Abstract
Branchio-oto-renal syndrome (BOR) is an autosomal dominant developmental disorder characterized by hearing loss, branchial arch defects, and renal anomalies. Recently, eight mutations in the SIX1 homeobox gene were discovered in BOR patients. To characterize the effect of SIX1 BOR mutations on the EYA-SIX1-DNA complex, we expressed and purified six of the eight mutants in Escherichia coli. We demonstrate that only the most N-terminal mutation in SIX1 (V17E) completely abolishes SIX1-EYA complex formation, whereas all of the other mutants are able to form a stable complex with EYA. We further show that only the V17E mutant fails to localize EYA to the nucleus and cannot be stabilized by EYA in the cell. The remaining five SIX1 mutants are instead all deficient in DNA binding. In contrast, V17E alone has a DNA binding affinity similar to that of wild type SIX1 in complex with the EYA co-factor. Finally, we show that all SIX1 BOR mutants are defective in transcriptional activation using luciferase reporter assays. Taken together, our experiments demonstrate that the SIX1 BOR mutations contribute to the pathology of the disease through at least two different mechanisms that involve: 1) abolishing the formation of the SIX1-EYA complex or 2) diminishing the ability of SIX1 to bind DNA. Furthermore, our data demonstrate for the first time that EYA: 1) requires the N-terminal region of the SIX1 Six domain for its interaction, 2) increases the level of the SIX1 protein within the cell, and 3) increases the DNA binding affinity of SIX1.
Branchio-oto-renal syndrome (BOR; Mendelian Inheritance in Man (MIM) 113650)5 is an autosomal dominant developmental disorder that is characterized by hearing loss, branchial fistulae, and renal anomalies. Although the penetrance of the syndrome is highly variable between and even within families (1), 70–93% of BOR patients exhibit hearing loss (1). This hearing loss can be conductive, sensorineural, or mixed and ranges in severity. In total, BOR affects an estimated 1 in 40,000 children and accounts for 2% of profoundly deaf children (2).
The most commonly mutated gene in BOR syndrome is EYA1 (3), with an estimated 40% of BOR patients exhibiting mutations in this gene (4). EYA1 belongs to the EYA gene family of transcriptional co-factors. There are four mammalian members (EYA1–4), each containing an N-terminal transactivation domain (5), and a highly conserved ∼270-amino acid C-terminal Eya domain (ED), also referred to as the eya homologous region. The ED possesses phosphatase activity (6–8) and is involved in protein-protein interactions with the SIX family of homeoproteins (9–12). The SIX family of homeoproteins are characterized by a DNA-binding homeodomain (HD) and the protein-interaction Six domain (SD), which binds directly to the ED of EYA (9). As a complex, the SIX and EYA proteins are believed to form a bipartite transcription factor where SIX confers DNA binding and EYA confers transactivation activity.
Recently, mutations in two SIX family members (SIX5 and SIX1) have also been identified in BOR syndrome (13–15). Four different heterozygous mis-sense mutations were found in SIX5 (13), and functional analysis revealed that two of the mutations affect SIX5-EYA1 complex formation and the ability of SIX5 or the SIX5/EYA1 complex to activate transcription. In an independent study, Ruf et al. (14, 15) identified three mutations in the SIX1 gene, which, like the mutations in the SIX5 gene, were argued to inhibit SIX1-EYA1 binding. In addition, two of the three identified mutations also interfere with SIX1-DNA binding. Five additional novel SIX1 mutations have been subsequently identified, although the effect of these mutations on SIX1 function have not yet been determined (16).
To better understand the molecular mechanism of BOR syndrome caused by SIX1 mutations, we attempted to analyze all eight of the SIX1 BOR mutations that have been identified to date. We report the effects of six of the eight SIX1 BOR mutations on a variety of biological functions including: protein expression, protein stability, protein-protein interaction, DNA binding, and transcriptional activation. Significantly, we found that only the most N-terminal mutation in SIX1 (V17E) is able to completely abolish the SIX1-EYA protein interaction, whereas the other five mutations appear to cause major deficiencies in the ability of SIX1 to bind DNA. Finally, we demonstrate that while residing in different regions of the protein, all six SIX1 mutations result in the inability of the complex to activate transcription.
MATERIALS AND METHODS
Molecular Cloning
Mutations within the human SIX1 cDNA were generated using the QuikChange site-directed mutagenesis kit (Stratagene), using WT human SIX1 in pcDNA-3.1 as a template, and the following primers: V17E (gcaagtggcgtgcgAgtgcgaggttctg), H73P (gatcctggagagccCccagttctcgcctc), V106G (ccctgggcgccgGgggcaaatatcg), R110Q (gtgggcaaatatcAggtgcgccgaaaa), R110W (gtgggcaaatatTgggtgcgccgaaaa), R112C (gcaaatatcgggtgTgccgaaaatttcc), Y129C (gaggagaccagctGctgcttcaaggagaag), and del133E (cagctactgcttcaagaagtcgaggggtgtc). Generation of all mutants was confirmed by sequencing. Bacterial expression plasmids were generated by subcloning the PCR products into the BamHI and XhoI sites of the pGEX-6P1 (GE Healthcare) vector and were sequenced.
Small Scale Expression Tests
pGEX-6P1 vectors were transformed into the Escherichia coli strain XA-90. Two independent clones for each mutant were used for small scale expression tests by inoculating 1 ml of LB medium with an overnight culture and grown at 37 °C for 2 h. Protein expression was induced by 1 mm isopropyl β-d-thiogalactopyranoside for 2 h at 37 °C. Protein expression was analyzed by electrophoresis of cell lysates on a 12% SDS-PAGE and visualized by Coomassie Blue staining.
Protein Expression and Purification
All SIX1 proteins were expressed in E. coli at room temperature by inducing protein expression for 4 h at an A600 value of 0.8–1.0 with 1 mm isopropyl β-d-thiogalactopyranoside. Cell pellets were lysed by sonication in precooled lysis buffer (50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 1 mm TCEP) containing the protease inhibitors phenylmethylsulfonyl fluoride, pepstatin, and leupeptin. The supernatant was then treated with 0.4% polyethyleneimine to precipitate out contaminating DNA, which was pelleted by centrifugation. The resultant supernatant was then purified on glutathione-Sepharose 4B resin (GE Healthcare), and the fusion proteins were cleaved with PreScission protease. The SIX1 protein was further purified by ion exchange using a RESOURCE S column (GE Healthcare) and the following buffer A (100 mm Tris-HCl, pH 8.8, 100 mm NaCl, 0.5 mm TCEP) and buffer B (same as buffer A but with 1 m NaCl). Purified proteins were adjusted to 5 mm TCEP, pH 8.0, and stored at −80 °C. For EYA2 ED, after PreScission protease cleavage, the protein preparation was purified using a Superdex 200 (GE Healthcare) size exclusion column in 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 0.5 mm TCEP. Purified protein was adjusted to 5 mm TCEP and stored at −80 °C.
Circular Dichroism Spectroscopy
The secondary structure contents of the purified proteins was assessed by CD spectroscopy using a Jasco 810 spectropolarimeter (Jasco, Inc). CD measurements were carried out in a 1-mm cuvette at 18 °C with a protein concentration of 0.25 mg/ml. Each spectrum was the average of six wavelength scans collected from 196–250 nm in 5 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.5 mm TCEP. Molar ellipticity was calculated using the following equation,
where 0obs is the observed ellipticity (mdeg), C is protein concentration (m), l is the cuvette path length (mm), and n is the number of residues in the protein.
Thermal Denaturation
Protein thermal stability was determined by monitoring the CD signal at 222 nm with increasing temperature. CD data points were obtained at a scan rate of 2 °C/min for the temperature range of 5–85 °C and plotted as the fraction of protein folded versus temperature.
Size Exclusion Complex Formation Assay
All SIX1 protein preparations were run alone or in the presence of the ED on an analytical Superdex 200 in 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 0.5 mm TCEP. The proteins were run alone at 30 μm or mixed at the same concentration with the ED in a 1:1 molar ratio and incubated together on ice for 15 min prior to loading on the column. The gel filtration profiles were overlaid to reveal the elution volumes of the proteins alone, and the complex. SDS-PAGE analysis and Coomassie staining was carried out on the “complex” peak fractions to demonstrate the presence of both proteins.
Reverse Phase HPLC Analysis of the Complex Peak
Reverse phase HPLC was used to confirm the presence of both proteins in the complex peak. Analytical reverse phase HPLC was performed with a linear gradient of eluents A (0.2% trifluoroacetic acid in water) and B (0.2% trifluoroacetic acid in acetonitrile) on a Zorbax SB300-C18 (2.1 × 150 mm) column. Prior to loading, the peak fraction was diluted 10-fold with eluent A.
Nuclear Localization Assay
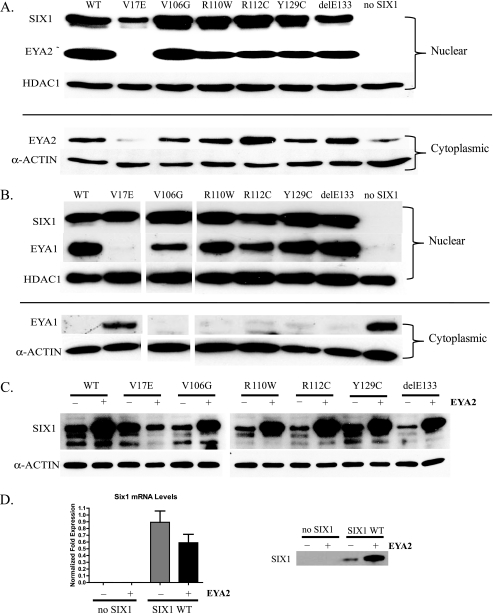
MCF7 cells were cultured onto 10-cm dishes and allowed to adhere overnight. The cells were co-transfected with 8 μg/dish of the indicated Six1 mutant construct and pcDNA3.1 FLAG-Eya2 using FuGENE 6 transfection reagent (Roche Applied Science). For the EYA1 nuclear localization experiments, the pCMVTag2-FLAG-Eya1 construct was used. After 48 h nuclear and cytoplasmic extracts were generated using the NE-PER nuclear/cytoplasmic kit (Thermo Scientific). Equal protein amounts were run out on 10% SDS-PAGE gels, transferred to polyvinylidene difluoride membranes, and probed with anti-FLAG M2 antibodies (Sigma). SIX1 expression levels were assessed by reprobing membranes with anti-SIX1 antibodies (Affinity Bioreagents). To demonstrate equal loading, the membranes were then probed with either anti-HDAC1 (Santa Cruz) or anti-α-actin antibodies (Sigma).
Intracellular Protein Stability Assay
8 × 104 MCF7 cells/well were plated in 12-well plates. The cells were transfected with 250 ng of pcDNA3.1-Six1 and 250 ng of FLAG vector or 250 ng of pcDNA3.1-Six1 and 250 ng of FLAG-Eya2 using the FuGENE transfection method. After 48 h the cell lysates and RNA were isolated. The cell lysates were obtained using lysis buffer PLB (Promega). 40 μg of lysate were electrophoresed on a 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membrane was probed with an anti-SIX1 antibody (Affinity Bioreagents) and to ensure that equal loading the membrane was reprobed with an anti-α-actin antibody (Sigma). To examine alterations in the message level, RNA was isolated using the TRIzol reagent (Invitrogen). 2 μg of RNA were used for cDNA synthesis using the SuperScript III First-Strand kit (Invitrogen). Real time PCR was carried out for Six1 and cyclophilin B (control) using TaqMan gene expression assays on demand (Applied Biosystems) (Six1 Hs00195590_m1, cyclophilin B Hs01018503_m1). The reactions were carried out in triplicate using the Bio-Rad CFX96 real time system. Six1 expression levels were normalized to cyclophilin B.
Electrophoretic Mobility Shift Assay
All of the oligonucleotides (Operon) were ordered HPLC-purified. The oligonucleotides used were a fragment of the myogenin promoter containing the SIX1 binding MEF3 site (GGGGGCTCAGGTTTCTGTGGC) and a second oligonucleotide with this MEF3 site scrambled (GGGGGCATGCTGCTCTGTGGC). The oligonucleotides were labeled using T4 polynucleotide kinase and [γ-32P]ATP and purified using CENTRI-SEP columns (Princeton Separations). The reactions were assembled on ice in 50 mm HEPES, pH 8.0, 75 mm KCl, 200 ng/μl bovine serum albumin, 1 mm TCEP, pH 7.0, 5% glycerol. 4 fmol of oligonucleotide was used along with 750 fmol of protein. For the reactions that include the ED, 750 fmol of the SIX1 protein was mixed with 750 fmol of the ED before the addition of DNA. For the Kd determination, varied quantities of SIX1 or SIX1 and ED were used (7500, 3750, 1500, 750, 375, 150, 75, 37.5, and 15 fmol) while holding the DNA quantity constant at 4 fmol/reaction. For competition reactions the wild type oligonucleotide or the scrambled version at 200× concentration (800 fmol/reaction) was used. Electrophoresis was carried out for 90 min at 60 volts at 4 °C using a 6% nondenaturing polyacrylamide gel. The gels were then dried and exposed to a phosphorimaging plate and visualized using a Storm phosphorimager (GE Healthcare).
Luciferase Assay
MCF7 cells were seeded onto 24-well plates at 25,000 cells/well and allowed to adhere overnight. The following day, the cells were transfected with 300 ng/well pGL3–6×MEF3, 70 ng/well Renilla luciferase, 100 ng/well of the indicated pcDNA3.1 Six1 mutant construct, and 150 ng/well of either pcDNA3.1 FLAG-Eya2 or empty vector using FuGENE 6 (Roche Applied Science) transfection reagent. The cells were lysed 48 h later with passive lysis buffer, and the resulting extracts were analyzed for luciferase and Renilla activities. To ensure expression of exogenous SIX1 protein, equal amounts of protein were run out on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes, which were probed with an anti-SIX1 antibody (Affinity Bioreagents), and to ensure equal loading the membranes were reprobed with anti-α-actin (Sigma).
RESULTS
Expression and Purification of SIX1 BOR Mutants
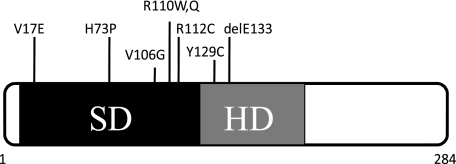
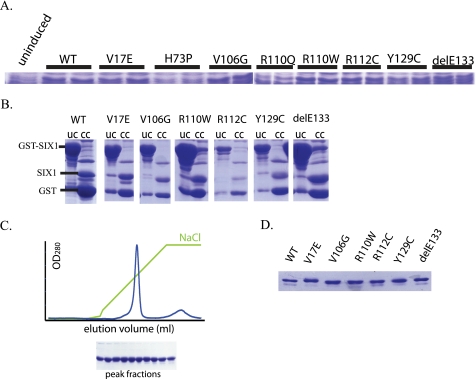
All eight identified SIX1 BOR mutations, including seven mis-sense mutations and one in-frame deletion, reside in either the SD or the HD (Fig. 1). To precisely define the molecular mechanism(s) by which these mutations compromise SIX1 function, we generated the eight mutations using site-directed mutagenesis. Wild type human SIX1 and the eight SIX1 mutants were then subcloned into a GST fusion vector in an effort to express these proteins in E. coli for biochemical analyses. Expression tests in E. coli were first performed using GST fusion proteins to determine whether all of the mutant proteins could be expressed effectively. Two independent clones for each mutant were treated with isopropyl β-d-thiogalactopyranoside to induce protein expression. Protein expression was analyzed by SDS-PAGE and Coomassie staining. The expression tests demonstrated that the H73P and R110Q mutants failed to express in E. coli, whereas all of the other six mutants were expressed efficiently (Fig. 2A). Therefore, all subsequent data presented will focus on wild type SIX1 and the six SIX1 mutants that were efficiently expressed in E. coli.
FIGURE 1.
Schematic representation of the functional domain structure of SIX1 and the location of BOR mutations.
FIGURE 2.
Expression and purification of human wild type SIX1 and BOR mutants. A, SDS-PAGE analysis of whole cell lysates from expression tests using two independent clones for each of the various mutants. The proteins were expressed as GST fusions. Note that H73P and R110Q were unable to be stably expressed. B, SDS-PAGE analysis of large scale preparations of the proteins purified by glutathione affinity chromatography with cleavage from the GST tag by PreScission protease. UC, uncut column, referring to the glutathione resin before PreScission digestion; CC, cut column, referring to the glutathione resin after PreScission digestion. C, further protein purification was achieved by ion exchange chromatography. A representative profile is shown along with a representative SDS-PAGE of the peak fractions (SIX1 WT). D, SDS-PAGE analysis of the purified protein preparations. 3 μg of protein was analyzed by SDS-PAGE to assure equal purity and quantities for biochemical characterization.
Subsequent large scale purification of wild type SIX1 and the six SIX1 mutants using glutathione affinity chromatography was carried out, and the GST moiety was removed using PreScission protease (Fig. 2B). The proteins were further purified by ion exchange chromatography to near homogeneity, as shown by SDS-PAGE and Coomassie analysis of the peak fractions (Fig. 2C). All of the purified protein preparations used for subsequent analyses were analyzed using SDS-PAGE and Coomassie staining, which demonstrated similar concentrations and purity (Fig. 2D).
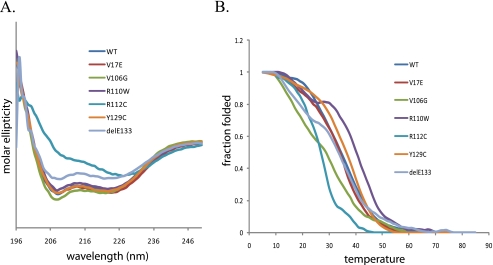
V106G and R112C Destabilize SIX1
To evaluate the impact of the mutations on overall protein structure, we first examined protein secondary structure using CD spectroscopy. In CD spectra, α-helical secondary structure is characterized by strong ellipticity minima at 222 and 208 nm. In highly α-helical proteins, the ellipticity minima at 222 and 208 nm are similar, as is observed with wild type SIX1 (Fig. 3A). Interestingly, the R112C and delE133 mutations cause a reduction in α-helical structure, whereas the rest of the mutations do not considerably alter the α-helical content of SIX1. To further measure protein stability, CD spectroscopy was used to record the thermal denaturation profiles of wild type SIX1 and the six SIX1 mutant proteins. Both the V106G and R112C mutants demonstrated a significantly decreased melting temperature (Tm) when compared with wild type SIX1 (Tm = 28.2 and 26.8 °C for V106G and R112C, respectively, versus Tm = 34.5 °C for wild type SIX1) (Fig. 3B). These data suggest that the V106G and R112C mutants are less stable than wild type SIX1 and the other SIX1 mutants. The decrease in protein stability is not surprising for R112C because it is consistent with the decrease in α-helical content. Although V106G did not disrupt the α-helical content noticeably, it nonetheless appears to have an overall destabilizing effect on the protein in vitro.
FIGURE 3.
Secondary structure and stability of wild type SIX1 and BOR mutants. A, CD spectra were obtained for wild type SIX1 and for the BOR mutants. Mean residue molar ellipticity is plotted versus wavelength. Note the significant decrease in α-helical structure for R112C and slight decrease for delE133. B, thermal stabilities of the proteins monitored by CD at 222 nm at a starting temperature of 5 °C. Fraction folded is plotted versus temperature in °C. Note the decrease in Tm for the mutants R112C and V106G.
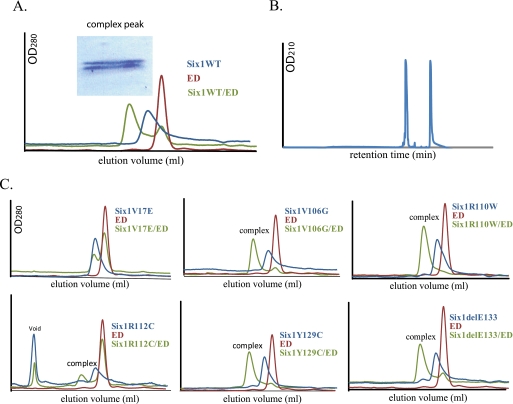
V17E Fails to Form a Complex with EYA2 ED in Vitro
Following our assessment of the stability of the SIX1 BOR mutants, we evaluated the ability of the same mutants to bind to the EYA co-factor using size exclusion gel filtration chromatography. Consistent with previous reports (8, 17), we had difficulty expressing functional recombinant EYA1 in E. coli. However, we were able to easily express and purify the ED of human EYA2. Both mammalian EYA1 and EYA2 can complement Drosophila eya mutations with comparable efficiency (18, 19), and the EYA2 ED, which contains the SIX1-binding region, shares over 90% sequence similarity with the ED of EYA1. Therefore, we used the EYA2 ED (hereafter referred to as ED), for all subsequent in vitro analyses.
The elution profiles of wild type SIX1 and the ED alone from an analytical S200 size exclusion column were very similar, with peaks in close proximity, consistent with their similar molecular mass. When wild type SIX1 and the ED were mixed and incubated to allow complex formation prior to analysis on the S200 column, a shifted elution peak corresponding to the SIX1-ED complex was observed, suggesting that the two proteins can form a stable complex. Indeed, examination of the protein content in the complex peak by SDS-PAGE and Coomassie staining revealed what appeared to be the presence of both wild type SIX1 and the ED (Fig. 4A). Because the molecular weights of the two proteins are similar, making it difficult to resolve them using SDS-PAGE, the complex peak fraction was also analyzed by analytical reverse phase chromatography to confirm the above finding. This analysis demonstrated that both SIX1 and the ED are present, and importantly, that they bind in a 1:1 molar ratio (Fig. 4B).
FIGURE 4.
The V17E mutation inhibits the ability of SIX1 to interact with EYA2 ED. A, analytical Superdex 200 gel filtration profiles of SIX1 WT (blue), ED (red), and SIX1 WT/ED (green) overlaid to show complex formation. The peak fraction from the complex peak was analyzed by SDS-PAGE and revealed the presence of both proteins. B, analysis of the Superdex 200 complex peak fraction by reverse phase HPLC confirms the presence of both proteins in a 1:1 molar ratio. C, analysis of the ability of the various SIX1 BOR mutants to complex with the ED by analytical gel filtration. Protein complex formation was analyzed as described in A. Note only V17E failed to complex with the ED, and a majority of R112C resides in the void volume, underscoring the unstable nature of this mutant.
We next analyzed the six SIX1 mutants for their ability to bind to the ED. Interestingly, we found that all of the mutants could successfully form a protein complex with the ED except the V17E mutant (Fig. 4C). Surprisingly, the R110W, Y129C, and delE133 mutants that were previously shown to be deficient in EYA1 binding using yeast two-hybrid analysis (15) were able to bind the ED in our assay. Although the R112C-ED complex formation appears reduced, this is likely due to the large amount of R112C protein that is lost in the void volume, which is consistent with its decreased secondary structure content and stability. Together, these results indicate that only the V17E mutation abolishes SIX1-ED binding in vitro.
V17E Fails to Localize Full-length EYA2 to the Nucleus
Previous reports have demonstrated that EYA, which is localized in the cytoplasm in the absence of SIX1, is translocated to the nucleus by SIX1 (20, 21). Thus, we analyzed the intracellular distribution of full-length FLAG-tagged EYA2 after transfection into MCF7 cells in the presence of either wild type SIX1 or each of the six SIX1 BOR mutants to assess whether proper complex formation is taking place within the cell. Using nuclear and cytoplasmic fractionation, we observed cytoplasmic EYA2 localization in the absence of SIX1 (Fig. 5A, no SIX1). In contrast, the addition of wild type SIX1 led to predominantly nuclear localization of EYA2 as expected (Fig. 5A, WT). Similarly, the V106G, R110W, R112C, Y129C, and delE133 mutants were all able to localize the majority of EYA2 in the nucleus (Fig. 5A, V106G, R110W, R112C, Y129C, and delE133). However, when the SIX1 V17E mutant was co-expressed with EYA2, no detectable level of EYA2 could be found in the nucleus, similar to the control lane that lacked SIX1 expression (Fig. 5a, V17E and no SIX1). Instead, all visible EYA2 was localized in the cytoplasm. These data further suggest that V17E is the only mutation that abolishes the interaction between SIX1 and EYA.
FIGURE 5.
The V17E mutant fails to localize EYA2 and EYA1 to the nucleus and to be stabilized by EYA2. A, analysis of the cellular localization of EYA2 in the presence of wild type SIX1, the various BOR mutants, or no SIX1 (control). SIX1 WT and the SIX1 BOR mutants were co-transfected with FLAG-tagged full-length EYA2, and nuclear and cytoplasmic lysates were isolated and analyzed by SDS-PAGE and Western blot analysis. HDAC1 and α-actin were used for nuclear and cytoplasmic loading controls, respectively. Note that transfection with V17E does not confer localization of EYA2 to the nucleus, similar to the control reaction that lacked SIX1 expression, whereas transfection with wild type SIX1, as well as all other BOR mutants, does enable nuclear localization of EYA2. B, analysis of the cellular localization of EYA1 in the presence of wild type SIX1, the various BOR mutants or no SIX1 (control). The experiment was carried out as in A, using EYA1 instead of EYA2. Note that transfection with V17E does not confer localization of EYA1 to the nucleus. C, EYA2 expression leads to an increase in SIX1 protein levels in the cell. Wild type SIX1 and SIX1 mutants were co-transfected with pcDNA-3.1 as a control (−) or with full-length EYA2 (+). Note that EYA2 is able to stabilize wild type SIX1 and all of the SIX1 mutants with the exception of V17E, again suggesting that this mutant is unable to bind EYA. D, analysis of the effect of EYA2 and SIX1-EYA2 expression on total SIX1 protein and mRNA levels. Western blot analysis (right panel) demonstrates that EYA2 expression alone does not increase the endogenous SIX1 protein to detectable levels, but when EYA2 is co-expressed with SIX1, it leads to a vast increase in SIX1 protein levels compared with when exogenous SIX1 is expressed alone. Quantitative reverse transcription-PCR analysis (left panel) reveals that when EYA2 is expressed with SIX1, EYA2 does not increase SIX1 transcript levels. The data are presented as normalized fold expression using data from three independent experiments.
V17E Fails to Localize Full-length EYA1 to the Nucleus
Because EYA1, not EYA2, is implicated in BOR syndrome and because previous studies had shown, in contrast to ours, that R110W, Y129C, and delE133 were deficient in EYA1 binding, we tested the ability of wild type SIX1 and the six SIX1 BOR mutants to specifically translocate full-length FLAG-tagged EYA1 to the nucleus. Again, using nuclear and cytoplasmic fractionation, we observed that in the absence of SIX1, EYA1 was localized in the cytoplasm (Fig. 5B, no SIX1). However, in the presence of wild type SIX1, EYA1 was localized in the nucleus (Fig. 5B, WT). Likewise, V106G, R110W, R112C, Y129C, and delE133 were all able to localize the majority of EYA1 in the nucleus (Fig. 5B, V106G, R110W, R112C, Y129C, and delE133). In contrast, V17E was unable to localize EYA1 in the nucleus (Fig. 5B, V17E). Because these results are consistent with the results obtained with EYA2, our data strongly suggest that V17E is the only mutation that abolishes the interaction between SIX1 and the EYA proteins.
V17E Fails to Be Stabilized by the Presence of EYA2 Protein
In the EYA2 nuclear localization experiments, we observed that co-transfection of EYA2 with wild type SIX1 increases the intracellular quantity of both the SIX1 and EYA2 proteins compared with levels when either protein is expressed alone (data not shown and Fig. 5A). We thus examined whether the levels of the various SIX1 BOR mutants were affected by the presence of EYA2. Interestingly, the levels of all the SIX1 BOR mutants as well as wild type SIX1 are increased with the addition of EYA2, with the exception, again, of the V17E mutant (Fig. 5C).
To test whether the increase in SIX1 protein was due to an increase in SIX1 transcription by the SIX1-EYA2 complex increasing SIX1 expression or EYA2 teaming up with endogenous SIX1 (or another SIX family member) to stimulate SIX1 expression, we performed a Western blot in conjunction with quantitative reverse transcription-PCR analysis of SIX1 transcript levels. When EYA2 is expressed alone, endogenous SIX1 protein levels are not increased to detectable levels (Fig. 5D, right panel). When SIX1 is exogenously expressed, the addition of EYA2 does not increase SIX1 mRNA levels (Fig. 5D, left panel); however, it does lead to an increase in SIX1 protein levels (Fig. 5D, right panel). These data suggest that EYA2 is able to stabilize SIX1 protein levels and further reinforce the finding that only the V17E mutation is able to abolish SIX1-EYA complex formation.
The Majority of SIX1 BOR Mutants Are Defective in DNA Binding
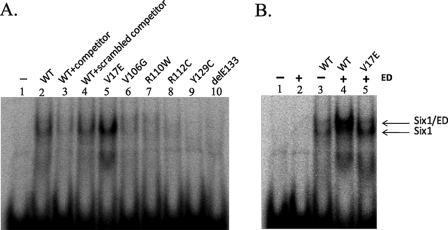
Because our data demonstrate that only the V17E mutant is completely deficient in binding EYA, we analyzed the ability of the SIX1 BOR mutants to bind to the MEF3 site of the myogenin promoter (TCAGGTT), a well characterized SIX1 target site (22). As expected, wild type SIX1 is able to efficiently and specifically bind the MEF3 oligonucleotide (Fig. 6A, lanes 1–4). Interestingly, the V17E mutation increased binding to the MEF3 site (Fig. 6A, lane 5). In contrast, all of the other SIX1 mutations inhibit the binding of SIX1 to this site (Fig. 6A, lanes 6–10).
FIGURE 6.
All SIX1 BOR mutants, with the exception of V17E, are unable to bind DNA efficiently. A, the ability of the BOR mutants to bind DNA was assessed by electrophoretic mobility shift assay utilizing an oligonucleotide containing a SIX1-binding site, MEF3 (TCAGGTT). B, the ability of the ED to enter the SIX1-DNA complex was assessed by electrophoretic mobility shift assay. Note that migration of the V17E-DNA complex is not altered by the addition of the ED, nor is DNA binding of V17E enhanced by the presence of the ED. In contrast, the addition of the ED into the wild type SIX1-DNA complex alters the mobility of the complex and increases the affinity of SIX1 for DNA.
We next investigated the role of the ED in SIX1-DNA binding. As anticipated, the ED alone is unable to bind DNA (Fig. 6B, lane 2). When the ED is added to a reaction containing SIX1 and DNA, a small but reproducible decrease in migration is observed, suggesting that a ternary complex is formed. In addition, the ED appears to increase SIX1-DNA binding activity (Fig. 6B, lanes 3 and 4). In contrast, when V17E and the ED are incubated with DNA, migration of the complex is not altered from the complex formed with V17E and DNA alone. These data demonstrate that the ED is unable to enter the V17E-DNA complex, further supporting the finding that V17E is unable to interact with EYA.
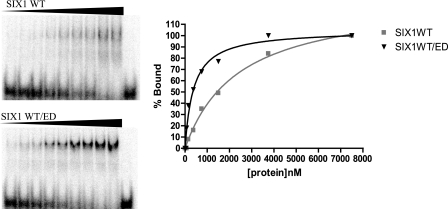
We then determined the DNA binding affinities of wild type SIX1 and the V17E mutant in the presence and absence of the ED. Interestingly, the binding affinity of wild type SIX1 is 10-fold higher when complexed with the ED (Kd = 2404 nm versus 383 nm). Surprisingly, V17E alone (Kd = 211 nm) has an affinity for DNA that is similar to the wild type SIX1-ED complex, and that affinity is not significantly changed in the presence of the ED (Kd = 336 nm) (Fig. 7 and Table 1).
FIGURE 7.
Determination of the SIX1-DNA binding affinity in the presence or absence of the ED. Left panels, electrophoretic mobility shift assays were carried out using an oligonucleotide containing the MEF3 SIX1-binding site, TCAGGTT. A fixed quantity of DNA (4 fmol) and increasing quantities of protein (15, 37.5, 75, 150, 375, 750, 1500, 3750, and 7500 fmol) were used to perform the experiment. Right panel, the data were analyzed by image quant, and Kd was determined by graphing the percentage of DNA bound versus protein concentration.
TABLE 1.
DNA binding affinity of wild type Six1 and V17E ± ED
| Kd | r2 | |
|---|---|---|
| nm | ||
| Six1 WT | 2404 ± 246 | 0.996 |
| Six1 WT + ED | 383 ± 64 | 0.966 |
| V17E | 211 ± 44 | 0.983 |
| V17E + ED | 336 ± 43 | 0.989 |
SIX1 BOR Mutants Are Deficient in Transcriptional Activation
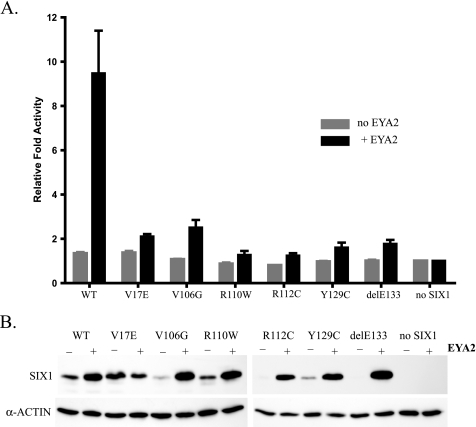
To analyze the effect of the various BOR mutations on the ability of SIX1 to stimulate transcription, we examined SIX1-mediated transcriptional activation using the pGL3–6×MEF3-luciferase construct, which contains six repeats of the SIX1-binding MEF3 site placed upstream of the luciferase gene (22). When wild type SIX1 was co-expressed with full-length EYA2, luciferase activity was increased ∼10-fold compared with SIX1 or EYA2 alone. However, all of the SIX1 BOR mutants were unable to stimulate luciferase activity when co-expressed with EYA2 (Fig. 8A).
FIGURE 8.
All BOR mutants are deficient in transcriptional activation of a SIX1-responsive promoter. A, Wild type SIX1 and the BOR mutants were either co-transfected with pcDNA-3.1 or full-length EYA2 and analyzed for their ability to stimulate transcription from the SIX1-responsive 6×MEF3 promoter-luciferase construct. B, representative Western blot analysis using extracts obtained in the luciferase experiments demonstrates the levels of wild type and mutant SIX1 protein within the luciferase assay. α-Actin was used as a loading control. Note the failure of V17E to be stabilized by EYA2.
To ensure proper transfection and expression of the mutants, equal quantities of the luciferase extracts were subjected to SDS-PAGE and Western blot analysis using an anti-SIX1 antibody. As previously observed, co-expression of EYA2 with all of the SIX1 BOR mutants, except for V17E, led to an increase in SIX1 protein levels (Fig. 8B).
These results demonstrate that all of the mutations, although occurring in different regions of the protein, functionally inactivate the complex by inhibiting the ability of the SIX1-EYA complex to activate transcription. It should be noted that although we were unable to express the two SIX1 mutants in bacteria, H73P and R110Q, we were able to express them in mammalian cells, and they both were capable of localizing EYA2 and EYA1 in the nucleus; their protein levels were stabilized by EYA2, and they failed to activate transcription (data not shown).
DISCUSSION
Biophysical, biochemical, and cell culture experiments presented here provide insight into the mechanism by which SIX1 BOR mutations affect the function of the SIX1-EYA transcriptional complex. We demonstrate that one of the mechanisms by which SIX1 BOR mutations affect SIX1 function is likely through an abolished interaction with its co-activator EYA. However, we find that the most prevalent mechanism by which the identified mutations inhibit the function of the SIX1 transcriptional complex is through decreasing the ability of SIX1 to bind DNA.
Using gel filtration chromatography we were able to analyze direct protein interactions between six SIX1 BOR mutants and the ED. Wild type SIX1 and the ED form a stable complex on the gel filtration column. Further analysis of the complex peak by reverse phase HPLC reveals, for the first time, that SIX1 and EYA bind in a 1:1 molar ratio. Surprisingly, all of the BOR mutants tested were able to form a stable complex with the ED with the exception of the most N-terminal mutant, V17E.
These gel filtration results are in contrast to the finding of Ruf et al. (15) that the R110W, Y129C, and delE133 SIX1 mutants are deficient in EYA1 binding. It is possible that the different observations could result from our use of a truncated form of EYA2 in our binding experiment as opposed to full-length EYA1, which was used in their yeast two-hybrid assays. In addition, we cannot rule out the possibility that the BOR mutants previously described to be deficient in EYA1 binding have different affinities for EYA than wild type SIX1, thus providing an explanation for the previous observation that the R110W, Y129C, and delE133 mutants are deficient in EYA1 binding. However, our cell culture experiments, in which full-length EYA1 and EYA2 cellular localization was examined in the presence of the different SIX1 mutants, strongly suggest that our in vitro biochemical results most likely reflect the in vivo situation, where only the V17E mutation abolishes binding to EYA within the cell. These data favor the model where a direct interaction between SIX1 and EYA rather than an indirect interaction between the proteins (21) is responsible for the nuclear localization of EYA because the only mutant that fails to bind EYA directly in vitro is the only mutant that fails to localize EYA to the nucleus.
Additionally, we found that EYA2 stabilizes SIX1 protein levels within the cell and that only the V17E mutant is not stabilized by the presence of EYA2. To our knowledge, this is the first demonstration that EYA2 leads to an increase in SIX1 protein levels. EYA2 protein levels also appear to be stabilized by SIX1 because similar levels of EYA2 are found in the V17E and no SIX1 lanes in the nuclear localization experiment, whereas EYA2 levels are increased in all other lanes where SIX1 and EYA2 proteins are able to interact (Fig. 5A).
We have previously reported that SIX1 is degraded through the anaphase-promoting complex (23). This degradation is facilitated by binding of the activating subunit, CDH1, to the SD of SIX1. The binding of EYA to the SD could potentially inhibit the ability of CDH1 to bind to SIX1, thus blocking/decreasing SIX1 degradation, although this remains to be proven. Thus, our protein stabilization finding further underscores the observation that only the V17E mutant is deficient in EYA binding and further provides insight into the mechanism by which SIX1 and EYA2 regulate each other.
It is important to note that the V17E mutation is the only SD mutation that resides in a predicted α-helix (the first α-helix of the SD) by secondary structure algorithms (data not shown). This suggests that the first α-helix of SIX1 is involved in its interaction with EYA. Indeed, this valine residue is conserved in all of the SIX family members, underscoring its importance in mediating SIX1 function.
We demonstrate that five of the six BOR mutants exhibit significantly diminished DNA binding, suggesting that this is the major mechanism by which SIX1 BOR mutants contribute to the syndrome. For the two mutations that reside in the DNA-binding HD (Y129C and delE133), this result is expected and is consistent with previous observations (15). The other mutations, V106G, R110W, and R112C all reside in an unstructured region of SIX1 (residues100–123) as predicted by secondary structure algorithms. This unstructured region links the last α-helix of the SD with the N-terminal arm and first α-helix of the HD. This region, N-terminal to the HD, is important in DNA binding specificity for the SIX family member, SIX4 (10). The integrity of this region may be crucial for the function of the N-terminal arm or for proper folding of the first α-helix of the HD, thus providing structural stability. Alternatively, this “linker” region may be involved directly in DNA binding, particularly in light of the observation that the N-terminal arm of the SIX family HD lacks important DNA contacting basic amino acids that are conserved in the homeodomain superfamily (24).
The N-terminal arm represents the first eight or nine amino acids of the homeodomain and is unstructured in the absence of DNA, but upon DNA binding forms direct contacts with bases in the minor groove (25). Canonical HD N-terminal arms have a stretch of basic amino acids including an arginine at position 5 that protrudes deep into the minor groove and makes extensive base and sugar contacts. Basic residues at positions 2 and 3 further stabilize the interaction by forming additional hydrogen bonds with the DNA backbone and give the N-terminal arm a net charge of +3. All three of these basic residues are absent in the SIX1 HD and in all of the SIX family members. In fact, two of these residues are replaced with acidic residues, resulting in an N-terminal arm with a net negative charge. Thus, the N-terminal arm of SIX1 most likely does not form these stabilizing DNA contacts. Given the DNA binding deficiencies of the mutants with mutations located in the linker region, and the fact that the region N-terminal of the HD (preceding the N-terminal arm) is important for DNA binding specificity for another family member SIX4, one could speculate that the SIX1 protein may contain a noncanonical extension of the N-terminal arm that is involved in DNA binding. Consistent with this hypothesis, the linker region of SIX1 has a net charge of +6, and three of the four BOR mutations in this linker target two arginines a single amino acid apart, amino acids that may be involved in forming direct DNA contacts, because they do not appear to significantly affect EYA binding. Thus, this linker region may help stabilize the DNA interaction by making direct DNA contacts, extending the DNA binding activity outside of the HD. Importantly, the residues in this linker region, Val106, Arg110, and Arg112 are conserved in all of the SIX family members, again suggesting that these residues play an important functional role.
Our observation that the V17E mutation in the SD enhances the DNA binding affinity of SIX1 leads to interesting hypotheses related to the function of the SD domain in DNA binding. It is possible that the SD may inhibit DNA binding through steric interference or by masking amino acids that can participate in DNA contacts. The V17E mutation may thus disrupt the structure and/or the positioning of the SD as to no longer sterically inhibit DNA binding. Alternatively, it may promote a structural change that allows more stable DNA contacts to be made. Consistent with these possibilities, SIX1-EYA complex formation leads to a ∼10-fold increase in DNA binding affinity, an increase that is similar to that reported for another SIX family member upon EYA binding, SIX2 (24). EYA binding to SIX1 may then disrupt this inhibition by the SD, either by relieving the steric interference or by exposing amino acids that can now take part in direct DNA binding. Further structural analysis will help answer these hypotheses.
In conclusion, our data demonstrate for the first time that the integrity of the first α-helix of the SD is critical for SIX1-EYA complex formation. We also show that SIX1-ED complex formation modulates the ability of SIX1 to bind DNA and that EYA2 expression leads to an increase in SIX1 protein levels within the cell. Furthermore, our data demonstrate that the SIX1 BOR mutations contribute to the pathology of the disease by at least two mechanisms; abolishing the formation of the SIX1-EYA complex (V17E) or, in the majority of cases, by diminishing the ability of SIX1 to bind DNA (V106G, R110W, R112C, Y129C, and delE133).
Acknowledgments
We thank Dr. Kiyoshi Kawakami for the pGL3–6×MEF3 construct. We thank Patrick Hutchins for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant 2RO1-CA095277 (to H. L. F.). This work was also supported by American Cancer Society Grant RSG-07-183-01-DDC (to H. L. F.), a V Scholar award from the V Foundation for Cancer Research, and a Kimmel scholar award from the Sidney Kimmel Foundation for Cancer Research (to R. Z.).
- BOR
- branchio-oto-renal syndrome
- ED
- Eya domain
- HD
- homeodomain
- SD
- Six domain
- WT
- wild type
- HPLC
- high pressure liquid chromatography
- GST
- glutathione S-transferase
- TCEP
- tris(2-carboxyethyl)phosphine.
REFERENCES
- 1.Kochhar A., Fischer S. M., Kimberling W. J., Smith R. J. H. (2007) Am. J. Med. Genet. A 143A, 1671–1678 [DOI] [PubMed] [Google Scholar]
- 2.Fraser F. C., Sproule J. R., Halal F. (1980) Am. J. Med. Genet. 7, 341–349 [DOI] [PubMed] [Google Scholar]
- 3.Abdelhak S., Kalatzis V., Heilig R., Compain S., Samson D., Vincent C., Levi-Acobas F., Cruaud C., Le Merrer M., Mathieu M., König R., Vigneron J., Weissenbach J., Petit C., Weil D. (1997) Hum. Mol. Genet 6, 2247–2255 [DOI] [PubMed] [Google Scholar]
- 4.Chang E. H., Menezes M., Meyer N. C., Cucci R. A., Vervoort V. S., Schwartz C. E., Smith R. J. (2004) Hum. Mutat. 23, 582–589 [DOI] [PubMed] [Google Scholar]
- 5.Xu P. X., Cheng J., Epstein J. A., Maas R. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11974–11979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayapureddi J. P., Kattamuri C., Steinmetz B. D., Frankfort B. J., Ostrin E. J., Mardon G., Hegde R. S. (2003) Nature 426, 295–298 [DOI] [PubMed] [Google Scholar]
- 7.Tootle T. L., Silver S. J., Davies E. L., Newman V., Latek R. R., Mills I. A., Selengut J. D., Parlikar B. E., Rebay I. (2003) Nature 426, 299–302 [DOI] [PubMed] [Google Scholar]
- 8.Li X., Oghi K. A., Zhang J., Krones A., Bush K. T., Glass C. K., Nigam S. K., Aggarwal A. K., Maas R., Rose D. W., Rosenfeld M. G. (2003) Nature 426, 247–254 [DOI] [PubMed] [Google Scholar]
- 9.Kawakami K., Sato S., Ozaki H., Ikeda K. (2000) Bioessays 22, 616–626 [DOI] [PubMed] [Google Scholar]
- 10.Kawakami K., Ohto H., Ikeda K., Roeder R. G. (1996) Nucleic Acids Res. 24, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R., Amoui M., Zhang Z., Mardon G. (1997) Cell 91, 893–903 [DOI] [PubMed] [Google Scholar]
- 12.Pignoni F., Hu B., Zavitz K. H., Xiao J., Garrity P. A., Zipursky S. L. (1997) Cell 91, 881–891 [DOI] [PubMed] [Google Scholar]
- 13.Hoskins B. E., Cramer C. H., Silvius D., Zou D., Raymond R. M., Orten D. J., Kimberling W. J., Smith R. J., Weil D., Petit C., Otto E. A., Xu P. X., Hildebrandt F. (2007) Am. J. Hum. Genet. 80, 800–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruf R. G., Berkman J., Wolf M. T., Nurnberg P., Gattas M., Ruf E. M., Hyland V., Kromberg J., Glass I., Macmillan J., Otto E., Nurnberg G., Lucke B., Hennies H. C., Hildebrandt F. (2003) J. Med. Genet 40, 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruf R. G., Xu P. X., Silvius D., Otto E. A., Beekmann F., Muerb U. T., Kumar S., Neuhaus T. J., Kemper M. J., Raymond R. M., Jr., Brophy P. D., Berkman J., Gattas M., Hyland V., Ruf E. M., Schwartz C., Chang E. H., Smith R. J., Stratakis C. A., Weil D., Petit C., Hildebrandt F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8090–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kochhar A., Orten D. J., Sorensen J. L., Fischer S. M., Cremers C. W., Kimberling W. J., Smith R. J. (2008) Hum. Mutat. 29, 565–576 [DOI] [PubMed] [Google Scholar]
- 17.Mutsuddi M., Chaffee B., Cassidy J., Silver S. J., Tootle T. L., Rebay I. (2005) Genetics 170, 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonini N. M., Bui Q. T., Gray-Board G. L., Warrick J. M. (1997) Development 124, 4819–4826 [DOI] [PubMed] [Google Scholar]
- 19.Bui Q. T., Zimmerman J. E., Liu H., Bonini N. M. (2000) Genetics 155, 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohto H., Kamada S., Tago K., Tominaga S. I., Ozaki H., Sato S., Kawakami K. (1999) Mol. Cell. Biol. 19, 6815–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buller C., Xu X., Marquis V., Schwanke R., Xu P. X. (2001) Hum. Mol. Genet. 10, 2775–2781 [DOI] [PubMed] [Google Scholar]
- 22.Spitz F., Demignon J., Porteu A., Kahn A., Concordet J.P., Daegelen D., Maire P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14220–14225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen K. L., Brennan J. D., Aldridge C. S., Ford H. L. (2007) Oncogene 26, 3406–3414 [DOI] [PubMed] [Google Scholar]
- 24.Hu S., Mamedova A., Hegde R. S. (2008) Biochemistry 47, 3586–3594 [DOI] [PubMed] [Google Scholar]
- 25.Ades S. E., Sauer R. T. (1994) Biochemistry 33, 9187–9194 [DOI] [PubMed] [Google Scholar]