Abstract
The structural characterization of environmentally-regulated lipid A derived from Francisella tularensis subspecies novicida (Fn) U112 is described using negative electrospray ionization with a linear ion trap Fourier transform ion cyclotron resonance (IT-FT-ICR) hybrid mass spectrometer. The results indicate that a unique profile of lipid A molecular structures are synthesized in response to Fn growth at 25 °C versus 37 °C. Molecular species were found to be tetra-acylated, sharing a conserved glucosamine disaccharide backbone, a galactosamine-1-phosphate linked to the reducing glucosamine, and multiple O- and N-linked fatty acyl groups. Deprotonated molecules were interrogated by MSn scanning techniques at both high and nominal mass resolution and were found to be complex heterogeneous mixtures where structures differed based on the positions and identities of the O- and N-linked fatty acyl substituents. For the dominant ion series, which consisted of five peaks, 30 unique lipid A structures were identified. Estimates for the relative abundance of each structure were derived from MS relative abundance ratios and fragment ion ratios from comparable dissociation pathways from MS2 through MS4 experiments. The results suggest a remodeling pathway in which the amide linked fatty acid of the reducing glucosamine favors a 3-hydroxyhexadecanoic acid substituent for growth conditions at 25 °C versus a 3-hydroxyoctadecanoic acid substituent for growth conditions at 37 °C.
Bacterial pathogens have evolved adaptive responses to environmental changes upon entering a host. These responses include remodeling of the bacterial outer membrane, which can alter the bacteria’s ability to cause disease or interact with the host’s innate immune defenses. Membrane remodeling is accomplished in a variety of ways and has been shown to play a role in virulence [1–3]. The cell envelope of gram-negative bacteria consists of two lipid bilayers: a symmetrical inner membrane composed primarily of phospholipids and an asymmetrical outer membrane containing primarily phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet. Lipid A, also know as endotoxin, is the hydrophobic membrane anchor of LPS [4, 5]. Tight packing of lipid A in the outer leaflet of the outer membrane creates a permeability barrier against environmental compounds and antimicrobial components of the host innate immune system [6]. Specific alteration of lipid A structure, including changes to the carbohydrate and fatty acid composition, can occur via a variety of environmental stimuli including divalent ion concentration, temperature, and other growth conditions [1, 7–10].
Francisella tularensis subspecies tularensis (Ft) is a highly infectious, gram-negative pathogen, and is the causative agent of tularemia in humans [11–13]. Human tularemia can occur in several forms depending on the route of entry, but in most cases is the result of a bite by an infected arthropod or inhalation of contaminated aerosols. Ft has been designated as a Class A select agent by the Centers for Disease Control and Prevention due to its potential use as a bioterrorism agent. The genus Francisella tularensis consists of four subspecies: tularensis (Type A), holartica (Type B), mediasiatica, and novicida. In humans, subspecies novicida (Fn) is only infectious in immunocompromised individuals [14]. However, in mice, both Ft and Fn cause a tularemia-like disease [15]. Previously, the structure of lipid A isolated from two type B Francisella tularensis subspecies holartica strains (1547–57 and LVS) after growth at 37 °C were elucidated using mass spectrometry, gas chromatography, and chemical methods [16, 17]. The base lipid A structure for both strains was similar and consisted of a β-1,6-linked diglucosamine disaccharide with a single phosphate moiety at the 1 position, amide-linked fatty acids at the 2 and 2′ positions, and ester-linked fatty acids at the 3, but not the 3′, positions. Interestingly, in the 1547–57 strain, the 1-phosphate moiety was determined to be substituted with galactosamine and defined a unique bacterial lipid A structure [16]. In studies of both strains, only the primary structures represented by the base peaks in the mass spectra were elucidated. In addition, matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) of lipid A isolated from clinical and environmental strains of F. tularensis subspecies tularensis, holartica, and mediasiatica after growth in rich medium at 37 °C were similar, and suggests a common lipid A structure for all F. tularensis subspecies [18].
The use of mass spectrometry for the structure elucidation of intact lipid A has been approached by both positive and negative ionization methods using fast atom bombardment [19–22], matrix-assisted laser desorption [23–25], and electrospray [26–29]. While these techniques often lead to complementary data, negative ionization is often necessary for acyl chain identification where collision induced dissociation (CID) of deprotonated molecules results in the observation of fatty acyl carboxylate anions [27, 28]. On quadrupoles, sectors, or time-of-flight instruments, these types of ions are readily observed, but on ion traps they are typically lost due to the low m/z cut-off imposed by the “Q” value of the ion trap and have to be inferred from parent ion neutral losses [30, 31]. More importantly, however, ion traps offer the advantage of multiple stages of CID (i.e., MSn) and provide a means to deconvolute and map the unimolecular fragmentation pathways for a given parent ion. With the advent of linear ion traps, this can be done with great sensitivity due to the increased ion storage capacity and dual electron multipliers found in the linear ion trap compared with previous three dimensional ion trap designs [32].
In this study, we report that lipid A isolated from Francisella tularensis subspecies novicida (Fn), strain U112, is a complex heterogeneous mixture of structures that display a unique abundance profile in response to a change in growth temperature (i.e., 25 °C versus 37 °C). This analysis was carried out using negative electrospray ionization (ESI) coupled to a linear ion trap (LTQ)-Fourier transform ion cyclotron resonance (FT-ICR) hybrid mass spectrometer (i.e., Thermo LTQ-FT). The approach utilized a combined advantage over previous methodologies: accurate mass measurement of deprotonated molecules and MS/MS fragments, the use of automatic gain control (AGC) that served to normalize the dynamic range between ions selected for MS/MS, and the use of MS and MSn fragment ion relative intensities to derive estimates for the relative abundance of each molecular species. The data indicate that the deprotonated molecules are heterogeneous mixtures, which differ as to the identities of the N- and O-linked fatty acyl substituents on the diglucosamine core. Further, analysis of the data support a membrane remodeling pathway in which the majority of lipid A structures from Fn grown at different temperatures results in alterations of the fatty acids linked to the diglucosamine core. In other gram-negative pathogens, temperature change induces expression of genes essential to their life cycle, such as transition between their natural reservoirs (e.g., arthropod to mammal) 33, [34]. Therefore, alteration of lipid A structure upon entry into the mammalian host may represent a pathogenesis strategy common to the Francisella species.
Experimental
Bacterial Strains and Growth Conditions
Francisella tularensis subspecies novicida strain U112, obtained from Francis Nano (University of Victoria, Victoria, Canada) was grown in tryptic soy broth (Gibco BRL, Grand Island, NY) supplemented with 0.1% cysteine (TSB-C; Sigma-Aldrich, St. Louis, MO) at 25 °C or 37 °C with aeration and harvested in the stationary phase.
LPS Purification and Lipid A Isolation
Fn LPS was extracted using a hot phenol/water extraction method [35]. Subsequently, LPS was treated with RNase A, DNase I, and proteinase K to ensure purity from contaminating nucleic acids and proteins [36]. Lipid A was isolated after hydrolysis in 1% SDS at pH 4.5 as described [37]. The yield of LPS was 0.9 to 10 mg dry cells.
Fatty Acid Analysis
LPS fatty acids were determined after conversion to methyl esters as previously described by methanolysis in 2 M methanolic HCl at 90 °C for 18 h 38, [39].
Electrospray Ionization IT-FT-ICR Mass Spectrometry
Lipid A was analyzed by electrospray ionization in the negative ion mode on an LTQ-FT linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (Thermo Electron Corp., San Jose, CA). Samples were diluted to ~0.3 to 1.0 mg/mL in chloroform/methanol (1:1) and infused using a syringe pump (Harvard Apparatus, Holliston, MA) at 0.5 to 1.0 μL/min via a fused silica capillary (75 μm i.d./360 μm o.d.) with a ~15 μm spray tip (New Objective, Woburn, MA). The electrospray voltage was applied via liquid junction using a gold wire inserted into a microtee union (Upchurch Scientific, Oak Harbor, WA). Ion source conditions were as follows (voltages are negative): ESI voltage, 1.4 to 1.8 kV; capillary temperature, 400 to 450 °C, capillary voltage, 16 to 44 V, and tube lens voltage, 200 to 220 V. Instrument calibration and all other tuning parameters were optimized using a solution of Ultramark 1621 (Lancaster Pharmaceuticals, PA).
Injection waveforms for the LTQ-FT linear ion trap and ICR cell were kept on for all acquisitions. For experiments analyzed in the ICR cell, resolution was set to 1.0e5 (m/z 400) and ion populations were held constant by automatic gain control (AGC) at 1.0e6 and 5.0e5 for MS and MS/MS, respectively. The maximum ion fill time for the ICR cell was set to 3 s. For MS/MS, the precursor ion selection window was set to 4 to 8 Da and the collision energy was set to 30%. Spectra were acquired over a 1 to 5 min period and averaged (typically, 30 to 150 scans depending on conditions). Measured mass accuracies following external calibration were typically +5 ppm. For MSn analysis in the linear ion trap, mass spectra were acquired using an ion population of 1.0e4 and a maximum ion fill time of 200 ms. The precursor ion selection window was set to 4 to 8 Da for MS2 and 2 Da for subsequent MS3 and MS4 events. The collision energy was set to 30% for MS2 and 25% for MS3 and MS4. For manual MSn acquisition, multiple scans were averaged with acquisition times of 1 to 2 min. For automated MS3 spectra using data dependent analysis of MS2 precursor ions, 20 scans were averaged for each spectrum. Data were acquired and processed using Xcalibur, Version 1.4 (Thermo) utilizing 7 point Gaussian smoothing.
Results and Discussion
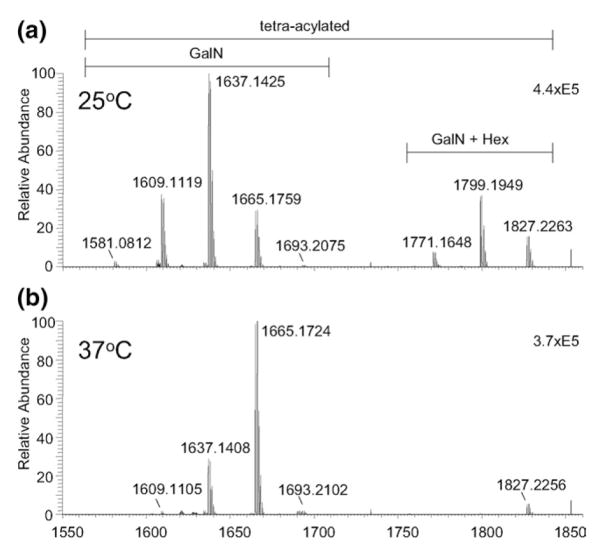
The negative electrospray ionization (ESI) Fourier transform ion cyclotron resonance (FT-ICR) mass spectra for LPS derived lipid A from Francisella tularensis subspecies novicida strain U112 (Fn) are shown in Figure 1. Dramatic shifts in the relative abundance of the major peaks (i.e., deprotonated molecules) are observed between the spectra for growth at 25 °C (Figure 1a) versus 37 °C (Figure 1b). The major ions for lipid A isolated after growth at 25 °C are found at nominal m/z values of 1609, 1637 (base), and 1665, with minor ions at m/z 1581 and 1693. A second set of ions arise at m/z 1771, 1799, and 1827, corresponding to a shift of 162.0 Da from the dominant ion series and indicate the presence of a hexose group (C6H12O6). For Fn grown at 37 °C, the major peaks are found at m/z 1637, 1665 (base), and 1827. Ions at m/z 1609 and 1799 are greatly reduced and ions at m/z 1581 and 1771 are absent. Comparison of the matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra to that produced by ESI was consistent (see Supplementary Material section which can be found in the online version of this article), resulting in the observation of the same ion series with similar relative abundances to that shown in Figure 1.
Figure 1.
Negative ion FT-ICR MS of Lipid A derived from Fn U112 grown at: (a) 25 °C; (b) 37 °C. The peak found at m/z 1853 arises from high-frequency noise.
Fatty acid analysis by gas chromatography with flame-ionization detection was used to confirm the fatty acids present in the LPS derived lipid A samples. Results indicate that the major fatty acids were 3-hydroxyhexadecanoic acid [(16:0)-3-OH], 3-hydroxyocta-decanoic acid [(18:0)-3-OH], tetradecanoic acid (14:0), hexadecanoic acid (16:0), and octadecanoic acid (18:0) (data not shown). Several other minor species were also observed that were not readily identified with the standard mixture used.
Characterization of the Base Peak at m/z 1665 Isolated After Growth at 37 °C
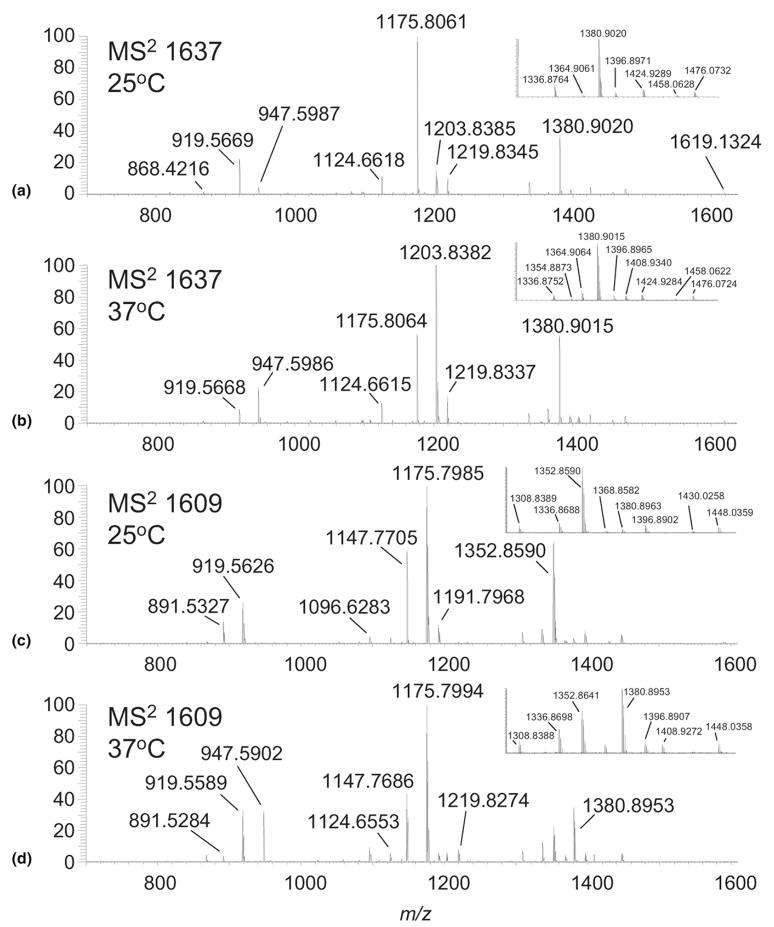
Lipid A structures were elucidated by linear ion trap tandem mass spectrometry with analysis of the fragment ions in either the ICR cell or linear ion trap. The base peak measured at m/z 1665.1724 (FT-ICR) from lipid A isolated from Fn grown at 37 °C (Figure 1b) had the same nominal mass and was determined to be the same structure to that reported for lipid A from Francisella tularensis subspecies holartica strain 1547–57 [16]. While we determined this peak to be a single lipid A structure, other peaks in the mass spectrum were determined to be isobaric mixtures of lipid A structures as described below. Thus, fragmentation described for m/z 1665 demonstrates the dissociation processes for a single lipid A entity and also serves to facilitate a method for the deconvolution of other lipid A structures found in the other peaks.
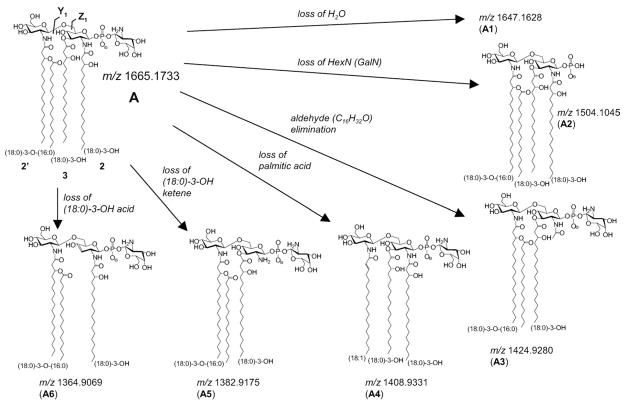
CID of m/z 1665 in the linear ion trap, followed by subsequent detection of the fragments in the ICR, gave rise to multiple fragmentation pathways illustrated in Scheme 1 and shown in Figure 2a. Primary dissociations of m/z 1665.1724 (−0.5 ppm), structure A, consist of neutral loss of H2O (m/z 1647.1594; −2.1 ppm; A1), hexosamine (m/z 1504.1031; −0.9 ppm; A2), C16H32O (m/z 1424.9286; 0.4 ppm; A3), palmitic acid (m/z 1408.9344; 0.9 ppm; A4), (18:0)-3-OH ketene (m/z 1382.9176; 0.1 ppm; A5), and (18:0)-3-OH fatty acid (m/z 1364.9065; −0.3 ppm; A6). The loss of the hexosamine (HexN) to give A2 was determined to be galactosamine (GalN), using a novel mass tagging LC-MS strategy for detection and quantitation of amino sugars (Ernst, manuscript in preparation). The ion at m/z 1424.9 (A3) arises from loss of C16H32O aldehyde, which is eliminated as the result of cleavage between the 2 and 3 position on (18:0)-3-OH. Ions at m/z 1408.9 (A4) and 1364.9 (A6) are due to losses of palmitic acid (16:0) and (18:0)-3-OH acid, respectively, and are indicative of O-linked fatty acids. The minor ion at m/z 1382.9 (A5) results from the loss of (18:0)-3-OH ketene from the N-linked (18:0)-3-OH group. Secondary dissociations lead to more abundant ions in the CID spectrum (Figure 2a) such as m/z 1247.8644 (loss of (16:0) and GalN; 0.1 ppm), m/z 1203.8381 (loss of (18:0)-3-OH acid and GalN; 0 ppm), m/z 1124.6614 (loss of (18:0)-3-OH acid and C16H32O aldehyde; −0.2 ppm), and m/z 947.5985 (loss of (18:0)-3-OH acid, palmitic acid, and GalN; 0.6 ppm).
Scheme 1.
Primary dissociations of m/z 1665 (monoisotopic m/z values given).
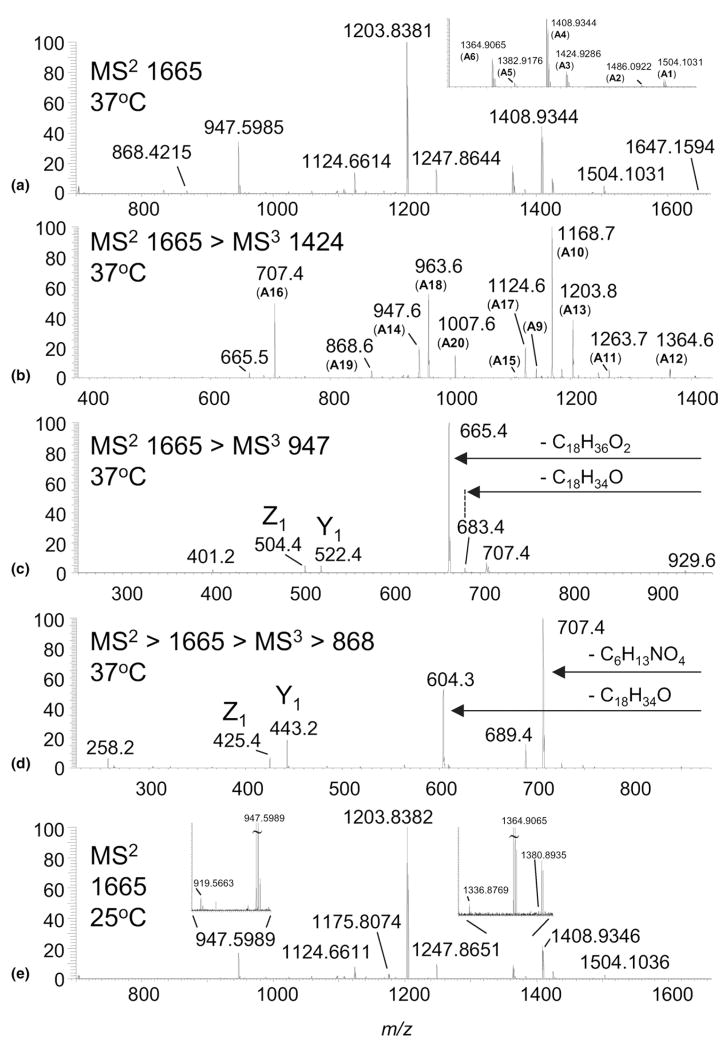
Figure 2.
(a) High-resolution MS/MS spectrum of m/z 1665 from Fn lipid A grown at 37 °C; (b) Ion trap (IT) MS3 spectrum of m/z 1424.9 derived from m/z 1665 (37 °C); (c) IT MS3 spectrum of m/z 947.6 derived from m/z 1665 (37 °C); (d) IT MS3 spectrum of m/z 868.4 derived from m/z 1665 (37 °C); (e) High-resolution MS/MS spectrum of m/z 1665 from Fn lipid A grown at 25 °C.
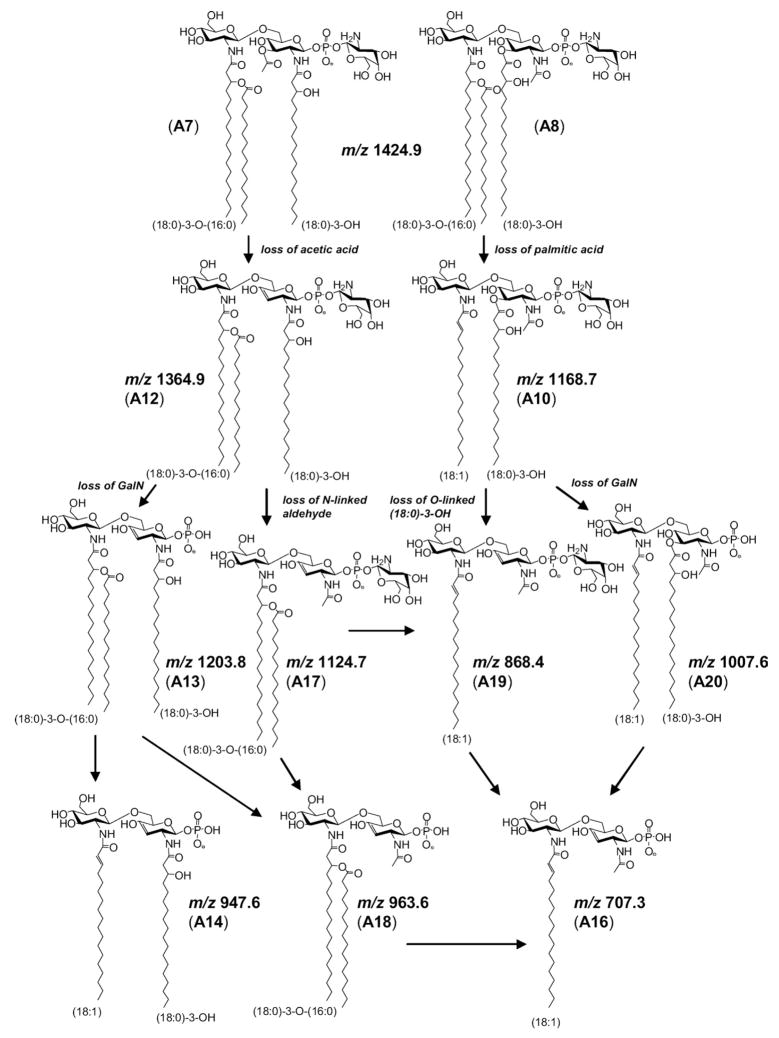
Previously, Gibson and coworkers showed that the ion at m/z 1424.9 in the MS/MS spectrum of A (Figure 2a), arises from elimination of C16H32O aldehyde, i.e., loss of 240.2 Da [16]. The authors hypothesized that this ion was formed from the N-linked (18:0)-3-OH acyl group of the reducing glucosamine, resulting from a McLafferty-like rearrangement initiated by the neighboring phosphate group. However, our study supports that the observed loss of 240.2 Da arises from C16H32O elimination from both N-linked and O-linked (18:0)-3-OH fatty acids of the reducing glucosamine. This is supported in the linear ion trap MS3 spectrum of m/z 1424.9 (Figure 2b), generated from a loss of 240.2 Da (C16H32O) from A. The ion at m/z 1424.9 can give rise to either ions A7 or A8 (Scheme 2), the latter depicted as the more stable ester and amide forms, respectively, rather than the corresponding enols [40–42]. Either A7 or A8 can fragment by eliminating a second C16H32O (m/z 1184.9, A9), palmitic acid (m/z 1168.7, A10), or GalN (m/z 1263.7; A11). However, only A7 is able to fragment by a loss of acetic acid to form the ion at m/z 1364.6 (A12). Subsequent loss of GalN (m/z 1203.8; A13) followed by palmitic acid leads to the ion at m/z 947.6 (A14). A14 is diagnostic for confirming C16H32O elimination from the O-linked (18:0)-3-OH fatty acid from A, the parent structure. This is because A8, with a stable acetamide group, undergoes multiple dissociations to form the stable ion at m/z 707.4 (A16), with none of the intermediate structures able to produce A14. Thus, with the presence of m/z 947.6 (A14) in the MS3 spectra of m/z 1424.9, the data support competitive losses of C16H32O aldehyde from both N- and O-linked fatty acyls on the reducing glucosamine. Further, this competitive fragmentation suggests that the formation of ions at m/z 1424.9 may be better described as initiating from a charge-remote mechanism [43, 44]. In this manner, fragmentation could proceed in a manner analogous to that described for the retro-aldol reaction [45].
Scheme 2.
MS3 fragmentation of m/z 1424.9 derived from A (m/z 1665).
Cleavage of the glycosidic bond of the glucosamine disaccharide was necessary for positional assignments of the N- and O-linked fatty acids, phosphate residues, and additional sugars, which cannot be inferred from the primary dissociations of A. As the CID spectrum of A (Figure 2a) is devoid of glycosidic bond cleavage, further interrogation was necessary. For example, the ion at m/z 947.6 (Figure 2a and Scheme 2 [A14]) results from the dissociation of GalN and both O-linked (18:0)-3-OH and (16:0) fatty acids, leaving the more stable amide linked fatty acids. MS3 of m/z 947.6 (Figure 2c) from A results in the formation of ions at m/z 522.4 and 504.4, Y1 and Z1 type fragment ions (Scheme 1), respectively, using the nomenclature described by Costello [46– 48]. These fragments facilitate assignment of the N-linked substituents to the distal (1′–6′ positions) and reducing (1–6 position) ends of the diglucosamine disaccharide as (18:1) and (18:0)-3-OH, respectively. The (18:1) fatty acid derives from the acyloxyacyl group, (18:0)-3-0-(16:0), following the loss of palmitic acid (16:0). Ions at m/z 683.4 and 665.4 (Figure 2b) further confirm the identities of the two N-linked substituents, (18:1) fatty acid at the 2′ position and (18:0)-3-OH acid at the 2 position, respectively. The Y1 and Z1 ions also assign the phosphate residue to the reducing end of the disaccharide. The MS3 spectrum is devoid of B or C type ions, which would only be observed for species carrying the charge on the distal end of the disaccharide.
Another important aspect in determining lipid A structure comes from identifying that the galactosamine moiety is linked through the phosphate residue on the reducing terminus of the disaccharide core. Following CID of A, there is little evidence for loss of a galac-tosamine-1-phosphate residue, which would tend to be lost as an ion rather than a neutral, and would fall below the m/z cut-off imposed by the Q value of the linear ion trap. Nonetheless, the MS3 spectrum of m/z 868.4 (A19, Scheme 2) from A clearly shows an ion at m/z 258.2, which supports the assignment of A with a galactosamine-1-phosphate residue (Figure 2d).
Determination of the Fatty Acid Structural Heterogeneity in the Dominant Ion Series
While only a single structure (A) was identified for the base peak at m/z 1665 isolated from Fn after growth at 37 °C, multiple structures were found for the same ion from the sample grown at 25 °C (Figure 1). This was evidenced initially in the MS2 spectrum of m/z 1665 where an ion at m/z 919.5663 (−0.3 ppm) was detected (Figure 2e). This ion is analogous in structure to the ion at m/z 947.6 (A14, Scheme 2) but possessing a shorter fatty acyl chain length (i.e., by C2H4). The other evidence was a small but discernable ion at m/z 1380.8935 (−6.0 ppm), presumably due to a loss of stearic acid (18:0) from the acyloxyacyl group. While the presence of additional trace isomers was evident in the MS2 spectrum, further experiments were necessary to identify all the possible isomers contained under the m/z 1665 peak.
To elucidate all the structures contained under m/z 1665 from lipid A isolated after growth at 25 °C, the MS2 fragment at m/z 1408.9 (Figure 2e), identified as the loss of palmitic acid from m/z 1665, was chosen for further interrogation and analysis in the linear ion trap. MS3 of m/z 1408.9 led to the formation of ions at m/z 947.6 and 919.6 (Table 1). These ions represent the combined loss of palmitic acid, GalN, and by deduction reveal the identity of the O-linked fatty acid residing on the 3 position of the reducing glucosamine as (18:0)-3-OH and (20:0)-3-OH, for ions at m/z 947.6 and 919.6, respectively (Table 2). As expected, MS4 of m/z 947.6 resulted in the formation of Y1 and Z1 type fragment ions, m/z 522 and 504, respectively, from A. Interestingly, an additional but minor Y1/Z1 ion series was observed at m/z 494 and 476, which facilitated assignment of the N-linked substituents to the distal and reducing ends of the glucosamine disaccharide as (20:1) and (16:0)-3-OH, respectively, and lead to the assignment of B (Table 2). Similarly, MS4 of m/z 919.6 from m/z 1408.9 also produced two Y1/Z1 ion pairs, 494/476 and 522/504, and led to the assignment of C and D. Finally, the MS2 fragment at m/z 1380.9, identified as the loss of stearic acid from m/z 1665, produced an ion at m/z 919.6 (Table 1). MS4 of m/z 919.6 produced ions at m/z 494 and 476, and led to the assignment of E. We conclude from the MS2 > MS4 spectra that m/z 1665 from Fn grown at 25 °C is an isobaric mixture whose relative abundance suggests that A ≫ B + C + D + E. Note that the absence of B–E from the sample grown at 37 °C cannot be due to a loss of MS dynamic range because of the use of AGC, which effectively normalizes the ion populations between the peaks selected for interrogation by MS/MS.
Table 1.
Fragment ion intensities from selected linear ion trap MSn spectra
| Temp. | MS2 Parent | MS2 Product | R.I. | MS3 Product | R.I. | Y1-/Z1-Ions | R.I. | σ | Struct.a |
|---|---|---|---|---|---|---|---|---|---|
| 25°C | 1665.1 | 1408.9-(16:0) | 100 | 947.6 | 100 | 522/504 | 100 | 1.0e6 | A |
| 494/476 | 2 | 2.0e4 | B | ||||||
| 919.6 | 8 | 494/476 | 100 | 8.0e4 | C | ||||
| 522/504 | 10 | 8.0e3 | D | ||||||
| 1380.9-(18:0) | 2 | 919.6 | 100 | 494/476 | 100 | 2.0e4 | E | ||
| 37°C | 1665.1 | 1408.9-(16:0) | 100 | 947.6 | 100 | 522/504 | 100 | 1.0e6 | A |
| 25°C | 1637.1 | 1380.9-(16:0) | 100 | 919.6 | 100 | 494/476 | 100 | 1.0e6 | F |
| 522/504 | 4 | 4.0e4 | G | ||||||
| 947.6 | 11 | 522/504 | 100 | 1.1e5 | H | ||||
| 37°C | 1637.1 | 1380.9-(16:0) | 100 | 947.6 | 100 | 522/504 | 100 | 1.0e6 | H |
| 919.6 | 80 | 494/476 | 100 | 8.0e5 | F | ||||
| 522/504 | 2 | 1.6e4 | G | ||||||
| 1408.9-(14:0)7 | 947.6 | 100 | 522/504 | 100 | 7.0e4 | I | |||
| 25°C | 1609.1 | 1352.9-(16:0) | 100 | 919.6 | 100 | 494/476 | 100 | 1.0e6 | J |
| 522/504 | 5 | 5.0e4 | K | ||||||
| 891.5 | 70 | 494/476 | 100 | 7.0e5 | L | ||||
| 466/448 | 3 | 2.1e4 | M | ||||||
| 1380.9-(14:0) | 6 | 919.6 | 100 | 494/476 | 100 | 6.0e4 | N | ||
| 947.6 | 12 | 522/504 | 100 | 7.2e3 | O | ||||
| 37°C | 1609.1 | 1380.9-(14:0) | 100 | 947.6 | 100 | 522/504 | 100 | 1.0e6 | O |
| 919.6 | 60 | 494/476 | 100 | 6.0e5 | N | ||||
| 1352.9-(16:0) | 80 | 919.6 | 100 | 494/476 | 100 | 8.0e5 | J | ||
| 522/504 | 11 | 8.8e4 | K | ||||||
| 891.5 | 20 | 494/476 | 100 | 1.6e5 | L | ||||
| 466/448 | 60 | 9.6e4 | M | ||||||
| 947.6 | 12 | 522/504 | 100 | 9.6e4 | P | ||||
| 1408.9-(12:0) | 16 | 947.6 | 100 | 522/504 | 100 | 1.6e5 | Q | ||
| 25°C | 1581.0 | 1324.8-(16:0) | 100 | 891.5 | 100 | 494/476 | 100 | 1.0e6 | R |
| 863.5 | 2 | 466/448 | 100 | 2.0e4 | S | ||||
| 919.6 | 1 | 494/476 | 100 | 1.0e4 | T | ||||
| 1352.9-(14:0) | 24 | 919.6 | 100 | 494/476 | 100 | 2.4e5 | U | ||
| 522/504 | 5 | 1.2e4 | V | ||||||
| 891.5 | 50 | 494/476 | 100 | 1.2e5 | W | ||||
| 25°C | 1693.2 | 1436.9-(16:0) | 100 | 947.6 | 100 | 522/504 | 100 | 1.0e6 | X |
| 975.6 | 50 | 522/504 | 100 | 5.0e5 | Y | ||||
| 550/532 | 100 | 5.0e5 | Z | ||||||
| 919.6 | 6 | 494/476 | 100 | 6.0e4 | AA | ||||
| 1408.9-(18:0) | 13 | 947.6 | 100 | 522/504 | 100 | 1.3e5 | BB | ||
| 494/476 | 3 | 3.9e3 | CC | ||||||
| 919.6 | 6 | 494/476 | 100 | 7.8e3 | DD | ||||
| 37°C | 1693.2 | 1436.9-(16:0) | 100 | 975.6 | 100 | 550/532 | 100 | 1.0e6 | Z |
| 522/504 | 60 | 6.0e5 | Y | ||||||
| 947.6 | 68 | 522/504 | 100 | 6.8e5 | X | ||||
| 1408.9-(18:0) | 16 | 947.6 | 100 | 504/522 | 100 | 1.6e5 | BB |
See Table 2. Relative intensities (R.I.) correspond to the dissociation pathways indicated (not the whole spectrum).
Table 2.
Lipid A structural isomers and fatty acyl positions for m/z 1581–1693
| (M – H)− | Structure | 2′ position | 2 position | 3 position |
|---|---|---|---|---|
| 1665.1733 | A | (18:0)-3-O-(16:0) | (18:0)-3-OH | (18:0)-3-OH |
| B | (20:0)-3-O-(16:0) | (16:0)-3-OH | (18:0)-3-OH | |
| C | (18:0)-3-O-(16:0) | (16:0)-3-OH | (20:0)-3-OH | |
| D | (16:0)-3-O-(16:0) | (18:0)-3-OH | (20:0)-3-OH | |
| E | (18:0)-3-O-(18:0) | (16:0)-3-OH | (18:0)-3-OH | |
| 1637.4250 | F | (18:0)-3-O-(16:0) | (16:0)-3-OH | (18:0)-3-OH |
| G | (16:0)-3-O-(16:0) | (18:0)-3-OH | (18:0)-3-OH | |
| H | (18:0)-3-O-(16:0) | (18:0)-3-OH | (16:0)-3-OH | |
| I | (18:0)-3-O-(14:0) | (18:0)-3-OH | (18:0)-3-OH | |
| 1609.1107 | J | (18:0)-3-O-(16:0) | (16:0)-3-OH | (16:0)-3-OH |
| K | (16:0)-3-O-(16:0) | (18:0)-3-OH | (16:0)-3-OH | |
| L | (16:0)-3-O-(16:0) | (16:0)-3-OH | (18:0)-3-OH | |
| M | (18:0)-3-O-(16:0) | (14:0)-3-OH | (18:0)-3-OH | |
| N | (18:0)-3-O-(14:0) | (16:0)-3-OH | (18:0)-3-OH | |
| O | (18:0)-3-O-(14:0) | (18:0)-3-OH | (16:0)-3-OH | |
| P | (18:0)-3-O-(16:0) | (18:0)-3-OH | (14:0)-3-OH | |
| Q | (18:0)-3-O-(12:0) | (18:0)-3-OH | (18:0)-3-OH | |
| 1581.0794 | R | (16:0)-3-O-(16:0) | (16:0)-3-OH | (16:0)-3-OH |
| S | (16:0)-3-O-(16:0) | (14:0)-3-OH | (18:0)-3-OH | |
| T | (18:0)-3-O-(16:0) | (16:0)-3-OH | (14:0)-3-OH | |
| U | (18:0)-3-O-(14:0) | (16:0)-3-OH | (16:0)-3-OH | |
| V | (16:0)-3-O-(14:0) | (18:0)-3-OH | (16:0)-3-OH | |
| W | (16:0)-3-O-(14:0) | (16:0)-3-OH | (18:0)-3-OH | |
| 1693.2046 | X | (18:0)-3-O-(16:0) | (18:0)-3-OH | (20:0)-3-OH |
| Y | (20:0)-3-O-(16:0) | (18:0)-3-OH | (18:0)-3-OH | |
| Z | (18:0)-3-O-(16:0) | (20:0)-3-OH | (18:0)-3-OH | |
| AA | (18:0)-3-O-(16:0) | (16:0)-3-OH | (22:0)-3-OH | |
| BB | (18:0)-3-O-(18:0) | (18:0)-3-OH | (18:0)-3-OH | |
| CC | (20:0)-3-O-(18:0) | (16:0)-3-OH | (18:0)-3-OH | |
| DD | (18:0)-3-O-(18:0) | (16:0)-3-OH | (20:0)-3-OH |
The ion at m/z 1637 was found to be a mixture of isomers whose percent composition changed dramatically between the two growth temperatures. The base peak in the ESI-MS spectrum of lipid A from Fn grown at 25 °C is m/z 1637.1425 (0.3 ppm; Figure 1a) and its MS/MS spectrum measured in the FT-ICR is shown in Figure 3a. Two distinct ions from the dissociation of the O-linked hydroxy fatty acid at the 3 position of the glucosamine disaccharide were observed (Figure 3a) at m/z 1336.8764 (0.6 ppm) and m/z 1364.9061 (−0.6 ppm) for the loss of (18:0)-3-OH and (16:0)-3-OH, respectively. Losses of C16H32O or C14H28O aldehyde from either the N- or O-linked fatty acyls of the reducing glucosamine were also observed at m/z 1396.8971 (0.3 ppm) and 1424.9289 (0.6 ppm), respectively. More evidence for the presence of a mixture was found in the ions at m/z 1175.8061 (−0.6 ppm) resulting from the combined loss of GalN and (18:0)-3-OH and m/z 1203.8385 (0.3 ppm) resulting from the loss of GalN and (16:0)-3-OH. Further dissociation of these ions by removal of the fatty acid esterified on the acyloxyacyl group of the 2-postion of the distal glucosamine leads lead to stable ions at m/z 919.5669 (0.3 ppm) and 947.5987 (0.8 ppm), ions that must derive from independent structures present in the parent ion.
Figure 3.
(a) High-resolution MS/MS spectrum of m/z 1637 from Fn lipid A grown at 25 °C; (b) at 37 °C; (c) High-resolution MS/MS spectrum of m/z 1609 from Fn lipid A grown at 25 °C; (d) at 37 °C.
While not observed in the high-resolution MS/MS spectrum from the 25 °C sample, MS/MS of m/z 1637 from the 37 °C sample produced an ion at m/z 1408.9340 (0.6 ppm) resulting from the loss of myristic acid (Figure 3b). This supports a structure in which myristate, rather than palmitate, is esterified to the amide linked fatty acyl on the distal glucosamine. By comparison with the ion for loss of palmitic acid (m/z 1380.9), the m/z 1408.9/1380.9 ratio was found to be ~0.08. Presumably, the abundance of this structure is small and comprises only a small fraction of the total ion abundance for m/z 1637. This estimate is further supported in secondary dissociations, where the combined loss of myristic acid (14:0) + GalN yield a small but discernable ion at m/z 1247.8645 (0.2 ppm). Comparison of its relative abundance with m/z 1219.8337 (0.6 ppm; loss of palmitic acid + GalN) produced a similar ratio of 0.04.
Utilizing the MSn strategy described above for m/z 1665, m/z 1637 was found to be a heterogeneous mixture of structures F, G, and H from Fn grown at 25 °C, and F, G, H, and I from Fn grown at 37 °C (Table 2). The components of the mixture changed dramatically between the two growth temperatures. To illustrate, F, G, and I can dissociate by loss of (18:0)-3-OH to form m/z 1336.9, while H can dissociate by loss of (16:0)-3-OH to produce m/z 1364.9. The measured m/z 1336.9/1364.9 ratio was ~6.6 and changed to ~0.65 for growth at 25 versus 37 °C, respectively. Further dissociation leads to ions at m/z 1175.8 [loss of (18:0)-3-OH and GalN] and 1203.8 [loss of (16:0)-3-OH and GalN]. Similarly, the 1175.8/1203.8 ratio was ~6.2 and changed to ~0.56 for growth at 25 °C versus 37 °C, respectively. Additionally, note that F and G are able to produce, upon successive dissociations, the ion at m/z 919.6, while H and I can produce the analogous ion at m/z 947.6. The m/z 919.6/947.6 ratio was found to be ~5.6 for the 25 °C sample and ~0.38 for the 37 °C sample. Since the abundance of I was found to be small, based on the minor loss of myristic acid, one can presume that most of the ion current from m/z 947 arises from structure H. Furthermore, Table 1 suggests that G is also a minor species in the mixture based on the MS4 spectra of m/z 919.6 isolated from m/z 1380.9 (loss of palmitic acid). Thus, most of the ion current from m/z 919.6 must arise from F. Taken collectively, the ion ratios suggest that F is the dominant structure at 25 °C, while H is dominant at 37 °C.
The peak at m/z 1609 was found to have the greatest fatty acid heterogeneity. This peak comprised ~37% relative intensity (m/z 1637 base) in the mass spectrum from lipid A isolated after growth at 25 °C, but was < 2% relative intensity (m/z 1665 base) in the mass spectrum from lipid A isolated after growth at 37 °C (Figure 1). The eight structures J–Q for m/z 1609 are found in Table 2. The structures differed by the identities of the O- and N-linked substituents of the reducing glucosamine that were identified as either (18:0)-3-OH, (16:0)-3-OH, or (14:0)-3-OH hydroxylated fatty acids. Structures can be grouped by the nature of the amide linked acyloxyacyl group of the distal glucosamine, identified as (18:0)-3-0-(16:0) (J, M, P), (16:0)-3-0-(16:0) (K, L), (18:0)-3-0-(14:0) (N, O), or (18:0)-3-0-(12:0) (Q). As with the other peaks investigated, the contributions of each structure that comprise m/z 1609 changed dramatically between the two growth temperatures studied. This was evidenced by examining the relative ion intensities for representing primary, secondary, and multiple dissociations from m/z 1609 in the MS/MS spectra for the two growth conditions (Figure 3c and d). As an example, the stable amide-linked fatty acid fragment ions at m/z 947.6, 919.6, and 891.5 were found in a ratio of ~0.96:1.0:0.12 for the 37 °C sample and ~0.01:1.0:0.54 ratio for the 25 °C sample. The dramatic changes in ion ratios, combined with the significant changes in relative ion abundance for the deprotonated molecules, indicate a massive lipid A remodeling effect from a change in growth temperature.
Modeling the Environmental Regulation of Lipid A
The use of precursor and fragment ion relative abundance ratios to quantitatively determine components of a mixture is problematic for several reasons. However, we chose this route in spite of two pitfalls. First, electrospray ionization efficiencies vary between structures, making their absolute abundances difficult to determine [49]. Second, collisional fragmentation efficiencies between similar structures can vary, even in the present case when comparing similar bond cleavages possessing different leaving groups. Both of these issues can be addressed with authentic analytical standards via a calibration response. However, commercial sources of lipid A are limited in availability and tend to be derived from natural sources where the molecular structure(s) are often unique to the corresponding species. Here we present a method for circumventing these problems.
Using an infusion tandem mass spectrometry approach, we used both precursor and fragment ion abundances to detect qualitative changes in Fn lipid A structures between different growth conditions. This was done by first utilizing eq 1.
| (1) |
where σ is a weighting coefficient for an individual structure (A-DD), RI MS2 is the relative intensity of the MS2 product ions following the loss of fatty acid from the acyloxyacyl group from the parent ions, RI MS3 is the relative intensity of the MS3 product ions following the combined loss of GalN and the hydroxy-fatty acid located at the 3 position, and RI MS4 is the relative intensity of the MS4 Y- and Z-type product ions following cleavage of the glycosidic bond of the diglucosamine core. The relative intensity values, RI MS2, RI MS3, and RI MS4 are found in Table 1. The tandem MS weighting coefficient σ was further manipulated to yield ω, using eq 2,
| (2) |
where %MS1 is the percent contribution for the deprotonated molecule ion in the MS spectra and ΣσMI is the sum of σ for all structures comprising a deprotonated molecule under a given set of sample conditions. Thus, ω represents a composite weighting factor for an individual structure (A-DD), derived from both the MS data (Figure 1) and the tandem MSn data (Table 1).
The value of ω can be used to estimate the relative abundance of a particular structure derived from a common precursor ion, and this in turn can be used to propose a rank order of abundance for lipid A structures A-DD under the two Fn growth conditions. Changes detected between the two growth conditions are indicative of remodeling and reflect changes in the expression of genes responsible for lipid A biosynthesis. This is also the case for monosaccharide addition, which by examination of the MS spectra in Figure 1, the hexose containing lipid A ions at m/z 1771, 1799, and 1827 comprised ~26% of the lipid A ion current from the 25 °C sample, but was <5% for the 37 °C sample.
Using Table 1 and Figure 1 to derive ω, the composite weighting factors were used to visualize the remodeling of all structures (A-DD) for each acyl position for the two growth temperatures studied using eq 3
| (3) |
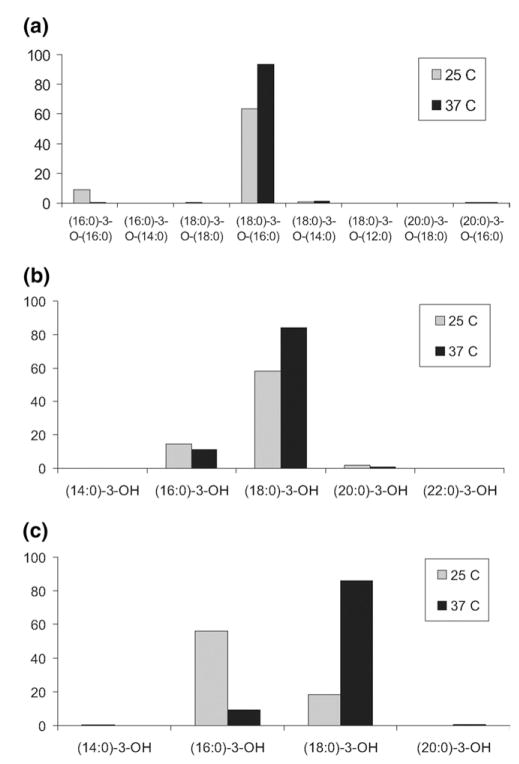
where τ represents the sum of all ω for all structures bearing a defined acyl group at a defined position on the diglucosamine core. Figure 4 displays τ, the frequency of acyl groups, for the 2, 3, and 3′ positions. The data indicate that (18:0)-3-O-(16:0) is the dominant substituent at the 3′ position and is enriched following growth at 37 °C (Figure 4a). By contrast, the (16:0)-3-O-(16:0) substituent is greatly reduced following growth at 37 °C. Further analysis of the esterified fatty acid component of the acyloxyacyl group showed that palmitic acid was the dominant substitution and was elevated at 37 °C relative to 25 °C (data not shown). For the 3 position, the most common substituent was (18:0)-3-OH; and was enriched at 37 °C relative to 25 °C (Figure 4b). For the 2 position, however, the most significant change was observed (Figure 4c). Growth of Fn at 25 °C indicates that (16:0)-3-OH is the dominant substituent but for growth at 37 °C the trend is reversed and favors (18:0)-3-OH. These observations implicate the 2 position of Fn lipid A as the major location for temperature-dependent remodeling of fatty acyl groups.
Figure 4.
Temperature-dependent lipid A remodeling as a function of acyl group frequency (τ) for: (a) the acyloxyacyl group (2′ position); (b) the 3 position; (c) the 2 position. Values derive from m/z 1581–1693 but ω was calculated using %MS1 values inclusive of the hexose modified ion series (i.e., m/z 1771–1827).
The data suggest that acyl chain remodeling favors (18:0)-3-OH at the higher temperature and is most pronounced at the 2 position but also occurs for the 3 and 2′ positions. This can help to explain the dramatic shift in ion intensities contained between the two samples in the ESI-MS spectra (Figure 1). For example, the ion at m/z 1581 (25 °C) is almost entirely comprised of R, U, and W (Table 1), all of which possess a (16:0)-3-OH at the 2 position (Table 2). It follows that if this position is preferentially remodeled to (18:0)-3-OH, then this ion would not be predicted to be observed in the MS spectrum from the lipid A sample isolated from Fn growth at 37 °C. Similarly, the ion at m/z 1609 (25 °C) is dominated by J and L; both of these possess a (16:0)-3-OH at the 2 position and both are greatly reduced in the MS spectrum from the 37 °C sample. The base peak in the MS spectrum at 25 °C, m/z 1637, consists primarily of F. Substitution of (16:0)-3-OH for (18:0)-3-OH at the 2 position in F would produce A (m/z 1665), the base peak (m/z 1665) in the MS spectrum at 37 °C. Note that A, having both the 2 and 3 positions occupied with (18:0)-3-OH would not be predicted to shift to higher m/z at the higher temperature; the weak increase of m/z 1693 (37 °C) supports this conclusion.
It follows that the measured changes in σ and ω between the sample sets reflect relative changes in sample composition. By comparison of similar dissociation pathways between all possible isomers, discrimination effects by ion activation and dissociation are minimized as much as possible as fragment ions are similar with respect to structure, degrees of freedom, and charge localization. Moreover, fragment ions are collisionally cooled before their isolation for subsequent activation and dissociation, so that energetic biases caused by CID would be minimized rather than be perpetuated through multiple stages of MSn. An advantage of the LTQ is the use of AGC, which sets the number of ions for both the linear ion trap and ICR cell [32]. If the maximum fill time is set high relative to the actual fill time needed, then equivalent numbers of ions are analyzed between samples. This has the effect of decoupling the incident ESI ion current on subsequent MSn events and allows measurement of fragment ion relative intensities (e.g., σ) that are less susceptible to diurnal variations in ESI ion current, ion source tuning parameters, and transmission dynamics through the pressure gradient to the LTQ analyzer. Moreover, given the normalization effect of AGC, the ability to detect the low abundance isomers contained within a peak is a direct measure of the methods dynamic range. For example, m/z 1693 (25 °C) gave a ratio of σ values of 250 (i.e., X/CC), thus providing an estimate of the dynamic range under the current set of ion population settings.
Measurements in both the LTQ and ICR cell were beneficial to the data interpretation. Comparison of ion ratios were similar between the two detectors at high signal intensity but tended to be dissimilar at low intensities. This discrepancy was due to the data reduction feature of the instrument control program, which subtracts out ~2.9 times the standard deviation of the noise from the FT transient following its conversion to a mass spectrum. Thus, from this standpoint, the use of ion ratios was favored from the LTQ data. The mass accuracy of the FT-ICR MS data were in agreement, but did little to authenticate the elemental compositions of the deprotonated molecules, given their high molecular weights. Further, most of the ions were mixtures of isobaric structures and deconvolution of the isomers had to come from MSn analysis. For MS/MS, the FT-ICR data were of great value as fragment ions and inferred neutral losses were realized with great accuracy (<5 ppm in most cases). Moreover, low intensity fragment ions that could be mistaken as noise in the LTQ could rapidly be identified in the FT-ICR by virtue of their mass defect and isotopic distribution.
Conclusions
The results show that a unique profile of Fn U112 lipid A molecular structures are produced in response to growth at 25 °C versus 37 °C. Deprotonated molecules were found to be heterogeneous mixtures, sharing a conserved glucosamine disaccharide backbone, a galac-tosamine-1-phosphate linked to the reducing glucosamine, but differed as to the identities of the N- and O-linked fatty acyl substituents. The extensive heterogeneity of structures observed has not been reported previously. Estimates for the relative abundance of each isomer were derived from MS relative abundance ratios and fragment ion ratios from comparable dissociation pathways. The results suggest a remodeling pathway in which the amide linked fatty acid of the reducing glucosamine favors a 3-hydroxyhexadecanoic acid substituent for growth conditions at 25 °C and a 3-hydroxyoctadecanoic acid substituent for growth conditions at 37 °C.
Supplementary Material
Acknowledgments
The authors thank the NCRR (1S10RR17262-01), NIAID (1U54AI57141-01), and NIEHS (P30ES07033) for generous funding and support. Additional thanks go to Professor František Tureček for helpful discussion and Thomas Kalhorn, Jace Jones, and Shawna Hengel for critical review of this manuscript.
References
- 1.Ernst RK, Guina T, Miller SI. Salmonella typhimurium Outer Membrane Remodeling: Role in Resistance to Host Innate Immunity. Microbes Infect. 2001;3(1415):1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 2.Sajbidor J. Effect of Some Environmental Factors on the Content and Composition of Microbial Membrane Lipids. Crit Rev Biotechnol. 1997;17(2):87–103. doi: 10.3109/07388559709146608. [DOI] [PubMed] [Google Scholar]
- 3.Suutari M, Laakso S. Microbial Fatty Acids and Thermal Adaptation. Crit Rev Microbiol. 1994;20(4):285–328. doi: 10.3109/10408419409113560. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CR, Whitfield C. Lipopolysaccharide Endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trent MS. Biosynthesis, Transport, and Modification of Lipid A. Biochem Cell Biol. 2004;82(1):71–86. doi: 10.1139/o03-070. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst RK, Guina T, Miller SI. How Intracellular Bacteria Survive: Surface Modifications that Promote Resistance to Host Innate Immune Responses. J Infect Dis. 1999;179(Suppl):2S326–2S330. doi: 10.1086/513850. [DOI] [PubMed] [Google Scholar]
- 8.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller S. Lipid A Acylation and Bacterial Resistance Against Vertebrate Antimicrobial Peptides. Cell. 1998;95(2):189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 9.Miller SI, Ernst RK, Bader MW. LPS, TLR4, and Infectious Disease Diversity. Nat Rev Microbiol. 2005;3(1):36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 10.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in Lipid A Structure in the Pathogenic . Tersiniae Mol Microbiol. 2004;52(5):1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 11.McLendon MK, Apicella MA, Allen LA. Francisella tularensis: Taxonomy, Genetics, and Immunopathogenesis of a Potential Agent of Biowarfare. Annu Rev Microbiol. 2006;60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyston PC, Sjostedt A, Titball RW. Tularaemia: Bioterrorism Defence Renews Interest in Francisella tularensis. Nat Rev Microbiol. 2004;2(12):967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 13.Santic M, Molmeret M, Klose KE, Abu Kwaik Y. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 2006;14(1):37–44. doi: 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15(4):631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieffer TL, Cowley S, Nano FE, Elkins KL. Francisella novicida LPS has Greater Immunobiological Activity in Mice than F. tularensis LPS and Contributes to F. novicida murine pathogenesis. Microbes Infect. 2003;5(5):397–403. doi: 10.1016/s1286-4579(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 16.Phillips NJ, Schilling B, McLendon MK, Apicella MA, Gibson BW. Novel modification of lipid A of Francisella tularensis. Infect Immun. 2004;72(9):5340–5348. doi: 10.1128/IAI.72.9.5340-5348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinogradov E, Perry MB, Conlan JW. Structural Analysis of Francisella tularensis lipopolysaccharide. Eur J Biochem. 2002;269(24):6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- 18.Hajjar AM, Harvey MD, Shaffer SA, Goodlett DR, Sjostedt A, Edebro H, Forsman M, Bystrom M, Pelletier M, Wilson CB, Miller SI, Skerrett SJ, Ernst RK. Lack of in Vitro and in Vivo Recognition of Francisella tularensis Subspecies Lipopolysaccharide by Toll-Like Receptors. Infect Immun. 2006;74(12):6730–6738. doi: 10.1128/IAI.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qureshi N, Takayama K, Heller D, Fenselau C. Position of Ester Groups in the Lipid A Backbone of Lipopolysaccharides Obtained from Salmonella typhimurium. J Biol Chem. 1983;258(21):12947–12951. [PubMed] [Google Scholar]
- 20.Qureshi N, Takayama K, Ribi E. Purification and Structural Determination of Nontoxic Lipid A Obtained from the Lipopolysaccharide of Salmonella typhimurium. J Biol Chem. 1982;257(19):11808–11815. [PubMed] [Google Scholar]
- 21.Seid RC, Jr, Bone WM, Phillips LR. Identification of Ester-Linked Fatty Acids of Bacterial Endotoxins by Negative Ion Fast Atom Bombardment Mass Spectrometry. Anal Biochem. 1986;155(1):168–176. doi: 10.1016/0003-2697(86)90242-3. [DOI] [PubMed] [Google Scholar]
- 22.Takayama K, Qureshi N, Ribi E, Cantrell JL. Separation and Characterization of Toxic and Nontoxic Forms of Lipid A. Rev Infect Dis. 1984;6(4):439–443. doi: 10.1093/clinids/6.4.439. [DOI] [PubMed] [Google Scholar]
- 23.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of Lipid A Modifications by Salmonella typhimurium Virulence Genes phoP-phoQ. Science. 1997;276(5310):250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 24.Kaltashov IA, Doroshenko V, Cotter RJ, Takayama K, Qureshi N. Confirmation of the Structure of Lipid A Derived from the Lipopolysaccharide of Rhodobacter sphaeroides by a Combination of MALDI, LS IMS, and Tandem Mass Spectrometry. Anal Chem. 1997;69(13):2317–2322. doi: 10.1021/ac9612943. [DOI] [PubMed] [Google Scholar]
- 25.Lindner B. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry of Lipopolysaccharides. Methods Mol Biol. 2000;145:311–325. doi: 10.1385/1-59259-052-7:311. [DOI] [PubMed] [Google Scholar]
- 26.Boue SM, Cole RB. Confirmation of the Structure of Lipid A from Enterobacter agglomerans by Electrospray Ionization Tandem Mass Spectrometry. J Mass Spectrom. 2000;35(3):361–368. doi: 10.1002/(SICI)1096-9888(200003)35:3<361::AID-JMS943>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 27.Chan S, Reinhold VN. Detailed Structural Characterization of Lipid A: Electrospray Ionization Coupled with Tandem Mass Spectrometry. Anal Biochem. 1994;218(1):63–73. doi: 10.1006/abio.1994.1141. [DOI] [PubMed] [Google Scholar]
- 28.Kussak A, Weintraub A. Quadrupole Ion-Trap Mass Spectrometry to Locate Fatty Acids on Lipid A from Gram-Negative Bacteria. Anal Biochem. 2002;307(1):131–137. doi: 10.1016/s0003-2697(02)00004-0. [DOI] [PubMed] [Google Scholar]
- 29.Sforza S, Silipo A, Molinaro A, Marchelli R, Parrilli M, Lanzetta R. Determination of Fatty Acid Positions in Native Lipid A by Positive and Negative Electrospray Ionization Mass Spectrometry. J Mass Spectrom. 2004;39(4):378–383. doi: 10.1002/jms.598. [DOI] [PubMed] [Google Scholar]
- 30.Douglas DJ, Frank AJ, Mao D. Linear Ion Traps in Mass Spectrometry. Mass Spectrom Rev. 2005;24(1):1–29. doi: 10.1002/mas.20004. [DOI] [PubMed] [Google Scholar]
- 31.March R. An Introduction to Quadrupole Trap Mass Spectrometry. J Mass Spectrom. 1997;32:351–369. [Google Scholar]
- 32.Schwartz JC, Senko MW, Syka JE. A Two-Dimensional Quadrupole Ion Trap Mass Spectrometer. J Am Soc Mass Spectrom. 2002;13(6):659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 33.Battisti JM, Sappington KN, Smitherman LS, Parrow NL, Minnick MF. Environmental Signals Generate a Differential and Coordinated Expression of the Heme Receptor Gene Family of Bartonella quintana. Infect Immun. 2006;74(6):3251–3261. doi: 10.1128/IAI.00245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinnebusch BJ. The Evolution of Flea-Borne Transmission in Yersinia pestis. Curr Issues Mol Biol. 2005;7(2):197–212. [PubMed] [Google Scholar]
- 35.Westphal O, Jann K. Bacterial Lipopolysaccharides: Extraction with Phenol-Water and Further Applications of the Procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 36.Fischer W, Koch HU, Haas R. Improved Preparation of Lipoteichoic Acids. Eur J Biochem. 1983;133(3):523–530. doi: 10.1111/j.1432-1033.1983.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 37.Caroff M, Tacken A, Szabo L. Detergent-Accelerated Hydrolysis of Bacterial Endotoxins and Determination of the Anomeric Configuration of the Glycosyl Phosphate Present in the “Isolated Lipid A” Fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175:273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- 38.Darveau RP, Cunningham MD, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, Dietsch M, Page RC, Aruffo A. The Ability of Bacteria Associated with Chronic Inflammatory Disease to Stimulate E-Selectin Expression and Neutrophil Adhesion. Prog Clin Biol Res. 1995;392:69–78. [PubMed] [Google Scholar]
- 39.Somerville JE, Jr, Cassian L, Bainbridge B, Cunningham MD, Darveau RP. A Novel Escherichia coli Lipid A Mutant that Produces an Anti-Inflammatory Lipopolysaccharide. J Clin Invest. 1996;97(2):359–365. doi: 10.1172/JCI118423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hegarty A, O’Neill P, Rappoport Z, editors. In The Chemistry of Enols. Wiley; Chichester, UK: 1990. p. 639. [Google Scholar]
- 41.Rappoport Z, Yamataka H. Calculated Amide/Enol of Amide Energy Differences for Several Interesting Amide Systems. Chem Commun. 2000;21:2101–2102. [Google Scholar]
- 42.Sklenak S, Apeloig Y, Rappoport Z. Calculated pKEnol Values for Enols of Carboxylic Acid Derivatives HC = C(OH)X, X = OH, NH2, NMe2, OMe, OCHO, F, Cl, Br. J Am Chem Soc. 1998;120:10359–10364. [Google Scholar]
- 43.Gross ML. Charge-Remote Fragmentation: An Account of Research on Mechanisms and Applications. Int J Mass Spectrom. 2000;200:611–624. [Google Scholar]
- 44.Jensen N, Tomer K, Gross M. Gas-Phase Ion Decomposition Occurring Remote to a Charged Site. J Am Chem Soc. 1985;107:1863–1868. [Google Scholar]
- 45.Mole T. Thermal Retro-Aldol Reaction. Chem Ind. 1960:1164–1165. [Google Scholar]
- 46.Costello CE, Vath JE. Tandem Mass Spectrometry of Glycolipids. Methods Enzymol. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. [DOI] [PubMed] [Google Scholar]
- 47.Domon B, Costello CE. A Systematic Nomenclature for Carbohydrate Fragmentations in FAB-MS/MS Spectra of Glycoconjugates. Glycoconjugate. 1988;5:397–409. [Google Scholar]
- 48.Domon B, Vath JE, Costello CE. Analysis of Derivatized Ceramides and Neutral Glycosphingolipids by High-Performance Tandem Mass Spectrometry. Anal Biochem. 1990;184(1):151–164. doi: 10.1016/0003-2697(90)90028-8. [DOI] [PubMed] [Google Scholar]
- 49.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative Determination of Phospholipid Compositions by ESIMS: Effects of Acyl Chain Length, Unsaturation, and Lipid Concentration on Instrument Response. J Lipid Res. 2001;42(4):663–672. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.