Abstract
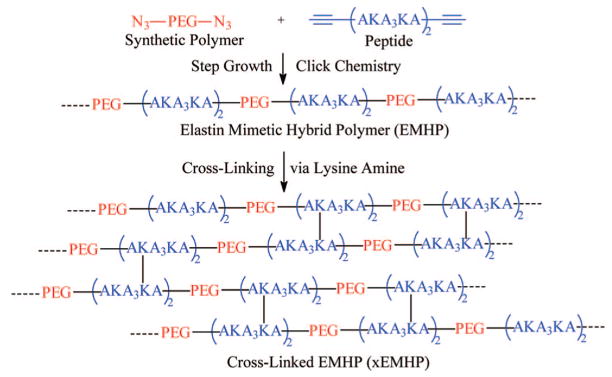
We are interested in developing elastin–mimetic hybrid polymers (EMHPs) that capture the multiblock molecular architecture of tropoelastin as well as the remarkable elasticity of mature elastin. In this study, multiblock EMHPs containing flexible synthetic segments based on poly(ethylene glycol) (PEG) alternating with alanine-rich, lysine-containing peptides were synthesized by step-growth polymerization using α,ω-azido-PEG and alkyne-terminated AKA3KA (K = lysine, A = alanine) peptide, employing orthogonal click chemistry. The resulting EMHPs contain an estimated three to five repeats of PEG and AKA3KA and have an average molecular weight of 34 kDa. While the peptide alone exhibited α-helical structures at high pH, the fractional helicity for EMHPs was reduced. Covalent cross-linking of EMHPs with hexamethylene diisocyanate (HMDI) through the lysine residue in the peptide domain afforded an elastomeric hydrogel (xEMHP) with a compressive modulus of 0.12 MPa when hydrated. The mechanical properties of xEMHP are comparable to a commercial polyurethane elastomer (Tecoflex SG80A) under the same conditions. In vitro toxicity studies showed that while the soluble EMHPs inhibited the growth of primary porcine vocal fold fibroblasts (PVFFs) at concentrations ≥0.2 mg/mL, the cross-linked hybrid elastomers did not leach out any toxic reagents and allowed PVFFs to grow and proliferate normally. The hybrid and modular approach provides a new strategy for developing elastomeric scaffolds for tissue engineering.
Introduction
Accumulated knowledge in cell biology provides new inspirations for designing biomimetic materials for tissue engineering. A close examination of mechanically active tissues (e.g., vocal folds, bladder, lungs, cardiac tissue, and blood vessels)1,2 indicates the presence of a relatively high concentration of elastin. A network of elastic fibers gives the tissue the required strength, flexibility, and resilience so that it can reversibly recoil after transient stretch up to 200%.3 Recent studies suggest that the cellular functions can be modulated by the stiffness of their substrates.4 Synthetic scaffolds that possess appropriate elasticity are required in order to effectively transmit the mechanical forces to the encapsulated cells.5 However, traditional polymeric scaffolds exhibit inferior mechanical properties, tending to fail catastrophically at high strain and/or demonstrating insufficient ability to recover from mechanical deformation. As a result, the development of biodegradable elastomers has been the focus of many recent studies.6,7
Tropoelastin, the soluble precursor of mature elastin, is synthesized by cells as a ~72 kDa protein8 with an alternating molecular architecture containing highly flexible segments rich in nonpolar amino acids and alanine-rich, lysine-containing peptides which form cross-links between adjacent molecules via the action of lysyl oxidase in the extracellular matrix (ECM).9 The multiblock, alternating structure not only fosters the hydrophobic aggregation but also synergistically aligns the lysine residues for subsequent covalent stabilization.10 The elastin elasticity has been explained in terms of the conformational entropy of the polypeptide chains (rubber elasticity11 and β-spiral12 theories) and the change in solvent (water) entropy resulting from the hydrophobic interaction between the polypeptide side chains and the water molecules.13 Consistent with the functions of the two types of domains, in the subsequent discussions, the nonpolar peptide sequences are referred to as the “entropic” segments, while the lysine-containing peptides are referred to as the “cross-linking” domains.
Despite these useful properties, native elastin is rarely used as a tissue engineering scaffold, and when it is used, only poorly defined preparations have been employed. Major challenges to the use of native elastin result from its substantial insolubility,14 which necessitates multistep purification15 or high-temperature hydrolysis that may lead to undesirable structural alteration.16 In addition, elastin isolated from natural tissues suffers from batch-to-batch variation and potential of disease transmission.17 Accordingly, there has been much recent work focusing on synthesizing elastin-like polypeptides (ELPs) that mimic the inherent assembly and elastomeric properties of native elastin. SPPS has been used to produce short elastin-like peptide sequences containing VPGVG peptide repeats that exhibit similar aggregation properties as elastin.18–20 High molecular weight ELPs with diblock, triblock, and multiblock structures and intriguing material properties and biological responses have been produced by recombinant DNA technology.21–23 Although variations exist in terms of their compositions and polymer architectures, these materials are based purely on amino acids, limiting the tunability of their overall material properties. Synthetic polymers, on the other hand, can be easily synthesized in large quantities with varying chemical compositions, controlled molecular weights, molecular architectures, and microscopic morphologies.24 Thus, strategies that combine peptide-and synthetic polymer-based approaches would offer significant advantages in the design of modular materials that permit the tuning of elastic and biological behavior.25–28
Our strategy of producing multiblock EMHPs involves replacing the dynamic, entropic peptide segments with appropriate synthetic polymer chains while retaining the structural component of the cross-linking domains of the natural elastin (Scheme 1). In a proof-of-concept demonstration described here, PEG was employed as the synthetic building block due to its inherent biocompatibility,29 water solubility, and simplicity in its chemical composition. Moreover, PEG not only exhibits distinct chain flexibility30 as compared to other synthetic polymers, it is also capable of associating with water molecules. In fact, molecular dynamics simulations31 suggest that water dramatically affects the elastic properties of PEG through its dynamic hydrogen bonding with the PEG main chain. Thus, the physical properties of PEG are reminiscent of the entropic segments of tropoelastin. On the other hand, alanine-rich, lysine-containing peptide sequences with a repeating unit of AKA3KA, found in the cross-linking regions of native elastin,32,33 were introduced for cross-linking and potential assembly purposes.
Scheme 1.
Schematic Representation of the Synthesis and Covalent Cross-Linking of Elastin–Mimetic Hybrid Polymer
We describe herein our initial efforts in synthesizing elastin–mimetic hybrid polymers (EMHPs) consisting of flexible synthetic segments based on poly(ethylene glycol) (PEG) alternating with AKA3KA peptides that have the potential to organize into defined structures. Nucleophilic substitution of mesylated hydroxyls of PEG with sodium azide gave rise to azide-functionalized, telechelic PEG (N3-PEG-N3). Separately, standard 9-fluorenylmethoxycarbonyl (Fmoc)-based solid phase peptide synthesis (SPPS) protocols were applied to prepare AKA3KA peptide containing alkyne groups at both C- and N-termini. Step growth polymerization of the functionalized PEG and peptide using copper-catalyzed 1,3-Huisgen cycloaddition afforded EMHPs with alternating structure and high molecular weight. Covalent cross-linking of EMHPs with hexamethylene diisocyanate (HMDI) afforded an elastomeric gel (xEMHP) that absorbed a large amount of water. The hydrated xEMHP was soft and compressible, exhibiting a compression modulus similar to that of a commercial polyurethane elastomer. Primary porcine vocal fold fibroblasts (PVFFs) were able to proliferate and adopt normal cell morphology without any detectable cell death when cultured in the presence of xEMHP in an indirect contact mode.
Experimental Methods72
Materials
All chemicals were used as received unless otherwise noted. Water was deionized and filtered through a NANOpure Diamond water purification system (Barnstead).
Characterization
1H NMR spectra were recorded in either D2O or CDCl3 on a Bruker AV400 NMR spectrometer under standard quantitative conditions and were analyzed with Bruker Topspin software. All chemical shift values were calibrated using the solvent peak (from proton impurities, 4.8 ppm for D2O, 7.24 ppm for CDCl3). FT-IR spectra of PEG and EMHPs were obtained with a Bruker Tensor 27 spectrometer and analyzed with OPUS software. Films were deposited on NaCl plates from methylene chloride solutions. Spectra were taken at a resolution of 4 cm−1 from 550 to 4000 cm−1, and an average of 256 scans was reported. A background of the atmosphere was subtracted from all sample spectra. FT-IR spectra of the peptide were obtained with a Thermo Nicolet Nexus 670 spectrometer equipped with a DuraSamplIR II ATR accessory (SensIR Technologies) and analyzed with OMNIC software. The solid peptide was deposited on the silicon ATR crystal and gently pressed down for the duration of the measurement. Spectra were taken at a resolution of 4 cm−1 from 550 to 4000 cm−1, and an average of 128 scans was reported. A background of the clean silicon crystal was subtracted from all sample spectra. Polymer molecular weights were determined by size exclusion chromatography (SEC) and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and SEC was conducted on a system comprised of a Waters 515 HPLC pump, two Waters Ultrahydrogel (7.8 × 300 mm) columns in series, a Waters 2414 refractive index detector, and a Waters 2996 photodiode array detector. The mobile phase was Dulbecco’s phosphate buffered saline (PBS). All samples (ca. 1–3 mg/mL) were run at a flow rate of 1 mL/min. Data were collected and analyzed with Waters Empower software. Molecular weight calibration was based on protein standards (trypsinogen and catalase (Sigma-Aldrich) and alkyne-functionalized peptide starting materials). SDS-PAGE analyses were carried out following standard procedures34 with Coomassie blue staining. High concentrations of PEG–peptide multiblock samples were required in order for them to be clearly visualized by Coomassie blue staining. For ESI-MS analyses, samples were dissolved in methanol at a concentration of ca. 0.1 mg/mL (with a few drops of water added for solubility, if necessary) and analyzed in positive ion mode on a Thermo Finnegan LCQ Advantage mass spectrometer with Surveyor MS pump. Circular dichroism (CD) spectra were recorded on a J-810 spectropolarimeter with Spectra Manager software in a 1 mm or 10 mm path length quartz cuvette. Background scans of buffers (PBS) were recorded and subtracted from the sample scans automatically using the software. Samples were prepared at a concentration of 50 μM, and the pH was adjusted using a NaOH solution. The temperature was held constant at 20 °C, and data were recorded from 190 to 260 nm. Three scans were recorded for each sample, and the average of the scans was reported. The molar ellipticity, [θ]MRW (deg cm2 dmol−1), was calculated using the molecular weight of the peptide and cell path length.35
End-Group Modification of PEG.36,37
The hydroxyl end groups of PEG were converted to azide groups (N3-PEG-N3) via the mesylateintermediate(CH3SO2O-PEG-OSO2CH3 orMsO-PEG-OMs).
MsO-PEG-OMs
PEG (Acros) (Mn = 1450 g/mol, 2.00 g, 2.76 mmol of OH) was dissolved in anhydrous dichloromethane (20 mL) containing triethylamine (Aldrich) (1.92 mL, 13.8 mmol). The flask was cooled to 0 °C in an ice bath, and 1.07 mL (13.8 mmol) of methanesulfonyl chloride (Acros) was added dropwise over 10 min under nitrogen. The mixture was stirred under nitrogen in the ice bath for about 1 h and then at room temperature for an additional 3.5 h. After the reaction, the mixture was washed twice with saturated sodium bicarbonate solution. The organic layer was collected, dried over magnesium sulfate, and precipitated twice into cold diethyl ether to obtain a pale yellow solid. Yield: 93%. 1H NMR (D2O) δ(ppm): 4.48 (4H, m, CH2OMs), 3.87 (4H, m, CH2CH2OMs), 3.65–3.75 (128H, m, CH2CH2O), 3.25 (6H, s, CH3SO2O).
N3-PEG-N3
MsO-PEG-OMs was dissolved in anhydrous DMF (Aldrich) and sodium azide (Acros) (1.1 equiv, based on the OMs end groups) was introduced. The mixture was stirred under nitrogen at 40 °C for 48 h. The product was precipitated two times in cold diethyl ether, with redissolution in dichloromethane after each precipitation. The solid precipitate was collected by vacuum filtration and dried in a vacuum oven. Yield: 70%. 1H NMR (D2O) δ(ppm): 3.65–3.75 (120H, m, CH2CH2O), 3.50 (4H, m, CH2N3).
Synthesis of KA3K Peptide
Fmoc SPPS was conducted with an automated peptide synthesizer (PS3, Protein Technologies Inc.) on a Rink amide MBHA resin (Novabiochem). The sequence of the peptide used in the PEG-peptide multiblock synthesis reported here was X(AKA3KA)2X, where A = Fmoc-L-alanine, K = Fmoc-L-lysine (Boc) (Protein Technologies Inc.), and X = Fmoc-L-propargylglycine (Anaspec). The N-terminus was acetylated by capping with acetic anhydride. After the synthesis was complete, the peptide was simultaneously cleaved and deprotected by stirring in a mixture of 9.5 mL/0.25 mL/0.25 mL trifluoroacetic acid/water/triisopropylsilane for 6–8 h at room temperature. The solution was precipitated twice into cold diethyl ether, and the solid precipitate was collected by centrifugation at 4000 rpm. The pellet was then dissolved in water, frozen at −80 °C, and lyophilized. The peptide was isolated as a fluffy, white solid. The mass of the peptide was confirmed via ESI mass spectrometry (m/z) 1473.4 [(M + H)+, calculated 1473.0], 737.1 [(M + H)2+, calculated 737.0], 491.9 [(M + H)3+, calculated 492]. Yield: 59%. 1H NMR (D2O) δ (ppm): 4.25–4.50 (m, 16H, NH–CH–CO), 3.02 (m, 8H, CH2NH2), 2.62 (m, 4H, CH2C≡CH), 2.35 (m, 2H, CH2C≡CH), 2.0 (m, 2H, CHCH2CH2CH2CH2NH2), 1.55–1.81 (m, 16H, CHCH2CH2CH2CH2-NH2), 1.22–1.50 (m, 30H, CHCH3).
Synthesis of Multiblock Hybrid Copolymers (EMHPs)
Ligand Synthesis
The ligand for click chemistry, tris(benzyltria-zolylmethyl)amine (TBTA), was synthesized and purified based on a previously reported method.38 Yield: 64%. 1H NMR (CDCl3) δ (ppm): 8.25 (s, 3H, triazole), 7.20–7.40 (m, 15H, Ph), 5.50 (s, 6H, CH2Ph), 4.21 (s, 6H, NCH2).
EMHPs by Step Growth, Click Coupling
N3-PEG-N3 and the peptide X(AKA3KA)2X were dissolved in a mixture of 300 μL of water and 300 μL of DMF to form a solution with an initial concentration of 0.05 M azide and 0.05 M alkyne. CuSO4 • 5H2O (0.1 equiv), sodium ascorbate (0.5 equiv), and TBTA (0.1 equiv) were added to the solution. The temperature was increased to 80 °C, and the reaction was allowed to proceed for 24 h. The azide-and alkyne-functionalized prepolymers (0.03 mmol each) were again added to the reaction flask along with ~600 μL of water, and the reaction was continued for another 24 h. At the end of the reaction, a brown insoluble byproduct was observed on the sides of the flask. The solid was removed by vacuum filtration, and the yellow filtrate was collected, diluted with water, and purified by dialysis against water in a Spectra/Por Biotech MWCO 2000 cellulose ester membrane (Spectrum Laboratories) for 3 days, frozen at −80 °C, and lyophilized to obtain a fluffy, pale yellow solid. Yield: 50%. 1H NMR (D2O) δ (ppm): 7.9 (s, 1 H, triazole C=CH), 4.20–4.40 (m, 8H, NH–CH–CO), 3.60–3.80 (m, 104H, OCH2CH2), 3.52 (m, 2H, OCH2CH2-triazole), 3.15–3.40 (m, 3H, NH2), 3.05 (m, 4H, CH2NH2), 2.04 (m, 2H, CH2C≡CH), 1.65–2.0 (m, 8H, CH2CH2CH2CH2NH2, CH2CH2CH2CH2NH2), 1.35–1.70 (m, 23H, CHCH2CH2CH2CH2NH2, CHCH3).
Covalent Cross-Linking of EMHPs (xEMHPs)
EMHP was dissolved in DMSO at a concentration of 100 mg/mL. HMDI (Fluka) was added at a molar ratio of 10:1 isocyanate:lysine functional groups. The solution was vortexed and pipetted to 10 ×10 × 1.5 mm3 wells in a PTFE mold. The wells were covered with a PTFE lid, and the mixture was allowed to cross-link overnight at room temperature. The cross-linked product (xEMHP) was dried by heating to ~80 °C for ~2 h and thoroughly rinsed with acetone/water. The final product was stored in water at 4 °C before use.
Water Uptake
xEMHPs (six repeats) prepared as described above were dried overnight in a vacuum oven at 40 °C. After the dry weights were measured, samples were immersed in water for 24 and 48 h at room temperature. The wet weights were measured after blotting the excess water. The swelling ratio was calculated as the wet weight divided by the dry weight.
Mechanical Analysis
Mechanical analysis was conducted on a dynamic mechanical analyzer (RSA III instrument, TA Instruments) using a parallel plate geometry that measures the deformation as a function of an applied force. The compression rate was maintained at 0.05 mm/s. Samples were prepared in PTFE molds and cut into cylindrical shape (height ≈ 1.5 mm, diameter = 5 mm) for the test. Both dry and hydrated xEMHPs, along with a Tecoflex SG80A (Noveon), were tested under the same conditions. The modulus was calculated using the initial linear portion of the stress–strain curve. Five cyclic loading and unloading tests were carried out on each sample, with 1 min recovery time between consecutive cycles.
Cytotoxicity Tests
MTT Assay
Primary porcine vocal fold fibroblasts (PVFFs) were isolated from 2–3 year old pigs using collagenase disaggregation39 and were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Confluent cells (passage 8) were trypsinized and seeded into 24-well plates at a density of 50 000 cells/well in 1 mL of culture medium. The sterile-filtered (0.2 μm) polymer solutions (N3-PEG-N3, X(AKA3KA)2X, and EMHPs) were added to each well so that their final concentrations were in the range of 0.002–2.0 mg/mL (five repeats for each sample). The relative toxicity of these polymers was assessed with an MTT cell proliferation assay kit (ATCC) after 3 days of culture without changing the medium.17 The absorbance was measured at 550 nm with a plate reader (PerkinElmer Universal Microplate Analyzer). Cells cultured under the same conditions in a pure medium were used as the control, and the results were expressed as the measured absorbance normalized to the absorbance of the control sample. Statistical analysis was performed using a two-tailed Student’s t test with a p value of 0.05 for statistical significance. All values are reported as the mean ± standard deviation of the mean.
Live/Dead Assay
Live/dead assay was used to evaluate the toxicity of xEMHPs in an indirect contact mode. Thin sheets of xEMHPs (dry weight 4.8 mg, thickness ~0.7 mm, three repeats) were sterilized by UV irradiation (254 nm, ~10 min) before being loaded into the Millicell cell culture inserts (Millipore). The inserts containing xEMHPs were subsequently submerged in DMEM without direct contact with the underlying cells that had been preincubated for 6 h. PVFFs cultured under the same condition in the absence of xEMHPs were used as the control. After 3 days of culture, xEMHPs were removed from the well, and the cells were rinsed with PBS before being stained with Syto 13 (Molecular Probes, 1:1000 in PBS) and propidium iodide (Molecular Probes, 1:2000 in PBS). The stained cells were visualized using a confocal microscope (Axiovert LSM 510 with Zeiss LSM image examiner).
Results and Discussion
Synthesis and Chemical Analysis of EMHPs
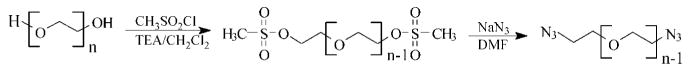
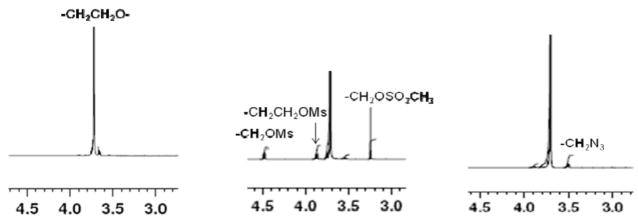
We employed copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) as a strategy for the production of multiblock architectures using telechelic azido-functionalized PEG and alkyne-derivatized peptide as the prepolymers. This reaction is known as one type of click chemistry and has been reported as an efficient reaction with tolerance to a wide variety of functional groups, few side reactions, and high reaction rates and efficiency in water as well as many organic solvents.24,40 The hydroxyl end groups of linear PEG were converted to mesylate groups using mesyl chloride and subsequently to azide groups using sodium azide (Scheme 2). The conversion of hydroxyls to mesylate end groups was confirmed by the appearance of CH2OMs and CH2CH2OMs peaks in the 1H NMR spectrum at 4.5 and 3.85 ppm, respectively (Figure 1). A 96% mesylate functionality was calculated from the ratio of the CH2OMs peak to the CH2CH2O repeat unit. The conversion of the mesylate to the azide end groups was confirmed by the disappearance of the CH2OMs, CH2CH2OMs, and OSO2CH3 peaks and the appearance of the CH2N3 peak at 3.55 ppm. Near 100% conversion from mesylate to azide was achieved. The overall end-group conversion was at least 96%, necessary for achieving high molecular weight products by the subsequent step-growth polymerization reactions.41 PEG is not expected to degrade under the experimental conditions employed for end-group modification.
Scheme 2.
Synthesis of α, ω-Azido-PEG
Figure 1.
1H NMR spectra of HO-PEG-OH (left), MsO-PEG-OMs (middle), and N3-PEG-N3 (right).
Alanine-rich, lysine-containing peptides with the sequence X(AKA3KA)2X, which mimics the hydrophilic/cross-linking domains of native elastin, were produced by standard Fmoc-based SPPS. The incorporation of terminal L-propargylglycine permitted the integration of the peptide into PEG–peptide multiblock copolymers via CuAAC. The peptide was analyzed by ESI-MS analysis (Figure S1, Supporting Information), and the measured molecular weight was in agreement with the calculated value of 1472 g/mol.
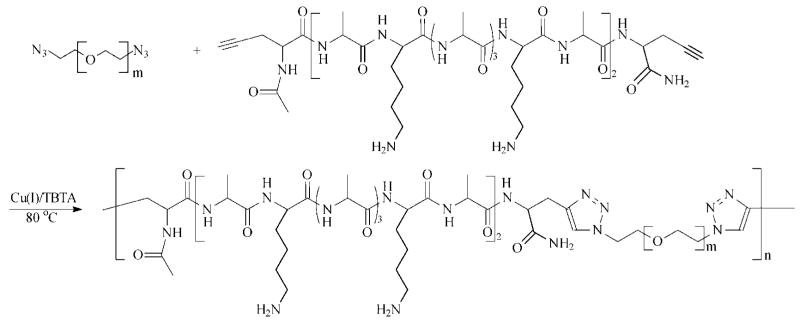
Hybrid multiblock copolymers with alternating PEG and peptide blocks were formed via step-growth polymerization by click coupling between the azide end groups of PEG with the alkyne end groups of the peptide (Scheme 3). These reactions were conducted in a water/DMF mixture with CuSO4 as the catalyst and sodium ascorbate as the reducing agent to generate copper(I) in situ. Stoichiometry for the complementary functional groups was maintained throughout the reaction. The mixture was heated to 80 °C to accelerate the reaction rate, and TBTA was used to stabilize the copper catalyst and to increase the polymerization rate. TBTA has been shown to be an efficient ligand due to the presence of both tertiary amine and 1,2,3-triazole functionalities that work in concert to increase the reaction rate.38 At the end of the polymerization, brown precipitates were observed. The byproducts were insoluble in common solvents such as DMF, DMSO, acetone, and water and are believed to be insoluble polymer aggregates resulting from copper coordinating to the triazole rings in the polymer products.42 The soluble fraction was isolated and purified for further chemical analyses and covalent cross-linking. Control experiments were conducted where peptide alone, dissolved in H2O/DMF (50/50), was heated at 80 °C for 24 h. SEC traces (data not shown) for peptide before and after heating showed no significant difference in terms of the elution time and the peak shape, suggesting the absence of peptide degradation during polymerization.
Scheme 3.
Synthesis of EMHP by Step-Growth Polymerization Using Azide-Functional PEG and Alkyne-Terminated Peptide
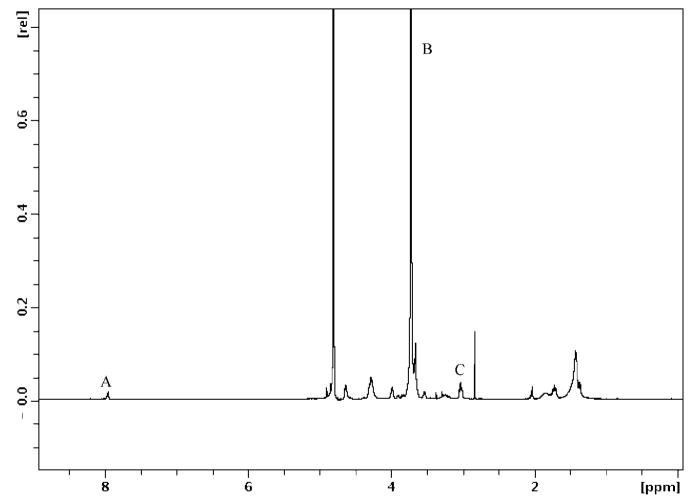
The formation of the multiblock product was confirmed collectively by 1H NMR, FT-IR, SEC, and SDS-PAGE. The 1H NMR spectrum of the EMHP is shown in Figure 2, where the ratio of the PEG, peptide, and the triazole linker per repeating unit in the multiblock copolymer was calculated as approximately 0.79:0.5:1 by comparing the integrations of the peaks at 3.7 ppm (PEG repeat unit), 3.0 ppm (lysine side chain CH2NH2), and 7.9 ppm (the triazole CH), labeled as B, C, and A in Figure 2, divided by the respective number of protons.
Figure 2.
1H NMR spectrum of EMHP. Peak A (7.9 ppm) = triazole CH, B (3.7 ppm) = CH2CH2O, and C (3.0 ppm) = CH2NH2.
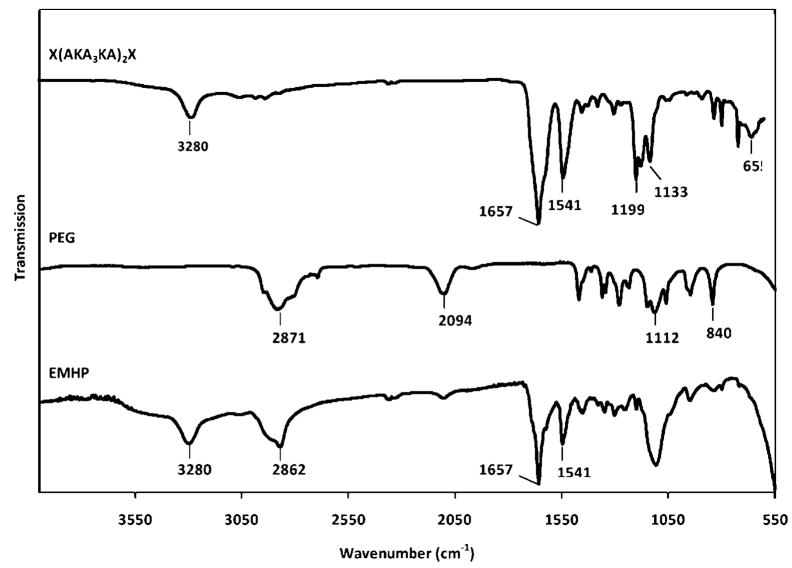
The FT-IR spectra of the alkyne-terminated peptide and the telechelic azido-PEG as well as the resulting EMHP are shown in Figure 3. The alkyne-terminated peptide exhibits characteristic N-H stretch (3280 cm−1), amide I (1657 cm−1), and amide II (1541 cm−1) peaks. The peaks at 1133–1199 cm−1 can be attributed to the C–N stretch of the lysine amine. The presence of the terminal alkyne is confirmed by the –C≡CH bend at 655 cm−1 although the =C–H stretch may have overlapped with the N–H stretch at 3280 cm−1. Telechelic azido-PEG shows distinct stretching bands at 2871 cm−1(CH2), 1112 cm−1 (C–O–C), and 840 cm−1 (C–C) that are characteristic of the PEG backbone. The strong band at 2094 cm−1 is indicative of the azide end groups in the starting PEG.43 The intensity of the azide peak decreased significantly in the spectrum for EMHPs. The additional peptide- and PEG-specific peaks are clearly visible in the spectrum for the EMHP, indicating the success of the coupling reaction. The characteristic triazole unsaturation (C=N stretching, 1380–1650 cm−1) could not be readily distinguished due to its overlapping with the amide bond.44,45
Figure 3.
FT-IR spectra of X(AKA3KA)2X peptide (top), N3-PEG-N3 (middle), and EMHP (bottom).
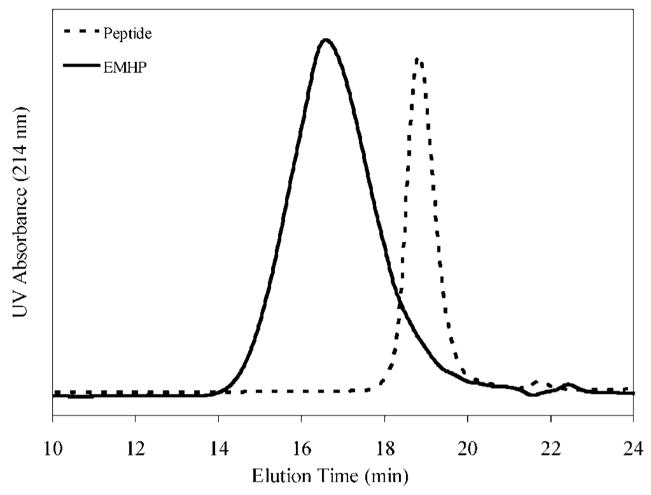
The change in molecular weight from the starting materials to the hybrid product was monitored by SEC and SDS-PAGE. SEC was run in PBS (pH = 7.4), and the traces shown in Figure 4 were collected using a UV detector at a wavelength of 214 nm. The starting PEG could not be detected due to the lack of a chromophore. The peptide appeared as a sharp peak at 18.9 min, while the multiblock copolymer showed a broad peak centered at a shorter elution time of ~16 min. A rough estimation using protein standards gave an Mw of 34 kg/mol for EMHP, with a polydispersity index of 4.3. Interestingly, although PEG and the peptide had comparable molecular weights (1450 g/mol for PEG and 1472 g/mol for peptide), their elution profiles were significantly different as indicated by their SEC traces using a refractive index detector (Figure S2, Supporting Information). While the peptide eluted at 18.9 min, the peak for PEG was centered at 17.7 min. Thus, PEG exhibited a higher “apparent” molecular weight than the peptide, suggesting the potential differences in their mobility and/or hydrodynamic volume in the mobile phase. It is also possible that the peptide interacts with the column packing material, delaying the elution time. The hybrid copolymer containing equal amounts of peptide and PEG is anticipated to elute at a later time (thus a lower apparent molecular weight) with a broader molecular weight distribution as compared to a linear, PEG-based polymer with similar molecular weight. Thus, the broad molecular weight distribution observed for the multiblock hybrid copolymers could be attributed to the different chemical and physical identities of the constituents in the multiblock copolymers.
Figure 4.
SEC analyses of the peptide and EMHP. Mobile phase: PBS, pH = 7.4. Detector: photodiode array (PDA) at 214 nm.
A broad molecular weight distribution is also expected from the step-growth polymerization due to the presence of polymerizing species of varying sizes. A theoretical polydispersity index (PDI) of 2 can be reached only at 100% conversion.41 Matyjaszewski and co-workers reported46 the step-growth click coupling between an azido-terminated polystyrene (Mn = 2020 g/mol) and propargyl ether. After 40 h of reaction, a polymer with a Mn of 16 700 g/mol and a PDI of 3.34 was obtained. Using a different chemistry, Guan and co-workers47 reported the synthesis of novel alternating saccharide–peptide hybrid copolymers for gene delivery. The polymers were produced via interfacial condensation polymerization between the amine from the oligolysines and the acid chloride derived from the protected monosaccharide. When the number of lysine residues in the starting oligomer increased from 2 to 4, the polydispersity of the resulting hybrid copolymers increased from 1.86 to 3.63. Larger monomers are likely to exhibit a reduced molecular dynamics and less efficient collisions with their partners, leading to slower polymerization and a broader molecular weight distribution. Our SEC results were not surprising considering the nature of the condensation polymerization, the relatively large size of the prepolymers, the possibility of competing side reactions, and the hybrid nature of the copolymer.
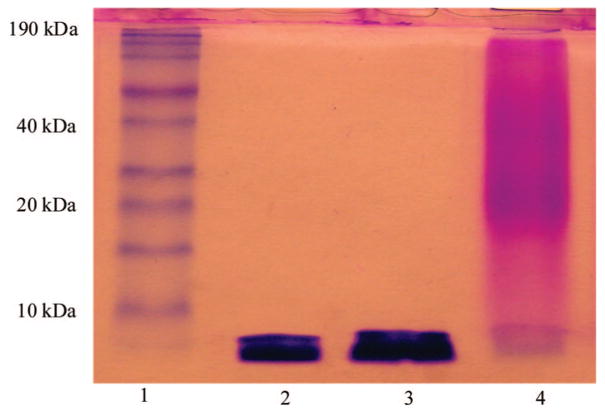
The SDS-PAGE results shown in Figure 5 agree with the SEC analyses. The alkyne-functionalized peptide appeared as a single band with an apparent molecular weight lower than 6 kDa as assessed by the protein standards (lane 2). When a physical mixture of the peptide and PEG diazide starting materials was run on the gel, a single band appeared at the same location (lane 3); the PEG is not stained by Coomassie blue and thus does not appear in the SDS-PAGE gel. The fact that the peptide alone and the peptide/PEG mixture appeared at the same location indicates that PEG did not interact with the peptide, nor did it carry the peptide along the gel when they were physically mixed. A clear shift to high molecular weight after the polymerization (lane 4) confirms the presence of covalent linkages between the peptide and PEG. Polymeric species with an apparent molecular weight of 20–40 kDa exhibit more intense staining than those with lower molecular weight, confirming the presence of a large population of polymers with such molecular weight in the product. The broad smear seen in the band for the hybrid copolymer is typical for protein/synthetic polymer bioconjugates as synthetic polymers usually exhibit a broader molecular weight distribution, even for well-defined polymers generated by living polymerization procedures.48,49 In SDS-PAGE, proteins are unfolded and rendered negatively charged via their interaction with the surfactant.50 In our case, SDS likely binds more strongly to the peptide due to the presence of both lysine and hydrophobic alanine residues, while its interaction with PEG may be limited because of PEG’s hydrophilicity and lack of charge. Therefore, the mobility of PEG along the gel is likely to be significantly lower than that of the peptide, this heterogeneity further contributing to the breadth of the smear. The 1H NMR and FT-IR spectra combined with SEC and SDS-PAGE analyses provided convincing evidence for the formation of hybrid copolymers containing approximately 3–5 PEG blocks in alternation with equal numbers of peptide in the backbone.
Figure 5.
SDS-PAGE of the peptide and PEG prepolymers as well as the multiblock copolymer: lane I, protein molecular weight ladder; lane 2, X(AKA3KA)2X; lane 3, a mixture of X(AKA3KA)2X and N3-PEG-N3; lane 4, multiblock hybrid polymer.
Structure Analyses of Peptide and EMHP
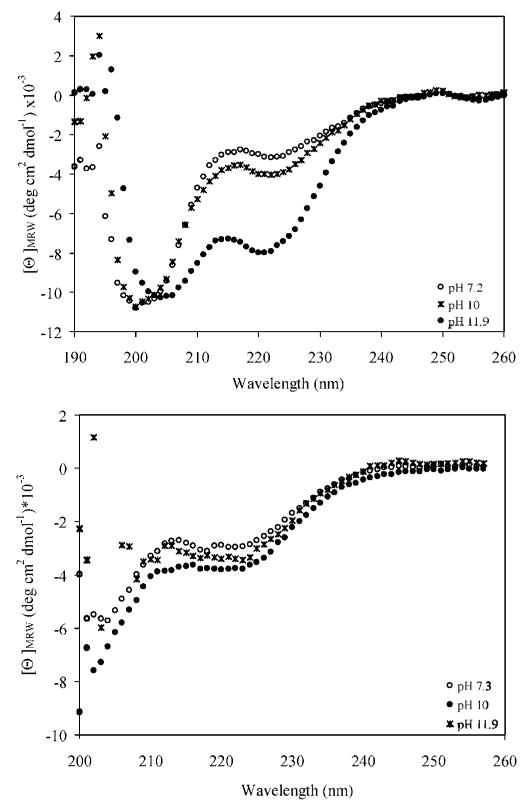
The secondary structures of the alkyne-functional peptide and EMHP were monitored by circular dichroic (CD) spectroscopy at various pH conditions (Figure 6). The positive charge of the lysine residues at pHs 7.4 and 10 causes the peptides to adopt a predominantly random coil conformation at room temperature, as evidenced by a single minimum at 198 nm in the spectra. The emergence of minima at 222 and 208 nm at pH 11.9 confirmed the increased helicity of the peptides under this condition. As the solution pH increased, there was a gradual increase in the peptide helical content (5.2% at pH = 7.2; 6.6% at pH = 10, 13% at pH = 11.9) due to the neutralization of the lysine amine above its pKa value (10.5). The relatively low helical content observed is expected for the short peptides under these conditions and is in good agreement with the structures of the cross-linking domains alone.
Figure 6.
CD spectra of KA3K peptide and EMHPs at pH 7.2 (circle), 10 (star), and 11.9 (filled circle).
The cross-linking domains in tropoelastin are generally believed to be predominantly α-helical in structure. However, several laboratories have observed that the α-helical content of the protein, measured by CD, is substantially less than that expected from the primary sequence prediction, suggesting the presence of less stable, frayed structures.33,51 Tamburro et al. studied the conformation of the individual cross-linking domains of human tropoelastin in both water and 2,2,2-trifluoroethanol (TFE). While these peptides mainly adopted flexible conformations such as random coil (and/or PPII helix) in water, they adopted stable α-helical conformations in TFE. In a separate study by Keeley and co-workers,51 ELPs derived from entropic exons 20 and 24 flanking a single cross-linking domain based on exons 21 and 23 were synthesized recombinantly. Approximately 15–18% α-helical content was observed when the ELP was dissolved in water, and a 28% helicity was obtained when the flexible “hinge” region was removed. In tropoelastin, the organized assembly appears to require a process of hydrophobically driven, temperature-induced coacervation, creating a highly hydrophobic microenvironment that is conducive to the helix formation in the cross-linking domains.
When the peptide was intercepted with PEG regularly along the hybrid polymer backbone, the fractional helicity at pH 11.9 was significantly reduced (5.6%) although similar helical content to that observed for the free peptide was found at lower pH conditions (Figure 6). The reduction in helicity is not surprising considering the hydrophilicity of PEG coupled with the covalent linkage between the peptide and PEG building blocks that may have introduced steric hindrance that inhibits peptide folding. Therefore, to further mimic the assembly properties of natural elastin, the hydrophobicity of the synthetic polymers, and the number, sequence, and arrangement of the cross-linking domains, will be systematically varied in future studies. Dry, solid films of the multiblock copolymer are suggested to contain α-helical conformation as evidenced by a strong peak at 1657 cm−1 in the FT-IR spectrum (Figure 3).52
Covalent Cross-Linking of EMHPs and Mechanical Evaluations
In nature, the preassembled amine groups from the lysine residues are cross-linked by the enzyme lysyl oxidase, which causes oxidative deamination of the lysine residues to form allysine (α-amino adipic δ-semialdehyde). The allysine residues then undergo spontaneous condensation reactions with each other or with lysine to form cross-links between tropoelastin chains.9 In our hybrid copolymers, the lysine amines were similarly used for cross-linking by hexamethylene diisocyanate (HMDI). HMDI has been previously used to cross-link recombinant ELPs53 and is less toxic than glutaraldehyde, another commonly used cross-linker for amine groups. Cross-linking was conducted in DMSO to prevent hydrolysis of the isocyanate groups. After cross-linking, DMSO and residual HMDI were removed by heating and thorough washing with acetone and water. The average swelling ratio for xEMHPs was measured as 3.6 ± 0.2 after 24 h swelling. Prolonged incubation (48 h) did not change the measured swelling ratio, indicating that the xEMHP has reached equilibrium swelling after 24 h. The hydrated xEMHPs visibly shrank and appeared more brittle after drying.
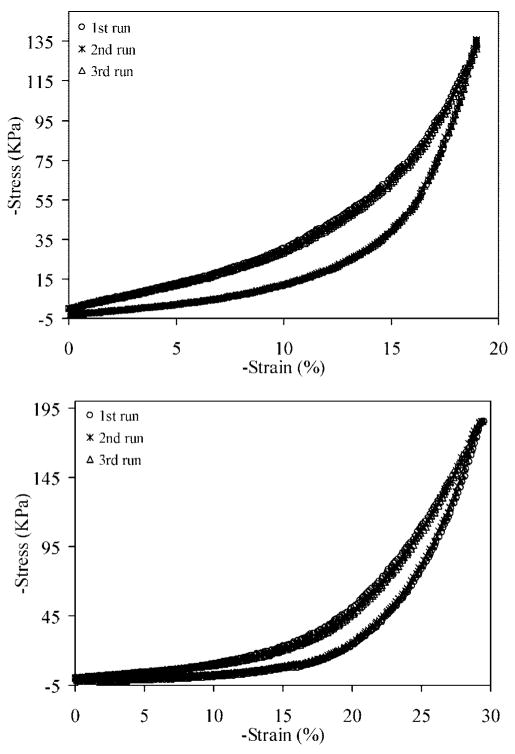
The mechanical properties of xEMHP (dry and wet) were analyzed by uniaxial compression testing. Dry xEMHP was soaked in water for 24 h, and the excess water was gently removed prior to the tests. Parallel measurements were carried out on Tecoflex SG80A, a soft, aliphatic polyurethane widely used as an elastomeric material for tissue engineering.54–56 The maximum strain applied to different samples (εmax; Tecoflex SG 80A, 35%; hydrated xEMHP, 30%; dehydrated xEMHP, 19%) was arbitrarily chosen to avoid reaching the upper transducer limit (2000 g applied force) of the instrument. At least five consecutive loading–unloading cycles (Figure 7; only three runs are shown for more facile interpretation) were carried out in order to assess the energy dissipation during cyclic compression, with 1 min recovery time before the next cycle was initiated. An examination of the loading curves for all three samples showed an initial compliant behavior up to a strain of 10–15% followed by stiffening behavior, which may be attributed to strain hardening and/or non-Gaussian behavior of polymer chains at high strains.57,58 The unloading paths typically show a hysteresis loop with a residual strain. Additional recovery occurred with time (1 min) after unloading; these behaviors are typical for elastomers.59,60 Remarkable overlapping for the loading–unloading curves was observed for all three types of materials, indicating the efficient recovery after loading and the absence of permanent deformation under our testing conditions.
Figure 7.
Representative loading–unloading curves (three runs) for dehydrated xEMHP (top) and hydrated xEMHP (bottom). Samples were allowed to recover for 1 min after each run.
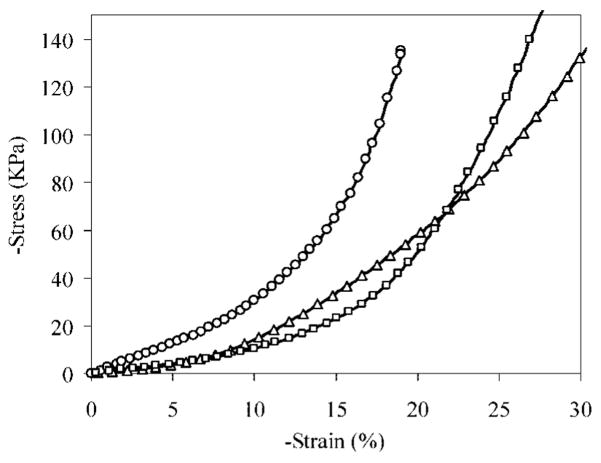
Representative stress–strain curves for hydrated and dry xEMHPs are plotted in Figure 8. The compressive modulus was calculated by taking the slope of the stress–strain curve in the linear region from 5–10% strain. The moduli of the dry and hydrated xEMHPs are 0.32 ± 0.008 and 0.12 ± 0.018 MPa, respectively. The material softens upon hydration, consistent with results observed for natural elastin, in which the modulus decreases with an increased amount of hydration due to the plasticizing effect of water and increased chain dynamics.61 For comparison purposes, the compression modulus of Tecoflex SG80A was measured as 0.214 ± 0.025 MPa. This modulus is comparable to that of the cross-linked EMHPs. Of note is that Tecoflex SG80A is a hydrophobic elastomer that does not absorb any significant amount of water, while the hydrated xEMHP contains ~70 wt % water and yet behaves as a compressible elastomer. Hydrated materials with desirable mechanical properties offer greater advantages over hydrophobic scaffolds for tissue engineering.
Figure 8.
Stress–strain curves for dehydrated xEMHP (circle), hydrated xEMHP (square), and Tecoflex SG80A (triangle).
Gosline and co-workers62 have done extensive investigation on the mechanical properties of elastin matrices. Using tensile tests, they found that pig aortic elastin had an elastic modulus of 0.81 MPa, stress at break of 1.02 MPa, strain at break of 103%, and a resilience of 77%. Using uniaxial tensile tests, Urry18,63 evaluated the mechanical properties of elastomeric polypentapeptides that had been prepared using recombinant DNA technology and cross-linked by γ-irradiation. The hydrated samples had a tensile modulus varying from 0.04 to 0.20 MPa depending on the polypeptide sequence and the irradiation dosage. Bellingham et al. prepared ELPs containing alternating hydrophobic domains and hydrophilic cross-linking domains by recombinant DNA technology.22 The resulting polypeptides were covalently cross-linked via the lysine residues using pyrroloquinoline quinine (PQQ) upon peptide coacervation. Under tension, these materials had an elastic modulus of ~0.25 MPa and could be extended to strains of ~100% before breaking. Repeated cycles of loading and unloading of these materials showed consistent and efficient recovery, with a resilience of ~80%. Although a similar range of modulus values was observed for xEMHP, a direct comparison of material properties under compression and tension is not practical. Other modes of deformations, such as shear, exist in compression tests,64 while in tensile tests, maximum deformation leads to catastrophic failure. On the other hand, the compressive modulus is expected to reach infinity at high strain due to the material densification, which is not directly materials related. While the area under loading–unloading curves can be used to calculate the energy dissipation (resilience), such exercises cannot be applied in compression.
Recently, Srokowski et al. used lysine diisocyanate (LDI)65 to cross-link genetically engineered, multiblock ELPs composed of five hydrophobic domains flanked by four cross-linking domains. Under unconfined compression, ELP foams had compression moduli of 0.14 MPa at 20% strain and 0.94 MPa at 80% strain. These values are similar to those obtained from our xEMHP under the same compression conditions. Similar loading–unloading curves between ELPs and xEMHPs were also observed. However, our hybrid and modular design offers the opportunities to finetune the material’s mechanical properties by systematically adjusting (1) the peptide sequence and sequence length, (2) the polymer molecular weight and architecture, (3) hydrophobicity of polymer and peptide building blocks, and (4) interchain covalent cross-linking.
Cytotoxicity Assays
As discussed above, cells actively probe and respond to mechanical cues in their surrounding environment, changing their adhesion structures, gene expression, and cell locomotion. In vitro cell cytotoxicity study is the first step in evaluating the potential of EMHPs for tissue engineering applications. Vocal fold fibroblasts were chosen for the toxicity studies for multiple reasons. First, the human vocal folds are the only tissue in the human body in which vibration occurs naturally and regularly at frequencies of 100–1000 Hz and amplitudes of about 1 mm.66 Second, vocal fold fibroblasts (VFFs) are the major type of cells that are responsible for maintaining the functional condition of the extracellular compartment in the vocal fold.67 Third, elastin plays a pivotal role in controlling the proper function of the vocal fold.68
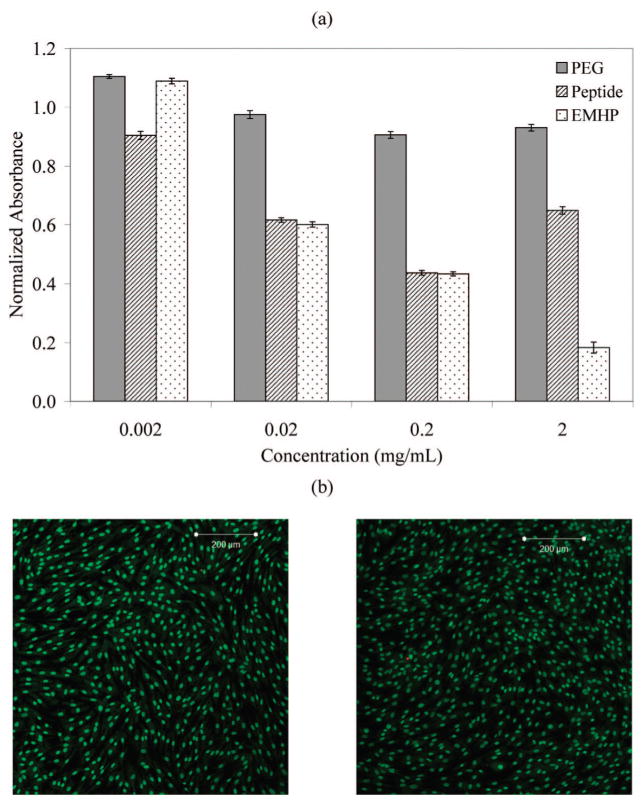
The toxicity of N3-PEG-N3 and the alkyne-functionalized peptide as well as the un-cross-linked multiblock copolymer were measured by an MTT assay using primary porcine vocal fold fibroblasts (PVFFs). The average absorbance shown in Figure 9a was normalized to a positive control of PVFFs cultured in unconditioned media. PEG diazide was not toxic to cells at concentrations of 0.002–2 mg/mL. Compared to PEG diazide, the peptide caused more cell death at all concentrations (p < 0.05), likely due to the abundant positive charges on the peptide69 (2 lysines every 5 alanines) and possible impurities from the SPPS process. The peptide and the hybrid copolymer showed similar toxicity at concentrations of 0.02 and 0.2 mg/mL, while reduced cell growth for the latter was observed at 2 mg/mL (p < 0.05). The observed toxicity is not likely to originate from the polymer degradation due to the stability of the amide and triazole bonds and the nondegradable nature of PEG. The presence of some residual copper catalyst that remained bound to the copolymers may have caused more cell death for EMHPs at 2 mg/mL.70 PVFFs cultured in the presence of 2 mg/mL peptide were more viable than those cultured at peptide concentrations of 0.2 and 0.02 mg/mL. The underlying mechanism for such observation is not clear.
Figure 9.
Cytotoxicity evaluations of the soluble polymers and the cross-linked EMHP using primary porcine vocal fold fibroblasts (PVFFs). (a) MTT assay of N3-PEG-N3, X(AKA3KA)2X, and EMHP at concentrations of 0.002, 0.02, 0.2, and 2 mg/mL. (b) Live/dead assay of xEMHP in an indirect contact mode. Left: PVFFs in the presence of xEMHP. Right: control cells in the absence of xEMHP. Live cells are stained green, and dead cells are stained red. Scale bar: 200 μm.
An indirect contact method was applied to test the toxicity of xEMHP. Swollen xEMHPs were suspended above the cells and fully immersed in the media using tissue culture inserts. PVFFs were cultured in the presence of xEMHPs for 3 days at 37 °C. A second well with PVFFs in unmodified media was used as a control. Figure 9b shows a representative live/dead staining. It is clear that cells exposed to xEMHPs were highly viable and capable of growing to confluence over the course of the culture. They showed the same cell morphology as the control cells, indicating that our purification procedures appear to adequately remove any toxic reagents (e.g., cross-linker, solvents). When PVFFs were directly seeded on top of the swollen gels (data not shown), they did not adhere well to the substrate, owing to the relative hydrophilicity of the xEMHPs and the lack of integrin-binding motifs in these proof-of-concept materials. Remarkably, cells in direct contact with the edges of the swollen gels reminded highly viable, further demonstrating the lack of toxicity of the xEMHPs. Thus, these types of materials may be promising candidates as elastic scaffolds for vocal fold tissue engineering.
This study represents our initial effort in synthesizing elastin mimetic hybrid copolymers with alternating molecular architecture. The modular nature of the design, coupled with the chemical nature of the synthesis, should permit facile adjustment of mechanical, morphological, and biological properties of the resulting polymers. The relatively broad molecular weight distribution for the EMHP indicates the structural heterogeneity of the cross-linked product, which may be useful if it can be controlled, as controlled heterogeneity at molecular and microscopic levels is necessary and has been shown to enhance the overall material properties.7,71 We are currently conducting mechanistic studies in order to elucidate the structure–function relationships between EMHP structure and mechanical properties. Insight gained from these studies will allow the engineering of hybrid elastomers with controlled heterogeneity and desired materials properties.
Conclusions
Elastin–mimetic hybrid polymers (EMHPs) with an alternating multiblock structure were synthesized by step-growth polymerization employing orthogonal click chemistry between an α,ω-azido-PEG and alkyne-terminated KA3K peptide. The formation of multiblock copolymers was confirmed collectively by 1H NMR, FT-IR, SEC, and SDS-PAGE. The resulting hybrid polymers contain PEG and peptide alternating along the polymer backbone, with an estimated molecular weight (Mw) of 34 kg/mol. Covalent cross-linking of EMHPs with HMDI through the lysine residue in the peptide domain afforded an elastomeric gel (xEMHPs) with a compressive modulus of 0.12 MPa when hydrated. The mechanical properties of xEMHP are comparable to a commercial polyurethane elastomer (Tecoflex SG80A) under the same conditions. Perfectly overlapping loading/unloading curves were obtained for xEMHP, both dry and wet, demonstrating their ability to recover from mechanical stress. The observed elasticity for xEMHPs was attributed to the hybrid multiblock molecular architecture, with PEG segments providing the chain flexibility and the peptide domains providing covalent cross-linking ability. Although the soluble multiblock copolymers exhibited some toxicity to primary porcine vocal fold fibroblasts (PVFFs) at concentrations ≥0.2 mg/mL, the cross-linked hybrid elastomer allowed PVFFs to grow and proliferate during 3 days of culture, suggesting that the cross-linked hybrid polymers do not contain any toxic impurities that leach during cell culture.
Supplementary Material
Acknowledgments
We thank Carl Giller for his help with acquiring the ATR-IR spectrum. The authors thank the NSF/DMR Biomaterials Program (Jia, Career: 0643226) and the NSF Integrative Graduate Education & Research Traineeship (IGERT) program for funding. This work was also supported in part by National Institute of Health (R01DC008965, 2P20RR017716) and the Center for Neutron Science at the University of Delaware (US Department of Commerce, #70NANB7H6178).
Footnotes
Supporting Information Available: ESI-MS characterization of X(AKA3KA)2X and SEC analyses of PEG and peptide prepolymers. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Sandberg LB, Soskel NT, Leslie JG. New Engl J Med. 1981;304:566–579. doi: 10.1056/NEJM198103053041004. [DOI] [PubMed] [Google Scholar]
- 2.Hammond TH, Gray SD, Butler J, Zhou R, Hammond E. Otolaryngol Head Neck Surg. 1998;119:314–322. doi: 10.1016/S0194-5998(98)70071-3. [DOI] [PubMed] [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York: 2002. [Google Scholar]
- 4.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 5.Ingber DE. FASEB J. 2006;20:811–827. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Ameer GA, Sheppard BJ, Langer R. Nat Biotechnol. 2002;20:602–606. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 7.Amsden B. Soft Matter. 2007;3:1335–1348. doi: 10.1039/b707472g. [DOI] [PubMed] [Google Scholar]
- 8.Daamen WF, Veerkamp JH, van Hest JCM, van Kuppevelt TH. Biomaterials. 2007;28:4378–4398. doi: 10.1016/j.biomaterials.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 9.Vrhovski B, Weiss AS. Eur J Biochem. 1998;258:1–18. doi: 10.1046/j.1432-1327.1998.2580001.x. [DOI] [PubMed] [Google Scholar]
- 10.Toonkool P, Regan DG, Kuchel PW, Morris MB, Weiss AS. J Biol Chem. 2001;276:28042–28050. doi: 10.1074/jbc.M103391200. [DOI] [PubMed] [Google Scholar]
- 11.Hoeve CAJ, Flory PJ. Biopolymers. 1974;13:677–686. doi: 10.1002/bip.1974.360130404. [DOI] [PubMed] [Google Scholar]
- 12.Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, Xu J, Parker T. Phil Trans R Soc London B. 2002;357:169–184. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosline JM. Biopolymers. 1978;17:677–695. doi: 10.1002/bip.1978.360170311. [DOI] [PubMed] [Google Scholar]
- 14.Daamen WF, Hafmans T, Veerkamp JH, van Kuppevelt TH. Biomaterials. 2001;22:1997–2005. doi: 10.1016/s0142-9612(00)00383-5. [DOI] [PubMed] [Google Scholar]
- 15.Smith DW, Brown DM, Carnes WH. J Biol Chem. 1972;247:2427–2432. [PubMed] [Google Scholar]
- 16.Singla A, Lee CH. J Biomed Mater Res. 2002;60:368–374. doi: 10.1002/jbm.10077. [DOI] [PubMed] [Google Scholar]
- 17.Jia X, Burdick JA, Kobler J, Clifton RJ, Rosowski JJ, Zeitels SM, Langer R. Macromolecules. 2004;37:3239–3248. [Google Scholar]
- 18.Lee J, Macosko CW, Urry DW. Biomacromolecules. 2001;2:170–179. doi: 10.1021/bm0000900. [DOI] [PubMed] [Google Scholar]
- 19.Reiersen H, Clarke AR, Rees AR. J Mol Biol. 1998;283:255–264. doi: 10.1006/jmbi.1998.2067. [DOI] [PubMed] [Google Scholar]
- 20.Schreiner E, Nicolini C, Ludolph B, Ravindra R, NO, Kohlmeyer A, Rousseau R, Winter R, Marx D. Phys Rev Lett. 2004;92:148101. doi: 10.1103/PhysRevLett.92.148101. [DOI] [PubMed] [Google Scholar]
- 21.Wright ER, Conticello VP. Adv Drug Delivery Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 22.Bellingham CM, Lillie MA, Gosline JM, Wright GM, Starcher BC, Bailey AJ, Woodhouse KA, Keeley FW. Biopolymers. 2003;70:445–455. doi: 10.1002/bip.10512. [DOI] [PubMed] [Google Scholar]
- 23.Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 24.Hawker CJ, Wooley KL. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- 25.Klok HA. J Polym Sci, Polym Chem. 2005;43:1–17. [Google Scholar]
- 26.Borner HG, Schlaad H. Soft Matter. 2007;3:394–408. doi: 10.1039/b615985k. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Stewart RJ, Kopecek J. Nature (London) 1999;397:417–420. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 28.Van Hest JCM. Polym Rev. 2007;47:63–92. [Google Scholar]
- 29.Rihova B. Adv Drug Delivery Rev. 1996;21:157–176. [Google Scholar]
- 30.Fromstein JD, Woodhouse KA. J Biomater Sci, Polym Ed. 2002;13:391–406. doi: 10.1163/156856202320253929. [DOI] [PubMed] [Google Scholar]
- 31.Heymann B, Grubmuller H. Chem Phys Lett. 1999;307:425–432. [Google Scholar]
- 32.Bellingham CM, Keeley FW. Curr Opin Solid State Mater Sci. 2004;8:135–139. [Google Scholar]
- 33.Tamburro AM, Pepe A, Bochicchio B. Biochemistry. 2006;45:9518–9530. doi: 10.1021/bi060289i. [DOI] [PubMed] [Google Scholar]
- 34.Ausubel FM, Kingston RE, Moore DM, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 4. John Wiley & Sons; New York: 1999. [Google Scholar]
- 35.Farmer RS, Kiick KL. Biomacromolecules. 2005;6:1531–1539. doi: 10.1021/bm049216+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polizzotti BD, Fairbanks BD, Anseth KS. Biomacromolecules. 2008;9:1084–1087. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 37.Malkoch M, Vestberg R, Gupta N, Mespouille L, Dubois P, Mason AF, Hedrick JL, Liao Q, Frank CW, Kingsbury K, Hawker CJ. Chem Commun. 2006:2774–2776. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- 38.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 39.Freshney RI. Culture of Animal Cells: A Manual of Basic Techniques. J. Wiley; New York: 2000. [Google Scholar]
- 40.Kolb HC, Finn MG, Sharpless KB. Angew Chem. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 41.Odian G. Principles of Polymerization. 4. John Wiley & Sons; Hoboken, NJ: 2004. [Google Scholar]
- 42.Meudtner RM, Hecht S. Macromol Rapid Commun. 2008;29:347–351. [Google Scholar]
- 43.Agut W, Agnaou R, Lecommandoux S, Taton D. Macromol Rapid Commun. 2008;29:1147–1155. [Google Scholar]
- 44.Jin YH, Zhu J, Zhang ZB, Cheng ZP, Zhang W, Zhu XL. Eur Polym J. 2008;44:1743–1751. [Google Scholar]
- 45.Lambert JB, Shurvell HF, Lightner DA, Cooks RG. Organic Structural Spectroscopy. Prentice Hall; Upper Saddle River, NJ: 1998. [Google Scholar]
- 46.Tsarevsky NV, Sumerlin BS, Matyjaszewski K. Macromolecules. 2005;38:3558–3561. [Google Scholar]
- 47.Metzke M, O’Connor N, Maiti S, Nelson E, Guan ZB. Angew Chem. 2005;44:6529–6533. doi: 10.1002/anie.200501944. [DOI] [PubMed] [Google Scholar]
- 48.Boyer C, Bulmus V, Liu JQ, Davis TP, Stenzel MH, Barner-Kowollik C. J Am Chem Soc. 2007;129:7145–7154. doi: 10.1021/ja070956a. [DOI] [PubMed] [Google Scholar]
- 49.Heredia KL, Bontempo D, Ly T, Byers JT, Halstenberg S, Maynard HD. J Am Chem Soc. 2005;127:16955–16960. doi: 10.1021/ja054482w. [DOI] [PubMed] [Google Scholar]
- 50.Hames BD. Gel Electropheresis of Proteins. 3. Oxford University Press; Oxford, NY: 1990. [Google Scholar]
- 51.Miao M, Cirulis JT, Lee S, Keeley FW. Biochemistry. 2005;44:14367–14375. doi: 10.1021/bi0510173. [DOI] [PubMed] [Google Scholar]
- 52.Creighton TE. Proteins: Structure and Molecular Properties. WH Freeman & Co; New York: 1993. [Google Scholar]
- 53.Nowatzki PJ, Tirrell DA. Biomaterials. 2004;25:1261–1267. doi: 10.1016/s0142-9612(03)00635-5. [DOI] [PubMed] [Google Scholar]
- 54.Liu YC, Webb K, Kirker KR, Bernshaw NJ, Tresco PA, Gray SD, Prestwich GD. Tissue Eng. 2004;10:1084–1092. doi: 10.1089/ten.2004.10.1084. [DOI] [PubMed] [Google Scholar]
- 55.Webb K, Li WH, Hitchcock RW, Smeal RM, Gray SD, Tresco PA. Biomaterials. 2003;24:4681–4690. doi: 10.1016/s0142-9612(03)00368-5. [DOI] [PubMed] [Google Scholar]
- 56.Mulder MM, Hitchcock RW, Tresco PA. J Biomater Sci, Polym Ed. 1998;9:731–748. doi: 10.1163/156856298x00118. [DOI] [PubMed] [Google Scholar]
- 57.Gent AN. J Rheol. 2005;49:271–275. [Google Scholar]
- 58.Miquelard-Garnier G, Creton C, Hourdet D. Soft Matter. 2008;4:1011–1023. doi: 10.1039/b717460h. [DOI] [PubMed] [Google Scholar]
- 59.Sarva SS, Deschanel S, Boyce MC, Chen WN. Polymer. 2007;48:2208–2213. [Google Scholar]
- 60.Qi HJ, Boyce MC. Mech Mater. 2005;37:817–839. [Google Scholar]
- 61.Lillie MA, Gosline JM. Biopolymers. 1990;29:1147–1160. doi: 10.1002/bip.360290805. [DOI] [PubMed] [Google Scholar]
- 62.Lillie MA, David GJ, Gosline JM. Connect Tissue Res. 1998;37:121–141. doi: 10.3109/03008209809028905. [DOI] [PubMed] [Google Scholar]
- 63.Lee J, Macosko CW, Urry DW. Macromolecules. 2001;34:5968–5974. [Google Scholar]
- 64.Treloar LRG. The Physics of Rubber Elasticity. Clarendon Press; Oxford: 1975. [Google Scholar]
- 65.Srokowski EM, Woodhouse KA. J Biomater Sci, Polym Ed. 2008;19:785–799. doi: 10.1163/156856208784522038. [DOI] [PubMed] [Google Scholar]
- 66.Titze IR. J Acoust Soc Am. 1989;85:901–906. doi: 10.1121/1.397562. [DOI] [PubMed] [Google Scholar]
- 67.Catten M, Gray SD, Hammond TH, Zhou R, Hammond E. Otolaryngol Head Neck Surg. 1998;118:663–667. doi: 10.1177/019459989811800516. [DOI] [PubMed] [Google Scholar]
- 68.Hammond TH, Zhou R, Hammond EH, Pawlak A, Gray SD. J Voice. 1997;11:59–66. doi: 10.1016/s0892-1997(97)80024-0. [DOI] [PubMed] [Google Scholar]
- 69.Cremers HFM, Lens JP, Seymour L, Feijen J. J Control Release. 1995;36:167–179. [Google Scholar]
- 70.Codelli JA, Baskin JM, Agard NJ, Berozzi CR. J Am Chem Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guelcher SA. Tissue Eng, Part B: Rev. 2008;14:3–17. doi: 10.1089/teb.2007.0133. [DOI] [PubMed] [Google Scholar]
- 72.Certain commercial materials and equipment are identified in this paper in order to specify adequately the experimental procedure. In no case does such identification imply recommendation by the National Institute of Standards and Technology nor does it imply that the material or equipment identified is necessarily the best available for this purpose
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.