Abstract
The homeodomain protein Cux1 is highly expressed in the nephrogenic zone of the developing kidney where it functions to regulate cell proliferation. Here we show that Cux1 directly interacts with the co-repressor Grg4 (Groucho 4), a known effector of Notch signaling. Promoter reporter based luciferase assays revealed enhanced repression of p27kip1 promoter activity by Cux1 in the presence of Grg4. Chromatin immunoprecipitation (ChIP) assays demonstrated the direct interaction of Cux1 with p27kip1 in newborn kidney tissue in vivo. ChIP assays also identified interactions of Cux1, Grg4, HDAC1, and HDAC3 with p27kip1 at two separate sites in the p27kip1 promoter. DNAse1 footprinting experiments revealed that Cux1 binds to the p27kip1 promoter on the sequence containing two Sp1 sites and a CCAAT box ~500 bp from the transcriptional start site, and to an AT rich sequence ~1.5 KB from the transcriptional start site. Taken together, these results identify Grg4 as an interacting partner for Cux1 and suggest a mechanism of p27kip1 repression by Cux1 during kidney development.
Introduction
Cux1 is the murine homologue of the Drosophila gene cut. Cux1 contains four potential DNA binding domains: three 60 amino acid repeats, termed cut repeats, and the homeodomain (1–3). Mammalian cut proteins function as cell cycle-dependent transcription factors that can function as activators or repressors (4–12). Targets of repression by Cux1 include γ-globin (13), c-myc (14), myosin heavy chain (15), NCAM (16), CD8a (17), c-mos (18), MMTV long terminal repeats (19), gp91-phox (20), and the cyclin kinase inhibitors p21waf1 and p27kip1 (21–22). The binding of Cux1 protein to the promoters of target genes appears to be limited to tissues or developmental stages in which the target genes are not expressed. Upon terminal differentiation, Cux1 is downregulated or loses the ability to bind to the promoters, permitting transcription of the target genes. Cux1 represses transcription by two mechanisms: 1) Competition for CCAAT or SP1 binding site occupancy, preventing activation by the corresponding transcription factors, or 2) Active repression via carboxy-terminal repression domain following binding at distance from transcription start site (23). It has been suggested that the mechanism of active repression involves the direct recruitment of HDAC1 deacetylase (24). In addition, Cux1 was reported to recruit G9a histone lysine methyltransferase to repress transcription of the p21waf1 promoter (7). During cell cycle progression, a nuclear isoform of the cysteine protease cathepsin L cleaves Cux1, forming the amino-truncated p110 Cux1 protein, which contains two cut repeats and the homeodomain (25). A shorter isoform, containing a single cut repeat and the homeodomain, is the result of transcription from an alternate promoter (26).
Mice carrying targeted deletions of Cux1 exhibit reduced growth, retarded differentiation of lung epithelia, hair follicle defects, reduced male fertility, and deficient T and B cell function (27– 29). In contrast, transgenic mice ectopically expressing Cux1 exhibit multiorgan hyperplasia including an increase in the size of the kidneys, heart, liver, and testis, apparently resulting from the repression of p27kip1 gene expression (22). These mice also develop glomerulosclerosis, renal interstitial fibrosis, and hepatic tumors (22, 30–31). Cux1 transgenic mice have a similar kidney phenotype as p27kip1 knockout mice (32–34), while p21waf1 knockout mice do not exhibit renal hyperplasia (35), suggesting that p27kip1, and not p21waf1, may be the primary target of Cux-1 repression in the kidney. Another Cux1 transgenic mouse ectopically expressing p75 Cux1 exhibits a myeloid leukemia like myeloproliferative disease with reduced p27kip1 expression in the spleen (36). Recently, p75 Cux1 transgenic mice have been reported to develop polycystic kidney disease, associated with reduced p27kip1 expression and upregulation of c-myc (11).
The TLE/ Groucho (Grg) proteins are members of a family of co-repressor proteins that do not themselves bind to DNA but are recruited to DNA by interactions with DNA binding proteins (37). Once recruited to a promoter, the Grg proteins can recruit histone deacetylases or directly interact with histones or the basal transcriptional machinery, resulting in short-term or long-term repression (38–40). In yeast, the Groucho related protein, Tup1, can repress transcription by interacting with subunits of RNA polymerase II holoenzyme (41–42). TLE/Grg proteins are broadly expressed in developing organs and interact with multiple transcription factors involved in patterning and differentiation (43).
In Drosophila, Cut functions as a downstream effector of the Notch signaling pathway (1, 44–46). The Notch pathway is highly conserved across most species. In mammals, there are four Notch receptors (Notch1–4) and five ligands (delta like 1, delta like 3, delta like 4, jagged 1, and jagged 2). Notch signaling is activated when jagged or delta ligands bind to Notch receptors, resulting in the proteolytic cleavage of the Notch receptor releasing the notch intracellular domain (NICD) (47–49). NICD translocates to the nucleus and associates with the RBP-jk transcription factor to activate the expression of Notch effector proteins, such as the HES or HERP family proteins (49). These proteins recruit Grg proteins as cofactors, and this complex functions to repress tissue specific genes such as Myo D and Mash during development (49–51). We have previously shown that Cux1 co-localizes with Notch pathway components in numerous tissues during embryogenesis and co-immunoprecipitates with Grg4 in a rat kidney epithelial cell line expressing a constitutively active Notch1, called RKE Notchic cells (52–53). The RKE Notchic cells exhibit significantly higher levels of Cux1 expression compared to vector only transfected cells, and show reduced p27kip1 expression (53). Here we show a direct interaction between Cux1 and the co-repressor protein Grg4, and show that Cux1 is part of a complex bound to the p27kip1 promoter in the native chromatin configuration in the developing kidney.
Materials and methods
Antibodies
Rabbit Anti-CDP (Cux1) (#sc-13024), goat anti-CDP (Cux1) (#sc-6327), rabbit anti-Grg4 (#sc-9125), rabbit anti-Grg1 (#sc-9121), and rabbit anti-p27 (#sc-528) antibodies were purchased from Santa Cruz Biotechnology. Rabbit anti-HDAC1 (#2062) and anti-HDAC3 (#2632) antibodies were purchased from Cell Signaling.
Immunohistochemistry
Kidneys were isolated from 3-day-old C57BL/6J mice and immersion fixed in 4% paraformaldehyde and embedded in paraffin. Five micron-thick tissue sections were de-paraffinized with xylene and hydrated with graded ethanols. Immunolabeling was performed as described previously (53).
Preparation of kidney lysates and western blot analysis
Kidneys isolated from 3-day-old mice were isolated and frozen immediately in dry ice and ethanol and stored at −80°C until used. Kidneys were thawed on ice and chopped with a sterile scalpel blade before use. The chopped kidneys were suspended in Ripa buffer (0.01M Sodium phosphate, pH 7.2; 125mM sodium chloride; 50mM sodium fluoride; 0.1% SDS; 1mM ethylenediaminetetra acetic acid; 1% sodium deoxycholate; 1%NP40; 4µg/ml aprotinin; 4µg/ml pepstatin; 4µg/ml leupeptin; 1mM PMSF (phenylmethylsulfonyl fluoride). The tissue was homogenized, using a dounce homogenizer, until no cellular clumps were visible, (~25 times up and down). This was incubated on ice for 20 minutes and centrifuged at 15000 rpm at 4°C for 30 minutes. The supernatant was collected and measured for protein concentration using BCA protein assay (Biorad). Western blotting was performed as described previously (53).
Co-immunoprecipitations
Coimmunoprecipitation assays were performed as previously described (53). Briefly, 60µl protein A-Sepharose beads were washed with RIPA buffer and were incubated with anti-Grg4, anti- Cux1, anti-HDAC1, or anti-HDAC3 antibodies. Immune complexes were washed and incubated with 500µg of protein (lysate) isolated from newborn kidney. Reactions were washed and analyzed by Western blot analysis.
Transient reporter assay
Transient luciferase assay was performed as described previously (22). Briefly, human embryonic kidney (293T) cells were cultured in Dulbecco’s modified Eagle’s medium (glucose concentration, 450mg/dl) supplemented with heat-inactivated 10% fetal bovine serum and 100U/ml penicillin and 100mg/ml streptomycin under humidified 5% CO2/95% air at 37°C. Cells were plated at a concentration of 6X105 cells per well of twelve well plates 18 hrs prior to the transfection. Transient transfections were performed using Fugene transfection reagent (Roche Applied Science), using 0.5 µg of luciferase reporter plasmid containing p27 promoter sequence (−1609 to +178), along with 0.1 µg renilla-expressing plasmid (to correct for transfection efficiencies), pCMV/Cux1 and pKW/Grg4 at the concentrations indicated, and pcDNA3.1 (to control for non-specific vector effects). After 48 hours, cells were lysed, and luciferase and renilla activities were determined by enzyme assay kits. Luciferase activity was normalized to renilla activity as an internal transfection control. Comparisons between luciferase activity were made using one-way ANOVA. P < 0.05 was considered statistically significant.
GST pull down assay
Full length GST fusion protein (Grg4-GST) and full-length Cux1 were expressed in Escherichia coli BL21 (DE-3) cells by induction with isopropyl-B-thiogalactopyranoside for 2 hours at 30°C and 37°C respectively. Cell pellets were suspended in NETN buffer (20 mM Tris, pH 8.0; 100mM NaCl; 1mM EDTA; 0.5% Nonidet P-40) and native purification buffer (40 mM NaH2 PO4 pH 8.0 and .5 M NaCl) at 4°C supplemented with protease inhibitors (Roche), followed by lysis. Cleared lysates were suspended with washed Glutathione beads (Amersham) or probond resin (Invitrogen) and Grg4 or Cux1 was immobilized on glutathione beads or probond Ni agarose beads followed by extensive washings with the respective buffers listed above. The Cux1 protein was eluted by 250mM imidazole supplemented in the native buffer, pH 8.0. The GST pulldown assay was performed by incubating eluted Cux1 with glutathione-sepharose-bound GST, or GST fusion protein Grg4 in the binding buffer (10mM Tris, pH 7.6; 50mM NaCl; 5mM EDTA; 1%Triton-X 100; protease inhibitor) at 4°C for 1 hour. After incubation, the beads were washed three times with binding buffer and boiled in 6X sample buffer. The eluted binding proteins were electrophoresed in 4–6% gradient SDS-acrylamide gels (Biorad) and transferred to polyvinylidene fluoride (PVDF) membranes, followed by Western blot analysis.
Chromatin Immunoprecipitations
ChIP assays were performed using the EZ ChIP kit (Upstate Corp. Lake Placid, New York) according to manufacturer’s directions, and as previously described (54). RKE cells or kidneys isolated from newborn mice were used for ChIP analysis. Briefly, equal aliquots of isolated chromatin were subjected to immunoprecipitation with an anti-Cux1, anti-Grg4, anti-HDAC1, anti- HDAC3, anti-RNA polymerase II, or IgG antibodies. DNA associated with immunoprecipitates was used as a template for PCR analysis with primers producing 249 bp (A), 317 bp (B), or 191 bp (C) fragments of the p27kip1 promoter spanning −1609 to −1360 (A), −1272 to −955 (B), or −687 to −496 (C), relative to the transcription start site. Primers used were: A) 5'- AGCATTTTCGCCCTTCAAGAG-3' and 5'-CCAAGTGGGAAAGCCATTGC-3', B) 5'-TGAAGAGGCTTGAGAGCACTG-3' and 5'-TGGCTTGTTTGGAGCCTCAGTG-3', and C) 5'-AATGTCCTGGCGGCGGT-3' and 5'-GGAGGCTGACGAAGAAGAAGATG-3'. For newborn kidneys, p27kip1 primers used were: 5'-CAGAGCAGGTTTGTTGGCAGTC-3' and 3'-GGCTGACGAAGAAGAAGATGATTG-5'. PCR conditions were determined to ensure that results were within linear range of the PCR. Results obtained from unrelated antibody controls combined with enrichment when specific antibodies were used confirmed they were in the linear range of product amplification and not a consequence of non-specifically immunoprecipitating chromatin.
DNase 1 footprint analysis
The p27kip1 −1609/-1360 (fragment A) and −687/-496 (fragment C) amplification products from the ChIP analysis were used for DNA footprinting. DNA fragments obtained from the ChIP analysis were cloned into PCR Topo TA cloning vector (Invitrogen). DNase1 footprinting was performed using a core footprinting System from Promega. Briefly, double-stranded, single 32P-end-labeled probe was obtained by digesting the plasmid with Spe1 and Not1 to obtain the double stranded 249bp and 191bp fragments. The 5’ ends were then dephosphorylated and radiolabeled, followed by restriction digestion with Pme1 and Pst1 for p27kip1 −1609/-1360 (fragment A) and p27kip1 −687/-496 (fragment C) respectively, so that only the 3’ end remained labeled. These probes were incubated with in vitro synthesized Cux1, which was generated using the Tnt quick coupled transcription/translation kit and transcend tRNA kit (Promega) according to manufacturer’s directions. Various concentrations of DNase 1 were added and samples were incubated for one minute, followed by the addition of stop solution. Following purification, the DNA samples were electrophoresed through a 6% polyacrylamide, 7M urea sequencing gel at 1500V for 2 hours. Gels were dried and visualized by autoradiography.
Results
Cux1 is the murine homologue of the Drosophila gene Cut. In Drosophila, Cut is regulated by the Notch signaling pathway. Previously we showed that Cux1 expression is upregulated by an activated Notch1 in rat kidney epithelial (RKE) cells (Notchic cells), while p27kip1, a target of repression by Cux1, is downregulated (53). Moreover, we identified Grg4, a mammalian homologue of the co-repressor groucho, as a potential interacting partner of Cux1 in RKE cells. Grg4 was significantly upregulated in Notchic cells and co-immunoprecipitated with Cux1. In addition, Cux1 and Grg4 are co-expressed in the nephrogenic zone of the developing kidney, with highest expression in the presumptive podocytes of capillary loop staged glomeruli. Taken together, our previous results raised the possibility that Cux1 interacts with Grg4 to regulate gene expression during kidney development.
Cux1 interacts with Grg4 in Vivo and in Vitro
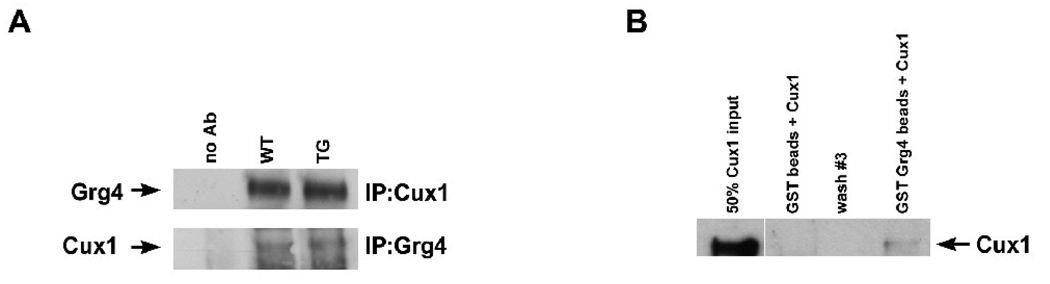
To determine whether Cux1 associates with Grg4 in the developing kidney, reciprocal co-immunoprecipitation assays were performed. Cux1 was immunoprecipitated from newborn kidney lysates from 3-day old wild type (WT) and Cux1 transgenic (TG) mice, followed by Western blotting for the presence of Grg4. Grg4 protein was coimmunoprecipitated with Cux1 from the kidney lysates, but not in control precipitations (Figure 1A). Similar immunoprecipitation experiments were performed showing that Cux1 protein was coimmunoprecipitated with Grg4 from newborn kidney lysates (Figure 1A). The presence of Groucho binding sites on Cux1 raised the possibility that Cux1 can physically interact with Grg4. To test this directly, we performed pulldown assays using bacterially expressed GST fused full-length Grg4 and full-length Cux1 protein. GST fused Grg4 protein was immobilized on glutathione agarose beads and incubated with bacterially expressed Cux1. This showed that Cux1 interacts directly with GST-Grg4 (Figure 1B). In contrast, the GST protein alone incubated with Cux1 showed no interaction. These results indicate a direct interaction between Grg4 and Cux1, and suggest that Cux1 and Grg4 form a complex during kidney development. The weaker interaction between Cux1 and Grg4 detected by the pulldown assay, compared to the co-immunoprecipitation results, suggests that direct interaction of Cux1 with Grg4 may be stabilized by other proteins present in a complex.
Figure 1. In vivo and in vitro interactions between Cux1 and Grg4.
A: Kidney lysates from 3-day-old wild type (WT) and Cux1 transgenic (TG) mice were immunoprecipitated with rabbit polyclonal anti-Cux1 (top panel) or rabbit polyclonal anti-Grg4 (bottom panel) as indicated. The immunoprecipitates were then blotted for Grg4 (top panel) or Cux1 (bottom panel) as indicated. Grg4 can be seen in WT and TG kidneys, after immunoprecipitation with Cux1 (right lanes, top panel). Cux1 can be seen in WT and TG kidneys, after immunoprecipitation with Grg4 (right lanes, bottom panel). No bands are observed when protein G beads alone are precipitated (no Ab). B: Bacterially expressed Cux1 protein was incubated with GST- Grg4 fusion protein immobilized on Glutathione agarose beads. Following washing, GST beads alone with Cux1, wash #3, and GST-Grg4 immobilized beads with Cux1 were transferred to a membrane and blotted with antibody directed against Cux1 protein. Only GST-Grg4 with Cux1 protein and the Cux1 input showed a positive band, indicating a direct physical interaction between Cux1 and Grg4.Lane 1 shows 50% of the total purified recombinant Cux1 protein used for the assay.
Grg4 enhances the ability of Cux1 to repress p27kip1 promoter activity
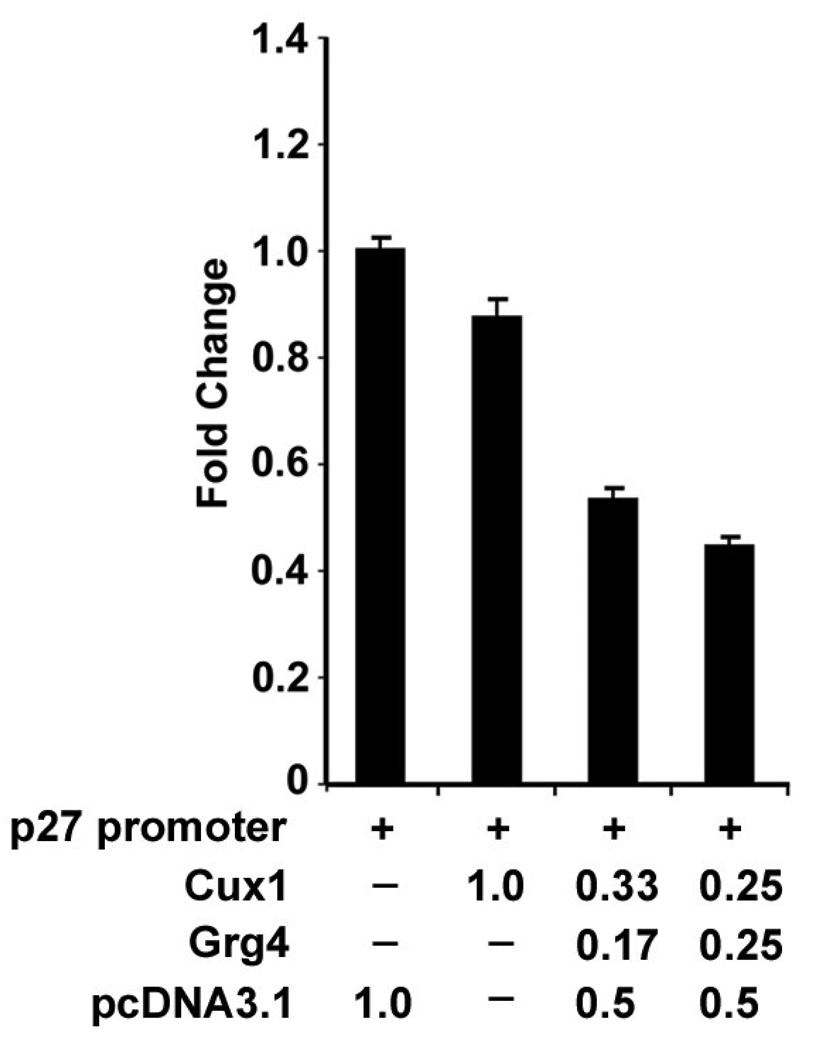
We previously showed that Cux1 represses p27kip1 promoter activity in a reporter assay. In addition, transgenic mice ectopically expressing Cux1 exhibit multiorgan hyperplasia, similar to p27kip1 knockout mice (22). Moreover, rat kidney epithelial cells stably transfected with the intracellular active form of Notch 1 (Notchic) showed increased Cux1 and Grg4 expression and decreased p27kip1 expression (53). To evaluate the functional importance of Grg4 on Cux1 mediated p27kip1 repression, we compared the ability of Cux1 to repress p27kip1 promoter activity with or without Grg4 (Fig 2). Unsynchronized 293T cells were cotransfected with a p27kip1/luciferase reporter construct and either the empty CMV vector, or the CMV/Cux1 expression construct, with or without a CMV/Grg4 expression construct. In three separate experiments, the luciferase activity was significantly reduced in the presence of Cux1 and Grg4, compared with Cux1 alone, suggesting that Cux1 repression of p27kip1 involves Grg4.
Figure 2. Repression of p27kip1 promoter activity by Cux1 and Grg4.
293T cells were transiently transfected with 1 µg of reporter construct containing p27 upstream sequences from −1609 to +178 fused to the luciferase reporter gene (+), together with different concentrations of the Cux1 and Grg4 expression vectors (amounts shown are in µg) or with empty pcDNA3.1 expression vector (amounts shown are in µg). In the presence of Grg4, significantly less Cux1 is required to repress p27 promoter activity (3rd and 4th bar). Promoter activity was plotted as fold change, normalized to the expression of a co-transfected renilla expression construct. Activity is expressed as the mean of three separate experiments performed in triplicate. Error bars indicate standard deviation. ANOVA showed a significant reduction in luciferase activity following cotransfection with Grg4 in a dose dependent manner (P< 0.002).
Co-expression and Co-immunoprecipitation of HDACs with Cux1
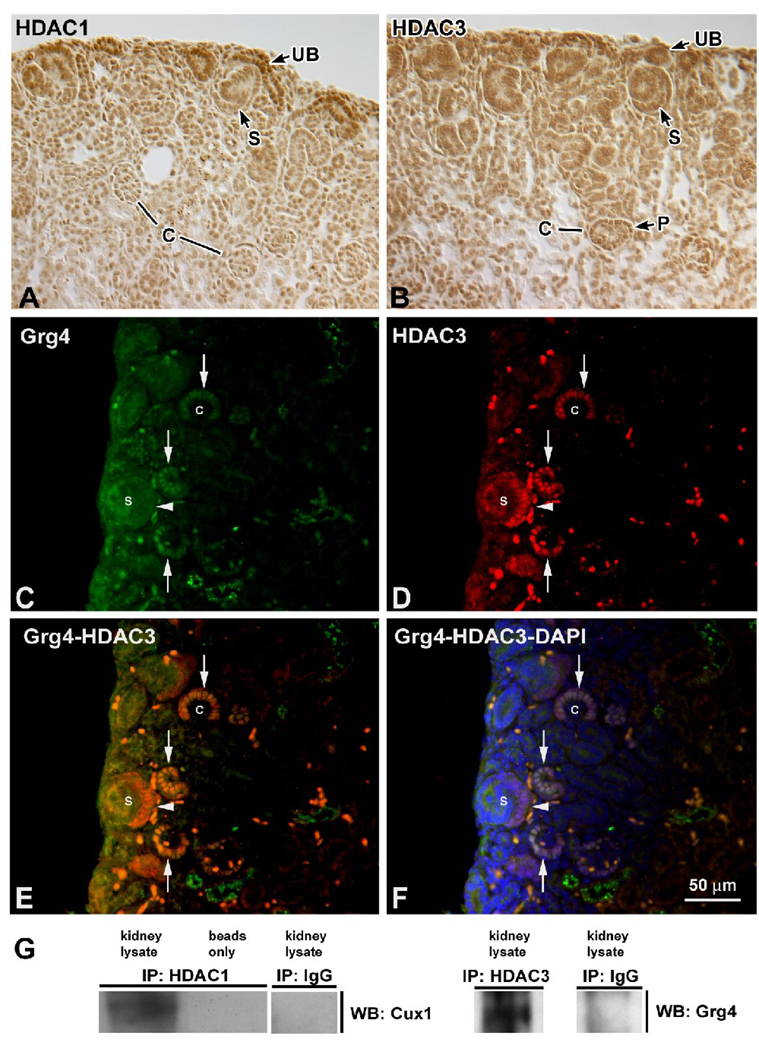
One mechanism of transcriptional repression by Cux1 involves the recruitment of HDAC1 (24). Groucho proteins can also act as cofactors either alone or by recruiting HDACs (55–57). To begin to determine whether Cux1 interacts with HDACs in the developing kidney, we evaluated the expression of HDAC1 and HDAC3. HDAC1 was expressed throughout the nephrogenic zone, with highest levels in the ureteric buds, and lower levels in the condensing mesenchyme and early nephric structures, including S-shaped bodies and capillary loop staged glomeruli (Fig. 3A). Similarly, HDAC3 was expressed in the nephrogenic zone, including the ureteric bud and S-shaped bodies. In capillary loop stage glomeruli, HDAC3 expression was observed in the presumptive podocytes (Fig. 3B). The expression of Cux1 (not shown) is broader and overlaps with the expression of both HDAC1 and HDAC3 (53). Grg4 was expressed at low levels in the nephrogenic zone, but was upregulated in the presumptive podocytes of capillary loop staged glomeruli (Fig. 3C), where it overlapped with the expression of HDAC3 (Fig. 3D–F). To begin to determine whether Cux1 repression involves the recruitment of HDACs, we asked whether an interaction between Cux1 and HDAC proteins could be detected in vivo. HDAC1 was immunoprecipitated from kidneys isolated from 3-day-old mice, followed by Western blotting for the presence of Cux1. Cux1 protein was coimmunoprecipitated with HDAC1 from the kidney lysates, but not in control preparations (Fig. 3G). Similar immunoprecipitation experiments were performed showing that Grg4 was coimmunoprecipitated with HDAC3 from the newborn kidney lysates (Fig. 3G).
Figure 3. Co-localization and interaction of Cux1, Grg4, and HDACs.
A: HDAC1 was expressed at highest levels in the nephrogenic zone, where it was localized to mesenchyme, ureteric buds (UB), and early nephric structures, including S-shaped bodies (S), but down regulated in capillary loop staged glomeruli (C). B: HDAC3 was also expressed in the nephrogenic zone, including the ureteric bud (UB) and S shaped bodies (S). In capillary loop stage glomeruli (C), HDAC3 expression was observed in the presumptive podocytes (P). C: Grg4 was expressed at low levels in the nephrogenic zone, including S-shaped bodies (arrowhead), and upregulated in the presumptive podocytes of capillary loop staged glomeruli (arrows). D: HDAC3 was similarly expressed in the nephrogenic zone, with highest levels in the presumptive podocytes (arrows) and in S-shaped bodies (arrowheads). E, F: Co-localization of HDAC3 and Grg4 without (E) or with (F) DAPI in capillary loop staged glomeruli (arrows). G: Newborn kidney lysates were immunoprecipitated (IP) with protein G beads alone (beads only), rabbit IgG (IgG), rabbit polyclonal anti-HDAC1 (left panel), or rabbit polyclonal anti-HDAC3 (right panel) as indicated. The immunoprecipitates were then blotted for Cux1 (left panel) or Grg4 (right panel) as indicated. Cux1 can be seen in newborn kidneys after immunoprecipitation with HDAC1 (left lane, left panel), and Grg4 can be seen in newborn kidneys after immunoprecipitation with HDAC3 (left lane, right panel).
Cux1, Grg4, HDAC1, and HDAC3 are present in complexes bound to the native promoter of p27kip1
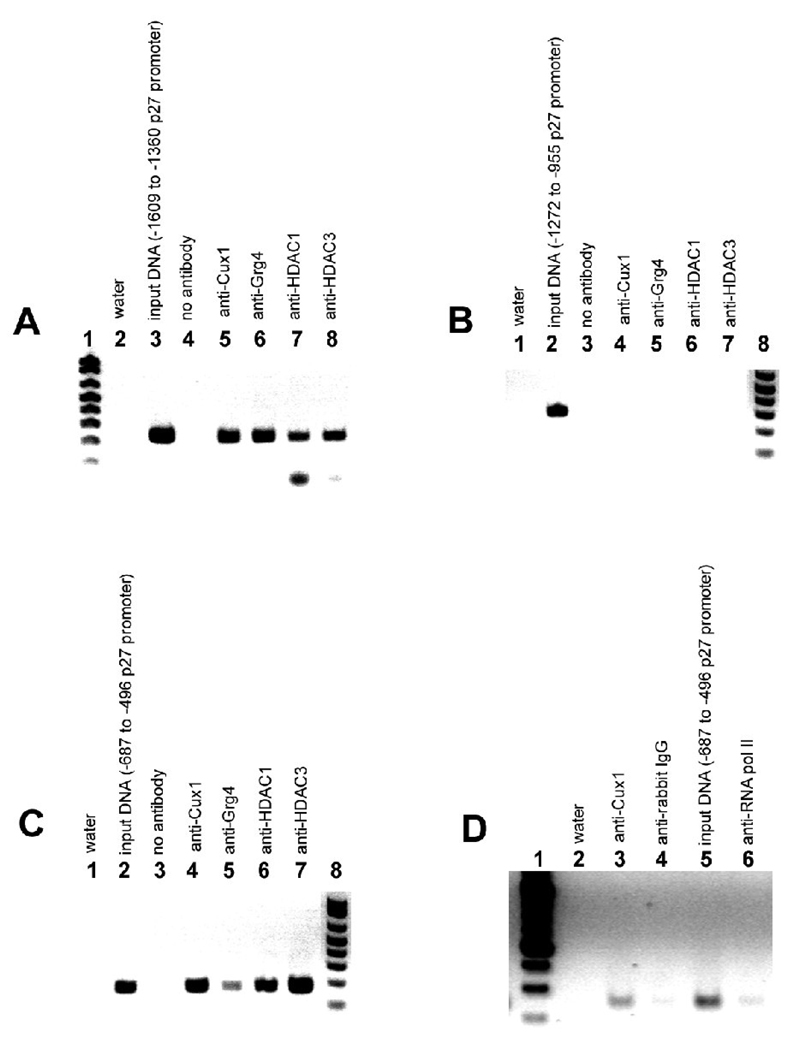
Our previous studies evaluating Cux1 transgenic mice revealed p27kip1 as a potential direct target of Cux1 during nephrogenesis. To directly determine whether Cux1 interacts with the native p27kip1 promoter, we performed chromatin immunoprecipitation (ChIP) analysis using chromatin isolated from Notchic cells or from newborn mouse kidneys. The ChIP assays were carried out using IgG (negative control), anti-Cux1, anti-Grg4, anti-HDAC1, anti-HDAC3, and anti-RNA polymerase II. Multiple primer sets were designed to amplify fragments of the p27kip1 promoter from 150–350 bp in length. Together, these primer sets spanned the entire length of the p27kip1 promoter. We found that two of the primer sets amplified the p27kip1 promoter following ChIP (panel A and C). The first positive primer set amplified a 191 bp fragment that contained two Sp1 sites and the CCAAT box at −543 to −522 relative to the transcription start site and is shown in panel C of Fig 4. The second primer set amplified a 249 bp fragment located at ~ −1.5 kb from the transcription start site and is shown in panel A of Fig 4. Amplification of sequences between regions A and C produced no Cux1 bound products. These results clearly show that Cux1 is part of a complex bound to two different regions of the p27kip1 promoter in its native chromatin configuration. Therefore, p27kip1 is a direct in vivo transcriptional target of Cux1, providing a mechanism for the regulation of cell cycle progression by Cux1 during kidney development. Moreover, our results show that Grg4, HDAC1 and HDAC3 are part of a complex bound to the same regions of the p27kip1 promoter in its native chromatin configuration.
Figure 4. p27Kip1 is a direct Cux1 target in the developing kidney.
Complexes isolated from Notchic cells (A–C) or from newborn kidneys (D) were immunoprecipitated with anti-Cux1, anti-Grg4, anti-HDAC1, anti-HDAC3, or anti-RNA polymerase II antibodies followed by reverse cross-linking of protein and DNA, and PCR amplification of a 249 bp fragment of the p27kip1 promoter spanning −1609 to −1360 (A), or a 191 bp fragment of the p27kip1 promoter spanning −687 to −496 (C, D), relative to the transcription start site. A: Inverse image of ethidium-stained agarose gel showing PCR amplification of a 249 bp p27kip1 product from input DNA (lane 3), following ChIP with antibody directed against Cux1 (lane 5), Grg4 (lane 6), HDAC1 (lane 7), and HDAC3 (lane 8), indicating that Cux1, Grg4, HDAC1, and HDAC3 interact with a site within the 5’ region of the p27kip1 promoter in vivo. B: To control for non-specific chromatin immunoprecipitation, a 317 bp product from −1272 to −955 of the p27kip1 promoter was amplified from input DNA (lane 2), following ChIP with antibody directed against Cux1 (lane 4), Grg4 (lane 5), HDAC1 (lane 6), and HDAC3 (lane 7). C: PCR amplification of a 191 bp p27kip1 product from input DNA (lane 2), following ChIP with antibody directed against Cux1 (lane 4), Grg4 (lane 5), HDAC1 (lane 6), and HDAC3 (lane 7), indicating that Cux1, Grg4, HDAC1, and HDAC3 interact with a site within the 3’ region of the p27kip1 promoter in vivo. D: PCR amplification of a 191 bp p27kip1 product from input DNA (lane 5), following ChIP with antibody directed against Cux1 (lane 3), and RNA polymerase II (lane 6), indicating that Cux1 interact with a site within the 3’ region of the p27kip1 promoter in newborn kidneys in vivo. In A controls were water (lane 2) and no antibody (lane 4). In both B and C, controls were water (lane 1) and no antibody (lane 3). In D controls were water (lane 2) and normal rabbit IgG (lane 4).
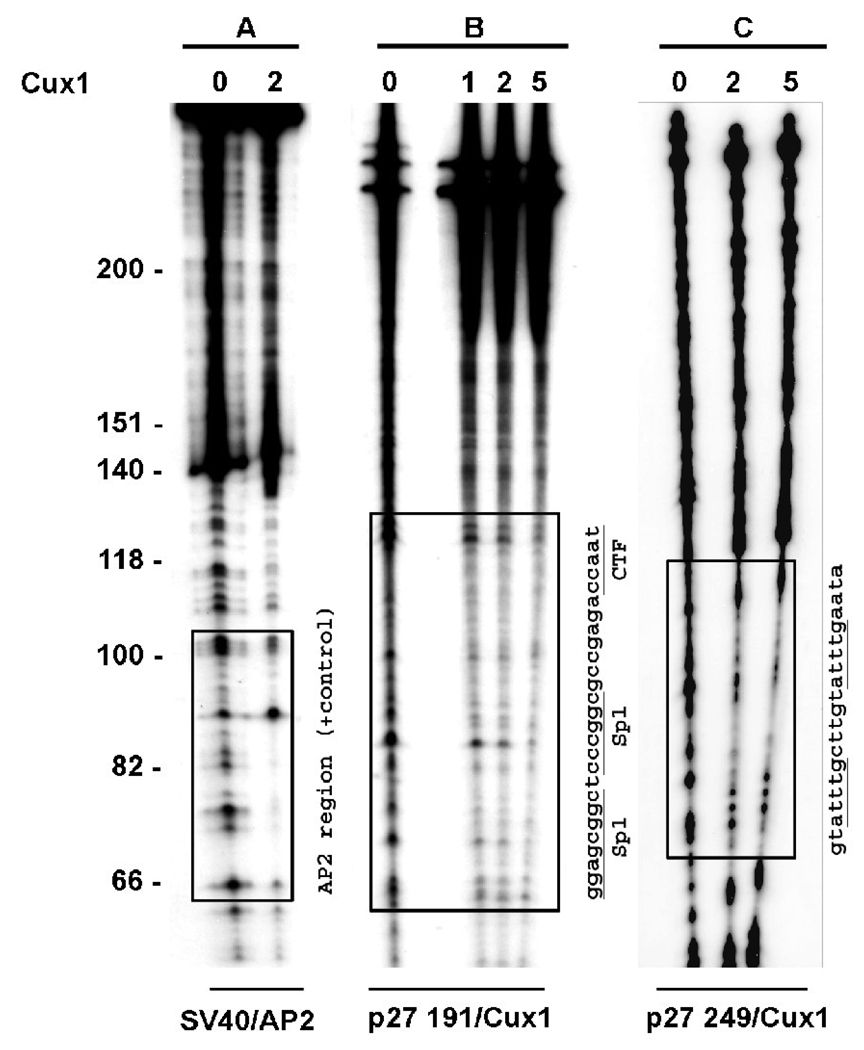
Identification of a novel region on the p27kip1 promoter bound by Cux1
To identify the DNA sequence in the p27kip1 promoter bound by Cux1 in vivo, we performed DNAse 1 footprinting analysis using the amplification products from the ChIP assays. The ChIP positive amplification products (p27/191) and (p27/229) were end-labeled and incubated with in vitro synthesized Cux1. We observed protection of a 38 bp region overlapping two SP1 sites and a CCAAT box within region B (Figure 5, panel B). A second protected region was identified in the p27kip1 promoter in region C (Figure 5, panel C). This novel site of interaction between Cux1 and the p27kip1 promoter is in a region located ~1.5 kb from the transcription start site and contains two ATTTG repeats, sequences previously shown to be bound by Cux1 (58).
Figure 5. Two separate Cux1 binding sites on the p27kip1 promoter.
DNAse I footprint analysis of the p27kip1 promoter regions bound by Cux1 in vivo. −687 to −496 (panel B) and −1609 to −1360 (panel C) PCR amplification products from ChIP assays were radiolabeled and incubated with purified in vitro synthesized Cux1 protein (amounts shown are in µl) and then digested with DNase1. Regions of DNA protected from DNase1 digestion are indicated by boxed regions. The corresponding sequence for each protected region is shown, with the previously identified Cux1 target sequences underlined. Panel A shows radiolabeled SV40 DNA following incubation with AP2 protein and DNase1 digestion as a positive control. Boxed region shows region of DNA protected from DNase1 digestion.
DISCUSSION
During kidney development, Cux1 is highly expressed in the nephrogenic zone, where it is associated with cell proliferation. Cux1 is down regulated at later stages of nephrogenesis, when cell proliferation is decreased (59). Transgenic mice ectopically expressing Cux1 develop renal hyperplasia resulting from the down regulation of p27kip1 (22). In addition, promoter reporter assays demonstrated that Cux1 repressed p27kip1 promoter activity in a concentration dependent manner. Cux1 can be regulated by the Notch signaling pathway suggesting that cell proliferation induced by Notch signaling may occur through the down regulation of p27kip1. To begin to determine the mechanism of p27kip1 repression by Cux1, we evaluated whether Cux1 interacts with the Notch effector protein groucho homologue Grg4 during kidney development
Two mechanisms of repression have been described for Cux1 protein: 1) Passive repression via competition for CCAAT or Sp1 binding site occupancy, preventing activation by the corresponding transcription factors, or 2) active repression via a carboxy terminal repression domain following binding at a distance from the transcription start site (23). The first mechanism is thought to involve rapid and unstable DNA binding activity and involves the first two cut repeats, while the second mechanism is thought to involve stable DNA binding activity and involves the third cut repeat and the homeodomain (2). Our results suggest that Cux1 can bind to the Sp1 and CCAAT sequences within the p27kip1 promoter. This site has previously been shown to be required for p27kip1 promoter activity, suggesting that Cux1 may engage in passive repression (60). However, we also identified a novel DNA binding site within the p27kip1 promoter that is bound by Cux1. The interaction between Cux1 and Grg4, both by co-immunoprecipitation and pull down assays, suggests that Cux1 repression of p27kip1 involves the recruitment of Grg4 to the promoter. This is supported by the observation that Grg4 significantly enhanced the repression of p27kip1 promoter activity by Cux1 in a reporter assay. Grg proteins cannot bind DNA by themselves, but interact with other transcription factors. One mechanism of repression by Grg proteins is the recruitment of HDACs to form a repression complex (37, 41–43). The chromatin immunoprecipitation of Cux1, in addition to Grg4 and HDAC1/3, on the native p27kip1 promoter, raises the possibility that p27kip1 repression by Cux1 involves the formation of a repression complex.
There are four domains in Cux1 that could potentially be involved in an interaction with Groucho proteins. The N-terminus of Cux1 protein contains the sequence FALNSLL that is similar to the Eh1 (Engrailed like homeodomain1 sequence) motif designated by the consensus sequence FXIXXIL (43). This sequence was originally identified in the Drosophila homeodomain proteins Engrailed (En) and Goosecoid and shown to mediate the interaction of these homeodomain proteins with groucho (61–62). In addition, the mammalian homeodomain proteins PRH and Six3 interact with groucho family members through the Eh1 domain (63–64). Three additional groucho interaction motifs exist in Cux1. These PKPW motifs are found in each of the cut repeats (CR) (65). This sequence is similar to the consensus sequence (W/F/Y)(K/R)P(WFY) used to recruit Groucho proteins to transcription factors (43). Further studies will be required to identify the specific sites of interaction between Cux1 and Grg4.
ACKNOWLEDGEMENTS
We thank Rosetta Barkley and Eileen Roach for expert technical assistance. We thank members of the Kidney Institute for many helpful discussions. MS was supported by a KUMC Biomedical Research Institute Award. GBVH was supported by K-INBRE award P20 RR16475 and by NIH grant DK58377.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- 2.Sansregret L, Nepveu A. The multiple roles of Cux1: Insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Alcalay NI, Vanden Heuvel GB. Pathophysiological roles of Cux1: Regulation of cell proliferation and differentiation in the kidney. Frontiers in Bioscience. doi: 10.2741/3582. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta S, Luong MX, Bleuming SA, Miele A, Luong M, Young D, Knudsen ES, Van Wijnen AJ, Stein JL, Stein GS. Tumor suppressor pRB functions as a co-repressor of the CCAAT displacement protein (CDP/cut) to regulate cell cycle controlled histone H4 transcription. J Cell Physiol. 2003;196:541–556. doi: 10.1002/jcp.10335. [DOI] [PubMed] [Google Scholar]
- 5.Lievens PM, Donady JJ, Tufarelli C, Neufeld EJ. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem. 1995;270:12745–12750. doi: 10.1074/jbc.270.21.12745. [DOI] [PubMed] [Google Scholar]
- 6.Nirodi C, Hart J, Dhawan P, Moon NS, Nepveu A, Richmond A. The role of CDP in the negative regulation of CXCL1 gene expression. J Biol Chem. 2001;276:26122–26131. doi: 10.1074/jbc.M102872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. EMBO J. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Q, Maitra U, Johnston D, Lozano M, Dudley JP. The homeodomain protein CDP regulates mammary-specific gene transcription and tumorigenesis. Mol Cell Biol. 2004;24:4810–4823. doi: 10.1128/MCB.24.11.4810-4823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truscott M, Raynal L, Premdas P, Goulet B, Leduy L, Bérubé G, Nepveu A. CDP/Cux stimulates transcription from the DNA polymerase alpha gene promoter. Mol Cell Biol. 2003;23:3013–3028. doi: 10.1128/MCB.23.8.3013-3028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon NS, Premdas P, Truscott M, Leduy L, Bérubé G, Nepveu A. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol Cell Biol. 2001;21:6332–6345. doi: 10.1128/MCB.21.18.6332-6345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadieux C, Harada R, Paquet M, Côté O, Trudel M, Nepveu A, Bouchard M. Polycystic kidneys caused by sustained expression of Cux1 isoform p75. J Biol Chem. doi: 10.1074/jbc.M709332200. in press. [DOI] [PubMed] [Google Scholar]
- 12.Truscott M, Harada R, Vadnais C, Robert F, Nepveu A. p110 Cux1 Cooperates with E2F Transcription Factors in the Transcriptional Activation of Cell Cycle-Regulated Genes. Mol Cell Biol. doi: 10.1128/MCB.02089-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Superti-Furga G, Barberis A, Schreiber E, Busslinger M. The protein CDP, but not CP1,footprints on the CCAAT region of the [gamma]-globin gene in unfractionated B-cell extracts. Biochimica et Biophysica Acta. 1989;1007:237–242. doi: 10.1016/0167-4781(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 14.Dufort D, Nepveu A. The human cut homeodomain protein represses transcription from the c-myc promoter. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- 16.Valarche I, Tissier-Seta JP, Hirsch MR, Martinez S, Goridis C, Brunet JF. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 17.Banan M, Rojas IC, Lee WH, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD. Interaction of the Nuclear Matrix-associated Region (MAR)-Binding Proteins, SATB1 and CDP/Cux, with a MAR Element (L2a) in an Upstream Regulatory Region of the Mouse CD8a Gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- 18.Higgy NA, Tarnasky HA, Valarche I, Nepveu A, van der Hoorn FA. Cux/CDP homeodomain protein binds to an enhancer in the rat c-mos locus and represses its activity. Biochimica et Biophysica Acta. 1997;1351:313–324. doi: 10.1016/s0167-4781(96)00221-7. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross SR, Dudley JP. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skalnik DG, Strauss EC, Orkin SH. CCAAT displacement protein as a repressor of the myelomonocytic- specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 21.Coqueret O, Berube G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ledford AW, Brantley JG, Kemeny G, Foreman TL, Quaggin SE, Igarashi P, Oberhaus SM, Rodova M, Calvet JP, Vanden Heuvel GB. Deregulated Expression of the Homeobox Gene Cux-1 in Transgenic Mice Results in Downregulation of p27kip1 Expression during Nephrogenesis, Glomerular Abnormalities, and Multiorgan Hyperplasia. Developmental Biology. 2002;245:157–171. doi: 10.1006/dbio.2002.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailly F, Berube G, Harada R, Mao PL, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, LeLeiko NS, Walsh MJ. Transcriptional Repression of the Cystic Fibrosis Transmembrane Conductance Regulator Gene,Mediated by CCAAT Displacement Protein/cut Homolog, Is Associated with Histone Deacetylation. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 25.Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 26.Goulet B, Watson P, Poirier M, Leduy L, Berube G, Meterissian S, Jolicoeur P, Nepveu A. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 2002;62:6625–6633. [PubMed] [Google Scholar]
- 27.Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic Ablation of the CDP/Cux Protein C Terminus Results in Hair Cycle Defects and Reduced Male Fertility. Mol Cell Biol. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, Tucker PW, Neufeld EJ, Scheuermann RH. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- 30.Brantley JG, Sharma M, Alcalay NI, Vanden Heuvel GB. Cux-1 transgenic mice develop glomerulosclerosis and interstitial fibrosis. Kidney Int. 2003;63:1240–1248. doi: 10.1046/j.1523-1755.2003.00889.x. [DOI] [PubMed] [Google Scholar]
- 31.Vanden Heuvel GB, Brantley JG, Alcalay NI, Sharma M, Kemeny G, Warolin J, Ledford AW, Pinson DM. Hepatomegaly in transgenic mice expressing the homeobox gene cux-1. Molecular Carcinogenesis. 2005;43:18–30. doi: 10.1002/mc.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 33.Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, Kaushansky K, Roberts JM. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia,retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 35.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 36.Cadieux C, Fournier S, Peterson AC, Bedard C, Bedell BJ, Nepveu A. Transgenic Mice Expressing the p75 CCAAT-Displacement Protein/Cut Homeobox Isoform Develop a Myeloproliferative Disease-Like Myeloid Leukemia. Cancer Res. 2006;66:9492–9501. doi: 10.1158/0008-5472.CAN-05-4230. [DOI] [PubMed] [Google Scholar]
- 37.Gasperowicz M, Otto F. Mammalian Groucho homologs: Redundancy or specificity? J. Cell. Biochem. 2005;95:670–687. doi: 10.1002/jcb.20476. [DOI] [PubMed] [Google Scholar]
- 38.Grbavec D, Lo R, Liu Y, Stifani S. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur J Biochem. 1998;258:339–349. doi: 10.1046/j.1432-1327.1998.2580339.x. [DOI] [PubMed] [Google Scholar]
- 39.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 40.Pinto M, Lobe CG. Products of the grg (Groucho-related gene) family can dimerize through the amino-terminal Q domain. J Biol Chem. 1996;271:33026–33031. doi: 10.1074/jbc.271.51.33026. [DOI] [PubMed] [Google Scholar]
- 41.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Du W, Stillman DJ, Roth SY. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buscarlet M, Stifani S. The 'Marx' of Groucho on development and disease. Trends Cell Biol. 2007;17:353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Jack J, DeLotto Y. Effect of wing scalloping mutations on cut expression and sense organ differentiation in the Drosophila wing margin. Genetics. 1992;131:353–363. doi: 10.1093/genetics/131.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Celis J, Garcia-Bellido A, Bray S. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- 46.Neumann C, Cohen S. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 1996;122:3477–3485. doi: 10.1242/dev.122.11.3477. [DOI] [PubMed] [Google Scholar]
- 47.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 48.McCright B. Notch signaling in kidney development. Curr Opin Nephrol Hypertens. 2003;12:5–10. doi: 10.1097/00041552-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 49.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the notch signaling pathway. Journal of Cellular Physiology. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 50.Sun J, Kamei CN, Layne MD, Jain MK, Liao JK, Lee ME, Chin MT. Regulation of Myogenic Terminal Differentiation by the Hairy-related Transcription Factor CHF2. J Biol Chem. 2001;276:18591–18596. doi: 10.1074/jbc.M101163200. [DOI] [PubMed] [Google Scholar]
- 51.Chen H, Thiagalingam A, Chopra H, Borges MW, Feder JN, Nelkin BD, Baylin SB, Ball DW. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: A hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. PNAS. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma M, Fopma A, Brantley JG, Vanden Heuvel GB. Coexpression of Cux-1 and notch signaling Coexpression of Cux-1 and notch signaling pathway components during kidney development. Developmental Dynamics. 2004;231:828–838. doi: 10.1002/dvdy.20175. [DOI] [PubMed] [Google Scholar]
- 54.Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development. 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi CY, Kim YH, Kwon HJC, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem. 1999;279:33194–33197. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- 56.Yochum GS, Ayer DE. Pf1, a Novel PHD Zinc Finger Protein That Links the TLE Corepressor to the mSin3A-Histone Deacetylase Complex. Mol Cell Biol. 2001;21:4110–4118. doi: 10.1128/MCB.21.13.4110-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 58.Erturk E, Ostapchuk P, Wells SI, Yang J, Gregg K, Nepveu A, Dudley JP, Hearing P. Binding of CCAAT Displacement Protein CDP to Adenovirus Packaging Sequences. J.Virol. 2003;77:6255–6264. doi: 10.1128/JVI.77.11.6255-6264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanden Heuvel GB, Bodmer R, McConnell KR, Nagami GT, Igarashi P. Expression of a cut-related homeobox gene in developing and polycystic mouse kidney. Kidney Int. 1996;50:453–461. doi: 10.1038/ki.1996.336. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Lin SC. Molecular characterization of the cyclin-dependent kinase inhibitor p27 promoter. Biochim Biophys Acta. 1997;1353:307–317. doi: 10.1016/s0167-4781(97)00063-8. [DOI] [PubMed] [Google Scholar]
- 61.Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;1997:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- 64.Swingler TE, Bess KL, Yao J, Stifani S, Jayaraman PS. The Proline-rich Homeodomain Protein Recruits Members of the Groucho/Transducin-like Enhancer of Split Protein Family to Co-repress Transcription in Hematopoietic Cells. J Biol Chem. 2004;279:34938–34947. doi: 10.1074/jbc.M404488200. [DOI] [PubMed] [Google Scholar]
- 65.Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]