SUMMARY
Bimodal targeting of the endoplasmic reticular protein, cytochrome P4501A1 (CYP1A1), to mitochondria involves activation of a cryptic mitochondrial targeting signal through endoprotease processing of the protein. Here, we characterized the endoprotease regulating CYP1A1 mitochondrial targeting. The endoprotease, which was induced by β-naphthoflavone, was a dimer of 90 kDa and 40 kDa subunits, each containing Ser protease domains. The purified protease processed CYP1A1 in a sequence-specific manner, leading to its mitochondrial import. The glucocorticoid receptor, retinoid X receptor, and p53 underwent similar processing-coupled mitochondrial transport. The inducible 90 kDa subunit was a limiting factor in many cells and some tissues, and thus, regulates the mitochondrial levels of these proteins. A number of other mitochondria-associated proteins with non-canonical targeting signals may also be substrates of this endoprotease. Our findings point to a novel mechanism of mitochondrial protein import that requires an inducible cytosolic endoprotease for activation of cryptic mitochondrial targeting signals.
INTRODUCTION
The targeting signals for proteins destined for mitochondria vary markedly in terms of sequence, structure, and location (Pfanner and Neupert, 1991; Haucke, et al., 1995). In most reported cases, the signal sequences contain multiple basic and hydroxylated residues, form amphiphilic helices, and are cleaved by matrix metalloproteases (Roise and Schatz, 1988; von Heijne, 1989). However, marked differences exist among many mitochondrial proteins in both the sequence and locations of these signals (Rapaport, 2003, Neupert and Hermann, 2007). All outer mitochondrial membrane proteins and most mitochondrial intermembrane space proteins are devoid of typical N-terminal presequences (Rapaport, 2003; Pfanner, N., et al., 2004; Mihara, 2000). Similarly, some of the inner mitochondrial membrane proteins, such as BCS1, Tim14, MDj2, DAKAP1 contain non-canonical and non-cleavable internal signals (Folsch et al., 1996, Neupert and Hermann, 2007, Ma and Taylor, 2008). The nature of the signals or the mechanism responsible for the mitochondrial translocation of the N-terminally processed proteins containing non-canonical signals, such as retinoid X receptor (RXR), glucocorticoid receptor (GR), estrogen receptor (EsR), and p53, remain unclear (Casas et al., 2003; Koufali et al., 2003; Chen and Yager, 2004).
Recently, comprehensive proteomic analyses of mitochondria from different cells and tissues have been reported (Mootha et al., 2003; Sickmann et al., 2003; Taylor et al., 2003; Prokisch et al., 2004). According to the MITOP database, yeast mitochondria contain up to 1000 different proteins, whereas their mammalian counterparts have approximately 2000 proteins. Proteomic data suggest that nearly half of nuclear-encoded proteins associated with mammalian mitochondria lack canonical N-terminal targeting signals (Mootha et al., 2003; Taylor et al., 2003). The mechanisms underlying the mitochondrial translocation of these proteins are likely to differ from those underlying translocation of proteins containing canonical unimodal targeting signals.
Over the past decade, studies in our laboratory have demonstrated that xenobiotic-inducible CYPs, which are widely known to be ER-associated proteins, as well as cytosolic glutathione S-transferases (GSTs) are present in the mitochondrial matrix (Addya et al., 1994; Addya et al. 1997, Anandatheerthavarada et al. 1999, Bhagwat et al. 1999, Boopathi et al. 2000; Robin et al., 2002:Raza et al., 2001; Sepuri et al. 2007). We have discovered that the multiple cellular locations of these proteins are attributable to a novel bimodal targeting mechanism involving the activation of cryptic N-terminal or C-terminal mitochondrial-targeting signals (Addya et al., 1997; Anandatheerthavarada et al., 1999; Robin et al., 2002; Robin et al., 2003). In the case of β-naphthoflavone (BNF)-inducible cytochrome P450 1A1 (CYP1A1), the mitochondrial-targeted species (mtCYP1A1) consisted mostly of N-terminally cleaved protein (+33/1A1) (Addya et al., 1997; Bhagwat 1999; Boopathi et al., 2000). Our results suggest that a fraction of CYP1A1 nascent chains (~ 25%) escape signal recognition particle (SRP) binding and are N-terminally clipped by a sequence-specific cytosolic endoprotease, leading to the exposure of a cryptic mitochondrial-targeting signal at positions +34 to +44 of the protein. We have observed that the cytosolic chaperones Hsp70 and Hsp90, peripheral TOM receptors, as well as the channel-forming TOM40 protein are required for the mitochondrial import of +33/1A1 and +5/1A1 (a minor processed product) (Anandatheerthavarada, H. K., Sepuri NB., Biswas, G. and Avadhani, N. G, unpublished observations). Here, we describe the purification a Ser protease family endoprotease from the liver cytosol of BNF-treated rats and demonstrate that this protease activates cryptic mitochondrial-targeting signals in CYP1A1, as well as in number of other proteins, through sequence-specific processing.
RESULTS
Endoprotease Processing of CYP1A1
To analyze processing of CYP1A1, we performed in vitro processing assays and cell transfection studies using a dihydrofolate reductase (DHFR) and CYP1A1 fusion protein (Figure 1A). This fusion protein was used for three reasons. First, the N-terminal fusion would block both mitochondrial and ER targeting signals. Second, the endoproteolytic nature of the putative protease could be ascertained by attaching an N-terminal extension. Finally, an N-terminal extension would improve detection of processed fragments, since CYP1A1 and its processed product have similar mobilities on polyacrylamide gels (Addya et al., 1997; Dasari et al., 2006).
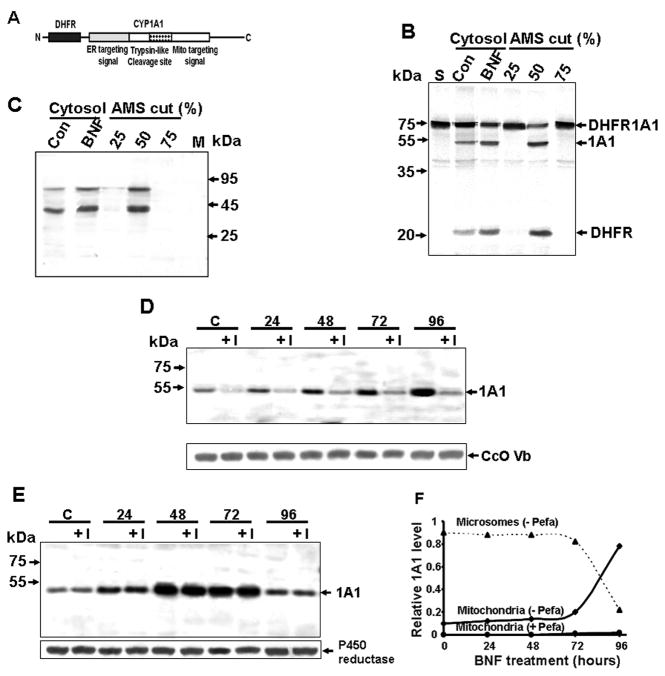
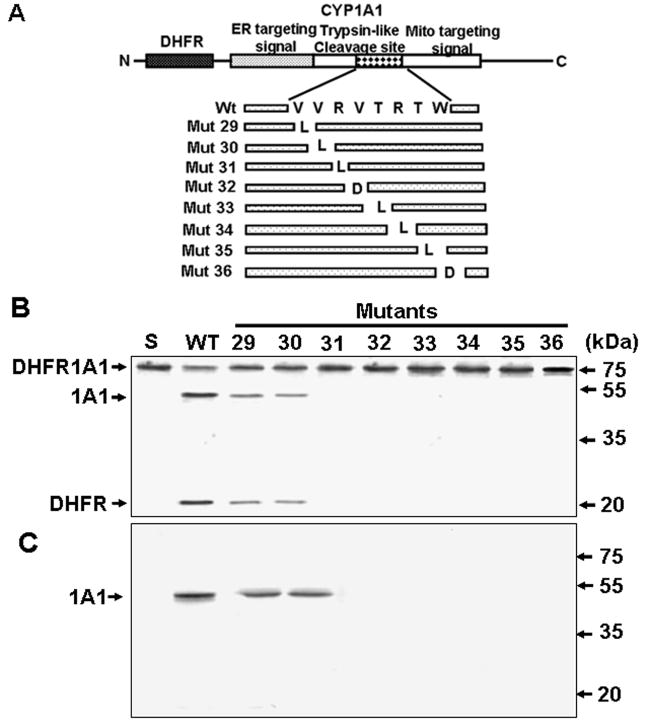
Figure 1. CYP1A1 Processing Activity of Rat Liver Cytosol.
(A) The DHFR-1A1 fusion construct used as a substrate for processing assays.
(B) DHFR-1A1 processing by liver cytosol from control and BNF-treated rats as well asby (NH4)2SO4 (AMS)-fractionated liver cytosol from the BNF group. The 1A1 (51 kDa) and DHFR (23 kDa) processed fragments are indicated. “S” indicates substrate alone.
(C) Far western analysis of cytosolic liver protein with aprotinin.
(D and E) Immunoblot analysis of 1A1 in trypsin treated (150 μ/ml, 30 min) mitochondrial (D) and microsomal fractions (E) from C6 glioma cells treated with 0.1% DMSO (“C”) or exposed to BNF (50 μM) ± pefabloc (“I”, 300 μM) for the indicated time.
(F) Relative distribution of mitochondrial and microsomal CYP1A1 in BNF treated C6 glioma cells based on a 2.5:1 ratio of microsomes and mitochondria recovered by cell fractionation.
Analysis of the processing activity of rat liver cytosol revealed that liver cytosol from untreated animals (200 μg) processed the 73 kDa DHFR-1A1 protein into a short 23 kDa fragment (mostly DHFR with a short N-terminal fragment of 1A1 attached) and a larger 50 kDa fragment of +33/1A1 (Figure 1B). Liver cytosol (30 μg) from rats treated with BNF, a known inducer of class I and class II drug-metabolizing enzymes, had a markedly higher processing activity, as seen by increased intensity of the 50 kDa band. Proteins precipitating at 25% and 75% (NH4)2SO4 saturation did not have any detectable activity, while the 50% (NH4)2SO4 precipitate showed a high processing activity. In control liver, BNF-treated liver, and the 50% (NH4)2SO4-precipitated fractions, a 90 kDa and 40 kDa protein species bound to aprotinin, suggesting that these factors may participate in CYP1A1 processing (Figure 1C). BNF induces the expression of CYP1A1 (Whitlock, 1986; Xu et al., 2005) in C6 glioma cells. In addition, we have previously shown that BNF treatment for > 72h induces an accumulation of mtCYP1A1 (+33/1A1), an effect accompanied by a sharp reduction in microsomal CYP1A1 (mcCYP1A1) most likely because of vastly different CYP1A1 turnover rates in these subcellular compartments (Boopathi et al., 2000). The effect of inhibition of cytosolic CYP1A1 processing on the time-dependent accumulation of mtCYP1A1 was investigated using the Ser protease inhibitor, Pefabloc (AEBSF-HCl). Pefabloc markedly inhibited the accumulation of mtCYP1A1 in C6 glioma cells, showing that processing by a Ser protease is required for mitochondrial targeting (Figures 1D and F). In contrast, accumulation of CYP1A1 up to 72h and a sharp decline after 72h of treatment was unaffected by Pefabloc (Figure E and F).
Purification and Characterization of the Processing Protease
Since BNF increased endoprotease activity and the levels of aprotinin-binding proteins (Figure 1B and C), we purified the processing protease from liver cytosol of BNF-treated rats. The endoproteolytic processing of DHFR-1A1 into putative 50 kDa and 23 kDa fragments was used as an index of purification. The 50% (NH4)2SO4 precipitate showing processing activity was subjected to chromatography on a DEAE-Sephacel column. The proteins eluted as several major and minor peaks (Figure S1A in the Supplemental Data), with only the peak IV fraction showing processing activity (Figure S1B). This processing activity was inhibited by the Ser protease inhibitor, aprotinin (250 μg/ml) (Figure S1C). Far western blot analysis of all fractions using biotinylated aprotinin and streptavidin-conjugated secondary antibodies revealed that only the 90 kDa and 40 kDa proteins from fraction IV interacted with aprotinin (Figures S1D-F).
The peak IV fractions were pooled and further purified using aprotinin-agarose affinity chromatography (Goldberg, 2000; Bicknell et al., 2001). Proteins bound to the aprotinin column were eluted with a step gradient of 1 M and 2 M NaCl (Figure 2A). SDS-PAGE of the eluted fractions revealed that the 1 M NaCl-eluted fractions contained multiple proteins ranging from about 20 kDa to greater than 100 kDa, while the 2 M NaCl-eluted proteins resolved into 40 kDa and 90 kDa bands (Figure 2B). The 2 M NaCl-eluted fraction contained processing activity, which was inhibited by Pefabloc (Figure 2C) or aprotinin (not shown). Additionally, both the 90 kDa and 40 kDa proteins interacted with biotinylated aprotinin, as determined by far western blot analysis (Figure 2D). As shown by silver-stained gels, successive (NH4)2SO4, DEAE-Sephacel, and aprotinin-agarose fractionation generated about 2 μg of protein (last lane) from 12 g of BNF-treated liver cytosolic protein.
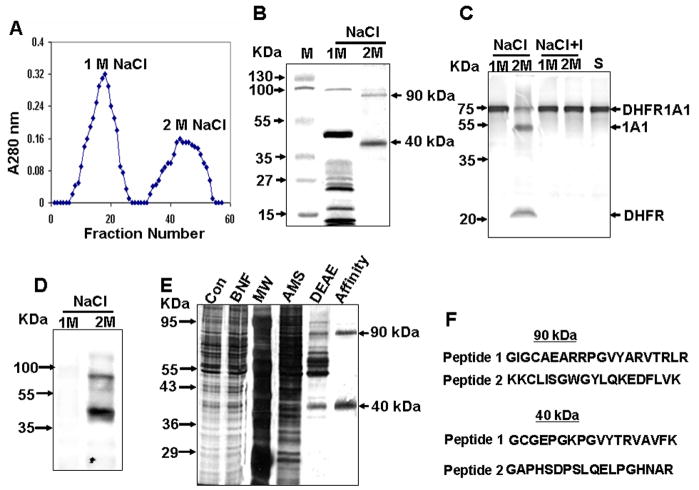
Figure 2. Endoprotease Purification by Aprotinin-Agarose Affinity Chromatography.
(A) Elution pattern of aprotinin-agarose column-bound proteins. The DEAE Sephacel-purified fraction (IV) was loaded onto the aprotinin-agarose column, and bound proteins were eluted using a NaCl step gradient.
(B) Cypro Ruby-stained protein fractions separated by SDS-PAGE.
(C) Proteolytic activity of protein fractions, as determined using 35S-labeled DHFR-1A1 as substrate. “S” indicates substrate alone, and “I” indicates reactions performed in the presence of Pefabloc.
(D) Far western blot analysis of fractions using aprotinin.
(E) Polyacrylamide gel electrophoresis (12%) of fractions at each progressive purification stage. Proteins shown, from left to right, are total liver cytosolic protein (Con, 10 μg protein), the liver cytosolic fraction from BNF-treated animals (BNF, 10 μg), molecular weight markers (MW), the 50% (NH4)2SO4 fraction (AMS, 10 μg), the peak IV DEAE sephacel-purified fraction (DEAE, 3 μg), and the aprotinin-agarose purified fraction (Affinity, 0.5 μg).
(F) Peptide sequences for p90 and p40.
Next, the 90 kDa and 40 kDa protein species, designated p90 and p40, were in-gel digested with trypsin and subjected to LC-MS using MALDI-QSTAR mass spectrometry. The 90 kDa protein yielded peptide fragments that matched the recently identified human transmembrane Ser protease 9 (also known as polyserase 1) and covered about 60% of length. Two peptides from p90 with mass values (m/z) of 2255.7 and 2201.61 as well as two peptides from p40 with mass values (m/z) of 1883.02 and 1865.19 were analyzed by MS/MS for amino acid sequencing (Figure 2F). Both peptides from p90 had sequences that matched the rat ortholog of human Ser protease 9 by 100%. The predicted nucleotide sequences of the p40 peptides matched a sequence entry in the rat genome that corresponded to a 1467 bp mRNA encoding a 42 kDa protein. More recently, a larger splice variant of this gene encoding a 63 kDa protein has also been reported (XM_001064945). This splice variant exhibits ~ 50% sequence identity with human transmembrane Ser protease 9. The LC-MS data for p40 matches well with this sequence. The mouse ortholog of this gene, which encodes a 85 kDa protein (XM_906114.3), has recently been characterized.
A partial cDNA for p90 hybridized to ~ 4.8 kb and 3.1 kb RNA species in BNF-treated rat liver and to a ~ 1.4 kb RNA species in control liver (Figure S2A). The identity of the shorter RNA in liver from control animals is unknown. The two closely migrating mRNAs in the BNF-treated group may represent splice variants. We also observed a 2.5 – 3 fold increase in the 4.8 kb mRNA in BNF-treated rat brain, although the 3.1 kb mRNA was not seen in this tissue. Immunoblot analysis revealed that p90 levels were increased more than 4-fold in the liver and brain of BNF-treated rats (Figure S2B). A partial cDNA for p40 hybridized to a considerable amount of 2.1 kb mRNA in the liver and brain of control rats. BNF treatment increased the levels of this mRNA by 2 – 3-fold in both tissues (Figure S2C). Immunoblot analysis using a peptide-specific antibody also showed a 2 – 3 fold increase in p40 levels following BNF treatment (Figure S2D). These results show that BNF induces the p90 by about 6-fold and p40 by about 2 – 3 fold.
Processing Activity Requires Both p40 and p90
To better understand the role of each subunit, we determined the apparent size of the active protease by resolving the DEAE column-purified protein (Figure S1) on a Sephacryl-S200 gel filtration column (Figure 3A). Only fractions 18 – 24 (size range 140–150 kDa) had processing activity (Figure 3B). The latter two factions (i.e., 33 – 39 and 45–51) had processing activity when combined, although they were unable to process DHFR-1A1 protein individually (Figure 3C). Immunoblot analysis revealed that fractions 18 – 24 contained both p90 and p40, fractions 33 – 39 contained p90, and fractions 45 – 51 contained p40 (Figures 3D–F). These proteins had a relative size of ~ 140 kDa, 90 kDa, and 40 kDa, based on the elution patterns of marker proteins (Figure 3A).
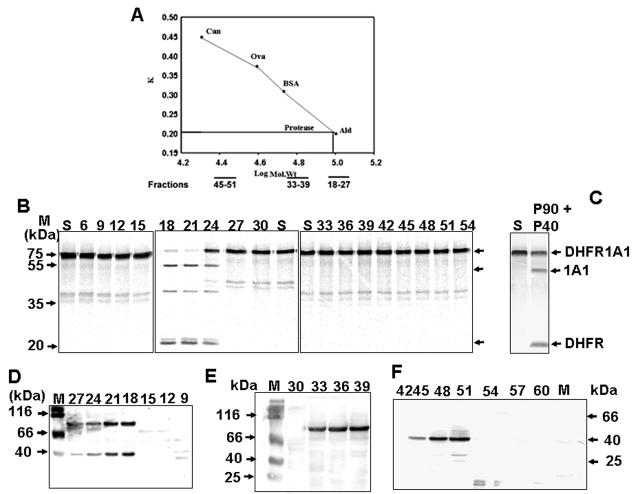
Figure 3. Size Exclusion Chromatography of the Processing Protease.
(A) Rates of elution of alcohol dehydrogenase (Ald), bovine serum albumin (BSA), ovalbumin (Ova), and carbonic anhydrase (Can) from the Sephacryl-200 column, based on apparent size.
(B) Resolution of 35S-DHFR-1A1 processing activity by size. Fractions 18–27 corresponded to 140 – 150 kDa, fractions 33 – 39 to 90 kDa, and fractions 45 – 51 to 40 kDa. 5μg protein each was used for assaying processing activity.
(C) Combined effects of proteins from fractions 33–39 and 45–51 in 35S DHFR-1A1 processing.
(D–F) Immunoblot analysis of Sephacryl S200-eluted fractions using p90 and p40 peptide antibodies.
Ser Protease Inhibition Blocks CYP1A1 Processing and Mitochondrial Targeting
The effects of different types of protease inhibitors on the in vitro processing of DHFR-1A1 were investigated. DHFR-1A1 processing was inhibited by Ser protease inhibitors (aprotinin, Pefabloc, and antipain), trypsin-inhibitor, or benzamidine (Figures 4A–C). However, calpain had no detectable effect (Figure 4C). In COS cells expressing DHFR-1A1, both DHFR-1A1 and processed 1A1 were detected in the cytosol (Figure 4D). Presennce of processed CYP1A1 in the cytosol may reflect a slow kinetic rate of mitochondrial targeting in this overexpression system. In accord with in vitro findings, incubation of these cells for 48 hr with increasing amounts of Pefabloc led to a progressive disappearance in cytosolic 1A1 (Figure 4D). The role of cytosolic processing on mitochondrial targeting of CYP1A1 was also investigated using the DHFR-1A1 fusion system. Since the leader sequence of DHFR lacks a targeting signal, we reasoned that only endoprotease-processed protein with an activated mitochondrial signal would enter mitochondria. The mitochondrial fraction of COS cells was treated with trypsin to eliminate cytosolic contamination (Figure 4E). Fully processed mitochondrial CYP1A1 (+33/1A1) was present in non Pefabloc-exposed cells, but decreased with increasing concentrations of Pefabloc. Although not shown, the mitochondrial targeting of CYPc27 was unaffected by Ser protease inhibitors aprotinin, Pefabloc, and antipain. These results confirm that a Ser protease activates the cryptic targeting signals through endoproteolytic processing and that its activity is essential for CYP1A1 mitochondrial targeting.
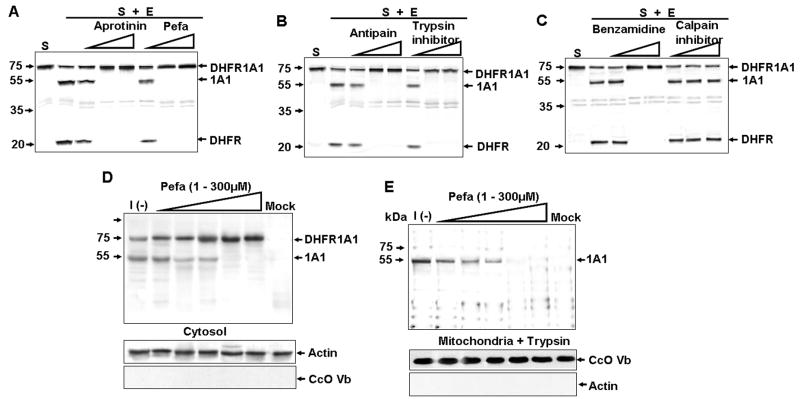
Figure 4. Effect of Protease Inhibitors on DHFR-1A1 Processing In Vitro and In Vivo.
(A–C) Processing of 35S-DHFR-1A1 by DEAE column-purified enzyme (1 μg) in the absence or presence of aprotinin (0.01, 0.1, or 0.5 mg/ml), Pefabloc (0.001, 0.01, or 0.1 mM), antipain (0.01, 0.1, or 10 μM), Soybean trypsin inhibitor (0.01, 0.1, or 10 μM), benzamidine (0.01, 0.1 or 1mM), or calpain inhibitor I (5, 25, or 50 μM). The enzyme was preincubated with the indicated inhibitor for 15 min at 37°C prior to the final incubation (30 min, 37°C) with DHFR-1A1. “S” indicates substrate alone.
(D and E) Immunoblot analysis of 1A1 in cytosolic (D) and trypsin-treated (150 μg/ml, 30 min) mitochondrial proteins (E) isolated from DHFR-1A1-expressing COS cells incubated without [I(−)] or with Pefabloc (1 – 300 μM).
Sequence Specificity of the Processing Protease
CYP1A1 is primarily processed between residues +32 and +33 (Addya et al., 1997; Dasari et al., 2006), while very minor processing occurs between residues +4 and +5. Val32 occupies the P1 site of the predicted Ser protease cleavage site, while the P’2 position is occupied by Thr, P’3 an Arg, P2 an Arg, and P3 a Val (Figure 5A). This cleavage site is conserved in CYP1A1 of all vertebrate species for which sequence information is available (Dasari et al., 2006). Point mutations were introduced in this site (Figure 5A), and the processing of the partially purified enzyme was investigated both in vitro and in transfected COS cells. Mutant proteins V29L and V30L were processed at a lower level than wild type protein (Figure 5B). However, the processing activity was nearly abolished by R31L, V32R, T33L, R34L, T35L, or W36D. In keeping with these findings, only the V29L and V30L proteins were detected in trypsin-treated mitochondrial fractions from COS cells (Figure 5C). These results indicate that CYP1A1 processing by cytosolic endoprotease is sequence specific, and they suggest that both the core cleavage site (+32/+33) and the flanking amino acids are important for processing.
Figure 5. Sequence Specificity of the Processing Protease.
(A) CYP1A1 signal domains and processing site mutations.
(B) Processing of wild type and mutant 35S-DHFR-1A1 by DEAE-Sephacel column purified enzyme.
(C) Processing of mutant DHFR-1A1 in COS cells, as shown by SDS-PAGE of trypsin-treated (150 μg/ml, 30 min) mitochondrial fractions (50 μg per lane).
AK131261 Overexpression in p90-Poor 3T3 Cells Increases CYP1A1 Processing and Mitochondrial Targeting
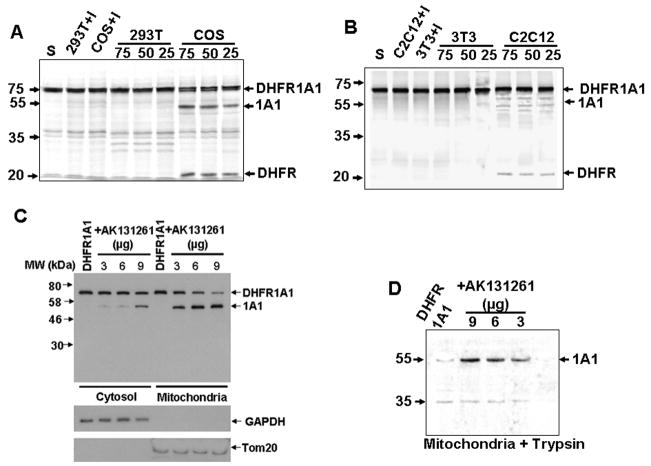
Both p40 mRNA (2.1 kb) and p90 mRNA (4.8 kb/3.1 kb) were moderately high in COS-7 cells (Figure S3). However, in 293-T and 3T3 cells, p40 mRNA was high, while p90 mRNA was very low. The opposite was true in C2C12 cells. Analysis of the in vitro processing activity of cytosolic fractions [25 – 50% (NH4)2SO4 fraction] from these cells revealed that COS cell cytosol exhibited high activity, while cytosol from all other three cell types had low to undetectable activity (Figures 6A and 6B).
Figure 6. Cell-Type Specific CYP1A1 Processing Activities.
(A and B) Processing of 35S-DHFR-1A1 by (NH4)2SO4-fractionated cytosolic extracts (50% fraction, 25 – 75 μg) from COS-7, 293T, C2C12, and NIH 3T3 cells. “I” indicates that the reaction was performed in the presence of Pefabloc (100μM).
(C and D) Immunoblot analysis of 1A1 in cytosolic and mitochondrial fractions isolated from AK 131261- and/or DHFR-1A1-overexpressing NIH3T3 cells. In (D), the mitochondrial fractions were subjected to trypsin digestion (150 μg/ml, 30 min) prior to immunoblot analysis.
Consistent with the low processing activity of 3T3 cytosol, mitochondrial targeting of DHFR-1A1 in these cells was very low (Figures 6C and 6D). DHFR-1A1 accumulated in the cytosol, with considerable amounts also being present in the mitochondrial fraction, possibly due to adventitious binding. However, no appreciable processed CYP1A1 was detected in mitochondrial fractions. To determine if reduced DHFR-1A1 mitochondrial targeting was due to low p90 expression, we transfected 3T3 cells with AK131261 cDNA, which encodes a 90 kDa variant containing intact second and third Ser protease domains, but lacking the N-terminal most Ser protease domain (Cal et al., 2003). The first and second domains of polyserase 1 are independently active in protein processing, and the third domain is relatively inactive (Cal et al., 2003). AK131261 expression increased the level of processed CYP1A1 in the mitochondrial fraction and reduced levels of full-length fusion protein in the cytosol (Figure 6C). Treatment of mitochondrial fractions with trypsin led to the disappearance of DHFR-1A1, confirming that it was externally adhered to mitochondria (Figure 6D). However, processed CYP1A1 protein was nearly completely protected against trypsin, suggesting that it is intramitochondrial. In experiments involving CYP1A1 cDNA transfection in 3T3 cells, increasing concentrations of AK131261 cDNA also increased levels of trypsin-resistant mtCYP1A1 (Figures S4A–C). Levels of mcCYP1A1 were not altered by AK131261, suggesting that the cytosolic Ser protease does not regulate this CYP pool. These results, along with the biochemical data in Figure 3, confirm that both subunits are necessary for the cytosolic processing and mitochondrial targeting of CYP 1A1.
Identification of Other Cytosolic Endoprotease Substrates That Are Bimodally Targeted to Mitochondria
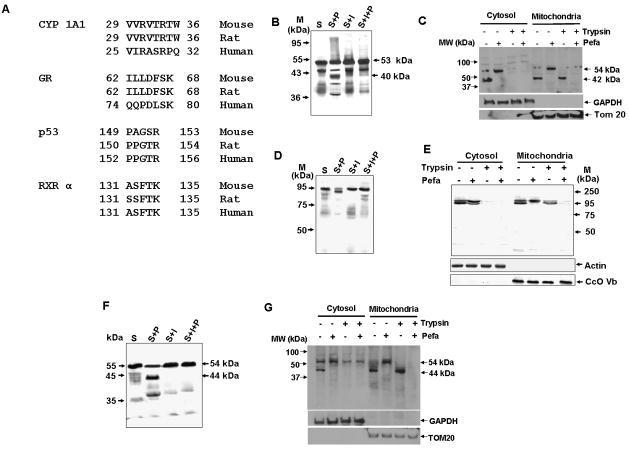
Using Fusspro algorithm (part of EMBOSS tool set from EMBL) we identified RXRα, GR and p53 as possible substrates for this protease. Search criteria included the presence of a Ser protease-like processing site, similar to that of +33/1A1 that was situated near the N-terminus of the protein. Ser-protease consensus site that is conserved among human, rat, and mouse sequences (Figure 7A) were found in all three proteins. Since these proteins are known to be imported into mitochondria we investigated their in vitro and in vivo processing by the cytosolic Ser protease. Under in vitro conditions, 35S-labeled p53 was processed into a number of fragments, including a major 40 kDa fragment, and this processing was inhibited by Pefabloc (Figure 7B). In COS cells, both the cytosolic and mitochondrial fractions contained the full-length protein as well as a major 42 kDa processed product. Pefabloc treatment blocked the appearance of this fragment in both fractions (Figure 7C). Only the ~ 40 kDa fragment was resistant to trypsin treatment of mitochondrial fractions, indicating that it is a mitochondrial imported species. GR was processing into a ~ 93 kDa species under the in vitro conditions (Figure 7D). A similar processing pattern was observed in COS cells transfected with GR cDNA (Figure 7E). Both the intact and 93 kDa fragment were completely degraded by the addition of trypsin to the cytosol. However, the 93 kDa fragment associated with the mitochondrial fraction was more resistant to trypsin treatment than the intact protein. Similar in vitro and in vivo processing patterns were observed for RXRα (Figures 7F and 7G), which had a mitochondrial-imported processed product of ~ 44 kDa. These results suggest that, as with CYP1A1, the cytosolic processing of GR, RXRα, and p53 is coupled to mitochondrial import. Although not shown, in all the three proteins, cryptic mitochondrial targeting signals were present immediately past the putative processing sites.
Figure 7. Processing of Other Substrates by the Purified Endoprotease.
(A) Putative protease processing sites on other mitochondrial-targeted proteins containing non-canonical targeting signals.
(B, D, F) In vitro processing of 35S-p53 (B), GR (D), or RXRα (F) by purified protease in the presence or absence of Pefabloc (100 μM). Processing was allowed to proceed for 30 min at 37°C.
(C, E, G) Processing of transfected p53 (C), GR (E), or RXRα (G) in COS cells treated with or without Pefabloc (300 μM), as determined by immunoblot analysis of cytosolic and mitochondrial fractions.
p90 or p40 siRNA-Induced Down Regulation of Processing Activity Inhibits CYP1A1 Mitochondrial Translocation
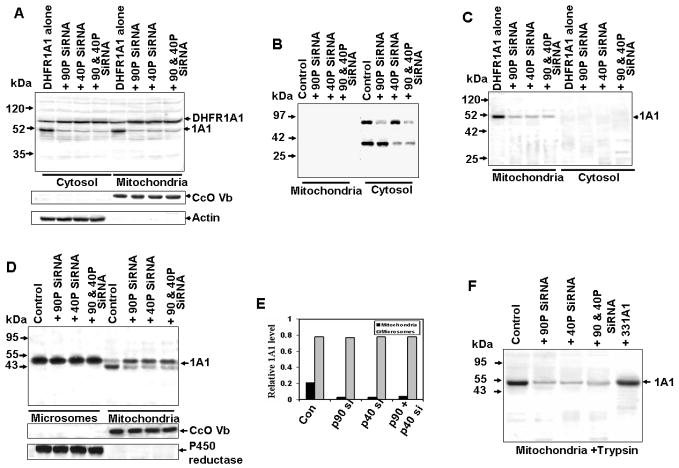
The role of p90 and p40 in the regulation of mitochondrial CYP1A1 biogenesis was further investigated using a siRNA approach. In COS cells expressing the p90 siRNA, p40 siRNA, or both siRNAs, cytosolic levels of processed DHFR-1A1 were reduced (Figure 8A). The mitochondrial level of processed CYP1A1 was also considerably reduced. Since the peptide antibodies raised against rat p90 and p40 did not cross react with the human orthologs, far western blot analysis was used to determine the steady-state levels of the two subunits in COS cell extracts. As shown in Figure 8B, the cytosolic levels of both proteins were significantly reduced in cells transfected with siRNAs targeted to the respective mRNAs (Figure 8B). Neither p90 nor p40 was detected in mitochondrial fraction of transfected cells, indicating that they were predominantly cytosolic. High levels of trypsin-resistant CYP1A1 were present in mitochondrial fractions from non siRNA-transfected cells (Figure 8C), and mtCYP1A1 was markedly reduced in cells transfected with one or both of the siRNA constructs. Trypsin completely degraded the cytosolic fusion protein and processed product.
Figure 8. Effect of p90 and p40 mRNA Depletion on DHFR1A1 Processing Activity and CYP1A1 targeting in COS Cells.
(A) Immunoblot analysis of 1A1 in cytosolic and mitochondrial fractions (50 μg protein per lane) from COS cells expressing p90 and/or p40 siRNA and cotransfected with DHFR-1A1.
(B) A duplicate blot was subjected to far western blotting using biotinylated aprotinin to determine the relative levels of p90 and p40.
(C) Samples in Figure 8A were reanalyzed following treatment with trypsin (150 μg/ml, 30 min).
(D) Immunoblot analysis of mitochondrial and microsomal proteins expressing p90 and or p40 siRNA, co-expressing CYP1A1 protein.
(F) Mitochondrial samples from (A) were reanalyzed following treatment with 150 μg/ml trypsin (30 min).
(E) Relative distribution of microsomal and mitochondrial CYP1A1 based on the 2.5:1 recovery of these membrane fractions by cell fractionation.
We also investigated the effect of silencing the cytosolic Ser protease (p90 and p40) on the microsomal and mitochondrial distribution of CYP1A1 in transfected COS cells. Immunoblot in Figure 8D and quantification in 8F show that microsomal CYP1A1 was unaffected by siRNA targeted silencing of p90, p40 or both. The level of mtCYP1A1 in trypsin treated mitochondria (Figure 8E and 8F) was vastly reduced in these cells. The siRNA-mediated knock down of Ser protease subunits also inhibited the mitochondrial targeting of GR (Figures S5A and S5B), RXRα (Figures S5C and S5D), and p53 (Figures S5E and S5F). These results provide direct proof that both p90 and p40 Ser protease subunits participate in the processing and mitochondrial biogenesis of CYP1A1, as well as of other proteins containing non canonical targeting signals.
DISCUSSION
Sequence-specific endoproteases regulate a myriad of cellular processes including activation of trypsin family proteases, regulation of clotting through activation of thrombin and fibrin, post-translational processing and activation of both endocrine and exocrine peptide hormones, and removal of signal peptides from the precursor proteins in the ER and mitochondria (Roise and Schatz, 1988, Rapaport, 2003). Here, we describe a new function for a cytosolic endoprotease in targeting bimodal proteins to the mitochondrial compartment. The endoprotease activates cryptic mitochondrial targeting signals in CYP1A1 and a number of other non canonical signal-containing proteins that are targeted to mitochondria. This is probably the first demonstration that a cytosolic Ser protease participates in mitochondrial biogenesis by regulating protein targeting.
We have previously shown that BNF, an inducer of family I CYPs and also cytosolic GSTs, stimulates the mitochondrial localization of CYP1A1 in C6 glioma cells and rat liver (Addya et al., 1997; Bhagwat et al., 1999; Boopathi et al., 2000; Dasari et al., 2006). The identity of mitochondrial CYP1A1 and the precise sites of processing were established by sequencing the purified protein (Addya et al., 1997). Here, we purified two polypeptide subunits of ~ 90 kDa and 40 kDa using a combination of DEAE-Sephacel and aprotinin-Sepharose affinity column chromatography. The overall processing activity and particularly, the steady-state level of the p90 subunit was induced by BNF (Figure S2). A key observation here is that the xenobiotic agent, BNF, not only affected the mitochondrial biogenesis by increasing protease activity, but also increased the mitochondrial drug metabolism/detoxification potential, possibly as part of the cellular defense system.
Both the p90 and p40 contain Ser-protease domains, as predicted by sequence analysis and/or direct biochemical assays (Cal et al., 2003). This is further supported by the finding that both of subunits bind to aprotinin, an active site inhibitor of Ser proteases (Figure 2). Surprisingly, both subunits were required for efficient processing of DHFR-1A1, both in vitro and in vivo. Both subunits were also required for processing-coupled mitochondrial translocation of p53, GR, and RXRα (Figure S5).
Multiple results demonstrate that both subunits are essential for processing activity. 1) Sephacryl S200 column-eluted fractions with an apparent size of 140 kDa were fully active, while those with sizes of 90 and 40 kDa were completely inactive. 2) Cells with low endogenous levels of p40 or p90 exhibited low in vitro and in vivo processing activity, as well as mitochondrial localization. 3) Overexpression of a portion of p90 (second and third Ser protease domains of Ser protease 9) in p40-rich 3T3 cells (Ser protease 9-like protein) restored processing activity. 4) siRNA-mediated silencing of either subunit markedly inhibited both in vitro and in vivo processing of DHFR-1A1 in COS cells. These results support the idea that the active enzyme is a heterodimeric complex. This is in line with studies showing that plasma membrane-anchored matrix proteases facing the exterior of the cells function as homo- or hetero-dimers (Bell et al., 2003; Cal et al., 2003).
Transmembrane Ser proteases containing trypsin-like catalytic domains are mostly targeted to the plasma membrane or secreted into the lumen of the ER via a signal peptide. In mammals, 13 members of this family, called transmembrane Ser proteases, have been identified. The complex mosaic architecture of Ser protease 9 (polyserase 1) is conserved in mouse and rat orthologs, although the functional significance of this multidomain architecture remains unclear. In the case of human liver polyserase 2, the activity of the first protease domain, as an independent unit, is greater than that of the entire protein, which includes the two additional Ser protease domains predicted to be catalytically inactive (Cal et al., 2005). A similar mosaic structure is present in Ser proteases from other vertebrates as well as invertebrates (Lindsay, et al., 1999; Hiyoshi et al., 2002; Krem and Cera 2001; Ross et al., 2003). Oviductin from Xenopus laevis or Bufo japonicus contains two Ser protease domains that also undergo proteolytic processing to generate independent, catalytically active units (Lindsay et al., 1999). The p90 subunit appears to be a post-translationally processed form of Ser protease 9, which is a 110 kDa plasma membrane-bound protein. Activity profiles of bacterially expressed individual domains show that the N-terminal most and the second Ser protease domains are active, while the C-terminal most domain is inactive. In COS cells overexpressing the full-length 110 kDa protein, both the 110 kDa membrane-bound form and the soluble 90 kDa form are present (Cal et al., 2003). However, the precise mechanisms underlying processing or the factors regulating this processing are unknown. The 3.1 kb mRNA species observed here may represent a splice variant encoding the 90 kDa soluble form.
Many Ser proteases undergo oligomerization, which is thought to facilitate alignment of canonical substrate sites in the substrate-binding pocket of the enzyme (Bell et al., 2003). The alpha helical structure of the substrate recognition site sterically blocks access to the peptide bond, and this steric hindrance is overcome through enzyme oligomerization (Bell et al., 2003; Batra et al., 2001). Oligomerization is also one of the strategies evolved by the cytosolic protease to facilitate substrate binding and orient the disulphide bridges. Therefore, we propose that the Ser protease characterized here is a functional dimer of p90 and p40, similar to Ser proteases found in other cellular compartments.
A previous analysis of the N-terminus of mitochondrial CYP1A1 revealed that processing occurs between V32 and T33. Mutational analysis in this study suggests that the flanking three amino acids are also important for processing activity. Ser proteases that cleave at P1 sites occupied by non-canonical amino acids are not uncommon. For example, kallikrein specifically cleaves low molecular weight kininogen between Met-Lys and Arg-Ser to generate the vasoactive kinin peptide (Muller-Esterl et al., 1986). Both mutational analysis and N-terminal sequencing (Addya et al., 1997) show that the Ser protease described here cleaves between a non-canonical Val-Thr to generate mtCYP1A1. N-terminal sequencing also revealed that minor processing occurs between Val4 and Ser5. Using DHFR-1A1, we were unable to detect the +5 processed species either in vitro or in vivo. A free N-terminus is likely required for the enzyme to gain access to the +4 residue for processing. Alternatively, a different enzyme may perform this minor processing.
Although the Ser protease inhibitor, Pefabloc, reduced mitochondrial CYP1A1, microsomal CYP1A1 levels were unaffected by this inhibitor or by AK131261 overexpression. A marked decline in the microsomal CYP1A1 and increase in mtCYP1A1 content in C6 glioma calls treated with BNF for >72h mostly reflects different turnover rates of CYP1A1 in these two subcellular compartments (Boopathi et al., 2000). An additional factor is the increased Ser-protease activity under prolonged treatment conditions which also contributes to increased mitochondrial targeting. In addition, DHFR-1A1, which was not targeted to the ER membrane, was efficiently processed and directed to mitochondria. These results suggest that the cytosolic pool of CYP1A1 is the substrate for the Ser protease. We previously reported that nascent CYP proteins have different SRP affinities and up to 30% of CYP1A1 is translated as free protein. Furthermore, CYP affinity for SRP is modulated by phosphorylation of nascent chains by PKA or PKC (Dasari et al., 2006). Thus, translation on free ribosomes and endoproteolytic processing at the N-terminus are important regulatory steps for the mitochondrial targeting of CYP1A1 and probably other CYP and non CYP proteins.
Purified protease processed not only 1A1, but also GR, RXRα, and p53, facilitating the mitochondrial import of each of these proteins. In each case, processing exposed a cryptic mitochondrial import signal at or very close to the N-terminus. The finding that a cytosolic Ser protease activates cryptic signals contained within CYP1A1 and other proteins provides direct support for the hypothesis that chimeric signals direct the bimodal targeting of proteins. Our findings also provide a rational basis for the import of non canonical signal-containing proteins into different submitochondrial compartments.
EXPERIMENTAL PROCEDURES
Expression Plasmids
DHFR was fused to the N-terminus of CYP1A1 (Yabusaki et al., 1984) through an EcoRI linker, and the fusion construct was cloned into pCMV4 or pGEM7zf plasmids. The cDNA constructs were engineered to contain a Kozak consensus sequence immediately upstream of the initiator ATG codon. Point mutations were introduced in the predicted trypsin-like cleavage site using the QuickChangeTM Site-Directed Mutagenesis System (Stratagene).
BNF Treatment of Animals and Isolation of Liver Cytosol
Male rats (~ 150 g, Harlan Sprague-Dawley Inc.) were subcutaneously injected with BNF (80 mg/kg body weight in corn oil) once daily for 6 – 10 days, as described elsewhere (Niranjan et al., 1984). Control rats received equivalent amounts of corn oil. All animal procedures, which were carried out as previously described (Niranjan et al., 1984), complied with NIH guidelines and were approved by the Institutional Animal Care and Use Committee. Liver cytosol was isolated in sucrose-mannitol buffer (2 mM HEPES, pH 7.4; 70 mM sucrose; 220 mM mannitol; 2 mM EDTA), as detailed previously. The cytosolic fraction (120,000 × g supernatant) was dialyzed against 50 mM phosphate buffer and subjected to (NH4)2SO4 fractionation using standard procedures. The (NH4)2SO4 precipitates were dissolved in 50 mM KH2PO4 buffer (pH 7.5) containing 5 mM NaCl and dialyzed against the same buffer. Insoluble proteins were collected by centrifugation at 10,000 × g for 10 min and stored at −80°C.
Aprotinin-Sepharose Affinity Chromatography
Aprotinin (Sigma) was biotinylated (Melrose et al., 1995), and biotinylated aprotinin (5 mg) was bound to an avidin monomer affinity column (10 ml bed volume) (Green and Toms 1973; Henrikson et al., 1979). The DEAE Sephacryl-fractionated proteins were further purified by aprotinin affinity column chromatography as detailed elsewhere (Melrose et al., 1995). The samples (50 – 200 μg protein) were applied to a 10 ml aprotinin-agarose affinity column in 50 mM Tris-HCl buffer (pH 7.4) containing 0.5 M NaCl. Unbound proteins were eluted with five bed volumes of the same buffer. Bound proteins were successively eluted with 1 ml of 50 mM sodium acetate buffer (pH 3.5) containing 1 and 2 M NaCl. Aliquots were collected from each fraction to monitor proteolytic activity using DHFR1A1 as substrate. The active fractions were pooled, dialyzed against 50 mM sodium phosphate buffer (pH 7.0), and stored at −80°C.
Size-Exclusion Chromatography
Gel filtration column chromatography was performed using Sephacryl S200. The column had a fractionation range of 10 to 500 kDa. The column (i.d.: 16 mm; h: 60 cm, 125 ml, Amersham Bioscience) was extensively washed and equilibrated with 50 mM phosphate buffer (pH 7.4) containing 200 mM NaCl. The column was calibrated with the following standards: carbonic anhydrase (Mr ~ 29,000), ovalbumin (43 kDa) bovine serum albumin (Mr ~ 76,000), and alcohol dehydrogenase (Mr ~ 150,000). The DEAE column-purified fraction (400 μg protein) was loaded at a flow rate of 0.5 ml/min. The fractions were tested for processing activity and subjected to immunoblot analysis using p90 and -p40 antibodies.
Assay for DHFR-1A1 Processing
DHFR1A 1 protein was synthesized in a coupled rabbit reticulocyte lysate translation system (Promega) in presence of 35S Met (40 mCi/200 μl, 1175 Ci/mmol, Amersham). The post ribosomal supernatant was prepared by centrifugation at 105,000 × g for 1 hr at 4°C. Processing reactions were carried out in a total volume of 50 – 100 μl with 50,000 cpm of translation product as well as with 200 μg cytosolic fraction, 50 μg (NH4)2SO4 fractions from cells or liver, or 2 μg DEAE-Sephacel column-purified fraction. Cytosolic extracts from COS-7, NIH 3T3, 293T, and C2C12 cells were fractionated and enriched using 50% (NH4)2SO4 cut for assaying DHFR-1A1 processing. The reactions were performed in 20 mM phosphate buffer (pH 7.6) containing 0.5 mM MgCl2 for 30min at 37°C and were terminated by addition of Pefabloc (100 μM final concentration). Inhibitors, aprotinin (250 μg/ml), or Pefabloc (100 μM), were added during the beginning of the reaction. The full-length substrate and processed products were analyzed by SDS-PAGE and the labeled proteins were imaged through a Molecular Dynamics STORM Phosphoimager system, and were quantified using ImageQuant 5.2 software.
Far western Blot Analysis using Biotinylated Aprotinin
Aprotinin immunoblotting was performed as described previously (Melrose et al., 1995). Proteins from polyacrylamide gels were transferred to nitrocellulose membrane and probed with horse radish peroxidase-conjugated avidin antibody. The blots were developed with the chemiluminiscence substrate, super signal femto (Pierce Biotechnology, Inc.).
Cell Culture, Transfection and Cell Fractionation
COS-7, NIH 3T3, 293T and C2C12 cells were grown and transiently transfected with various cDNA constructs as described elsewhere (Addya et al., 1997; Guha et al., 2007). Cultures were maintained in Dulbecco’s modified Eagle’s medium containing 10% (v/v) fetal bovine serum and gentamycin (50 μg/ml). Cells were transfected for 66–72 hr with column-purified plasmid DNA (2 – 10 μg/plate) using the lipophilic transfection reagent, fugene-6 (Quiagen). Cells were disrupted using a Teflon-fitted glass homogenizer, and homogenates were subjected to differential centrifugation to isolate mitochondria (Addya et al., 1997). Mitochondrial fractions were gently resuspended in sucrose/mannitol and purified by banding through a discontinuous sucrose gradient (Addya et al., 1997). Mitochondrial fractions migrating to the 1.35 and 1.6 M sucrose interface were recovered, washed twice with sucrose/mannitol buffer, and analyzed by immunoblotting. In specified experiments, mitochondria in sucrose/mannitol buffer were treated with trypsin (150 μg/ml) for 15–45 min at 4°C.
Northern Blot Analysis
Nylon filters containing total RNA (30 μg per lane) were prehybridized at 42°C for 3 hr in 50% formamide, 5 × saline/sodium phosphate/EDTA, 10 × Denhardt’s solution, 2% SDS, and 100 μg/ml denatured salmon sperm DNA. The partial cDNAs of EST clones for the 90 and 40 kDa proteases were 5’ labeled by nick translation and then used for hybridization, which was performed at 65 °C for 20 h. The filters were washed with 5 × SSC and 0.1% SDS at room temperature for 20 min, washed with 0.1 × SSC and 0.1% SDS at 50°C for an additional 20 min, and then autoradiographed.
Immunoblot Analysis
The mitochondrial, microsomal and the cytosolic proteins were dissociated in Laemmli’s sample buffer at 65°C for 15 min and subjected to immunoblot analysis by standard conditions. The membranes were probed with polyclonal antibody to CYP1A1, stripped with glycine solution (100 mM, pH 3), and reprobed with Actin, cytochrome P450 reductase or CcO Vb antibodies. The latter served as loading controls for the cytosolic, microsomal and mitochondrial fractions, respectively. The blots were developed using Pierce Femto Super signal reagent. The band intensity was determined using the VersaDoc Imaging System Model 3000 (Bio-Rad).
Supplementary Material
Acknowledgments
We thank the Avadhani lab members for their suggestions and useful comments. We also thank Dr. Anil Rustgi for providing the p53 cDNA used in this study. This work was supported, in part, by the National Institutes of Health (GM-34883).
References
- Addya S, Anandatheerthavarada HK, Biswas G, Bhagwat SV, Mullick J, Avadhani NG. Targeting of NH2-terminal-processed Microsomal Protein to Mitochondria: A Novel Pathway for the Biogenesis of Hepatic Mitochondrial P450MT2. J Cell Biol. 1997;139:589–599. doi: 10.1083/jcb.139.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, Vijayasarathy C, Bhagwat SV, Biswas G, Mullick J, Avadhani NG. Physiological role of the N-terminal processed P4501A1 targeted to mitochondria in erythromycin metabolism and reversal of erythromycin-mediated inhibition of mitochondrial protein synthesis. J Biol Chem. 1999;274:6617–6625. doi: 10.1074/jbc.274.10.6617. [DOI] [PubMed] [Google Scholar]
- Batra R, Khayat R, Tong L. Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease. Nature Structural Biology. 2001;8:810–817. doi: 10.1038/nsb0901-810. [DOI] [PubMed] [Google Scholar]
- Bell JK, Goetz DH, Mahrus S, Harris JL, Fletterick RJ, Craik CS. The oligomeric structure of human granzyme A is a determinant of its extended substrate specificity. 2003;10:527–534. doi: 10.1038/nsb944. [DOI] [PubMed] [Google Scholar]
- Bhagwat SV, Biswas G, Anandatheerthavarada HK, Addya S, Pandak W, Avadhani NG. Dual Targeting Property of the N-terminal Signal Sequence of P4501A1. Targeting of heterologous proteins to endoplasmic reticulum and mitochondria. J Biol Chem. 1999;274:24014–24022. doi: 10.1074/jbc.274.34.24014. [DOI] [PubMed] [Google Scholar]
- Bicknell AB, Lomthaisong K, Woods RJ, Hutchinson EG, Bennett HP, Gladwell RT, Lowry PJ. Characterization of a Ser protease that cleaves pro-gamma-melanotropin at the adrenal to stimulate growth. Cell. 2001;105:903–912. doi: 10.1016/s0092-8674(01)00403-2. [DOI] [PubMed] [Google Scholar]
- Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol. 1994;14:2298–2306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi E, Anandatheerthavarada HK, Bhagwat SV, Biswas G, Fang JK, Avadhani NG. Accumulation of mitochondrial P450MT2, NH(2)-terminal truncated cytochrome P4501A1 in rat brain during chronic treatment with beta-naphthoflavone. A role in the metabolism of neuroactive drugs. J Biol Chem. 2000;275:34415–34423. doi: 10.1074/jbc.M004431200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cal S, Quesada V, Llamazares M, Díaz-Perales A, Garabaya C, López-Otín C. Human polyserase-2, a novel enzyme with three tandem Ser protease domains in a single polypeptide chain. J Biol Chem. 2005;280:1953–1961. doi: 10.1074/jbc.M409139200. [DOI] [PubMed] [Google Scholar]
- Cal S, Quesada V, Garabaya C, Lopez-Otin C. Polyserase-I, a human polyprotease with the ability to generate independent Ser protease domains from a single translation product. Proc Natl Acad Sci U S A. 2003;100:9185–9190. doi: 10.1073/pnas.1633392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C. Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins. FASEB J. 2003;17:426–436. doi: 10.1096/fj.02-0732com. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Yager JD. Estrogen’s effects on mitochondrial gene expression: mechanisms and potential contributions to estrogen carcinogenesis. Ann N Y Acad Sci. 2004;1028:258–72. doi: 10.1196/annals.1322.030. [DOI] [PubMed] [Google Scholar]
- Dasari VR, Anandatheerthavarada HK, Robin MA, Boopathi E, Biswas G, Fang JK, Nebert DW, Avadhani NG. Role of protein kinase C-mediated protein phosphorylation in mitochondrial translocation of mouse CYP1A1, which contains a non-canonical targeting signal. J Biol Chem. 2006;281:30834–30847. doi: 10.1074/jbc.M510725200. [DOI] [PubMed] [Google Scholar]
- Fölsch H, Guiard B, Neupert W, Stuart RA. Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J. 1996;15:479–487. [PMC free article] [PubMed] [Google Scholar]
- Goldberg DM. Proteases in the evaluation of pancreatic function and pancreatic disease. Clin Chim Acta. 2000;291:201–221. doi: 10.1016/s0009-8981(99)00229-6. [DOI] [PubMed] [Google Scholar]
- Green NM, Toms EJ. The properties of subunits of avidin coupled to sepharose. Biochem J. 1973;133:687–700. doi: 10.1042/bj1330687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V, Lithgow T, Rospert S, Hahne K, Schatz G. The yeast mitochondrial protein import receptor Mas20p binds precursor proteins through electrostatic interaction with the positively charged presequence. J Biol Chem. 1995;270:5565–5570. doi: 10.1074/jbc.270.10.5565. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W. Mitochondrial protein import: Identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Henrikson KP, Allen S, Maloy HWL. An avidin monomer affinity column for the purification of biotin-containing enzymes. Anal Biochem. 1979;94:366–370. doi: 10.1016/0003-2697(79)90374-9. [DOI] [PubMed] [Google Scholar]
- Hiyoshi M, Takamune K, Mita K, Kubo H, Sugimoto Y, Katagiri C. Oviductin, the Oviductal Protease That Mediates Gamete Interaction by Affecting the Vitelline Coat in Bufo japonicus: Its Molecular Cloning and Analyses of Expression and Posttranslational Activation. Developmental Biology. 2002;243:176–184. doi: 10.1006/dbio.2001.0558. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosom. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Koufali MM, Moutsatsou P, Sekeris CE, Breen KC. The dynamic localization of the glucocorticoid receptor in rat C6 glioma cell mitochondria. Mol Cell Endocrinol. 2003;209(1–2):51–60. doi: 10.1016/j.mce.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Krem MM, Cera DE. Molecular markers of Ser protease evolution. EMBO J. 2001;20:3036–3045. doi: 10.1093/emboj/20.12.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunii D, Shimoji M, Nakama S, Ikebe M, Hachiman T, Sato I, Tamaki A, Yamazaki K, Aniya Y. Purification of liver Ser protease which activates microsomal glutathione S-transferase: possible involvement of hepsin. Biol Pharm Bull. 2006;29:868–874. doi: 10.1248/bpb.29.868. [DOI] [PubMed] [Google Scholar]
- Lindsay LL, Yang JC, Hedrick JL. Ovochymase, a Xenopus laevis egg extracellular protease, is translated as part of an unusual polyprotease. Proc Natl Acad Sci U S A. 1999;96:11253–11258. doi: 10.1073/pnas.96.20.11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Taylor SS. A molecular switch for targeting between endoplasmic reticulum (ER) and mitochondria: conversion of a mitochondria-targeted element into ER-Targeting signal in DAKAP1. J Biol Chem. 2008;283:11743–11751. doi: 10.1074/jbc.M710494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melrose J, Ghosh P, Patel M. Biotinylated aprotinin: a versatile probe for the detection of Ser proteinases on western blots. Int J Biochem & Cell Biol. 1995;27:891–904. doi: 10.1016/1357-2725(95)00068-z. [DOI] [PubMed] [Google Scholar]
- Mihara K. Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. BioEssays. 2000;22:364–391. doi: 10.1002/(SICI)1521-1878(200004)22:4<364::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Müller-Esterl W, Hock H, Rauth G, Kellermann J, Lottspeich F, Henschen A. Limited proteolysis of HMW kininogen by plasma kallikrein in man--evidence for a processing mechanism different from the bovine system. Adv Exp Med Biol. 1986;198(Pt A):97–103. doi: 10.1007/978-1-4684-5143-6_14. [DOI] [PubMed] [Google Scholar]
- Neurath H. Proteolytic enzymes, past and future. Proc Natl Acad Sci USA. 1999;96:10962–10973. doi: 10.1073/pnas.96.20.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjan BG, Wilson NM, Jefcoate CR. Hepatic mitochondrial cytochrome P-450 system. Distinctive features of cytochrome P-450 involved in the activation of aflatoxin B1 and benzo(a)pyrene. J Biol Chem. 1984;259:12495–12501. [PubMed] [Google Scholar]
- Pfanner N, Söllner T, Neupert W. Mitochondrial import receptors for precursor proteins. Trends Biochem Sci. 1991;16:63–67. doi: 10.1016/0968-0004(91)90026-r. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedmann N, Meisinger C, Lithgow T. Assembling the mitochondrial outer membrane. Nat Struct Mol Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- Perona JJ, Craik CS. Structural basis of substrate specificity in the Ser proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokisch H, Scharfe C, Camp DG, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, Hixson KK, Mottaz HM, Zischka H, Ueffing M, Herman ZS, Davis RW, Meitinger T, Oefner PJ, Smith RD, Steinmetz LM. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2004;2:0795–0804. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D. Finding the right organelle. EMBO reports. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin MA, Anandatheerthavarada HK, Biswas G, Sepuri NP, Cordon DM, Pain D, Avadhani NG. Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J Bio chem. 2002;277:40583–40593. doi: 10.1074/jbc.M203292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin MA, Prabu SK, Raza H, Anandatheerthavarada HK, Avadhani NG. Phosphorylation enhances mitochondrial targeting of GSTA4-4 through increased affinity for binding to cytosolic Hsp70. J Biol Chem. 2003;278:18960–189670. doi: 10.1074/jbc.M301807200. [DOI] [PubMed] [Google Scholar]
- Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- Ross J, Jiang H, Kanost MR, Wang Y. Ser proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:239–240. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- Whitlock JP. The regulation of Cytochrome P-450 gene expression, Ann. Rev Pharmacol Toxicol. 1986;26:333–369. doi: 10.1146/annurev.pa.26.040186.002001. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28(3):249–68. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Yabusaki Y, Shimizu M, Murakami H, Nakamura K, Oeda K, Ohkawa H. Nucleotide sequence of a full-length cDNA coding for 3-methylcholanthrene-induced rat liver cytochrome P-450MC. Nucleic Acid Res. 1984;12:2929–2938. doi: 10.1093/nar/12.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.