Abstract
There is increasing evidence that noncoding RNAs play a functional role in the nucleus. We previously reported that the microRNA (miRNA), miR-206, is concentrated in the nucleolus of rat myoblasts, as well as in the cytoplasm as expected. Here we have extended this finding. We show by cell/nuclear fractionation followed by microarray analysis that a number of miRNAs can be detected within the nucleolus of rat myoblasts, some of which are significantly concentrated there. Pronounced nucleolar localization is a specific phenomenon since other miRNAs are present at only very low levels in the nucleolus and occur at much higher levels in the nucleoplasm and/or the cytoplasm. We have further characterized a subset of these miRNAs using RT-qPCR and in situ hybridization, and the results suggest that some miRNAs are present in the nucleolus in precursor form while others are present as mature species. Furthermore, we have found that these miRNAs are clustered in specific sites within the nucleolus that correspond to the classical granular component. One of these miRNAs is completely homologous to a portion of a snoRNA, suggesting that it may be processed from it. In contrast, the other nucleolar-concentrated miRNAs do not show homology with any annotated rat snoRNAs and thus appear to be present in the nucleolus for other reasons, such as modification/processing, or to play roles in the late stages of ribosome biosynthesis or in nonribosomal functions that have recently been ascribed to the granular component of the nucleolus.
Keywords: microRNAs, nucleolus, nucleus, muscle, nuclear RNAs, in situ nucleic acid hybridization
INTRODUCTION
MicroRNAs (miRNA) have been known and studied for a number of years now, but their spatial disposition within the cell has sometimes been difficult to determine. Many miRNAs are transcribed as large primary transcripts, sometimes containing more than one embedded miRNA element, which are commonly processed into precursor “hairpin” miRNAs by Drosha/DGCR8 in the nucleus (Zeng et al. 2005; Yeom et al. 2006; Carthew and Sontheimer 2009). This processing can occur cotranscriptionally (Morlando et al. 2008; Pawlicki and Steitz 2008) and is subject to regulation (Thomson et al. 2006; Lee et al. 2008). For example, lin-28 can bind to pri-let-7 at some stages during neuronal development and inhibit its processing to the mature form (Wulczyn et al. 2007; Newman et al. 2008). Drosha-generated precursors are transported to the cytoplasm where Dicer further processes them into small double-stranded RNAs that interact with Argonaute proteins (Matranga and Zamore 2007; Carthew and Sontheimer 2009). Like Drosha processing, this step is also subject to regulation (Lee et al. 2008). Additionally, some miRNA transcripts are also subject to A→I editing (Luciano et al. 2004; Blow et al. 2006; Habig et al. 2007; Kawahara et al. 2008), and such editing can interfere with processing (Yang et al. 2006; Kawahara et al. 2007). miRNAs that have been diced and assembled into an Argonaute complex then interact via short “seed” sequences with mRNAs in the cytoplasm to regulate translation (Du and Zamore 2005; Filipowicz et al. 2008), and in some cases, miRNAs have been isolated in polysomal complexes (Kim et al. 2004; see Wang et al. 2008).
There is also mounting evidence that small RNAs, including miRNAs, play important nuclear roles and that some miRNAs, once exported to the cytoplasm, may revisit the nucleus (Hwang et al. 2007; Guang et al. 2008; Kim et al. 2008; Marcon et al. 2008; Ohrt et al. 2008; Place et al. 2008). The nuclear site(s) and the functional significance of these returning miRNAs are not known.
During a study of miRNAs expressed during muscle cell differentiation (Politz et al. 2006), we observed that miR-206, a microRNA implicated in muscle cell differentiation and maintenance (Sempere et al. 2004; Kim et al. 2006; McCarthy 2008; van Rooij et al. 2008), is concentrated in the nucleoli of a rat myoblast cell line. We decided to follow up on this observation to determine whether this nucleolar localization was specific to miR-206 or muscle-specific miRNAs or whether many miRNAs might visit the nucleolus. The nucleolus is a tripartite structure in terms of known functions and corresponding fiduciary molecular markers (Huang 2002; Raska et al. 2006; Boisvert et al. 2007). Preribosomal RNA is transcribed at so-called fibrillar centers, and initially processed in a surrounding domain known as the nucleolar dense fibrillar component (DFC), where fibrillarin and GAR1, among other proteins, are complexed with small nucleolar RNAs (snoRNAs) with the resulting snoRNPs modifying rRNA at multiple sites (Ochs et al. 1985; Matera et al. 1994, 2007; Pogacic et al. 2000; Gerbi et al. 2003). A third nucleolar domain, the granular component (GC), harbors the late stages of ribosome biosynthesis, where some rRNA modification still occurs and snoRNPs also accumulate. Although the nucleolus has classically been viewed solely as the site of ribosome biosynthesis, more recently it has been found that other proteins and RNAs also localize therein, including components involved in cell cycle control (Pederson 1998; Visintin and Amon 2000; Raska et al. 2006; Ma and Pederson 2008b; Pederson and Tsai 2009). We earlier determined that miR-206 colocalizes with 28S rRNA in the GC, where some of the newly identified proteins with nonribosomal functions also concentrate (Karayan et al. 2001; Politz et al. 2002, 2005; Ma and Pederson 2008a).
We have expanded our studies and now report that a number of miRNAs associate with the nucleolus in rat myoblasts. Some of these are significantly concentrated in the nucleolus relative to their prevalence in the nucleoplasm and/or the cytoplasm. We have also defined the intranucleolar localization of three of the most highly nucleolus-concentrated miRNAs and find that they are clustered, like miR-206, in the GC, raising the possibility that they may be involved in common functions at these particular sites.
RESULTS
To systematically determine which miRNAs might be present in the nucleolus, nuclei were isolated from L6 rat myoblasts using a protocol optimized specifically for this cell type (Mellon and Bhorjee 1982). We found that this method exactly replicated the high nuclear purity initially reported. The cytoplasmic fraction was saved, and the nuclear fraction was subjected to a nucleolus:nucleoplasm separation protocol (Maggio et al. 1963; Muramatsu et al. 1963) modified for cultured mammalian cells by our laboratory (Bhorjee and Pederson 1972, 1973). Total RNA was then isolated from each fraction and analyzed by microarray profiling.
Because mature miRNAs are so small, their detection by hybridization represents a technical challenge. Accordingly, we employed a system that uses locked nucleic acid (LNA) probes, which are phosphodiester backbone oligodeoxynucleotides containing at one or more internal sites a bicyclonucleoside that has a methylene group bonded to the ribose 2′-OH and C′4. This locks the ribose into the C3′-endo pucker and confers exceptional stability when the LNA is hybridized to a complementary target in either microarray assays or during in situ hybridization (Thomsen et al. 2005; Ason et al. 2006; Castoldi et al. 2008). Each LNA monomer raises the Tm of the probe:RNA hybrid by 2°C–8°C, and the specificity of the hybridization increases such that single nucleotide differences between miRNAs can often be distinguished. We used the LNA-based microRNA profiling analysis miChip (Castoldi et al. 2008) to screen for the full complement of rat miRNAs recorded in miRBase (version 9.2, Wellcome Trust Sanger Institute). The LNAs on these microarray chips are designed to detect mature, primary, and precursor forms of miRNAs, which allowed us to screen for nucleolar miRNAs that concentrate in the nucleolus in any form.
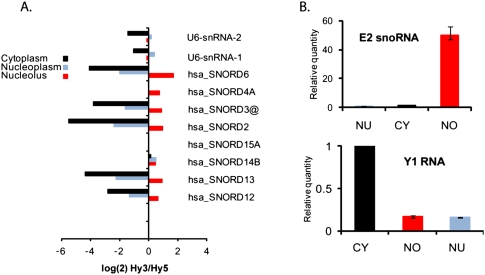
The purity of each cellular fraction was assayed by microscopy during purification (Supplemental Fig. 1) and was subsequently corroborated by the relative abundance of compartment-specific RNAs as detected by both the microarray and RT-qPCR (Fig. 1). Known snoRNAs were present at high levels in the nucleolus-enriched fractions and at much lower levels in nucleoplasm- and cytoplasm-enriched fractions. The U6 small spliceosomal RNA was present at similar levels in the nucleoplasm, where it functions, and the nucleolus, where it traffics for ribose modification (Fig. 1A; Ganot et al. 1999). Since most nucleoplasm-concentrated small RNAs traffic through the nucleolus, we were unable to test a marker completely specific for the nucleoplasm. The fractionation was further substantiated by RT-qPCR analysis of marker RNAs. Thus, a known snoRNA, E2 (Mishra and Eliceiri 1997), was present at highest levels in the nucleolus-enriched fraction, while a Ro small RNA family member, Y1 RNA, was detected at highest levels in the cytoplasmic fraction, as expected for this known cytoplasm-concentrated RNA species (Fig. 1B; Hendrick et al. 1981).
FIGURE 1.
Purity of nucleolar fraction. (A) Normalized log signal in nucleolar, nucleoplasmic, and cytoplasmic fractions from L6 rat myoblasts to human U6 and snoRNA probes (provided as controls on Exiqon miRCURY chips). Signal (Hy3) was normalized using total RNA (Hy5), which was an equimolar mixture of nucleoplasmic, nucleolar, and cytoplasmic RNA. The sequence of human and rat U6 are identical at 104/106 nt. Human snoRNAs have varying homology with rat snoRNAs, but both homologous and nonhomologous human snoRNAs are shown to demonstrate specificity and purity of the rat cell fractions used in this study. Cross-species homology is as follows: hsa_SNORD6, 4-nt difference; hsa_SNORD4A, expression not verified in rat; hsa_SNORD3@, 3-nt difference; hsa_SNORD2, 4-nt difference; hsa_SNORD15A, 18-nt difference; hsa_SNORD14B, not identified in rat; hsa_SNORD13, 19-nt difference; and hsa_SNORD12, 9-nt difference. (B) RT-qPCR of rat Y1 RNA and rat snoRNA E2 showing relative quantities (RQ) in nucleolar, nucleoplasmic, and cytoplasmic fractions from L6 rat myoblasts.
The microarray results revealed that at least one third of the 280 rat miRNAs probed on the microarray were expressed in L6 myoblasts, and surprisingly, most of these showed a significant presence in the nucleolus (Supplemental Table 1). Indeed, one third of the detected miRNAs exhibited nucleolar levels at least as high as those observed in the cytoplasm (Supplemental Table 2). The majority of these miRNAs are not known to be involved specifically in muscle differentiation, so these results suggest that nucleolar localization is not unique to muscle-specific miRNAs but rather is a more general phenomenon. As expected, many miRNAs showed cytoplasmic levels higher than the nucleoplasmic and nucleolar signals, and not all expressed miRNAs were detectable in the nucleolus. About 7% of the miRNAs showed nucleolar levels far below their respective levels in the nucleoplasm and/or the cytoplasm, thus indicating that presence in the nucleolus is not an obligatory step in miRNA biogenesis and pointing to the specificity of this finding.
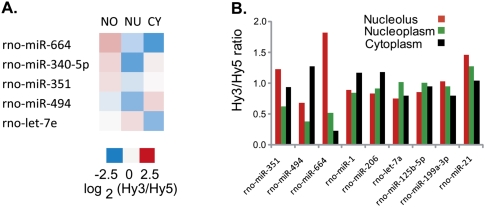
Five miRNAs, miR-340-5p, miR-351, miR-494, miR-664, and let-7e, were significantly concentrated (two- to eightfold) in the nucleolus compared with the nucleoplasm and/or the cytoplasm (Fig. 2A; Supplemental Table 1). None of these are muscle-specific miRNAs (i.e., miR-1, miR-133, miR-206, and perhaps miR-95, miR-128a, miR-499) (Lee et al. 2008; McCarthy 2008), although one of these miRNAs has been linked to muscle differentiation, viz. miR-494 is expressed at elevated levels in some muscle diseases (van Rooij et al. 2006; Eisenberg et al. 2007). miR-340-5p and miR-351 were originally identified as part of a large miRNA group in neurons (Kim et al. 2004). miR-351, along with many other stress response miRNAs, has been shown to be up-regulated in cardiohypertrophy (van Rooij et al. 2006). Neither let-7e nor miR-664 has reported links to muscle differentiation.
FIGURE 2.
Microarray analysis of nucleolar, nucleoplasmic, and cytoplasmic subcellular fractions from L6 rat myoblasts. miCURY LNA miChips were used for analysis. (A) Heat map showing relative levels of five significantly nucleolus-concentrated miRNAs. NO indicates nucleolus; NU, nucleus; and CY, cytoplasm. (B) Normalized Hy3/Hy5 ratios of selected miRNAs showing relative signal detected in each fraction. Hy3 indicates Hy3-labeled RNA from single fraction; Hy5, Hy5-labeled pooled RNA used as normalization control, containing equal amounts of nucleolar, nucleoplasmic, and cytoplasmic RNA.
Three of these five miRNAs, miR-351, miR-494, and miR-664, were chosen for further characterization (Fig. 2B). Let-7e was not pursued because although its nucleolar levels were significantly higher than its cytoplasmic levels, its even higher nucleoplasmic levels (see Supplemental Tables 1, 2) might have complicated analysis. miR-340-5p was also not pursued further because its sequence is not annotated in the rat genome to date (although it is in other species). We labeled LNA in situ hybridization probes to miR-351, miR-494, and miR-664 as well as the muscle-specific miR-206 and its family member miR-1, which is found at elevated levels in both cardiac and skeletal muscle (Kim et al. 2006; Rosenberg et al. 2006; McCarthy 2008; Sweetman et al. 2008). Both miR-1 and miR-206 were detected at high levels in the cytoplasm in the microarray analysis as expected. But somewhat surprisingly in view of our earlier results (Politz et al. 2006), the microarray assay showed nucleolar/nucleoplasmic ratios closer to one for both of these miRNAs (Fig. 2B). To further explore this observation, we made probes to an additional four miRNAs that also showed nucleolar/nucleoplasmic levels close to 1, miR-199a-3p, miR-21, miR-125a-5p, and let-7a (Fig. 2B), and additionally, exhibited high affinity LNA hybridization during the microarray profiling (data not shown). We were also interested in miR-199a-3p and miR-21 because they had been shown to increase during cardiac hypertrophy (van Rooij et al. 2006; Thum et al. 2008) and Duchenne muscular dystrophy (Eisenberg et al. 2007). miR-125a-5p and let-7a are both expressed ubiquitously (Lee et al. 2008).
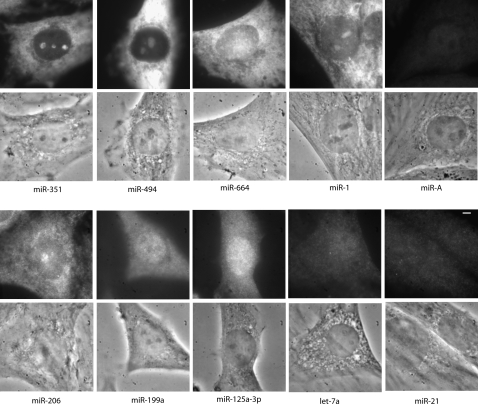
LNA probes to these various miRNAs (Fig. 2b) were hybridized to fixed rat myoblasts using previously established hybridization conditions (Politz et al. 2006). Figure 3 shows that all three of the miRNAs indicated to be nucleolus-concentrated in the microarray assays also showed nucleolar concentration as detected by in situ hybridization. Both miR-351 and miR-494 were present at higher levels in the nucleolus compared with the nucleoplasm as determined by in situ hybridization, correlating well with the microarray findings. Indeed, the nucleolar level of miR-351 determined by in situ hybridization was higher than that in either the cytoplasm or the nucleoplasm, as in the microarray analysis. Moreover, miR-494 nucleolar levels were higher than in the nucleoplasm but not higher than in the cytoplasm, also as observed in the microarray data. We also detected high levels of miR-664 in the nucleolus using in situ hybridization, but miR-664 did not always appear more concentrated in the nucleolus than in the nucleoplasm.
FIGURE 3.
miRNA LNA in situ hybridization in L6 myoblasts. Both fluorescence (top) and phase (bottom) images of typical cells are shown for each miRNA tested, with the name of the miRNA listed below the pair. Images are scaled to allow visualization of intranuclear localization patterns and should not be used to compare relative quantities between microRNAs. Exposure times were 3 sec for miR-125, miR-199, let-7a, miR-206, and miR-21; 500 msec for miR-351 and miR-664; 200 msec for miR-1 and miR-A; and 100 msec for miR-494. Bar, 3 μm.
As mentioned, we had earlier observed (Politz et al. 2006), and show here for comparison (Fig. 3), that miR-206 was concentrated in the nucleolus compared with the nucleoplasm in about half the cells in any given population, with an additional, more substantial presence in the cytoplasm. In the present study miR-1, like miR-206, showed concentrated nucleolar signal in some but not all cells. Figure 3 shows an example of a cell with strong miR-1 nucleolar hybridization. Such inter-cell differences during in situ hybridization can reflect true differences in nucleolar concentration between cells or, alternatively, be due to variations in probe accessibility among cells. However, if miR-1 and miR-206 were actually concentrated in all nucleoli, we would have expected to see high nucleolus/nucleoplasm ratios of miR-206 and miR-1 in the microarray profiles (where miRNAs are deproteinized and uniformly available for hybridization). Instead, the microarray data showed statistically similar levels of miR-206 and miR-1 in both the nucleolus and nucleoplasm. Taken together, these results suggest that these muscle cell-expressed miRNAs are actually concentrated in some nucleoli and not in others in a given rat myoblast population. The overall moderate levels of these miRNAs in the nucleolus-enriched biochemical fraction used for the microarray analysis thus more likely reflect the fact that the RNAs are derived from a mixed population of cells with higher versus lower nucleolar levels of miR-206 and miR-1.
The remainder of the miRNAs selected for further characterization did not show nucleolar concentration in the in situ hybridization experiments, although it should be stressed that, as in the microarray results, this does not mean there was no nucleolar presence at all. miR-199a-3p and miR-125a-5p appeared more concentrated in the nucleoplasm and cytoplasm compared with the nucleolus; their hybridization pattern more closely resembled that of let-7a, a microRNA that earlier in situ hybridization experiments had shown not to be concentrated in the nucleolus (Fig. 3; Politz et al. 2006). These results corroborated the microarray results, which indicated that these miRNAs were not significantly nucleolus-concentrated, although they have somewhat lower nucleolus/nucleoplasm ratios in the in situ hybridization study than predicted by the microarray data (data not shown). Importantly, although these three miRNAs, miR-199a-3p, miR-125a-5p, and let-7a, showed microarray nucleolus/nucleoplasm ratios similar to miR-206 and miR-1, we did not observe concentrated nucleolar signal in any cells hybridized with probes to these miRNAs. This indicates that the group of miRNAs showing similar levels of nucleolar and nucleoplasmic signal in the array represents miRNAs that show nucleolar localization in some cells (e.g., miR-1 and miR-206) as well as those that do not (e.g., miR-199a-3p, miR-125a-5p, let-7a). Thus, it appears that microarray analysis alone gives a low estimate of the number of nucleolus-concentrated miRNAs in a population.
Figure 3 also shows that miR-A, a variant of rat miR-1 that contains a G-to-A substitution, did not display substantial nucleolar hybridization, verifying the specificity of the LNA probe hybridization. All hybridization signals shown in Figure 3, except for miR-21 (vide infra), were RNase sensitive, indicating that the LNA probes are targeting RNA (Supplemental Fig. 2; Politz et al. 2006).
As just mentioned, miR-21 unexpectedly showed no significant RNase-sensitive in situ hybridization signal (Fig. 3; Supplemental Fig. 2) even though high levels of this microRNA were detected in all cell compartments in the microarray analysis (Fig. 1). miR-21 is expressed in many cell types besides muscle and its deregulated expression has been linked to neoplasia (Calin et al. 2005; Tong and Nemunaitis 2008). It is possible that this particular miRNA interacts with a protein (or RNA or other molecule) in rat myoblasts that blocks its in situ hybridization. This interpretation was substantiated by RT-qPCR analyses below.
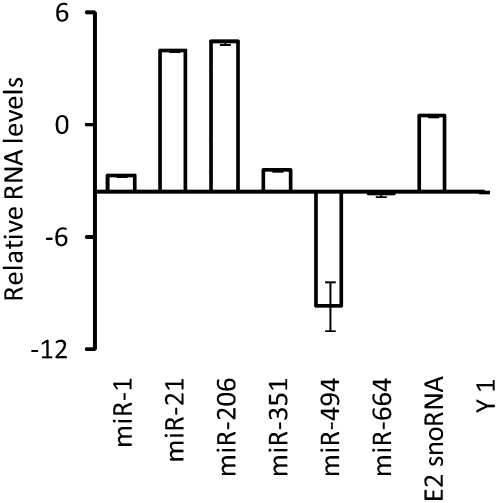
Since the above results demonstrated that a number of miRNAs were concentrated in the nucleolus, we wished to learn whether these miRNAs were present in precursor or mature form and also determine their relative amounts compared with one another. This information cannot be gleaned from the microarray and in situ analyses because the LNA probes were designed to detect all miRNA forms and possessed potentially different affinities for their target miRNAs. Therefore, we used TaqMan RT-qPCR to compare the relative amounts of each mature miRNA in the nucleolus-enriched RNA fractions isolated from L6 myoblasts. TaqMan RT-qPCR selectively amplifies mature miRNAs because the primers do not hybridize appreciably or at all to miRNA precursor forms (Chen et al. 2005; Schmittgen et al. 2008). Figure 4 shows the relative amounts of the nucleolus-concentrated miRNAs detected using this method. Levels of the E2 snoRNA and Y1 Ro RNA controls provide an estimation of the relative levels of a nucleolus-concentrated and a nucleolus-depleted small RNA, respectively (see also Fig. 1C).
FIGURE 4.
Detection of mature miRNAs in the nucleolus. TaqMan RT-qPCR microRNA assays were employed to characterize and quantify miRNAs identified in the nucleolus-enriched fraction of L6 myoblasts. In this method, a hairpin primer selectively hybridizes to a particular mature miRNA target and primes reverse transcription of only that mature miRNA and not its precursor or primary transcript within an RNA fraction. The RT product is then amplified using miRNA-specific PCR primers, and the number of copies is measured by hybridization of a specific fluorescent probe as real time PCR proceeds (Chen et al. 2005; Schmittgen et al. 2008). Relative copy numbers were normalized between two experiments using parallel RT reactions that were spiked with a control RNA, IPC (see Materials and Methods). The graph illustrates the levels of mature miRNAs relative to one another (in arbitrary units expressed as ΔCt in log base 2) in nucleolus-enriched fractions. The y intercept for the x axis has been set at the value for Y1 (a prevalent cytoplasmic small RNA), which represents background. Also shown are results for snoRNA E2, a small RNA that is highly concentrated in the nucleolus. Error bars, SE between the two experiments.
Both miR-206 and miR-21 were detected in the nucleolus-enriched fraction at levels higher even than the E2 snoRNA, which strongly suggests that high levels of the mature form of these two miRNAs are present in the nucleolus. These results substantiate our earlier findings for miR-206 (Politz et al. 2006). As discussed above, miR-21 was not detected in situ using the LNA probe, but it was detected at high levels in the microarray assay (in all compartments) (Fig. 2B; data not shown), which is consistent with these RT-qPCR results. Taken together, a simple interpretation of these results is that miR-21 is present as a mature miRNA in all three compartments but is somehow masked from detection by in situ hybridization. miR-1 and miR-351 were detected by RT-qPCR at lower levels than the nucleolus-abundant snoRNA but, nevertheless, were present at levels higher than Y1, the predominantly cytoplasmic RNA species that represents the “background” in the nucleolar preparation. The RT-qPCR thus confirms that these two miRNAs are present in significant amounts in the nucleolus, although the levels of the mature forms of these miRNAs are considerably lower than miR-206 and miR-21. Finally, miR-494 and miR-664 were detected by RT-qPCR of nucleolar RNA at levels similar to or below that of Y1 RNA, which indicates that the mature forms of these two miRNAs are not concentrated in the nucleolus. Since both these miRNAs were observed at significant levels in the nucleolus in the microarray analysis and by in situ hybridization (where the LNA probes detect both mature and precursor forms), taken together these results suggest that these miRNAs are present in precursor form in the nucleolus.
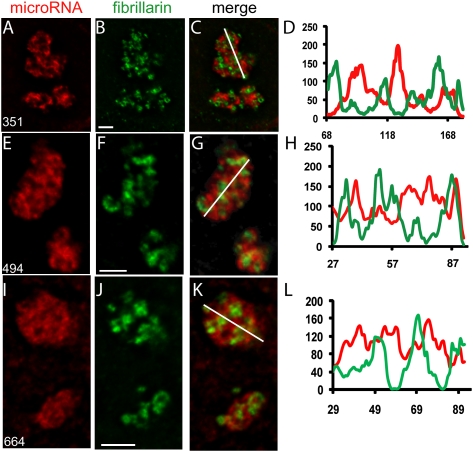
As mentioned in the Introduction, the nucleolus has a tripartite organization consisting of fibrillar centers, the DFC and the GC (Huang 2002; Raska et al. 2006). From the results in Figure 3 and as shown for miR-206 earlier (Politz et al. 2006), we knew that the newly identified nucleolar miRNAs did not localize exclusively to the small, focal fibrillar centers. In order to determine the distribution of these miRNAs with respect to the DFC and GC, we first immunostained cells for fibrillarin, a fiduciary marker of the DFC, and then performed in situ hybridization with LNA probes for miR-351, miR-494, or miR-664. The distribution of each miRNA within the nucleolus was compared to that of fibrillarin after deconvolution of optical stacks (Fig. 5). All three miRNAs showed highest signals at regions that did not overlap with the DFC and thus were primarily concentrated in the GC. The miR-351 signal was most restricted to the GC, while miR-494 and especially miR-664 showed additional signal in the DFC (note intensity line scans in Fig. 5). Therefore, as previously reported for miR-206 (Politz et al. 2006), the most concentrated signal for all three nucleolar-concentrated miRNAs was localized in the GC, the compartment in which other nonribosomal components congress (Ma and Pederson 2008b).
FIGURE 5.
Subnucleolar mapping of miRNAs. L6 myoblasts were grown to ∼70% confluence; fixed and subjected to immunostaining for fibrillarin, a fiduciary marker for the nucleolar DFC; followed by detection of miRNAs by in situ hybridization. Optical stacks were then captured, deconvolved, and color-merged. Images show magnified nucleoli cropped from a central plane of deconvolved stacks. (A) miR-351; (E) miR-494; (I) miR-664. (B,F,J) Fibrillarin. (C,G,K) Color merge of two images in row. The white line across the nucleolus indicates the position of the densitometric line scans shown in D, H, and L, respectively. Line scans indicate the relative intensities of fibrillarin (green) and the miRNA of interest (red). Scale bars, 0.5 μm.
DISCUSSION
We show here that many miRNAs are present in the nucleolus of rat myoblasts and that some of these miRNAs are concentrated therein. Our data suggest that some miRNAs are present in the nucleolus in precursor form (miR-494 and miR-664), while others are present as mature miRNAs (miR-206, miR-21, miR-1, and miR-351), and that members of both groups concentrate in the same subnucleolar compartment, the GC. This compartment also harbors various nonribosomal nucleolar components discovered recently (Karayan et al. 2001; Politz et al. 2002, 2005; Ma and Pederson 2008a).
It is still early days in the microRNA field, and there are many possible roles that nucleolar association might play in the life cycle of a miRNA. Our results allow a clearer delineation of the possibilities. First, none of the five miRNAs that showed the highest nucleolar concentration using the microarray assay, miR-340-5p, miR-351, miR-494, miR-664, or let-7e, are thought to be skeletal muscle-specific miRNAs (i.e., miR-1, miR-133, and miR-206 and perhaps miR-95, miR-128a, and miR-499) (Lee et al. 2008; McCarthy 2008). It is thus clear that non-muscle-specific miRNAs concentrate in the nucleolus of L6 myoblasts. Furthermore, we found that the majority of the miRNAs expressed in myoblasts (which includes ubiquitously expressed miRNAs) can be detected in all three subcellular compartments (see Supplemental Table 1), so our data do not support a model in which myoblast-enriched miRNAs alone exhibit a nucleolar presence. It also does not appear that there is a class of “nucleolar” miRNAs that persists across tissue types because in the myoblast cells we used, we do not see a significant nucleolar presence of some miRNAs shown (by others concurrently with this study) to be nucleolar in testes, e.g., miR-127 (Robertus et al. 2009) and miR-214 (Marcon et al. 2008).
Other small RNAs are present in the nucleolus for processing/modification or assembly into ribonucleoprotein complexes or are active in the nucleolus as regulatory modification/processing components themselves (Politz et al. 2002; Kiss et al. 2006; Raska et al. 2006; Boisvert et al. 2007; Matera et al. 2007). A nucleolar role in pri-miRNA processing or modification is possible, although it has been shown recently that the processing of at least some pri-miRNAs is linked to their transcription and thus occurs at sites of pol II activity and not the nucleolus (Morlando et al. 2008; Pawlicki and Steitz 2008). However, it has also been reported that Drosha associates with the nucleolus (Shiohama et al. 2007), and moreover, a noncoding RNA that is processed by Drosha has been shown to associate with the nucleolus (Ganesan and Rao 2008). Furthermore, some miRNAs are processed from mirtrons in a Drosha-independent manner (Berezikov et al. 2007), and most intriguingly, there is very recent evidence that snoRNAs may be precursors to some miRNAs and/or small RNAs (Ender et al. 2008; Saraiya and Wang 2008; Taft et al. 2009). Indeed, it has been reported that the small nucleolar RNA, SNORA36B/ACA36b, which was identified as a GAR1-binding snoRNA that is predicted to guide pseudouridylation of 18S rRNA in the nucleolus (Kiss et al. 2004), is a precursor to miR-664 (Ender et al. 2008). Nothing is known about the mechanism of miRNA processing from a snoRNA, although it is certainly possible that processing occurs in the nucleolus, since snoRNAs accumulate there. Our results suggest that miR-664 is present in the nucleolus as a precursor, which is consistent with this model. Since we did not see the most concentrated miR-664 signal in the DFC, where it might be expected if the LNA probe were hybridizing to a SNORA36B/ACA36b-GAR1 complex, more work will be necessary to determine the compartment and mechanism of this putative processing step. However, no matter how inviting the hypothesis may be that all nucleolus-localized miRNAs have snoRNA precursors, we have found no evidence for complete identity between the sequences of the other miRNAs we have characterized here and any rat snoRNA or rRNA sequences. (Homology searches were performed using the Toulouse human snoRNA database [http://www-snorna.biotoul.fr/index.php] in conjunction with Ensembl [http://www.ensembl.org/ and ncRNA BLAT at http://www.ensembl.org/Multi/blastview] and USCS [http://genome.ucsc.edu/] genome browsers to test human/rat alignments). It should be pointed out here that cross-hybridization of the miR-664 LNA probe to SNORA36B would occur regardless of the precursor nature of this molecule, but we observe, at most, only partial homology of other miRNAs to snoRNAs, which can occur randomly and would not support cross-hybridization of the highly specific LNA probes. Thus, our data do not currently support a model that all nucleolar-localized miRNAs are processed from snoRNA precursors.
It is also possible that some miRNAs traffic to the nucleolus for modification. Significantly, we note that A→I editing of non-miRNAs by the adenosine deaminase, ADAR2, has been shown to occur in the nucleolus (Vitali et al. 2005), and it is known that some miRNA precursors are subject to editing (Luciano et al. 2004; Blow et al. 2006; Habig et al. 2007; Kawahara et al. 2008). miRNA editing can also interfere with processing (Yang et al. 2006; Kawahara et al. 2007), which conceivably could cause sequestration of unprocessed miRNA precursors in the nucleolus. The spatiotemporal aspects of small RNA editing requires more study, however, since another adenosine deaminase, ADAR1, has been shown to bind (but not modify) dsRNA and inhibit RNAi in the cytoplasm in mammals (Yang et al. 2005).
Rather than RNA processing or modification, it may be that certain miRNAs visit the nucleolus to assemble with the appropriate proteins and/or RNAs necessary for transport of the active miRNP from the nucleus. The signal recognition particle RNA traffics through the nucleolus to assemble with specific proteins (Jacobson and Pederson 1998; Politz et al. 2000; Sommerville et al. 2005) and, in the process, accumulates in the GC (Politz et al. 2002), just as do the nucleolus-concentrated miRNAs. We also speculate that some miRNAs could combine with target messenger RNAs in the nucleolus to be exported as “presuppressed” mRNAs, a somatic cell analogy with masked maternal mRNAs in oocytes. Consistent with this idea, there is evidence that the nucleolus is involved in mRNA nucleocytoplasmic transport (Schneiter et al. 1995; Thomsen et al. 2008), and some mRNAs have been shown to associate with the nucleolus (John et al. 1977; Bond and Wold 1993; Bártová et al. 2008; Kim et al. 2009).
Finally, miRNAs may concentrate in the nucleolus because they are active as regulatory modification/processing components themselves. For example, small noncoding RNAs have been implicated in the control of ribosomal DNA transcription (Mayer et al. 2008), and snoRNAs guide modification of rRNA and some snRNAs in the nucleolus (Boisvert et al. 2007; Matera et al. 2007), as well as perhaps modulating the activity of ADAR2 (Vitali et al. 2005). However, neither miR-206 nor the nucleolus-enriched miRNAs identified in the present study were localized at the rDNA transcription sites (fibrillar centers) or to the DFC, where most snoRNA-guided rRNA modifications are thought to occur, but rather were clustered in the GC, a region that is known to be the site of late stages of rRNA processing. Furthermore, homology searches revealed that, except for miR-664, the sequences of these miRNAs do not share identity with any annotated snoRNA-like sequences necessary to guide 18S or 28S rRNA modifications, although we did find that miRNA-199a-3p and miR-494 possess 9–10 nucleotide (nt) sequences that are complementary to regions immediately adjacent to the guide sequences of SNORA61 and SNORA60, respectively (data not shown). If miRNAs hybridized to these regions, this could interfere with snoRNA-guided modification of target RNAs. However, our results suggest that miR-494 is present as a precursor in the nucleolus and we did not observe similar complementarity for any other miRNAs with any other snoRNAs, although we point out that the rat snoRNAs are not as well annotated as the human/yeast snoRNAs so this search is presently incomplete. If miRNAs do perform this and/or other functions in the nucleolus, they presumably would do this in addition to their function in gene silencing at the translational level in the cytoplasm.
In summary, we find it unlikely that only myoblast-specific miRNAs accumulate in nucleoli and likely that at least some miRNAs arise from snoRNA precursors. It remains possible that some miRNAs visit the nucleolus for modification, assembly with RNA/protein partners, or regulation of snoRNA activity. In these conceptions of the possible functional significance of the presence—and in some cases a concentration of—these miRNAs in the nucleolus, we again note that mature miRNAs and other small RNAs can return to the nucleus from the cytoplasm (Hwang et al. 2007; Guang et al. 2008; Kim et al. 2008; Marcon et al. 2008; Ohrt et al. 2008; Place et al. 2008). The fact that even the first translational RNA discovered, transfer RNA, also is known to display this beguiling “retrograde” transit in both yeast (Shaheen and Hopper 2005; Takano et al. 2005) and mammalian cells (Shaheen et al. 2007) invites the consideration of broader, evolutionary possibilities for the nuclear and nucleolar presence of microRNAs. We have demonstrated that a large number of miRNAs are present in the nucleolus, rather than a select few, so any models to explain a role for nucleolar miRNAs must apply to a general miRNA population. We have also carried out preliminary miRNA LNA in situ hybridization experiments in HeLa cells and have found that some of the miRNAs that concentrated in the nucleoli of rat myoblasts also concentrate in the nucleolus of these cancer cells (not shown), giving further weight to the notion that this localization is a general phenomenon. Further work is now necessary to define the role that nucleolar transit plays in the life cycle of miRNAs.
MATERIALS AND METHODS
Cell fractionation
L6 myoblasts were grown in DMEM with 10% fetal bovine serum at 37°C with 5% CO2 to 80% subconfluence in ten 225-cm2 flasks (yielding ∼108 cells), harvested using trypsin, and then washed three times with DMEM salts. Cells were resuspended in a hypotonic buffer (Mellon and Bhorjee 1982), allowed to swell, and then broken using a Dounce homogenizer with a clearance of ∼50 μm (Penman et al. 1966). Nuclei were purified through a sucrose cushion (Andersen et al. 2002) and sonicated (Pederson 1974), and the nucleoli were pelleted (Andersen et al. 2002). During isolation and washing, purity of nucleoli was judged by light microscopy. Total RNA was isolated from the nucleolar, nucleoplasmic, and cytoplasmic fractions using a Qiagen miRNAeasy kit. The resulting RNA is enriched in all the cell's small RNA species, including miRNAs <200 nt.
Microarray analysis
Two-color microarray analysis was performed using an LNA-based miCHIP (miRCURY microarrays, Exiqon) to screen for the full complement of rat miRNAs recorded in miRBase (version 9.2, Wellcome Trust Sanger Institute). Five to eight micrograms of total RNA from each subcellular fraction described above was labeled with Hy3 (a proprietary Cy3-like dye of Exiqon), and additionally, equal aliquots of all three fractions were pooled and labeled with Hy5 (Exiqon). This pooled “total RNA” was used for normalization in all runs. Four replicate two-color runs were performed for each fraction, and the results were background subtracted and subjected to a Loess and quantile normalization by Exiqon's in house profiling service. miRNAs showing significant difference in nucleolar level were determined as those having a SD ≥ 0.5 (see Supplemental Table 1).
In situ hybridization and immunostaining
In situ hybridization using specific miRNA LNA probes, image capture, and analysis was as described (Politz et al. 2006). RNase treatment was also as described (Politz et al. 2006). Immunostaining followed by in situ hybridization was performed with a fibrillarin monoclonal antibody as detailed previously (Politz et al. 2002), except optical stacks were captured using a Deltavision workstation and deconvolved using Softworx. Control cells subjected to either immunostaining or in situ hybridization alone showed no bleed-through of signal into the other channel. Images were adjusted for size and contrast and color merged using Image J. The Image J plug-in, color profiler, was used to obtain line intensity plots.
Quantitative RT-PCR
The two-step TaqMan miRNA RT-qPCR system (Applied Biosystems) was used to follow miRNA amplification in real time (Chen et al. 2005; Schmittgen et al. 2008). Reverse transcription and PCR reactions were performed according to the manufacturer's protocol using TaqMan rat stem-loop RT and PCR primers and probes specific for each miRNA under study. A synthetic exogenous internal positive control (IPC, Applied Biosystems no. 4308323) was spiked into parallel samples of total nucleolar RNA, amplified using IPC-specific primers and TaqMan probes, and used for normalization between PCR reactions. snoRNA E2 (U64702) (Selvamurugan et al. 1997) and the Ro family Y1 RNA controls were purchased from Applied Biosystems. Fluorescence was quantitated using an Applied Biosystems 7500 Fast Real-Time PCR System.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by grant MCB-0445841 from the National Science Foundation. We thank Michael Hansen at Exiqon technical services (Vedbaek, Denmark) for help with microarray data analysis and Mark Groudine and the Scientific Imaging Group at the Fred Hutchinson Cancer Research Center for the use of the Deltavision workstation. We also thank Paul Gardner (University of Massachusetts Medical School) for the use of his RT-qPCR thermal cycler, Hanhui Ma for help with cell culture, and Denise Maclachlan for assistance in manuscript preparation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1470409.
REFERENCES
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártová E, Harnicarová A, Krejcí J, Strasák L, Kozubek S. Single-cell c-myc gene expression in relationship to nuclear domains. Chromosome Res. 2008;16:325–343. doi: 10.1007/s10577-007-1196-0. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhorjee JS, Pederson T. Nonhistone chromosomal proteins in synchronized HeLa cells. Proc Natl Acad Sci. 1972;69:3345–3349. doi: 10.1073/pnas.69.11.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhorjee JS, Pederson T. Chromatin: Its isolation from cultured mammalian cells with particular reference to contamination by nuclear ribonucleoprotein particles. Biochemistry. 1973;12:2766–2773. doi: 10.1021/bi00738a033. [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27.1–R27.8. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bond VC, Wold B. Nucleolar localization of myc transcripts. Mol Cell Biol. 1993;13:3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Schmidt S, Benes V, Hentze MW, Muckenthaler MU. miChip: An array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat Protocols. 2008;3:321–329. doi: 10.1038/nprot.2008.4. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Ganesan G, Rao SM. A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA. 2008;14:1399–1410. doi: 10.1261/rna.838308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol. 1999;19:6906–6917. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA, Borovjagin AV, Lange TS. The nucleolus: A site of ribonucleoprotein maturation. Curr Opin Cell Biol. 2003;15:318–325. doi: 10.1016/s0955-0674(03)00049-8. [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig JW, Dale T, Bass BL. miRNA editing—we should have inosine this coming. Mol Cell. 2007;25:792–793. doi: 10.1016/j.molcel.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: Further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Building an efficient factory: Where is pre-rRNA synthesized in the nucleolus? J Cell Biol. 2002;157:739–741. doi: 10.1083/jcb.200204159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John HA, Patrinou-Georgoulas M, Jones KW. Detection of myosin heavy chain mRNA during myogenesis in tissue culture by in vitro and in situ hybridization. Cell. 1977;12:501–508. doi: 10.1016/0092-8674(77)90126-x. [DOI] [PubMed] [Google Scholar]
- Karayan L, Riou JF, Seite P, Migeon J, Cantereau A, Larsen CJ. Human ARF protein interacts with topoisomerase I and stimulates its activity. Oncogene. 2001;20:836–848. doi: 10.1038/sj.onc.1204170. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JWS. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 2009;21 doi: 10.1105/tpc.109.067736. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Pederson T. Nucleophosmin is a binding partner of nucleostemin in human osteosarcoma cells. Mol Biol Cell. 2008a;19:2870–2875. doi: 10.1091/mbc.E08-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Pederson T. Nucleostemin: A multiplex regulator of cell-cycle progression. Trends Cell Biol. 2008b;18:575–579. doi: 10.1016/j.tcb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Maggio R, Siekevitz P, Palade GE. Studies in isolated nuclei. I. Isolation and chemical characterization of a nuclear fraction from guinea pig liver. J Cell Biol. 1963;18:267–291. doi: 10.1083/jcb.18.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Babak T, Chua G, Hughes T, Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- Matera AG, Tycowski KT, Steitz JA, Ward DC. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–1299. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Matranga C, Zamore P. Small silencing RNAs. Curr Biol. 2007;17:R789–R793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ. MicroRNA-206: The skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Bhorjee J. Isolation and characterization of nuclei and purification of chromatin from differentiating cultures of rat skeletal muscle. Exp Cell Res. 1982;137:141–154. doi: 10.1016/0014-4827(82)90016-7. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Eliceiri GL. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Natl Acad Sci. 1997;94:4972–4977. doi: 10.1073/pnas.94.10.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Smetana K, Busch H. Quantitative aspects of isolation of nucleoli of Walker carcinosarcoma and liver of the rat. Cancer Res. 1963;23:510–518. [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: A new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Proteins associated with heterogeneous nuclear RNA in eukaryotic cells. J Mol Biol. 1974;83:163–183. doi: 10.1016/0022-2836(74)90386-6. [DOI] [PubMed] [Google Scholar]
- Pederson T. Growth factors in the nucleolus? J Cell Biol. 1998;143:279–281. doi: 10.1083/jcb.143.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penman S, Smith I, Holtzman E. Ribosomal RNA synthesis and processing in a particulate site in the HeLa cell nucleus. Science. 1966;154:786–789. doi: 10.1126/science.154.3750.786. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J, Yarovoi S, Kilroy S, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J, Lewandowski L, Pederson T. Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J Cell Biol. 2002;159:411–418. doi: 10.1083/jcb.200208037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J, Polena I, Trask I, Bazett-Jones D, Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell. 2005;16:3401–3410. doi: 10.1091/mbc.E05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- Robertus JL, Harms G, Blokzijl T, Booman M, de Jong D, van Imhoff G, Rosati S, Schuuring E, Kluin P, van den Berg A. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod Pathol. 2009;22:547–555. doi: 10.1038/modpathol.2009.10. [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Kadowski T, Tartakoff AM. mRNA transport in yeast: Time to reinvestigate the functions of the nucleolus. Mol Biol Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamurugan N, Joost OH, Haas ES, Brown JW, Galvin NJ, Eliceiri GL. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997;25:1591–1596. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13.1–R13.11. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci. 2007;104:8845–8850. doi: 10.1073/pnas.0700765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Sommerville J, Brumwell C, Politz J, Pederson T. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J Cell Sci. 2005;118:1299–1307. doi: 10.1242/jcs.01726. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206, and miR-133. Dev Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- Thomsen R, Nielsen PS, Jensen TH. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005;11:1745–1748. doi: 10.1261/rna.2139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R, Saguez C, Nasser T, Jensen TH. General, rapid, and transcription-dependent fragmentation of nucleolar antigens in S. cerevisiae mRNA export mutants. RNA. 2008;14:706–716. doi: 10.1261/rna.718708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A. The nucleolus: The magician's hat for cell cycle tricks. Curr Opin Cell Biol. 2000;12:752. doi: 10.1016/s0955-0674(00)00165-4. [DOI] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc Natl Acad Sci. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yang W, Wang Q, Howell KL, Lee JT, Cho DS, Murray JM, Nishikura K. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]