against MYC (5), and MYC (6).
DLX5 Accelerates Jurkat Cell Proliferation by Regulating the Expression of MYC
We next tested whether expression of DLX5 can promote cell proliferation by directly regulating the expression of MYC. Transient transfection of DLX5 into Jurkat cells resulted in a ∼2.5-fold or 6-fold increase in cell proliferation at 24 or 48 h, respectively. However, when cells were transfected with both DLX5 and siRNAs against MYC, cell proliferation increased only about 1.5- or 3.5-fold at 24 or 48 h, respectively, which was similar to that of cells transfected with empty vector alone. On the other hand, when the cells were transfected with both DLX5 and non-targeting siRNA, the cell proliferation rate was similar to that observed with DLX5 alone. Transient transfection of MYC into Jurkat cells led to a similar enhancement in cell proliferation (Fig. 4E).
Immunoblot analysis demonstrated that MYC expression can be significantly up-regulated by transient transfection of DLX5 in Jurkat cells. When Jurkat cells were transfected with both DLX5 and siRNA against MYC, the expression level of MYC decreased to about the same level seen in cells transfected with vector alone, whereas in cells co-transfected with DLX5 and non-targeting siRNA, expression of MYC was comparable with that observed in cells transfected with DLX5 alone (Fig. 5F). Thus, these data strongly suggest that DLX5 accelerates cell proliferation by regulating the expression of MYC.
FIGURE 5.
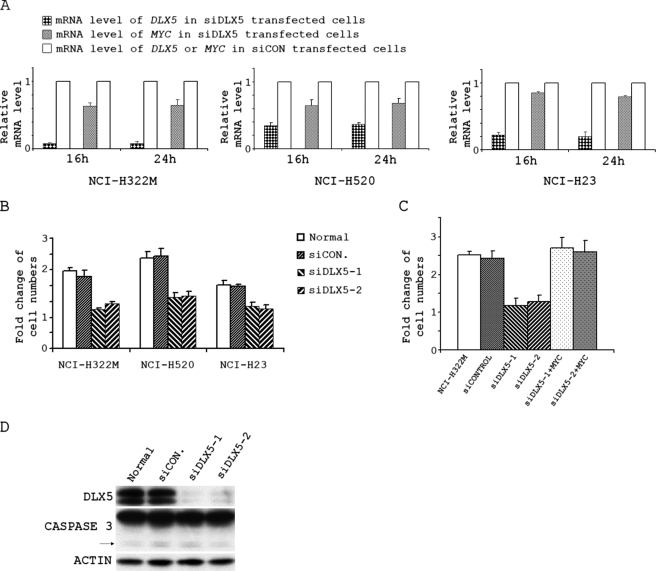
DLX5 promotes proliferation of lung cancer cells by regulating the expression of MYC. A, real time RT-PCR analyses of -fold change of DLX5 and MYC mRNA levels in NCI-H322M, NCI-H520, or NCI-H23 lung cancer cells transfected with siRNA against DLX5 (siDLX5; Invitrogen Oligo ID, HSS102810) or non-targeting siRNA (siCON) for 16 h or 24 h. The DLX5 and MYC mRNA levels were normalized against the respective DLX5 and MYC mRNA levels observed in cells transfected with control non-targeting siRNA. B, the same number of cells were transfected with buffer (Normal), non-targeting siRNA (siCON), siRNA1 against DLX5 (siDLX5-1; Invitrogen Oligo ID, HSS102808), or siRNA2 against DLX5 (siDLX5-2; Invitrogen oligo ID, HSS102810) and counted 48 h after transfection. C, cell numbers counted at 48 h in NCI-H322M cells transfected with buffer (NCI-H322M), non-targeting siRNA (siCONTROL), siRNA1 against DLX5 (siDLX5-1), siRNA2 against DLX5 (siDLX5-2), siRNA1 against DLX5 + pcDNA3-MYC (siDLX5-1 + MYC), or siRNA2 against DLX5 + pcDNA3-MYC (siDLX5-2 + MYC). D, expression of DLX5, CASPASE 3, and β-ACTIN in NCI-H322M cells transfected with buffer (Normal), non-targeting siRNA (siCON), siRNA1 against DLX5 (siDLX5-1), or siRNA2 against DLX5 (siDLX5-2). The arrow points to the bands representing the cleaved form of active CASPASE 3.
DLX5 Regulates Cell Proliferation and Expression of MYC in Lung Cancer Cells
NCI-H322M, NCI-H520, and NCI-H23 human lung cancer cells express high levels of DLX5. Stealth select siRNAs against DLX5 or non-targeting siRNA from Invitrogen were transfected into these cells by using the Amaxa Nucleofector System. To test the transfection efficiency, siRNA conjugated with fluorescein was introduced into cells. After transfection for 24 h, the cells with positive fluorescence were counted under a fluorescence microscope, which revealed a transfection efficiency of nearly 100% (data not shown).
To test whether inhibition of the expression of DLX5 diminishes the expression of MYC, we performed real time RT-PCR analysis on NCI-H322M, NCI-H520, and NCI-H23 cells transfected with siRNA against DLX5. To minimize the possibility that expression of MYC could be affected by cell cycle progression, we analyzed early time points (i.e. 16 and 24 h) after transfection (Fig. 5A). Interestingly, MYC transcription was rapidly inhibited following transfection of siRNA against DLX5 in all of the three lung cancer cell lines. As shown in Fig. 5A, transfection of NCI-H322M cells with stealth siRNA against DLX5 resulted in ∼90% knockdown of DLX5 16 or 24 h after transfection, whereas MYC mRNA expression was decreased ∼40%. In NCI-H520 cells, the expression of DLX5 was diminished about 70% after transfection with siRNA against DLX5, whereas MYC mRNA expression was decreased by nearly 40%. About 80% knockdown of DLX5 and 20% decrease of MYC mRNA were observed in NCI-H23 cells.
Furthermore, knockdown of DLX5 in lung cancer cells also resulted in decreased cell proliferation (Fig. 5B). Cell proliferation was reduced by ∼50% in NCI-H322M and NCI-H520 cells following knockdown of DLX5 compared with ∼40% in NCI-H23 cells. Importantly, the reduction in cell proliferation caused by knockdown of DLX5 was rescued by overexpression of MYC (Fig. 5C). Taken together, these findings suggest that DLX5 regulates cell proliferation, at least in part, by reducing MYC expression.
DISCUSSION
The Dlx5 and Dlx6 homeobox genes are co-expressed in all skeletal structures found in midgestation embryos after the initial formation of cartilage. Subsequent studies revealed that Dlx5 can be induced by BMP and is involved in osteoblast differentiation; furthermore, knock-out studies have confirmed that Dlx5 plays an important role in bone formation (24). Recently, overexpression of DLX5 was reported in human lymphomas and lung cancers and has been proposed to act as an oncogene (36, 37). For example, we showed that Dlx5 can enhance the proliferation of JML-5 lymphoma cells as well as promote anchorage-independent growth of Rat-1 fibroblasts. Furthermore, Dlx5 was found to cooperate with activated Akt2 kinase to promote increased soft agar colony formation in Rat-1 fibroblasts (36). In non-small cell lung cancer (NSCLC) patients, overexpression of DLX5 in lung tumors correlated with a poor prognosis. Proliferation of NSCLC cells in vitro was inhibited by specific siRNA against DLX5 (37).
MYC plays pivotal roles in controlling various cellular functions, such as cell growth, proliferation, differentiation, and senescence, by regulating the expression of numerous target genes (1). Deregulated MYC expression resulting from upstream oncogenic signals leads to constitutive activation of MYC in a variety of cancers to promote tumorigenesis.
In an earlier study of thymic lymphomas from Akt2 transgenic mice, we reported overexpression of Dlx5 due to a rearrangement that places the Dlx5 locus near the Tcrb enhancer (36). An analysis of global gene expression in these tumors revealed that Myc was consistently up-regulated compared with that observed in wild-type T cells. To confirm overexpression of Myc in the thymic lymphomas, real time RT-PCR analysis was performed on fresh (uncultured) T-cell tumors and control T cells harvested from wild-type mice at the same age (Fig. 1). Since numerous oncogenic transcription factors appear to regulate the expression of Myc (12–14), we hypothesized that overexpression of Dlx5 might up-regulate Myc and accelerate lymphoma progression in our mouse model. Here, we have used luciferase reporter assays to demonstrate that DLX5 can indeed regulate MYC promoter activity (Fig. 2). The fact that DLX5 binds to the MYC promoter both in vitro (Fig. 3A) and in vivo (Fig. 3B) suggests that DLX5 regulates MYC transcriptional activity. In addition, we have identified two DLX5 binding sites, D1 and D4, which appear to be important in regulating MYC expression. As shown in Fig. 4, A and B, transient transfection of DLX5 in Jurkat results in significant up-regulation of MYC. Furthermore, we observed that in Jurkat cells, expression of the ornithine decarboxylase gene, ODC1, a transcriptional target of MYC, was also up-regulated by DLX5 (data not shown). Moreover, DLX5 regulates the expression of MYC in a dose-dependent manner (Fig. 4, C and D). Therefore, our findings indicate that DLX5 can specifically bind to the MYC promoter, resulting in elevated expression of MYC.
Interestingly, immunohistochemical analysis of NSCLCs showed that positive staining for DLX5 correlates with increased tumor size and poor prognosis and was an independent prognostic factor (37). Moreover, treatment of NSCLC cells with siRNAs against DLX5 effectively knocked down its expression and suppressed cell proliferation. Intriguingly, it is reported that overexpression of MYC is very common in NSCLC (40). We identified three NSCLC cell lines, NCI-H322M, NCI-H520, and NCI-H23, with high expression of DLX5, and knockdown of DLX5 in these cells resulted in a decreased expression of MYC and decreased cell proliferation (Fig. 5, A and B). To exclude the possibility that decreased cell proliferations in siRNA-treated cells were due to apoptosis, we analyzed caspase 3 in those cells and found that the expressions of cleaved caspase 3 showed no significant difference between siRNA-transfected cells and normal cells (Fig. 5D). Since it is well known that MYC controls cellular proliferation, we had hypothesized that DLX5 might regulate cell proliferation via transcriptional regulation of MYC. Indeed, inhibition of MYC by siRNA inhibited the proliferative effects of DLX5 expression in Jurkat cells. Moreover, ectopic overexpression of MYC in Jurkat cells resulted in a similar enhancement of proliferation as DLX5 in this assay (Fig. 4, E and F). However, MYC may not be the only target of DLX5 responsible for enhanced proliferation connected with oncogenesis, because knockdown of DLX5 by 80–90% resulted in only a 20–40% decrease in MYC transcription and a ∼50% decrease in cell proliferation (Fig. 5, A and B). It is noteworthy that a growing number of homeobox genes are deregulated in cancers (25–27). Since homeobox genes share the same DNA binding motif and can have multiple targets, it is likely that the number of targets of DLX5 involved in tumorigenesis will increase. Thus, in future investigations, we plan to use ChIP-on-chip assays to identify other potential targets of DLX5 that may contribute to increased cell proliferation.
Because of its involvement in many different types of cancer, including lymphoma, lung cancer, osteogenic sarcoma, and pancreatic cancer, many investigators have conducted preclinical studies to explore the possibility of targeting Myc (15–19). However, small molecule inhibitors of MYC have had limited effectiveness in human cancer cells (20, 21). Unlike MYC, DLX5 has a restricted pattern of expression in adult tissues (37), and thus targeting DLX5 may prove to be more efficacious in the treatment of a subset of tumors that co-express elevated levels of these transcription factors.
Acknowledgments
We thank Dr. A. Bellacosa (Fox Chase Cancer Center) for helpful comments about the manuscript. We thank Dr. A. Klein-Szanto (Fox Chase Cancer Center) for providing the NCI-H520 cell line. The following Fox Chase Cancer Center shared facilities were used in the course of this work: Genomics, Cell Biology, and Biochemistry and Biotechnology.
This work was supported, in whole or in part, by National Institutes of Health Grants CA77429 and CA06927. This work was also supported by an appropriation from the Commonwealth of Pennsylvania.
- Lck-Myr-Akt2
- transgene encoding myristoylated (active) form of Akt2 kinase, driven by the Lck (lymphocyte-specific protein-tyrosine kinase) gene promoter
- NSCLC
- non-small cell lung cancer
- siRNA
- small interfering RNA
- RT
- reverse transcription
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Dang C. V. (1999) Mol. Cell Biol. 19, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelengaris S., Khan M. (2003) Arch. Biochem. Biophys. 416, 129–136 [DOI] [PubMed] [Google Scholar]
- 3.Sikora K., Evan G., Stewart J., Watson J. V. (1985) Br. J. Cancer 52, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada H., Sakamoto H., Taira M., Nishimura S., Shimosato Y., Terada M., Sugimura T. (1986) Jpn. J. Cancer Res. 77, 370–375 [PubMed] [Google Scholar]
- 5.Sikora K., Chan S., Evan G., Gabra H., Markham N., Stewart J., Watson J. (1987) Cancer 59, 1289–1295 [DOI] [PubMed] [Google Scholar]
- 6.Dubik D., Dembinski T. C., Shiu R. P. (1987) Cancer Res. 47, 6517–6521 [PubMed] [Google Scholar]
- 7.Masters J. R., Vesey S. G., Munn C. F., Evan G. I., Watson J. V. (1988) Urol. Res. 16, 341–344 [DOI] [PubMed] [Google Scholar]
- 8.Nowell P., Finan J., Dalla-Favera R., Gallo R. C., ar-Rushdi A., Romanczuk H., Selden J. R., Emanuel B. S., Rovera G., Croce C. M. (1983) Nature 306, 494–497 [DOI] [PubMed] [Google Scholar]
- 9.Popescu N. C., Zimonjic D. B. (2002) J. Cell Mol. Med. 6, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis M., Malcolm S., Rabbitts T. H. (1984) Nature 308, 286–288 [DOI] [PubMed] [Google Scholar]
- 11.Kubokura H., Koizumi K., Yamamoto M., Tanaka S. (1999) Nippon Ika Daigaku Zasshi 66, 107–112 [DOI] [PubMed] [Google Scholar]
- 12.Han Y., San-Marina S., Liu J., Minden M. D. (2004) Oncogene 23, 6933–6941 [DOI] [PubMed] [Google Scholar]
- 13.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 14.Wierstra I., Alves J. (2006) FEBS J. 273, 4645–4667 [DOI] [PubMed] [Google Scholar]
- 15.Pelengaris S., Khan M., Evan G. I. (2002) Cell 109, 321–334 [DOI] [PubMed] [Google Scholar]
- 16.Felsher D. W., Bishop J. M. (1999) Mol. Cell 4, 199–207 [DOI] [PubMed] [Google Scholar]
- 17.Marinkovic D., Marinkovic T., Mahr B., Hess J., Wirth T. (2004) Int. J. Cancer 110, 336–342 [DOI] [PubMed] [Google Scholar]
- 18.Soucek L., Whitfield J., Martins C. P., Finch A. J., Murphy D. J., Sodir N. M., Karnezis A. N., Swigart L. B., Nasi S., Evan G. I. (2008) Nature 455, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain M., Arvanitis C., Chu K., Dewey W., Leonhardt E., Trinh M., Sundberg C. D., Bishop J. M., Felsher D. W. (2002) Science 297, 102–104 [DOI] [PubMed] [Google Scholar]
- 20.Huang M. J., Cheng Y. C., Liu C. R., Lin S., Liu H. E. (2006) Exp. Hematol. 34, 1480–1489 [DOI] [PubMed] [Google Scholar]
- 21.Lin C. P., Liu J. D., Chow J. M., Liu C. R., Liu H. E. (2007) Anticancer Drugs 18, 161–170 [DOI] [PubMed] [Google Scholar]
- 22.Tupler R., Perini G., Green M. R. (2001) Nature 409, 832–833 [DOI] [PubMed] [Google Scholar]
- 23.Kraus P., Lufkin T. (2006) Am. J. Med. Genet. A 140, 1366–1374 [DOI] [PubMed] [Google Scholar]
- 24.Robledo R. F., Rajan L., Li X., Lufkin T. (2002) Genes Dev. 16, 1089–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abate-Shen C. (2002) Nat. Rev. Cancer 2, 777–785 [DOI] [PubMed] [Google Scholar]
- 26.Samuel S., Naora H. (2005) Eur. J. Cancer 41, 2428–2437 [DOI] [PubMed] [Google Scholar]
- 27.Hennessy B. T., Mills G. B. (2006) Int. J. Biochem. Cell Biol. 38, 1450–1456 [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K., Haraguchi R., Ogata T., Barbieri O., Alegria O., Vieux-Rochas M., Nakagata N., Ito M., Mills A. A., Kurita T., Levi G., Yamada G. (2008) Eur. J. Hum. Genet. 16, 36–44 [DOI] [PubMed] [Google Scholar]
- 29.Scherer S. W., Poorkaj P., Massa H., Soder S., Allen T., Nunes M., Geshuri D., Wong E., Belloni E., Little S., Zhou L., Becker D., Kere J., Ignatius J., Niikawa N., Fukushima Y., Hasegawa T., Weissenbach J., Boncinelli E., Trask B., Tsui L. C., Evans J. P. (1994) Hum. Mol. Genet. 3, 1345–1354 [DOI] [PubMed] [Google Scholar]
- 30.Depew M. J., Lufkin T., Rubenstein J. L. (2002) Science 298, 381–385 [DOI] [PubMed] [Google Scholar]
- 31.Miyama K., Yamada G., Yamamoto T. S., Takagi C., Miyado K., Sakai M., Ueno N., Shibuya H. (1999) Dev. Biol. 208, 123–133 [DOI] [PubMed] [Google Scholar]
- 32.Newberry E. P., Latifi T., Towler D. A. (1998) Biochemistry 37, 16360–16368 [DOI] [PubMed] [Google Scholar]
- 33.Benson M. D., Bargeon J. L., Xiao G., Thomas P. E., Kim A., Cui Y., Franceschi R. T. (2000) J. Biol. Chem. 275, 13907–13917 [DOI] [PubMed] [Google Scholar]
- 34.Kim Y. J., Lee M. H., Wozney J. M., Cho J. Y., Ryoo H. M. (2004) J. Biol. Chem. 279, 50773–50780 [DOI] [PubMed] [Google Scholar]
- 35.Lee M. H., Kim Y. J., Yoon W. J., Kim J. I., Kim B. G., Hwang Y. S., Wozney J. M., Chi X. Z., Bae S. C., Choi K. Y., Cho J. Y., Choi J. Y., Ryoo H. M. (2005) J. Biol. Chem. 280, 35579–35587 [DOI] [PubMed] [Google Scholar]
- 36.Tan Y., Timakhov R. A., Rao M., Altomare D. A., Xu J., Liu Z., Gao Q., Jhanwar S. C., Di Cristofano A., Wiest D. L., Knepper J. E., Testa J. R. (2008) Cancer Res. 68, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 37.Kato T., Sato N., Takano A., Miyamoto M., Nishimura H., Tsuchiya E., Kondo S., Nakamura Y., Daigo Y. (2008) Clin. Cancer Res. 14, 2363–2370 [DOI] [PubMed] [Google Scholar]
- 38.Shimamoto T., Nakamura S., Bollekens J., Ruddle F. H., Takeshita K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3245–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hay N., Bishop J. M., Levens D. (1987) Genes Dev. 1, 659–671 [DOI] [PubMed] [Google Scholar]
- 40.Cline M. J., Battifora H. (1987) Cancer 60, 2669–2674 [DOI] [PubMed] [Google Scholar]