Abstract
Our previous studies have implicated CHIP (carboxyl terminus of Hsp70-interacting protein) as a co-chaperone/ubiquitin ligase whose activities yield protection against stress-induced apoptotic events. In this report, we demonstrate a stress-dependent interaction between CHIP and Daxx (death domain-associated protein). This interaction interferes with the stress-dependent association of HIPK2 with Daxx, blocking phosphorylation of serine 46 in p53 and inhibiting the p53-dependent apoptotic program. Microarray analysis confirmed suppression of the p53-dependent transcriptional portrait in CHIP+/+ but not in CHIP−/− heat shocked mouse embryonic fibroblasts. The interaction between CHIP and Daxx results in ubiquitination of Daxx, which is then partitioned to an insoluble compartment of the cell. In vitro ubiquitination of Daxx by CHIP revealed that ubiquitin chain formation utilizes non-canonical lysine linkages associated with resistance to proteasomal degradation. The ubiquitination of Daxx by CHIP utilizes lysines 630 and 631 and competes with the sumoylation machinery of the cell at these residues. These studies implicate CHIP as a stress-dependent regulator of Daxx that counters the pro-apoptotic influence of Daxx in the cell. By abrogating p53-dependent apoptotic pathways and by ubiquitination competitive with Daxx sumoylation, CHIP integrates the proteotoxic stress response of the cell with cell cycle pathways that influence cell survival.
Death domain-associated protein (Daxx)3 is a nuclear protein active in a number of apoptotic pathways (1). In the nucleus, Daxx is found in promyelocytic leukemia oncogenic domains and in heterochromatic domains of the chromatin (2). The interaction of Daxx with a number of transcription factors generally has a repressive influence on transcriptional activity, tipping cells toward suppressed growth and enhanced apoptosis (3). Under certain stress conditions, Daxx translocates to the cytoplasm, promoting the c-Jun amino-terminal kinase (JNK)-dependent pathway of apoptosis through activation of the mitogen-activated protein (MAP) kinase kinase kinase, apoptosis signal-regulating kinase (ASK1) (4, 5).
However, other studies have indicated that Daxx has anti-apoptotic functions. The increased apoptosis found after depletion of endogenous Daxx (6) and the ability of overexpressed Daxx to enhance the heat shock transcription factor-1 (HSF1)-dependent transcriptional program (7) both support an anti-apoptotic role for this protein. That Daxx knockouts are embryonic lethals attests to its importance in normal cellular growth and development (8). Because the physiological state of the cell affects Daxx localization and Daxx binding partners, the exact role and the regulation of this molecule has been difficult to discern.
Carboxyl terminus of Hsp70-interacting protein (CHIP) is a ubiquitin ligase that is a critical regulator of proteotoxic stress through its ability to mediate degradation of misfolded proteins (9–11). CHIP provides protection against apoptosis under some circumstances (12, 13). Primarily a cytoplasmic molecule, CHIP inhibits the JNK-dependent apoptotic pathway by ubiquitinating and targeting ASK1 for proteasomal degradation (5). In the nuclear compartment, CHIP is part of the HSF1 transcriptional complex that up-regulates the production of heat shock proteins (12), which in turn interact with and suppress many apoptotic signaling molecules (14).
The counter-regulation of ASK1 by both Daxx and CHIP, the effect of CHIP on soluble Daxx distribution and steady-state levels (5), and the transcriptional regulation of HSF1 by both CHIP and Daxx (7) suggest intersections between Daxx signaling and CHIP-governed proteotoxic stress responses. These observations led us to examine more carefully the molecular relationship between CHIP and Daxx in the setting of cell stress. In this study, we describe a stress-induced interaction between Daxx and CHIP that results in reduced homeodomain-interacting protein kinase 2 (HIPK2) binding to Daxx, ubiquitination and reduced steady-state levels of cytoplasmic Daxx, and reduced stress-dependent sumoylation of Daxx. These stress-induced effects of CHIP on Daxx shift the cellular stress response toward survival pathways and away from apoptotic pathways.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture Conditions, and Transfections
COS-7 or HEK-293 cells were grown at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). CHIP+/+ and CHIP−/− primary mouse embryonic fibroblasts (MEFs) isolated from day 13.5 embryos of C57BL6 CHIP+/− crosses were grown under similar conditions with the addition of β-mercaptoethanol. Cells were transfected with FuGENE 6 reagent (Roche Applied Science) using pcDNA3 to equalize DNA. Transfection efficiencies were ≥95% as estimated by cells transfected with green fluorescent protein. Cells were treated and harvested 24 h after transfection.
Site-directed Mutagenesis
To create CHIP deletion mutants, site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis protocol (Stratagene) using appropriate oligonucleotides. The sequences of the mutated deletions were verified by DNA sequencing.
Western Blotting
For whole cell lysates, cells were harvested, washed twice in PBS, and then lysed in lysis buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 2 mm EGTA, 1 mm Na3VO4, 1× protease inhibitor mixture, 20 mm NaH2PO4, pH 7.2, 25 mm NaF, 10% glycerol, 0.50% Triton X-100, 1 mm phenylmethylsulfonyl fluoride). Cell lysates were centrifuged at 12,000 rpm for 10 min, and the supernatants were removed as the soluble fraction. The pellets were solubilized by adding lysis buffer and 10% DNase and incubating for 30 min at room temperature with mechanical trituration with a pipette. Pellet samples were then boiled for 30 min in SDS sample buffer (0.5 m Tris-HCl, pH 6.8, 30% glycerol, 10% SDS, 0.035% bromphenol blue, 7.5% β-mercaptoethanol). For cytoplasmic nuclear separation, the NER-PERTM nuclear and cytoplasmic extraction kit from Pierce was utilized. After the cytoplasmic and nuclear fractions had been removed, the remaining insoluble pellet was solubilized with radioimmune precipitation buffer and 10% DNase with mechanical trituration. 30–50 μg of protein were loaded and separated by SDS-PAGE gel electrophoresis. To preserve sumoylated species, cells were lysed in an “anti-desumoylation” buffer containing 1% SDS, 1% Nonidet P-40, 5 mm EDTA, and 1× protease inhibitor mixture in PBS heated to 95 °C and followed by sonication. Blots were probed with the appropriate dilutions of the primary antibodies: anti-FLAG (Sigma), anti-Myc (Santa Cruz Biotechnology), anti-Daxx (Santa Cruz Biotechnology), anti-CHIP (9), anti-β-actin (Sigma), anti-SUMO-1 (Upstate Cell Signaling), anti-p53 (Santa Cruz Biotechnology), anti-phospho p53 Ser-46 (Cell Signaling), anti-HIPK2 (Santa Cruz Biotechnology), anti-HA (Roche Applied Science), and anti-ubiquitin (Chemicon). All Western blots included in Figs. 1–7 are representative of at least three blots from three independent experiments with β-actin as the standard loading control.
FIGURE 1.
Endogenous levels of Daxx are higher in CHIP−/−MEFs than in CHIP+/+MEFs. A, freshly isolated MEFs from CHIP−/− or CHIP+/+ embryos were plated on coverslips. 24 h after plating, cells were left untreated or heat shocked at 43 °C for 30 min, fixed, and stained for Daxx. The rhodamine-conjugated secondary antibody labels Daxx in red, and 4′,6-diamidino-2-phenylindole stains the nuclei in blue. B, the number of Daxx speckles per nucleus was counted in 10 randomly chosen fields. CHIP−/− n = 118 nuclei, CHIP+/+ n = 158 nuclei. Student's t test p = 0.000006. C, Western blot (IB) of CHIP−/− and CHIP+/+ lysates with no treatment and immunoblotted with anti-Daxx.
FIGURE 2.
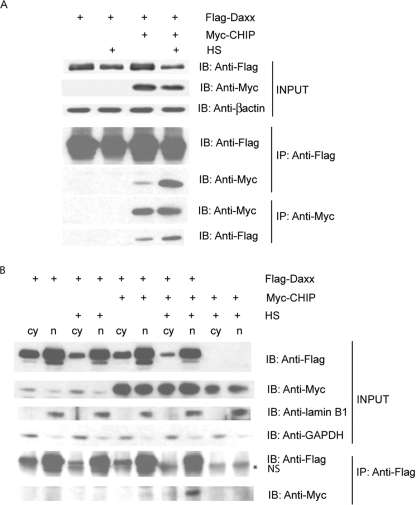
Co-expression of CHIP reduces Daxx under stress of heat shock. A, COS-7 cells were transiently transfected with FLAG-Daxx or FLAG-Daxx and Myc-CHIP. 24 h after transfection, cells were left untreated or heat shocked at 43 °C for 30 min. The cell lysates were immunoprecipitated (IP) with anti-FLAG resin for immunoprecipitating Daxx, or conversely, immunoprecipitated with anti-Myc resin for immunoprecipitating CHIP. Lysates were analyzed on Western blot (IB) with β-actin as a loading control and probed for Daxx and CHIP with anti-FLAG and anti-Myc respectively. B, COS-7 cells were transiently transfected as in A and then separated into cytoplasmic (cy) and nuclear (n) fractions. These fractions were then immunoprecipitated with anti-FLAG resin for Daxx and immunoblotted with anti-FLAG for Daxx and anti-Myc for CHIP. Immunoblotting for lamin B1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as controls for the nuclear and cytoplasmic compartments respectively. HS, heat shocked.
FIGURE 3.
The interaction between CHIP and Daxx requires specific domains. A, HEK-293 cells were transiently transfected with FLAG-Daxx WT and Myc-CHIP WT or one of the indicated Myc-CHIP deletion mutants. 24 h after transfection, cells were left untreated or heat shocked at 43 °C for 30 min, lysed, and immunoprecipitated (IP) with anti-FLAG resin for Daxx and then immunoblotted (IB) with anti-Myc for CHIP and anti-FLAG for Daxx. TPR, tetratricopeptide. B, COS-7 cells were transiently transfected with Myc-CHIP WT and FLAG-Daxx WT or the indicated FLAG-Daxx deletion mutants. 24 h after transfection, cells were treated as in A. PML, promyelocytic leukemia protein; FAS, transforming growth factor-β receptor. C, COS-7 cells were transiently transfected with FLAG-CHIP WT and the indicated Myc-Daxx deletion mutants. 24 h after transfection, the cells were treated as in A except for immunoprecipitating with anti-Myc agarose conjugate for Daxx, immunoblotting with anti-Myc for Daxx, and immunoblotting with anti-FLAG for CHIP.
FIGURE 4.
CHIP ubiquitinates Daxx under the stress of heat shock. A, FLAG-Daxx and increasing amounts of Myc-CHIP were co-expressed in COS-7 cells. 24 h after transfection, cells were left untreated or heat shocked at 43 °C for 30 min and then lysed and separated into soluble (s) and insoluble (p) fractions. IB, immunoblot. B, left panel, in vitro ubiquitination was performed with immunopurified FLAG-Daxx as substrate and the indicated components of the ubiquitination reaction system. E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; Ub, ubiquitin; 3KTR, K29R,K48R,K63R. B, right panel, in vitro ubiquitination was performed using the indicated WT and mutant ubiquitin proteins with immunopurified FLAG-Daxx as the substrate.
FIGURE 5.
Endogenous CHIP and Daxx co-immunoprecipitate with the stress of heat shock (HS). A, CHIP+/+ MEFs were left untreated or heat shocked at 43 °C for 30 min and then lysed. The lysates were immunoprecipitated with anti-Daxx, and the co-immunoprecipitating (IP) proteins were then subjected to Western blot (IB) analysis and immunoblotting with anti-Daxx, anti-CHIP, and anti-β-actin as loading control. B, CHIP+/+ MEFs were heat shocked at 43 °C, and samples were collected at 0, 10, 20, 30, 40, and 150 min from the start of heat shock. The nuclear fraction of the lysates was immunoprecipitated with agarose-conjugated anti-Daxx (Santa Cruz Biotechnology). A Western blot of the immunoprecipitated proteins was probed with anti-Daxx (Santa Cruz Biotechnology) and anti-CHIP (Chemicon). C, CHIP+/+ and CHIP−/− MEFs were treated as in A and separated into soluble (SOL) and insoluble (INSOL) fractions. The fractions were immunoblotted with anti-Daxx, anti-CHIP, and anti-β-actin as loading control. The * indicates possible sumoylated Daxx. D, CHIP+/+ and CHIP−/− MEFs treated as in A. The lysates were separated into soluble and insoluble fractions and immunoprecipitated with agarose conjugated anti-Daxx (Santa Cruz Biotechnology). A Western blot of the two fractions was probed with anti-Daxx and anti-ubiquitin (Chemicon). E, CHIP+/+ and CHIP−/− MEFs were untreated or heat shocked at 43 °C and allowed to recover for the indicated times before lysis. The lysates were immunoprecipitated with anti-Daxx resin (Santa Cruz Biotechnology), and the co-immunoprecipitating proteins were analyzed by Western blot and immunoblotted with anti-Sumo-1 (Cell Signaling), anti-Daxx and anti-β-actin.
FIGURE 6.
CHIP interferes with the stress-induced sumoylation of Daxx. A, COS-7 cells were transiently transfected with combinations of Myc-CHIP, FLAG-Daxx, and HA-SUMO-1. 24 h after transfection, the cells were left untreated or heat shocked at 43 °C for 30 min and then lysed and immunoprecipitated (IP) for 3 h with anti-FLAG-conjugated resin (Santa Cruz Biotechnology). The co-immunoprecipitating proteins were analyzed by Western blot (IB) with the indicated probes. B, COS-7 cells were transfected with FLAG-Daxx WT or FLAG-Daxx-K630A/K631A (K630/1A, a sumoylation mutant) and with Myc-CHIP. 24 h after transfection, the cells were left untreated or heat shocked at 43 °C for 30 min and then lysed. Both the soluble (s) and the insoluble (p) fractions were run on an 8% SDS gel and immunoblotted for Daxx with anti-FLAG, for CHIP with anti-Myc, and for β-actin as loading control. C, COS-7 cells treated as in B, and the soluble and insoluble fractions were immunoprecipitated with anti-FLAG for Daxx. The recovered proteins were run on an 8% SDS gel and immunoblotted for ubiquitin (Ub) and CHIP (anti-Myc).
FIGURE 7.
Under conditions of heat shock stress, CHIP interferes with HIPK2 phosphorylation of Ser-46 of p53. A, COS-7 cells were transiently transfected with FLAG-Daxx alone or with Myc-CHIP. 24 h after transfection, the cells were left untreated or heat shocked at 43 °C for 30 min. The cells were then lysed, separated into soluble and insoluble fractions, and immunoprecipitated (IP) for Daxx with anti-FLAG resin. The immunoprecipitated proteins were then run on an 8% SDS gel and probed with anti-FLAG for FLAG-Daxx, anti-HIPK2, anti-phospho p53 Ser-46, and anti-Myc for Myc-CHIP. IB, immunoblot. B, COS-7 cells were plated on slides and then transiently transfected with WT FLAG-Daxx alone or with Myc-CHIP and with the HIPK2 binding mutant FLAG-Daxx 154–740, alone or with Myc-CHIP. The cells were heat shocked for 30 min at 45 °C and then allowed to recover at 37 °C for 16 h. They were then stained, and 10 random fields were analyzed for TUNEL-positive apoptotic cells. Student's t test assuming equal variance gave p = 0.0002 for * and p = 0.003 for **. C, CHIP+/+ and CHIP−/− MEFs were heat shocked at 43 °C for 30 min and allowed to recover for the indicated times. The cell lysates were then run on a 12% SDS gel and immunoblotted for the indicated proteins.
Immunoprecipitation Analysis
Cell extracts were incubated for 3 h at 4 °C with the appropriate agarose-conjugated antibody: EZ view anti-FLAG-AG (Sigma), anti-Myc-AG (Santa Cruz Biotechnology), or anti-Daxx-AG (Santa Cruz Biotechnology). After three washes, the immunoprecipitated proteins were recovered by boiling in SDS sample buffer and analyzed by Western blot using the appropriate antibodies.
Immunofluorescence Analysis
MEFs were grown on coverslips, fixed with 3.7% formaldehyde, rinsed with PBS, blocked, and permeabilized with 1% bovine serum albumin, 0.5% Triton X-100 in PBS. They were then incubated with anti-Daxx (Santa Cruz Biotechnology) for 1.5 h, washed twice with PBS, 1% bovine serum albumin, 0.05% Triton X-100, and then incubated in rhodamine-conjugated goat anti-rabbit (Molecular Probes) for 45 min, washed three times in PBS and then washed for 5 min in 4′,6-diamidino-2-phenylindole (100 ng/ml) to stain the nuclei. The cells were then dehydrated, mounted in Prolong Gold antifade mounting medium (Invitrogen), and viewed using a Nikon Eclipse E800 upright fluorescent microscope utilizing Qcapture (QImaging Corp.) and IPLab (Scanalytics Inc.) software. Nuclei from cells in 10 random fields were scored for the number of Daxx speckles.
Daxx Purification and in Vitro Ubiquitination Analysis
COS-7 cells were transiently transfected with FLAG-Daxx. Cells were lysed (50 mm Tris, pH 7.4 with 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and protease inhibitor mixture) and then incubated with anti-FLAG resin for 1.5 h at 4 °C. After three lysis buffer washes, the resin was incubated with FLAG peptide (100 μg/ml) to chase off FLAG-Daxx. Excess FLAG peptide was cleared from the supernatant by centrifugation through a Centricon YM-30 (Millipore). In vitro ubiquitination assays were performed as described previously (15). Briefly, the immunopurified FLAG-Daxx was incubated for 2 h at 30 °C in the presence of 4 μm CHIP, 0.1 μm purified human ubiquitin-activating enzyme (E1), 2 μm UbcH5a, 50 μm ubiquitin, 2 mm Mg2+, and 5 mm ATP. The reactions were quenched by the addition of 1× loading buffer.
TUNEL Staining for Analysis of Apoptosis
COS-7 cells that had been plated on slides and then transiently transfected were analyzed for TUNEL-positive cells using the Promega DeadEnd fluorometric TUNEL system following the manufacturer's instructions. Ten randomly chosen fields were analyzed for each transfection condition.
RNA Extraction and Microarray Analysis
Total RNA was extracted from MEFs using the Qiagen RNeasy Plus mini kit. RNA integrity was verified by assay on an Agilent Bioanalyzer 2100. 500 ng of MEF total RNA were labeled with cyanine-5 CTP in a T7 transcription reaction using the Agilent low input linear RNA amplification/labeling system. Labeled cRNA from test samples was hybridized to Agilent G4122F mouse 4 × 44,000 microarray slides in the presence of equimolar concentrations of cyanine-3 CTP-labeled mouse reference RNA prepared from pools of 1-day-old mouse pups (16). Microarray data (n = 24 arrays) were loess-normalized (17, 18), and probes were filtered for features having a normalized intensity of <30 arbitrary fluorescence units in either channel. A probe was removed if <70% of the data were present across all samples. Missing data points were imputed using the k nearest neighbors algorithm (k = 3). 41,174 probes passed these filters and were subsequently used for analysis. Scripts written in the R Statistical Language and Environment (R; version 2.2.1, build r36812, release date 12/20/2005) and Perl (ActiveState Perl 5.8.1, build 807, release date 11/6/2003) were used to standardize (μ = 0, σ = 1) the data set.
Lists of differentially expressed genes were identified using the statistical analysis of microarray algorithm (17, 19–22) with a typical false discovery rate of 10% and custom R scripts written in our laboratory. Unsupervised, semisupervised, and supervised clustering analysis was performed on gene lists essentially as described (23) using Cluster (version 2.11, available from the Eisen laboratory). Heat maps of cluster analyses were visualized with Java TreeView (version 1.0.12, release date 3/14/2005). High level pathway analysis and mapping to gene ontology (Gene Ontology (GO) Home) categories and identification of predicted transcription factor binding sites were performed on gene lists using GATHER (Gene Annotation Tool to Help Explain Relationships) (24). The Chilibot (25) contextual data mining algorithm was used for text mining of the PubMed data base with selected genes and keywords. Differential expression values as a function of time were calculated, and the significance analysis of microarray (10% false discovery rate threshold) algorithm was used to identify genes that were significantly differentially expressed between 30 and 240 min in the heat shocked CHIP−/− when compared with the CHIP+/+ cells. The 30 and 240 min after heat shock treatments were then normalized by their respective unshocked controls. This gene list was subsequently subjected to higher order analysis using the GATHER algorithm to associate the differentially expressed genes with significantly over-represented predicted transcription factor binding sites (26). The complete data set is available online through the Gene Expression Omnibus (record GSE14339) at www.ncbi.nlm.nih.gov/geo.
RESULTS
Regulation of Daxx Nuclear Localization and Steady-state Soluble Daxx Levels by CHIP in MEFs
Having observed changes in Daxx localization and steady-state levels coincident with the ectopic expression of CHIP (5), we compared Daxx levels in CHIP+/+ MEFs and CHIP−/− MEFs to examine the relationship of these proteins in a more physiological setting. MEFs were grown on coverslips and left untreated or exposed to heat shock for 30 min at 43 °C and then fixed and stained for Daxx. Although the well documented speckle appearance of Daxx (27) was observed in both cell strains, there were significantly more (Student's t test p < 0.0001) and larger speckles in the CHIP−/− MEFs when compared with the CHIP+/+ MEFs (Fig. 1, A and B). Western blotting demonstrated similar differences in endogenous soluble Daxx levels between CHIP+/+ MEFs and CHIP−/− MEFs (Fig. 1C). Following heat shock, the CHIP−/− MEFs displayed much brighter Daxx fluorescence in both the nuclei and the cytoplasm when compared with CHIP+/+ MEFs. Collectively, these data suggest a CHIP-dependent regulation of distribution and abundance of endogenous Daxx under both steady-state and stress conditions.
Heat Stress-dependent Molecular Interaction between Daxx and CHIP
Having observed a decrease in soluble Daxx coincident with CHIP expression, we addressed whether a direct stress-dependent interaction occurred between Daxx and CHIP. COS-7 cells were transiently transfected either with FLAG-tagged Daxx (FLAG-Daxx) alone or with FLAG-Daxx in combination with Myc-tagged CHIP (Myc-CHIP). 24 h after transfection, the cells were left untreated or exposed to heat shock. Western blot analysis for Daxx and CHIP expression demonstrated that soluble Daxx levels were reduced in the setting of heat shock, but this effect was markedly enhanced when CHIP was co-expressed (Fig. 2A). Immunoprecipitating FLAG to pull down Daxx demonstrated that the amount of CHIP that co-immunoprecipitated with Daxx increased dramatically under the condition of heat shock when compared with that co-immunoprecipitated under control conditions. The reverse immunoprecipitation demonstrated the same increased association of Daxx with CHIP under the conditions of heat shock stress. These data indicate a stress-induced association between Daxx and CHIP correlated with a CHIP-dependent reduction in soluble Daxx. Because both CHIP and Daxx undergo subcellular relocalization in the setting of stress (12, 28, 29), the Daxx-CHIP interaction seen in the previous studies could occur in the nucleus, in the cytoplasm, or in both compartments. To explore this, we transfected cells with FLAG-Daxx and Myc-CHIP, treating them as above and then separating cellular lysates into nuclear and cytoplasmic fractions. These experiments indicated that the stress-induced Daxx-CHIP interaction occurred primarily in the nucleus (Fig. 2B), which is consistent with previous studies observing nuclear localization of CHIP after heat shock (12).
Specific Domains Required for Daxx-CHIP Interaction
We utilized both CHIP and Daxx deletion mutants to determine the specific domains necessary for the Daxx-CHIP heat shock stress interaction. CHIP has three characterized functional domains: the tetratricopeptide repeats (see Fig. 3A, TPR) at the amino terminus, required for its interaction with Hsp/Hsc70, the charged domain required for CHIP homodimerization (30), and the U-box domain at its carboxyl terminus, required for its ubiquitin ligase activity (9). HEK-293 cells were transiently transfected with full-length Daxx and either wild-type CHIP or CHIP deletions spanning each of these domains and then left untreated or heat shocked. Immunoblotting for CHIP after Daxx immunoprecipitation indicated that the charged domain of CHIP was required for the interaction with Daxx (Fig. 3A). Interestingly, deletion of the U-box of CHIP stabilized the Daxx-CHIP association under non-stressed conditions.
When cells were transfected with full-length CHIP and with wild-type Daxx or Daxx deletion mutants that isolate specific protein-protein interaction motifs, CHIP could be co-immunoprecipitated with the amino terminus of Daxx but not with the 240-amino acid stretch at the carboxyl terminus (Fig. 3B). To further define the interaction of CHIP with the amino terminus of Daxx, full-length CHIP was co-expressed with three additional Daxx deletion mutants cotransfected under the same conditions. These experiments revealed that CHIP can bind the HIPK2 interaction motif (Fig. 3C), Daxx residues 1–240, with high affinity, and residues 241–492, with lower affinity. The requirement of definite domains of both molecules for the Daxx-CHIP interaction supports a specific regulatory interaction between the two.
Daxx Is a Substrate for the Ubiquitin Ligase Activity of CHIP
The specific heat stress Daxx-CHIP interaction correlated with decreased soluble Daxx (Fig. 2A), suggested Daxx as a substrate for the ubiquitin ligase activity of CHIP. To test this possibility, we co-expressed Daxx and increasing amounts of CHIP, separating lysates into soluble and insoluble fractions (Fig. 4A). With increasing CHIP, we observed decreasing levels of soluble Daxx and increasing levels of a high molecular weight Daxx species in the insoluble fraction, changes noticeably accentuated by heat shock. Importantly, CHIP also moved into the insoluble fraction with the stress of heat shock. These observations suggested CHIP-dependent ubiquitination of Daxx. To determine whether CHIP could directly target Daxx for ubiquitination, we utilized an in vitro system to recapitulate Daxx ubiquitination. High molecular weight species of Daxx were detected only when all in vitro ubiquitin reaction components were present (Fig. 4B, left panel). These data demonstrate that Daxx can be a substrate for the ubiquitin ligase activity of CHIP. Surprisingly, we did not observe convincing recovery of soluble Daxx in our in vivo studies when heat shock was carried out in the presence of the proteasome inhibitor MG132 (data not shown), suggesting that CHIP may not be targeting Daxx to the proteasome via Lys-48-linked ubiquitination. Polyubiquitin chains with isopeptide linkages utilizing lysines other than lysine 48 have been shown to be resistant to proteasomal degradation, and CHIP can synthesize such non-canonical lysine linkages (11, 31). To determine whether Daxx could therefore be a CHIP substrate for non-canonical ubiquitination, single ubiquitin mutants K29R, K48R, K63R, and a triple mutant K29R,K48R,K63R were tested in an in vitro ubiquitination system (Fig. 4B, right panel). High molecular weight species of Daxx were found with the K29R, the K48R, and the K63R ubiquitin mutants. However, these species were completely absent when reactions were carried out with the K29R,K48R,K63R ubiquitin mutant. These results indicate that CHIP can assemble ubiquitin chains on Daxx via linkages other than Lys-48 and that Daxx can indeed be a substrate for CHIP-dependent non-canonical ubiquitination. Although such non-canonical linkages have been shown to resist proteasomal degradation (31), the biological significance of these linkages is yet to be fully elucidated. Conceivably, these linkages could result in transient regulation of the solubility and localization of Daxx.
Endogenous Daxx and CHIP Interact in Heat Shocked MEFs
To address whether or not endogenous proteins behave similarly, we first looked in MEFs to ascertain whether or not CHIP co-immunoprecipitated with Daxx under the stress of heat shock. CHIP+/+ MEFs were untreated or heat shocked for 30 min. at 43 °C. The lysate was then immunoprecipitated with anti-Daxx resin and analyzed by Western blot, revealing that endogenous CHIP co-immunoprecipitated with Daxx under the conditions of heat shock (Fig. 5A). Immunoprecipitating Daxx from the nuclear fraction of CHIP+/+ MEFs and immunoblotting for CHIP demonstrated that the Daxx-CHIP nuclear interaction commences shortly after the start of heat shock and terminates by 2 h after heat shock. This interaction caused a transient reduction of soluble Daxx that was coincident with the Daxx-CHIP interaction (Fig. 5B). Additionally, whole cell lysate soluble protein levels of Daxx were notably reduced in the CHIP+/+ MEFs after heat shock when compared with the CHIP−/− MEFs (Fig. 5C). This reduction of soluble Daxx in the CHIP+/+ MEFs correlated with a large increase of Daxx in the insoluble fraction. Notably, CHIP also moved into the insoluble fraction after heat shock. Such an increase in insoluble Daxx did not occur in the CHIP−/− MEFs. Also, the smear of high molecular weight Daxx species, visible in the insoluble fraction of the CHIP+/+ MEFs, was not seen in the CHIP−/− MEFs (Fig. 5C). To determine whether or not the high molecular weight Daxx species could be ubiquitinated Daxx, lysates from CHIP+/+ and CHIP−/− MEFs, treated as in Fig. 5C, were immunoprecipitated with agarose-conjugated anti-Daxx and analyzed by Western blotting. A ubiquitin-positive, high molecular weight species was observed after heat shock in the CHIP+/+ insoluble fraction, indicating that endogenous Daxx is ubiquitinated by CHIP after heat shock (Fig. 5D). A small amount of ubiquitin staining was present in the CHIP−/− heat shock fraction, possibly due to Cullin 3-dependent ubiquitination of Daxx (32). However, the presence of CHIP greatly increased Daxx ubiquitination after heat shock stress in these studies.
Because the CHIP−/− MEFs have a distinct Daxx-positive band not seen in the CHIP+/+ MEFs (indicated in Fig. 5C by the *), we looked to determine whether this band could be sumoylated Daxx. Because the sumoylated species of Daxx is associated with transcriptional repression that is correlated with suppressed growth and apoptosis (3, 33), we were interested in ascertaining whether or not CHIP might interfere with this modification. CHIP+/+ and CHIP−/− MEFs were heat shocked, and the lysates were immunoprecipitated with anti-Daxx resin and then immunoblotted for small ubiquitin-like modifier (SUMO-1). In the CHIP−/− MEFs, the immunoprecipitation for Daxx co-immunoprecipitated SUMO-1-positive species after heat shock (Fig. 5E, see the bar in the CHIP (−/−) 30/0 lane). These SUMO-1-positive bands were not seen in the comparable CHIP+/+ lane (Fig. 5E, compare the CHIP (+/+) and CHIP (−/−) lanes at the 30/0 time point). The time line of the appearance of these bands coincided with the nuclear Daxx-CHIP interaction portrayed in Fig. 5B, suggesting that in the CHIP+/+ MEFs, the Daxx-CHIP interaction prevents the formation of this sumoylated Daxx species. Collectively, these data suggest that CHIP ubiquitination of Daxx is competitive with Daxx sumoylation. We next investigated whether the residues known to be sumoylated were the same residues ubiquitinated by CHIP.
CHIP Competes with Sumoylation Machinery for Daxx Lysines 630 and 631
We speculated that the ubiquitination of Daxx by CHIP could compete with Daxx sumoylation as these two modifications sometimes compete for the same lysine residues (34). To test for this possibility, COS-7 cells were transiently transfected with FLAG-Daxx, with or without Myc-CHIP, and with or without HA-SUMO-1. The cells were heat shocked and allowed to recover at 37 °C for 1 h and then lysed and immunoprecipitated for Daxx with anti-FLAG-conjugated agarose. When CHIP was co-expressed with Daxx and SUMO-1, substantially less SUMO-1 co-immunoprecipitated with Daxx in the setting of heat shock (Fig. 6A), indicating that CHIP suppresses heat shock-induced Daxx sumoylation. To further investigate the interference of CHIP with stress-induced Daxx sumoylation, FLAG-Daxx WT and FLAG-Daxx K630A/K631A (a mutant that cannot be sumoylated (33, 35)) were each transiently transfected into COS-7 cells. 1 h after heat shock, the cell lysates were separated into soluble and insoluble fractions and subjected to Western blot analysis. Immunoblotting for FLAG to detect Daxx revealed a high molecular weight species in the insoluble fraction after heat shock in cells transfected with wild-type Daxx but not for the sumoylation-defective mutant (Fig. 6B). Next, lysates from COS-7 cells, treated as in Fig. 6B, were immunoprecipitated with anti-FLAG to immunoprecipitate Daxx and subsequently immunoblotted for CHIP and for ubiquitin. Although both wild-type Daxx and the K630A/K31A sumoylation mutant co-immunoprecipitated CHIP, only cells transfected with wild-type Daxx and then heat shocked stained positive for ubiquitin (Fig. 6C). These experiments support the hypothesis that the same lysines that are necessary for Daxx sumoylation are the primary residues ubiquitinated by CHIP and that in the setting of stress, CHIP and the Daxx SUMO ligase compete for these residues.
CHIP Blocks HIPK2 Binding with Daxx
Our results demonstrated that CHIP binds to Daxx at its amino terminus (Fig. 3), where HIPK2 also binds (36). Because phosphorylation of serine 46 on p53 by HIPK2 is facilitated by Daxx and leads to activation of the p53-dependent apoptotic program (37), we sought to determine what effect the Daxx-CHIP interaction might have on HIPK2 binding to Daxx and p53 serine 46 phosphorylation. COS-7 cells were transiently transfected with FLAG-Daxx or with FLAG-Daxx and Myc-CHIP. The cell lysates were collected after heat shock, separated into the soluble and insoluble fractions, and immunoprecipitated with anti-FLAG resin for 3 h to immunoprecipitate Daxx. Western blot analysis revealed that endogenous HIPK2 and p53 phospho-Ser-46 co-immunoprecipitated with Daxx after heat shock. The co-expression of CHIP reduced this co-immunoprecipitation of HIPK2 and p53 phospho-Ser-46 by Daxx (Fig. 7A). These observations demonstrated that the heat shock-induced Daxx-CHIP interaction reduces the binding of HIPK2 to Daxx and thus reduces the phosphorylation of p53, abrogating this pathway toward apoptosis. That this interaction occurred in the insoluble fraction of the cell lysate suggests that this fraction of the cell has biological relevance that deserves more investigation. Furthermore, knowing that both Daxx and p53 phospho-Ser-46 are transcriptional effectors, the possibility that the interaction occurred in the context of heterochromatin domains known to be insoluble seems possible.
To corroborate that the absence of HIPK2 binding to Daxx would have an effect on apoptosis, COS-7 cells grown on slides were transiently transfected with WT FLAG-Daxx or with a mutant that lacks the HIPK2 binding domain, FLAG-Daxx 154–740. Each of these constructs was transfected alone or with WT Myc-CHIP. The transfected cells were then heat shocked at 45 °C for 30 min and allowed to recover for 16 h. The cells were analyzed for TUNEL positivity in 10 randomly chosen fields. A significantly lower percentage of apoptotic cells was seen in the case of cells transfected with the HIPK2 binding mutant when compared with the WT Daxx transfected cells. The same significant reduction of apoptotic cells was observed when CHIP was cotransfected with Daxx (Fig. 7B). This observation supports the hypothesis that the stress-induced association of CHIP with Daxx abrogates the pathway to apoptosis by interfering with HIPK2 binding to Daxx and interfering with the apoptosis-promoting downstream effects.
Observations of endogenous molecules further support this hypothesis. We had observed less p53 serine 46 phosphorylation in CHIP+/+ MEFs when compared with CHIP−/− MEFs after UV irradiation where apoptosis is known to be mediated by Daxx (38) (data not shown). In heat shocked cells, at the 30/0 and the 30/1 time points, immunoblotting for p53 revealed more slowly migrating p53-positive bands, which we interpret to be the phosphorylated species of p53 that are formed after heat shock. Only in the CHIP−/− MEFs did one of these bands prove to stain for the phospho-Ser-46 species of p53. Additionally, at the later time points, the CHIP−/− MEFs had stronger induction of p53 than that seen in CHIP+/+ MEFs (Fig. 7C), which would be consistent with the known transcriptional up-regulation of p53 that is associated with the Ser-46 phosphorylated species. The reduced or abrogated phosphorylation of p53 serine 46 and much lower p53 induction after heat shock in the CHIP+/+ MEFs when compared with the CHIP−/− MEFs are consistent with the hypothesis that CHIP blocks the binding of HIPK2 to Daxx and therefore its effects on p53 phosphorylation. We also observed increased apoptosis in the CHIP−/− MEFS when compared with the CHIP+/+ MEFs as has been previously reported (12). Collectively, these data support a Daxx-CHIP interaction that, by blocking the HIPK2/Daxx association, prevents the phosphorylation of Ser-46 on p53 and thereby abrogates the apoptotic pathway this species supports.
Transcriptional Profiling Analysis of Heat Shocked MEFs Reveals Up-regulation of p53-responsive Genes in CHIP−/− MEFs When Compared with CHIP+/+ MEFs
Because the phospho-serine 46 species of p53 leads to sustained activation of p53, we predicted that p53-responsive genes would remain active longer in CHIP−/− MEFs than in wild-type cells. Therefore, we measured the global patterns of gene transcription in CHIP+/+ and CHIP−/− MEFs before, 30 min after, and 240 min after heat shock. Genes that were significantly and differentially changed between 30 and 240 min after heat shock between CHIP+/+ and CHIP−/− MEFs were selected for further analysis. The genes chosen were up-regulated in the CHIP−/− cells and concurrently down-regulated in the CHIP+/+ cells, or conversely, they were down-regulated in the CHIP−/− cells and concurrently up-regulated in the CHIP+/+ cells (Fig. 8A). Clustering of the predicted transcription factor binding sites demonstrated common upstream binding sites among the selected genes (Fig. 8B and supplemental Fig. 1). A number of these transcription factors are known to activate apoptotic pathways (e.g. C/EBP-homologous protein (CHOP), Cdx2, and Bach2 (39–41)), whereas AP1 and Myc/Max, which activate both proliferation and apoptotic pathways, promote apoptotic pathways under conditions of stress (42, 43). Interestingly, AP1 and the Myc/Max dimer are required for activation of the p53 promoter (44). The overall transcription binding site portrait of this gene set provides evidence of coordinated control of apoptotic pathways through multiple transcription factors, with up-regulation occurring in the CHIP−/− heat shock cells and down-regulation occurring in the CHIP+/+ heat shock cells.
FIGURE 8.
Microarray analysis of control and heat shocked MEFs revealed up-regulation of p53-responsive genes after heat shock (HS) in CHIP−/− MEFs when compared with CHIP+/+ MEFs. A, CHIP−/− and CHIP+/+ MEFs were heat shocked for 30 min at 43 °C and then allowed to recover for 30 or 240 min prior to microarray analysis. Gene expression values were normalized to untreated controls, and -fold change at 240 min relative (240R) to 30 min (30R) was calculated for CHIP+/+ and CHIP−/− MEFs. Of the genes selected, those with a false discovery rate of 10% were further filtered to select those genes that had a false discovery rate of 10% in both genotypes, and then only those genes that were differentially regulated between the two genotypes (up in knock out (KO) and down in WT or down in knock out and up in WT) were selected for further analysis. B, hypecluster heat map of differentially expressed genes. The selected genes were hierarchically clustered with transcription factor binding site data. CHOP, C/EBP-homologous protein.
Chilibot analysis of the clustered genes correlated gene expression patterns with two primary pathways, apoptosis and survival, with apoptotic pathways up-regulated in the CHIP−/− heat shocked MEFs and survival pathways down-regulated in this genotype (Fig. 8B and supplemental Fig. 1 and supplemental Data Sheets 1 and 2). Two of the most highly up-regulated genes, Bad and Hzf, are both p53-regulated genes and promote apoptosis through their effect on mitochondrial membrane permeability (45, 46). BNIP1, another up-regulated gene, also affects mitochondrial membrane permeability. Because mitochondria have been described as the central control point for apoptosis, the coordinate up-regulation of these three genes is noteworthy (47). Down-regulated genes in CHIP−/− cells are associated with cell migration and proliferation (Map4K4 (48) and Has1 (49)) and cell survival (Il6ra (50)), and some are classified as oncogenes (Pparbp (51)). The down-regulation of such genes would reduce the ability of the cell to mount an anti-apoptotic response. The Chilibot pathway analysis combined with the TRANSFAC promoter binding site analysis present a striking picture of coordinated gene regulation that promotes apoptotic pathways in the CHIP−/− cells and inhibits these pathways in the CHIP+/+ cells. That two of the most highly up-regulated genes in this list are p53-regulated genes supports our hypothesis that the absence of CHIP in the CHIP−/− cells leads to increased and sustained p53 activation and therefore increased apoptosis pathways that are normally kept in check by the stress-regulated ubiquitin ligase CHIP through its stress-dependent Daxx association.
DISCUSSION
The data presented here demonstrate that CHIP is a stress-induced regulator of Daxx. We have shown that, in the setting of heat shock, CHIP ubiquitinates Daxx, resulting in removal of Daxx from the soluble compartments of the cell. Interestingly, this reduction in soluble Daxx is transient in nature, and soluble Daxx levels return within a time period that seems too short to be newly synthesized Daxx; i.e. less than 30 min (Fig. 5B). The ability of CHIP to form non-canonical ubiquitin chain linkages on Daxx could provide a mechanism for removal of Daxx from current cellular events without targeting it for proteasomal degradation. Through the action of a deubiquitinase, Daxx could then return to the soluble compartment of the cell. A candidate Daxx deubiquitinase exists in a complex described by Tang et al. (52). This complex, comprised of Daxx, murine double minute 2 (MDM2), p53, and herpesvirus-associated ubiquitin-specific protease (HAUSP), has specificity and agility, quickly changing which components are ubiquitinated and which are deubiquitinated. The deubiquitinating enzymes in the cell have been shown to have an “editing” function that, by removing ubiquitin chains, spares proteins from proteasomal degradation and allows them to return to their normal cellular function (53–55). Non-canonical ubiquitin chain linkages that are resistant to proteasomal degradation are more susceptible to the action of deubiquitinases. Our data support a model in which CHIP enters the Daxx-MDM2-p53 complex under conditions of stress, replacing MDM2 and ubiquitinating Daxx. Upon the departure of CHIP from the complex, herpesvirus-associated ubiquitin-specific protease may deubiquitinate Daxx, allowing a return to the soluble fraction. Interestingly, previous studies have shown that p53 can be a substrate for CHIP (56, 57). Indeed, when comparing CHIP+/+ with CHIP−/− cells, in the CHIP+/+ cells, we have observed reduced p53 levels coincident with reduced Daxx levels after the stress of heat shock, after UV irradiation, and after the oxidative stress of H2O2 (data not shown). Our observation that, in the setting of heat shock-induced stress, CHIP and Daxx form a complex raises the possibility that this Daxx-CHIP interaction places CHIP in a position to ubiquitinate p53. Our data support CHIP-dependent abrogation of the Daxx-facilitated phosphorylation of serine 46 on p53 as well as the apoptotic gene program this species of p53 subsequently activates (Figs. 7 and 8). The interference of CHIP with Daxx sumoylation reveals a mechanism by which cells could escape the transcriptional repression and subsequent growth suppression that is associated with sumoylated Daxx. Our microarray heat shock data indicate up-regulation of genes in wild type cells that are pro-proliferation and pro-migration, whereas this same transcriptional program is down-regulated in the CHIP−/− cells (supplemental Fig. 1 and supplemental Data Sheets 1 and 2). These observations lead us to propose the following sequence of events: upon stress, CHIP enters the nucleus and binds to Daxx. This interaction prevents or reduces the movement of Daxx to the cytoplasm. Additionally, this interaction prevents HIPK2 binding to Daxx and phosphorylating p53, preventing the creation of a “super p53” (58). CHIP decorates Daxx with ubiquitin molecules at lysines 630 and 631, interfering with the sumoylation of Daxx and its transcriptional repressive effect. As stress ends, CHIP separates from Daxx, Daxx is deubiquitinated and returns to the soluble fraction of the cell, and CHIP returns to the cytoplasmic compartment. Our observations add to the data that place CHIP as a unique sensor of proteotoxic stress and as a key player in mounting the cellular response to stress, not only through its role in Hsf1 activation (12) but also as a key participant in the abrogation of an apoptotic response to cell stress.
Supplementary Material
Acknowledgments
We thank Dr. Tadashi Matsuda for the generous gift of the Myc-tagged Daxx deletion constructs and the FLAG-tagged WT and K630A/K631A Daxx constructs and Dr. Frank Boellmann for the generous gift of the FLAG-tagged Daxx deletion constructs. We also thank Dr. Xiaolu Yang for the WT and K630A/K631A FLAG-Daxx constructs.
This work was supported, in whole or in part, by National Institutes of Health Grants AG024282 and GM61728.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and supplemental Data Sheets 1 and 2.
The data set reported in this study has been submitted to the Gene Expression Omnibus under accession number GSE14339.
- Daxx
- death domain-associated protein
- CHIP
- carboxyl terminus of Hsc/Hsp 70-interacting protein
- ASK1
- apoptosis signal-regulating kinase 1
- HIPK2
- homeodomain-interacting protein kinase 2
- HSF1
- heat shock factor 1
- JNK
- c-Jun amino-terminal kinase
- MDM2
- murine double minute 2
- MEF
- mouse embryonic fibroblast
- SUMO-1
- small ubiquitin-like modifier 1
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling
- WT
- wild type
- C/EBP
- CCAAT/enhancer-binding protein.
REFERENCES
- 1.Salomoni P., Khelifi A. F. (2006) Trends Cell Biol. 16, 97–104 [DOI] [PubMed] [Google Scholar]
- 2.Ishov A. M., Vladimirova O. V., Maul G. G. (2004) J. Cell Sci. 117, 3807–3820 [DOI] [PubMed] [Google Scholar]
- 3.Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 4.Chang H. Y., Nishitoh H., Yang X., Ichijo H., Baltimore D. (1998) Science 281, 1860–1863 [DOI] [PubMed] [Google Scholar]
- 5.Hwang J. R., Zhang C., Patterson C. (2005) Cell Stress Chaperones 10, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L. Y., Chen J. D. (2003) Mol. Cell. Biol. 23, 7108–7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4100–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaelson J. S., Bader D., Kuo F., Kozak C., Leder P. (1999) Genes Dev. 13, 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Höhfeld J., Patterson C. (2001) Nat Cell Biol. 3, 93–96 [DOI] [PubMed] [Google Scholar]
- 11.Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J., Patterson C. (2001) J. Biol. Chem. 276, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 12.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C., Xu Z., He X. R., Michael L. H., Patterson C. (2005) Am. J. Physiol. Heart Circ Physiol 288, H2836–2842 [DOI] [PubMed] [Google Scholar]
- 14.Beere H. M. (2005) J. Clin. Invest. 115, 2633–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian S. B., McDonough H., Boellmann F., Cyr D. M., Patterson C. (2006) Nature 440, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X. R., Zhang C., Patterson C. (2004) BioTechniques 37, 464–468 [DOI] [PubMed] [Google Scholar]
- 17.Riva A., Carpentier A. S., Torrésani B., Hénaut A. (2005) Comput Biol. Chem. 29, 319–336 [DOI] [PubMed] [Google Scholar]
- 18.Smyth G. K., Speed T. (2003) Methods 31, 265–273 [DOI] [PubMed] [Google Scholar]
- 19.Storey J. D., Tibshirani R. (2003) Methods Mol. Biol. 224, 149–157 [DOI] [PubMed] [Google Scholar]
- 20.Storey J. D., Tibshirani R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H., Gao L., Tu K., Guo Z. (2005) Gene 352, 75–81 [DOI] [PubMed] [Google Scholar]
- 23.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang J. T., Nevins J. R. (2006) Bioinformatics 22, 2926–2933 [DOI] [PubMed] [Google Scholar]
- 25.Chen H., Sharp B. M. (2004) BMC Bioinformatics 5, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al'Qteishat A., Gaffney J., Krupinski J., Rubio F., West D., Kumar S., Kumar P., Mitsios N., Slevin M. (2006) Brain 129, 2158–2176 [DOI] [PubMed] [Google Scholar]
- 27.Nefkens I., Negorev D. G., Ishov A. M., Michaelson J. S., Yeh E. T., Tanguay R. M., Müller W. E., Maul G. G. (2003) J. Cell Sci. 116, 513–524 [DOI] [PubMed] [Google Scholar]
- 28.Ecsedy J. A., Michaelson J. S., Leder P. (2003) Mol. Cell. Biol. 23, 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song J. J., Lee Y. J. (2003) J. Biol. Chem. 278, 47245–47252 [DOI] [PubMed] [Google Scholar]
- 30.Xu Z., Devlin K. I., Ford M. G., Nix J. C., Qin J., Misra S. (2006) Biochemistry 45, 4749–4759 [DOI] [PubMed] [Google Scholar]
- 31.Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 32.Kwon J. E., La M., Oh K. H., Oh Y. M., Kim G. R., Seol J. H., Baek S. H., Chiba T., Tanaka K., Bang O. S., Joe C. O., Chung C. H. (2006) J. Biol. Chem. 281, 12664–12672 [DOI] [PubMed] [Google Scholar]
- 33.Muromoto R., Ishida M., Sugiyama K., Sekine Y., Oritani K., Shimoda K., Matsuda T. (2006) J. Immunol. 177, 1160–1170 [DOI] [PubMed] [Google Scholar]
- 34.Müller S., Hoege C., Pyrowolakis G., Jentsch S. (2001) Nat. Rev. Mol. Cell Biol. 2, 202–210 [DOI] [PubMed] [Google Scholar]
- 35.Jang M. S., Ryu S. W., Kim E. (2002) Biochem. Biophys. Res. Commun. 295, 495–500 [DOI] [PubMed] [Google Scholar]
- 36.Li Q., Wang X., Wu X., Rui Y., Liu W., Wang J., Wang X., Liou Y. C., Ye Z., Lin S. C. (2007) Cancer Res. 67, 66–74 [DOI] [PubMed] [Google Scholar]
- 37.Mayo L. D., Seo Y. R., Jackson M. W., Smith M. L., Rivera Guzman J., Korgaonkar C. K., Donner D. B. (2005) J. Biol. Chem. 280, 25953–25959 [DOI] [PubMed] [Google Scholar]
- 38.Wu S., Loke H. N., Rehemtulla A. (2002) Neoplasia 4, 486–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonhomme C., Duluc I., Martin E., Chawengsaksophak K., Chenard M. P., Kedinger M., Beck F., Freund J. N., Domon-Dell C. (2003) Gut 52, 1465–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Copanaki E., Schürmann T., Eckert A., Leuner K., Müller W. E., Prehn J. H., Kögel D. (2007) Biochim. Biophys. Acta 1773, 157–165 [DOI] [PubMed] [Google Scholar]
- 41.Muto A., Tashiro S., Tsuchiya H., Kume A., Kanno M., Ito E., Yamamoto M., Igarashi K. (2002) J. Biol. Chem. 277, 20724–20733 [DOI] [PubMed] [Google Scholar]
- 42.Mohammed K. A., Nasreen N., Antony V. B. (2007) Lung 185, 355–365 [DOI] [PubMed] [Google Scholar]
- 43.Nesbit C. E., Fan S., Zhang H., Prochownik E. V. (1998) Blood 92, 1003–1010 [PubMed] [Google Scholar]
- 44.Kirch H. C., Flaswinkel S., Rumpf H., Brockmann D., Esche H. (1999) Oncogene 18, 2728–2738 [DOI] [PubMed] [Google Scholar]
- 45.Jiang P., Du W., Heese K., Wu M. (2006) Mol. Cell. Biol. 26, 9071–9082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugimoto M., Gromley A., Sherr C. J. (2006) Mol. Cell. Biol. 26, 502–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desagher S., Martinou J. C. (2000) Trends Cell Biol. 10, 369–377 [DOI] [PubMed] [Google Scholar]
- 48.Collins C. S., Hong J., Sapinoso L., Zhou Y., Liu Z., Micklash K., Schultz P. G., Hampton G. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3775–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourguignon L. Y., Gilad E., Peyrollier K. (2007) J. Biol. Chem. 282, 19426–19441 [DOI] [PubMed] [Google Scholar]
- 50.Grivennikov S., Karin M. (2008) Cancer Cell 13, 7–9 [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto K., Yu S., Jia Y., Ahmed M. R., Viswakarma N., Sarkar J., Kashireddy P. V., Rao M. S., Karpus W., Gonzalez F. J., Reddy J. K. (2007) J. Biol. Chem. 282, 17053–17060 [DOI] [PubMed] [Google Scholar]
- 52.Tang J., Qu L. K., Zhang J., Wang W., Michaelson J. S., Degenhardt Y. Y., El-Deiry W. S., Yang X. (2006) Nat. Cell Biol. 8, 855–862 [DOI] [PubMed] [Google Scholar]
- 53.Amerik A. Y., Hochstrasser M. (2004) Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 54.Meray R. K., Lansbury P. T., Jr. (2007) J. Biol. Chem. 282, 10567–10575 [DOI] [PubMed] [Google Scholar]
- 55.Schmitz C., Kinner A., Kölling R. (2005) Mol. Biol. Cell 16, 1319–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esser C., Scheffner M., Höhfeld J. (2005) J. Biol. Chem. 280, 27443–27448 [DOI] [PubMed] [Google Scholar]
- 57.Tripathi V., Ali A., Bhat R., Pati U. (2007) J. Biol. Chem. 282, 28441–28454 [DOI] [PubMed] [Google Scholar]
- 58.Nakamura Y., Futamura M., Kamino H., Yoshida K., Nakamura Y., Arakawa H. (2006) Cancer Sci. 97, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.