Abstract
Many of the currently established human embryonic stem cell lines have been characterized extensively in terms of their gene expression profiles and genetic stability in culture. Recent studies have indicated that miRNA, a class of non-coding small RNA that participate in the regulation of gene expression, may play a key role in stem cell self renewal and differentiation. Using both microarrays and quantitative PCR, we report here the differences in miRNA expression between undifferentiated human embryonic stem cells (hESC) and their corresponding differentiated cells that underwent differentiation in vitro over a period of two weeks. Our results confirm the identity of a signature miRNA profile in pluripotent cells, comprising a small subset of differentially expressed miRNAs in hESCs. Examining both mRNA and miRNA profiles under multiple conditions using cross-correlation, we find clusters of miRNAs grouped with specific, biologically-interpretable mRNAs. We identify patterns of expression in the progression from hESC to differentiated cells that suggest a role for selected miRNAs in maintenance of the undifferentiated, pluripotent state. Profiling of the hESC “miRNA-ome” provides an insight into molecules that control cellular differentiation and maintenance of the pluripotent state, findings that have broad implications in development, homeostasis and human disease states.
Keywords: miRNA, gene expression, correlation, human embryonic stem cells, differentiation
Introduction
Embryonic stem cells (ESCs) share several unique features which include unlimited self renewal and the ability to differentiate into any of the three embryonal lineages, ectoderm, endoderm and mesoderm. For the cell fate decision to be made in response to internal and/or niche-specific signals, a complex set of dynamic feedback loops and cross regulation of pathways is required (1, 2). In addition to feedback loops, regulation by methylation of CPG islands has been proposed (3, 4). This is critical in X chromosome inactivation, imprinting of specific gene loci, and maybe important in regulating expression of ESC specific genes (5, 6). Additional miRNA-directed regulatory pathways have been proposed including those regulating the timing of differentiation. None of these pathways have been rigorously explored, although a recent convergence of technologies allowing isolation of sufficient amounts of purified cells for analysis and various large scale analytical methods have made these types of studies more feasible.
For instance, hESCs can now be maintained in feeder-independent cultures in an undifferentiated state and be used to obtain robust differentiation via the addition of exogenous reagents to the culture media (7, 8). While cell culture techniques and media additives have contributed much to our knowledge of stem cell differentiation and maintenance of pluripotency, significant work needs to be done to understand the molecular mechanisms involved in these processes. Global mRNA expression and methylation profiling of various hESC lines have been well characterized (3, 9-14). More recently, microRNA (miRNA) expression in stem cells has been shown to differ significantly from other cell types tested to date (15-17).
MicroRNAs are short (20-24 nucleotide), non-coding RNAs that have been identified in various organisms including mammalian cells. The sequence of most microRNAs are highly conserved across species with nearly 90% of the currently sequenced human microRNAs identical to mouse and rat and at least 30% homologous to microRNAs from C. elegans (18). microRNAs are thought to negatively regulate gene expression by direct mRNA cleavage (19-23); mRNA decay by deadenylation (24, 25) or via translational repression (26). To complicate the specific mapping of microRNA binding sites in the transcriptome, it has been determined that, at least in animal cells, translational repression occurs by annealing of microRNA to mRNA at sites with imperfect complementarity (27). Due to this complexity and the lack of a clear understanding of the mode of action of microRNA function, the identification of target mRNAs regulated by microRNA has been difficult (28). Nevertheless, the importance of microRNA in several biological processes such as cell growth and apoptosis (29), viral infection (30) and human cancer (31-33) is well documented. Based on several studies, it has been suggested that microRNAs regulate gene expression of more than 30% of protein coding genes in humans (34). The role of microRNA-mediated regulation of stem cell division (35), as well as adipocyte (36), cardiac (37), neural (28, 38) and hematopoeitic lineage differentiation (21, 39) is well known. More recently, a unique set of microRNAs has been shown to be associated with mouse ESC and EB (embryoid body) formation (15, 17, 40-42). Using northern blot analysis and cloning, several microRNAs were identified in hESCs, of which several were identical to microRNAs previously reported in mouse ESCs (16). Consistent with this observation a mouse ESC knockout lacking Dicer (40) and DGC8 (43), two key processing enzymes in microRNA biosynthesis, exhibits a failure to undergo differentiation, further implicating their importance as key regulators during this process.
Analytical methods for gene expression analysis have been available for some time and are now widely used in the field. Recently, tools for systematic analysis of epigenetic changes in cells have become available opening the door for broad-scale analysis on another level of transcriptional and translational regulation. In this study, NCode™ microRNA arrays (44) and qPCR were used to analyze microRNA profiles of various hESC lines and their differentiated cells derivatives. We show here that although there are some informative variations in the microRNA profiles between hESC lines, there are also several markers that are highly expressed across all hESC lines tested in this study. Furthermore, as these cells differentiate, the microRNA profiles change significantly. Using a semi-quantitative assay, microRNA copy numbers were estimated across pluripotent hESC, differentiating cells, and adult human brain, a representative sample of terminally differentiated adult tissue. Finally, gene expression and microRNA expression were correlated to identify potential regulators of key pluripotent genes. The results of this study will form the basis for further perturbation studies to study epigenetic regulation of microRNA to determine stem cell fate.
Methods
Embryonic Stem Cell culture
hESC lines CyT25 and CyT203, cultured and differentiated as previously described (45), were kindly provided by Melissa Carpenter, Novocell. hESC lines (HES2, HES3, and HES4) were from ES Cell International, http://stemcells.nih.gov/research/registry/esci.asp) at passage numbers ranging between 75-125 and with a normal karyotype were cultured and differentiated as described previously (46, 47). In short, hESC were cultured on a mitotically inactive in-house derived mouse embryonic fibroblast feeder layer using gelatin (Sigma) coated culture dishes (Falcon). Culture media was changed daily and was composed of Dulbecco's modified eagle medium (DMEM; with or without glucose and sodium pyruvate respectively; Invitrogen), supplemented with 20% fetal bovine serum, 0.1 mM β-mercaptoethanol (Invitrogen), 1% non-essential amino acids (Invitrogen), 2 mM L-glutamine (Invitrogen), 1% insulin-transferrin-selenium (Invitrogen), and 50 IU/mL penicillin and 50 μg/mL streptomycin (Invitrogen). Cells were subcultured every seven days by mechanical microdissection. For differentiation, cells were washed once with PBS and treated with Collagenase IV (1 mg/ml) for 3-4 min at 37°C. Collagenase was replaced by serum-free (SF) medium (DMEM medium supplemented with 1% MEM non-essential amino acids, 2 mM L-Glutamine, 1× ITS, 0.1 mM β-mercaptoethanol and Penicillin/Streptomycin and culture plates were scored with a 10 μl pipette tip (Eppendorf). The entire adherent cell layer was scraped off using a cell scraper (Iwaki) and the cell suspension transferred to a 50 ml tube (Falcon) and allowed to settle. The cell pellet was then resuspended in fresh SF medium and briefly triturated before an equal volume of cell suspension was transferred to ultra low attachment 6 well plates (Costar). Media changes of EBs were performed every 3 days for a period of 12 days prior to harvesting EBs for analysis.
RNA Isolation
Total RNA was isolated using Trizol Reagent (Invitrogen) according to manufacturer's instructions. Contaminating genomic DNA was removed from the isolated RNA by treatment with amplification grade DNase I (Invitrogen) for 2 h at 37°C. RNA was precipitated and quantified spectrophotometrically and its purity assessed by electrophoresis on a 15% Nupage urea-TBE gel (Invitrogen).
Gene Expression using Microarrays
For Illumina BeadArray, total RNA was amplified and labeled as reported earlier (11). Labeled, amplified material (∼700 ng per array) was hybridized to the Illumina HumanRef-8 v2 BeadChip according to the manufacturer's instructions (containing >22,000 probes based on the Human RefSeq database, Illumina, Inc., San Diego, CA). Array data processing and analysis was performed using Illumina BeadStudio software.
Enrichment of micro RNA
microRNA was isolated from Trizol extracted total RNA using Purelink microRNA Isolation Kit (Invitrogen) according to recommended protocol. The amount of microRNA was quantified spectrophotometrically and its purity assessed by electrophoresis on a 15% Nupage urea-TBE gel (Invitrogen).
Ncode™ microRNA array
Experimental Design
To compare global microRNA expression between hESC and their EB, a heterotypic dye swap experiment was carried out using the NCode™ microRNA array. Five hundred nanograms of the enriched microRNA was labeled with the Ncode™ direct labeling system and hybridized to replicate NCode™ multispecies microRNA arrays (44) as described earlier (48). Briefly, hESC microRNA fraction was labeled with Alexa 3 dye (Green) and the corresponding differentiated cell microRNA fraction was labeled with Alexa 5 (Red). A second slide reversed the dyes, with hESC labeled with Alexa 5 (Red) and the differentiated sample with Alexa3 (Green). Since the microRNA probes are printed in duplicate on the NCode array, a total of 4 data points for each microRNA can be obtained with minimal sample and slides. The identified markers were subsequently validated by the more sensitive and quantitative method of microRNA qPCR using the unamplified total RNA from all the five hESCs and their differentiated cells.
NCode™ qPCR
qPCR was performed on unamplified total RNA using the NCode™ qPCR kit (Invitrogen) as described earlier (48). The differences in microRNA expression between samples were determined using the relative quantification method. Briefly, the Ct values of the samples were normalized to the Ct values of GAPDH, a house keeping gene. The resulting values were further normalized to 2102Ep human embryonic carcinoma cells which were used as a reference cell line. Fold-difference in gene expression of the sample from the reference 2102Ep cells was calculated using the equation 2-ΔΔCt. To assign microRNA copy numbers, a standard curve was generated from a pure synthetic template diluted over several logs. The Ct values of the reference 2102Ep cell line were converted to copy number based on fold differences of each microRNA obtained using the relative quantification method. The copy number generated therefore is a relative approximation and not absolute numbers.
Correlation of mRNA and microRNA expression
Samples of hESC cultures were classified a priori into three biological groups; embryonic stem (ES) cells, differentiated ES cells (EB), or embryonic carcinoma (EC) cells. Illumina data were quantile normalized (49) and filtered for genes with a detection threshold of at least 0.99 under one or more conditions. NCode and Illumina array data were normalized and corrected for experimental effects using a linear model (50). Briefly, the following linear model was fit to the log-expression profile of each gene:
where A,D,V, and G represent the additive effect of the ith array, jth dye, kth biological group, and gth gene respectively, AG represents the combinatorial effect of the ith array with the gth gene, effectively modeling any chip or spot artifacts, εijkg describes the random error associated with each measurement, and VG represents the combinatorial effect of the kth biological group with the gth gene. This last effect, VG, is used to interpret any gene expression differences between the biological groups. All additive effects were estimated via least squares. An F-test was performed on the modeled data, with or without VGkg effects, to select differentially expressed genes. The p-values were obtained via bootstrap, and adjusted for multiple comparisons using the Benjamini-Hochberg method (51), with a tolerated false discovery rate of 5%. The microRNA/mRNA data were jointly examined by computing pairwise correlations between the estimated VGkg effect profiles. Heatmap displays were constructed from these correlations using BioConductor (http://www.bioconductor) and R (http://www.r-project.org). Target predictions were identified from a downloaded database of RNA22 predictions (52), using ENSEMBL transcript identifiers to link Illumina probes as predicted microRNA targets.
Results
Global mRNA expression profiles of ESC and EB
As a first step, we sought to characterize the hESC lines and their differentiated cells used in the study. Gene expression analysis was carried out using Illumina bead array as described previously (53). Prior to consideration of the data, the quality of each array was confirmed by comparing the signal intensity distribution obtained for each sample. The entire set of gene expression data and a table showing the signal intensity distribution is provided in Supplemental Table 1 (http://cord.rutgers.edu/appendix/Supplemental_Table_1.xls). Samples were then compared pair-wise by creating a scatter plot of expressed genes with a detection level greater than 0.99. Pair-wise comparison of two samples with a correlation coefficient (R2) value closer to 1.0 indicates similarity in transcript expression between the two cell types while a variation of transcript expression by over two fold is reflected by a lower correlation value. For example, the R2 value between the two hESC lines CyT25 and CyT203 is 0.938 (Figure 1A). This value is however decreased 0.839 when the hESC line CyT203 is compared to its corresponding differentiated cells (Figure 1B). A similar decrease in the correlation value is observed for all the five hESC upon differentiation indicating a change in the expression level of a significant number of transcripts between hESC and their corresponding differentiated cells (not shown). It is interesting to note here that HES2, HES3 and HES4 show a correlation R2 value close to 1. However, the R2 value between HES2 and CyT203 is 0.858, indicating that the HES lines are more similar to each other than compared to CyT203. This may be largely due to cell culture conditions since the cell maintenance and differentiation protocols were different for these two sets of samples. Nevertheless, comparison of the ES with their corresponding differentiated cells does indicate a change in transcript expression reflected by the decrease in the correlation (R2) value. Based on global gene expression profiles, the relatedness of samples is plotted as a dendrogram (Figure 1C).
Figure 1. Representative global mRNA expression profiles of hESC and differentiated cells.
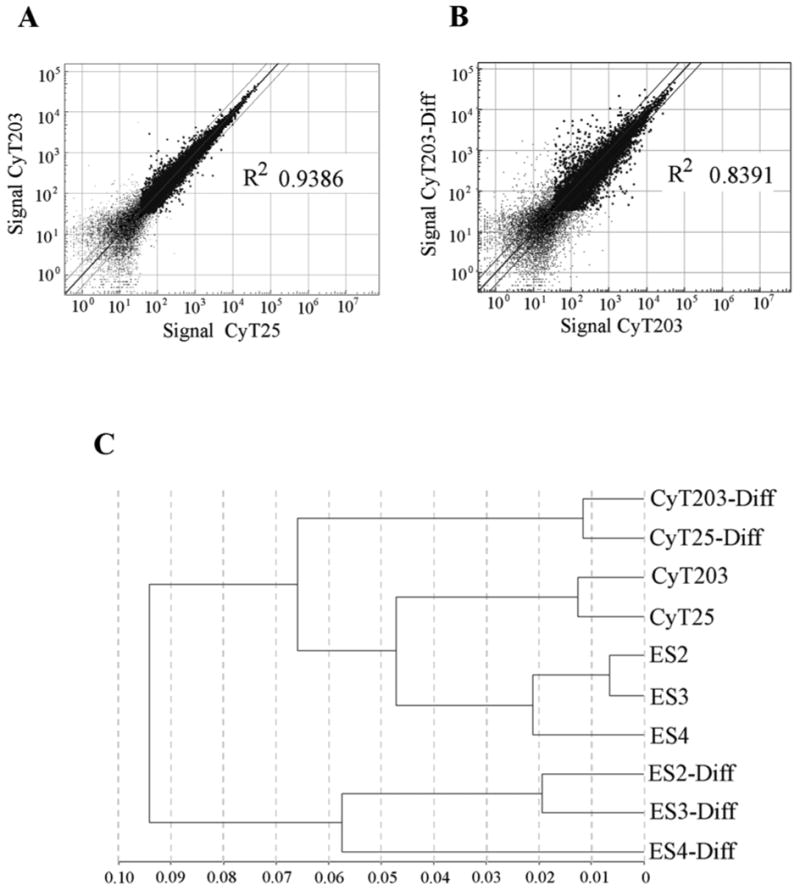
Total RNA isolated from the three hESC cell lines HES2, HES3 and HES4 and their corresponding cells after 12 days of differentiation were used for mRNA expression analysis using the Illumina bead array. Data were analyzed using Bead Studio software and data points representing greater than a 0.99 detection threshold are represented as points in the scatter plot. Points lying outside the lines represent genes with greater than 2 fold difference and R2 value closer to 1 suggest similar gene expression pattern.
A. Comparison of global gene expression between two hESC lines CyT25 and CyT203 with a correlation (R2) value of 0.938.
B. Comparison of global gene expression between CyT203 and its CyT203-differentiated cell for two weeks with a correlation (R2) value of 0.839.
C. Dendrogram showing relatedness in global gene expression of the hESC and differentiated samples.
While there is some variation in the gene expression profiles between different hESC lines, there are sets of genes that are known to be regulated during the very early stages of differentiation. To confirm uniform differentiation of all the hESC lines used in this study, the expression levels (expressed as arbitrary signal units) of pluripotency markers [Oct4 (POU5F1), Nanog, Rex1 (ZFP42), UTF1 and TDGF1] and markers for general differentiation [HAND 1, AFP and OTX1; specific for mesoderm, endoderm and ectoderm respectively] (53), were measured. The expression of Oct4 decreased after differentiation more prominently (>10 fold) for HES2, HES3 and HES4 compared to CyT25 and CyT203 (2 -3 fold). Consequently, an increase in the signal for the differentiation markers is apparent in differentiated cells relative to the parent hESC line (Table 1). The decrease in pluripotency associated genes together with an increase in general differentiation marker expression indicates differentiation of all the hESC lines after differentiation.
Table 1.
Signal intensity of key pluripotent and differentiation markers in ESC and differentiated cells samples obtained on Illumina gene analysis chip. The intensity, represented as arbitrary units, is represented in bold when the value is greater between ESC and their corresponding EB.
| hESC | Differentiated Cells | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Specificity | Genes | CyT25 | CyT203 | ES2 | ES3 | ES3 | CyT25-Diff | CyT203-Diff | ES2-Diff | ES3-Diff | ES3-Diff |
| Pluripotent | POU5F1 | 701 | 763 | 408 | 486 | 369 | 343 | 480 | 36 | 5 | 8 |
| NANOG | 89 | 133 | 141 | 158 | 66 | 75 | 107 | -12 | -13 | 5 | |
| ZFP42 | 2048 | 2715 | 1168 | 1218 | 2 | 925 | 950 | 144 | 141 | 27 | |
| UTF1 | 1208 | 1880 | 614 | 402 | 362 | 117 | 134 | 38 | 38 | 31 | |
| TDGF1 | 8387 | 10858 | 5967 | 5390 | 5767 | 10881 | 9530 | 92 | 63 | 64 | |
| Differentiation | HAND1 | 158 | 217 | 138 | 3017 | 27 | 1233 | 1748 | 15147 | 11743 | 3206 |
| AFP | 297 | 83 | 187 | 292 | 148 | 714 | 684 | 27189 | 22294 | 9497 | |
| OTX1 | -1 | 11 | 20 | 12 | 2 | 17 | 31 | 292 | 143 | 168 | |
Global microRNA expression profiles of ESC and EB
The expression profiles of 55 human microRNAs determined to be significantly regulated between ES, EB, and EC biological groups are sufficient to separate samples into biologically interpretable groups (Figure 2). Biological replicates are appropriately grouped indicating technical proficiency. Interestingly, in contrast to the relationships deduced from mRNA expression profiles, the Cyt25 and Cyt203 samples are inter-dispersed among the hESC samples suggesting a closer relationship to the ES cells when comparing microRNA profiles across all cell samples, including carcinomas (Ntera and 2102Ep samples). The differences between the differentiated Cyt25 samples and the undifferentiated Cyt25 samples are less drastic than those observed between the undifferentiated and differentiated Cyt203. Ntera2 and 2102Ep samples, while both classified in this study as embryonic carcinoma cells, cluster with distinct subsets of the ES group, suggesting that microRNA profiles classify samples based on an unexpected relatedness between these cell types.
Figure 2. Representative global microRNA expression heatmap of hESC and differentiated cells (day 12).
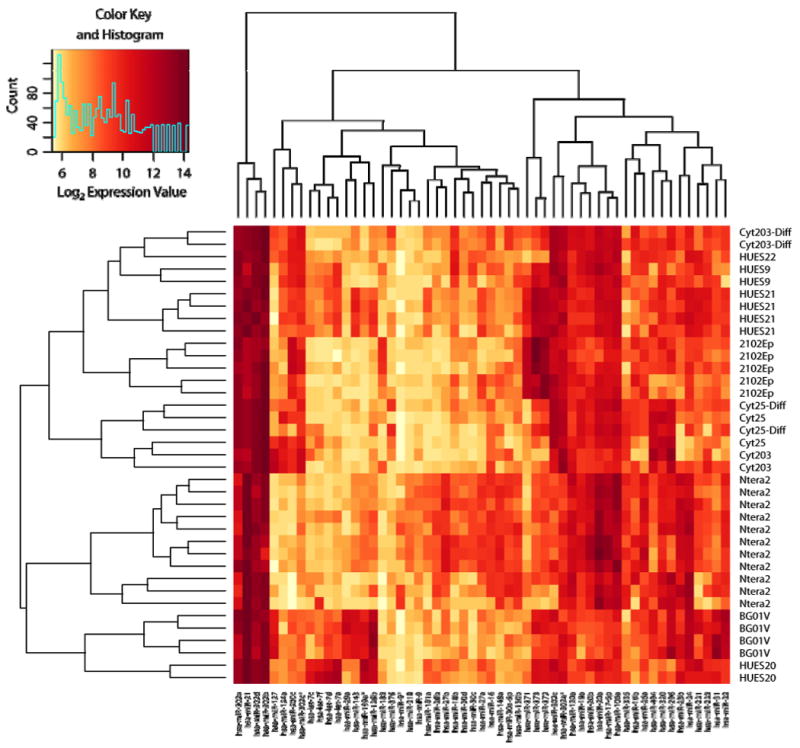
Significantly-regulated microRNAs were examined for expression levels in all samples by drawing a hierarchically clustered heatmap with associated dendrograms. Colors represent log2 expression values as depicted in the color key (inset). In general, technical replicates clustered together tightly as expected. The two embryonic carcinoma (EC) lines (Ntera and 2102Ep) cluster with distinct ES cell lines suggesting that microRNAs identify a range of cell phenotypes, potentially including the potential for tumorigenesis and/or differentiation.
Select microRNAs or groups of microRNAs can readily be identified within the significance list, and their relationships among samples illustrate several interpretations. For example, miR-17 cluster members (33) appear to be enriched in Ntera2 and differentiated EB sample, as well as select ES samples, suggesting they may be further along the differentiation path. MiR-302, a previously described ES cell-specific marker (15, 16) appears enriched in all samples, although slightly less so in the Ntera2 group. let-7 family members are more highly enriched in all of the HUES samples as well as the BGOV1. miR-21, a microRNA previously associated with tumorigenesis (54, 55) is elevated in Ntera2, BG0V1, and HUES20 cell lines. miR-9, a microRNA associated with the neuronal phenotype (56), is elevated in the Ntera2 cell line. In several other cases, tissue specific/enriched microRNAs are restricted to subsets of ES/EB/EC cell types. Clearly, microRNA expression patterns are informative and allow a novel classification of samples based on this set of parameters.
The entire NCode microRNA expression data and signal intensity distribution is provided as Supplemental Table 2 (http://cord.rutgers.edu/appendix/Supplemental_Table_2.xls). To further quantitatively measure the differences in the microRNA levels between ES and EB samples, a list of microRNA candidates differentially expressed between ES and EB (with P values less than 0.02) along with markers expressed similarly in ES and EB and earlier reported microRNAs associated with hESC (16) were chosen for further validation by quantitative PCR (qPCR) of all the five hESC samples and their corresponding EBs.
microRNAs expression between hESC and differentiated cells
To determine if a subset of significantly regulated microRNAs are useful to classify hESC and their differentiated products, we assayed three new hESC preparations (ES2, ES3, and ES4) by qPCR. Analysis of the chosen group of microRNAs differentially expressed between the three hESC lines and their corresponding differentiated cells was carried out using total cellular RNA fractions. As no specific microRNAs have been identified to be expressed at consistent levels across all cell lines (i.e. potential housekeeping microRNAs), microRNA qPCR values in this study were normalized to the GAPDH mRNA transcript. These values were further normalized to microRNA levels in reference to the nullipotent human EC line 2102Ep (57). We chose this cell line to serve as a reference standard for pluripotent cells to allow for normalization between data sets obtained during multiple qPCR runs (Supplemental Table 3, http://cord.rutgers.edu/appendix/Supplemental_Table_3.xls). Data are presented as a heat map of the fold change relative to 2102Ep cells with higher expression levels represented as red and lower expression levels represented as green (Figure 3). With the three newly-tested ES lines (ES2, ES3, and ES4), the selected microRNAs correctly cluster the samples by differentiation status. The Cyt25 and Cyt203 ES and EB samples were also clustered by cell phenotype. The qPCR microRNA expression patterns correctly distinguished ES from EB states for each cell line.
Figure 3. Heat map of microRNA expression in hESC and their corresponding differentiated cells.
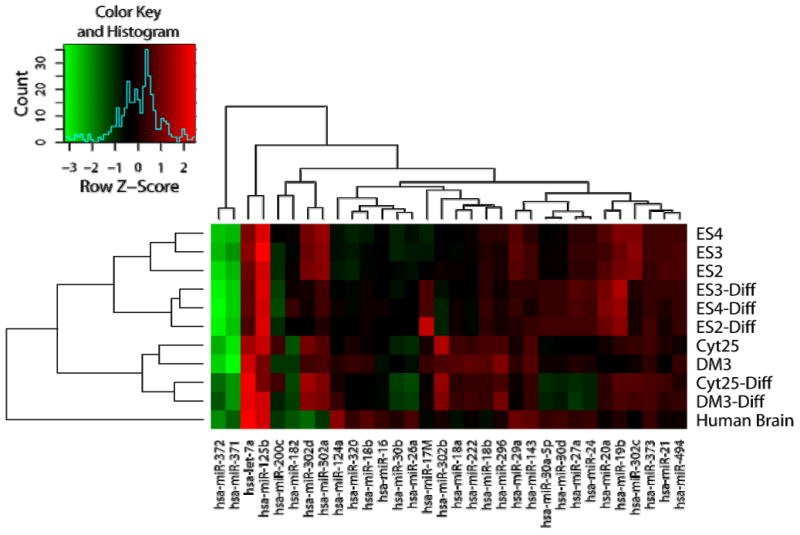
qPCR of statistically significant microRNA candidates was performed on three independent hESC lines and their corresponding differentiated cells. Expression of microRNA relative to the reference 2102Ep human EC cell line was determined and the log2 fold change depicted as a heat map with red representing higher and green lower levels of microRNA relative to the reference 2102Ep cell line (see color key, inset). Results were scaled by row (sample) and represented as the Z-score. Results confirm the ability to distinguish ES cells from their differentiated EB samples using this limited set of microRNAs.
While relative expression values provide a clear indication of differential expression, the absolute level of expression (copy number) of each microRNA itself is a more useful measure of microRNA concentration differences between multiple samples. The fold change values for hESC and differentiated samples were therefore converted to copy numbers to reflect the abundance of candidate microRNAs in each sample. microRNA copy numbers were generated using a standard curve of a known microRNA and compared the microRNA qPCR values from the 2102Ep sample against this standard. The fold-change (from 2102Ep) values of all cell specific microRNA were then converted to approximate copy numbers by multiplication against the copy number value of each microRNA measured in the 2102Ep sample. Adult human brain tissue was included in the analysis as a representative of a terminally differentiated cell type.
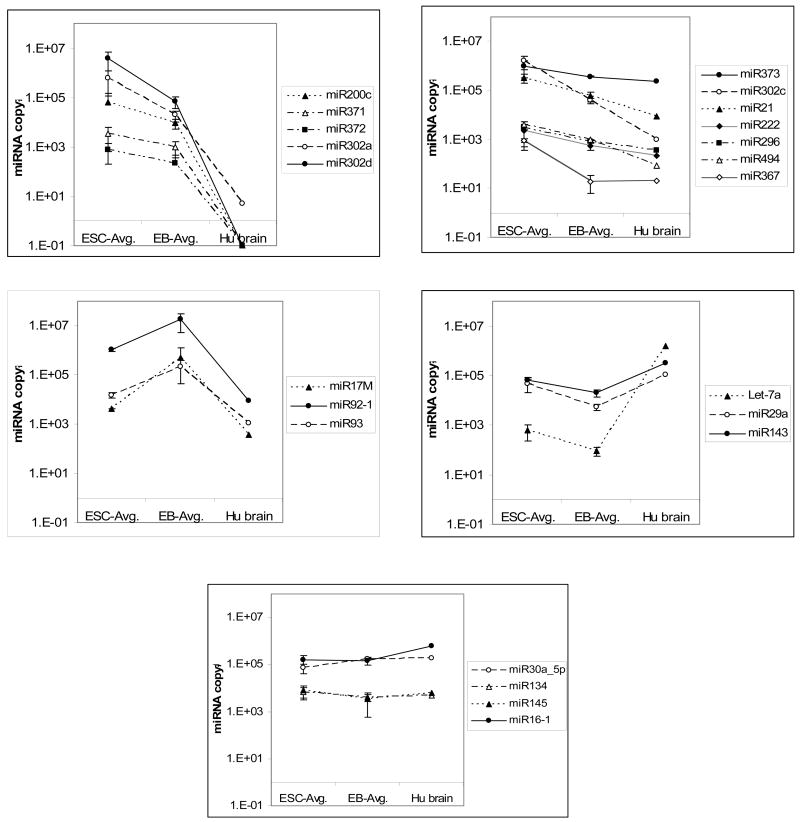
Findings from this calculation show that the pattern of several microRNA was consistent across the undifferentiated ESC and differentiated cell sample pairs. The average copy number of selected microRNAs in the hESC lines, their corresponding differentiated cells, and adult tissue is shown as a plot in Figure 4 with the error bars representing the standard deviation between the samples. Based on microRNA expression differences between hESC, differentiated cells, and adult tissue, candidate microRNAs can be divided into four main groups (Table 2). The first group of microRNA (group 1a) is highly expressed in hESC with expression levels decreasing after differentiation and being undetectable in adult brain tissue (Figure 4, Panel A). microRNAs in this group (302a, 302d, 371, 372 and 200c) have been reported to be associated with ESCs and EC cells in previous studies (15, 16). A class of microRNAs under the first group is present at very high levels in hESC with lower levels of expression in differentiated cells and adult tissue (Figure 4, Panel B). In this group a significant decrease in miR21, 222, 296 and 494 was noted changing from hESC to differentiated cells. These microRNAs, while present, are significantly lower in adult tissue compared to hESC and differentiated samples. In the second group, microRNAs are consistently expressed at higher levels in differentiated cells when compared with ESC or adult tissue (Figure 4, Panel C). Three microRNAs from a genomic cluster (33), 17M, 92 and 93, fall under this category which may be indicative that these microRNAs play a role during the differentiation process. The third group of microRNAs is expressed at relatively low levels in hESC and differentiated cells but has increased expression in the adult brain tissue control. (Figure 4, Panel D). Interestingly, Let7a, which is thought to be rare in pluripotent cells, was detected at varying levels in all the hESC and differentiated cells. However, the level of Let-7 expression in hESC was 3 logs lower than the adult tissue control. A fourth group consisting of microRNAs such as miR16, 134, 246 and 30a_5p were found to be expressed at relatively comparable levels in hESC, differentiated cells and adult brain tissue (Figure 4, Panel E) suggesting that they may be involved in general cellular function.
Figure 4. Classification of differentially expressed markers between hESC, differentiated cells and adult tissue (human brain).
Relative expression of microRNAs compared to the standard 2102Ep control cells was calculated as fold change as described in Materials and Methods. A standard curve of a pure βAct fragment generated over 7 logs was used to deduce the copy number of microRNA in 2102Ep cells based on Ct values. These values were then used to determine the relative copy numbers in other samples using fold change values.
A. microRNAs highly expressed in hESC decline with differentiation and are absent or present in very low levels in adult tissue.
B. microRNAs highly expressed in hESC decline with differentiation and are present in relatively low levels in adult tissue.
C. microRNAs expressed in both hESC and adult tissue with increasing levels in differentiated cells. Only mir-17M was significantly regulated but two other members of the mir-17 cluster (33) were assayed for comparison.
D. microRNAs with relatively low expression in hESC and differentiated cells compared to terminally differentiated adult tissue.
E. microRNAs expressed at comparable levels in hESC, differentiated cells and adult tissue.
Table 2.
microRNAs specific to hESC or differentiated cells based on NCode™ qPCR profiles.
| microRNA | ESC | Diff | Adult Tissue | Known Function |
|---|---|---|---|---|
| miR200c, 371, 372, 302a, 320d | High | Low | Absent | Present in hESC and EC (16) |
| miR373, 302c, 21, 222, 296, 494, 367 | High | Low | Lower | Inhibits erythroblast formation; Antiapoptotic factor (72, 73) |
| miR154, 29a, 143, 29c, Let7a | High | Low | High | Present in MEF/NIH3T3,; Promotes adipocyte diff (15, 36) |
| miR17M, 92, 93 | Low | Higher | Low | Reported in single mESC (17) |
| miR16, 134, 145, 30a_5p | Same | Same | Same | General Cellular Function (?) |
Cross-correlation of mRNA and microRNA
To determine potential correlation of mRNA expression and microRNA expression, the data generated from this study were combined with data reported in an earlier study, including several hESC lines and hEC lines (57), and modeled for cross-correlation. We reasoned that microRNAs and mRNAs that are coordinately regulated or mRNAs that are targeted for destruction by specific microRNAs ought to exhibit distinct patterns of correlated levels over specific cell types or conditions. Therefore we modeled the data in order to most appropriately examine these correlations.
A linear model was fit to the log-expression profile of each mRNA or microRNA, where additive effects for each experiment group were estimated via least squares. Undifferentiated hESC lines (HUES9, HUES20, HUES22, BG0V1, CyT25 and CyT203) comprised the baseline group, while differentiated CyT25 and differentiated CyT203 comprised of two separate groups. Finally, Ntera and 2102Ep formed the last two groups, for a total of 5 groups. An F-test was used to select differentially expressed genes (that differ between at least two of the biological groups). The p-values were adjusted for multiple comparisons using the Benjamini-Hochberg method at 5% FDR, resulting in 55 microRNAs and 2,678 mRNAs. The microRNA/mRNA data were jointly examined by computing pairwise correlations between the estimated additive effect profiles (results shown in Supplemental Table 4 http://cord.rutgers.edu/appendix/Supplemental_Table_4.xls). A heatmap depicting the cross-correlation of the entire set of significant genes is shown in Supplemental Figure 1. While the number of genes depicted makes it impossible to label specific mRNAs, it is clear that identifiable clusters of mRNAs and microRNA emerge from the analysis as regions of positive correlations (green) and negative correlations (red). Perhaps these relationships will be simpler to examine if we focus on an understandable subset of the significant mRNAs.
Using a list of pluripotency, germline-specific, and general differentiation markers (53), we selected 40 genes from the significant mRNA list and re-drew the heatmap using correlations to all 55 microRNAs (Figure 5). Again, clear patterns of positive and negative correlations were identified (green and red regions, respectively). Using this cross-clustering technique, mRNAs are clustered according to their cell-specific expression pattern (53) nearly perfectly (see the color coding to the left of the heatmap in Fig. 5). Similarly, microRNAs are divided into two major clusters. Within these groups, polycistronic family members are clustered together, microRNAs derived from the same hairpin precursor cluster together, and the ES-specific mir-302 family is both positively correlated with ES-specific mRNA markers and negatively correlated with EB mRNA markers.
Figure 5. Cross-correlation heatmap of selected mRNA and microRNA expression patterns.
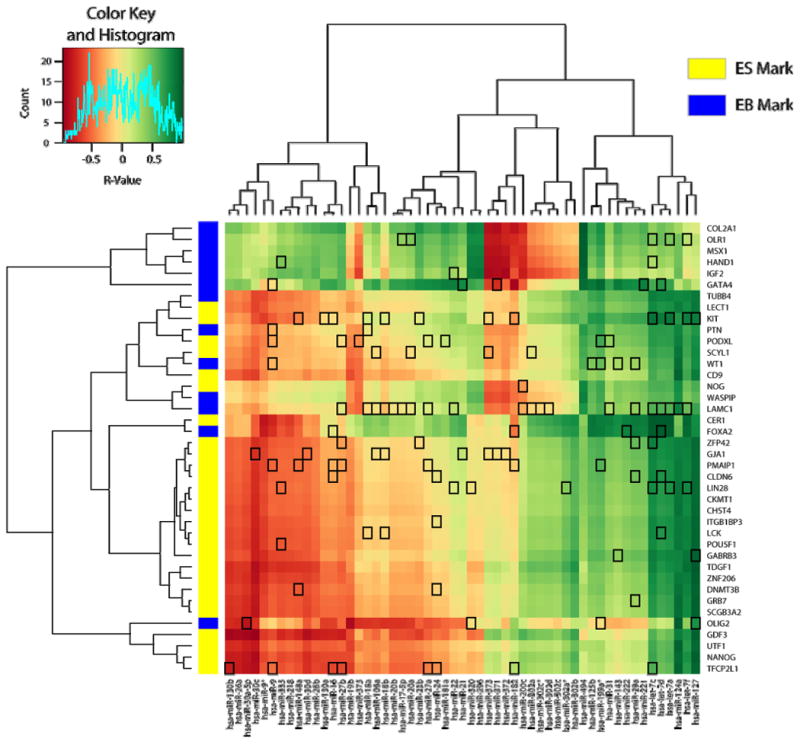
A cross-correlation table (Supplemental Table 4) was constructed for all 2,678 selected mRNAs and 55 microRNAs and expressed as a heatmap (Supplemental Figure 1). Here, a subset of mRNAs specific for pluripotency, germline-specific, and general differentiation markers was selected and used to construct a heatmap, colored by the Pearson correlation coefficient (R-value, see color key, inset). Specific mRNAs were color coded (left) as being ES-specific (yellow) or EB-specific (blue) based on previous studies (53). Target predictions computed using the RNA22 algorithm (52) are identified by drawing a box around the correlation cell at the intersection of the microRNA and the predicted mRNA target.
If expression correlations imply the possibility of a functional interaction, we might predict that microRNAs within clusters of negative correlation (red) should have a preponderance of predicted mRNA targets. Using the target predictions from the RNA22 algorithm (52), we do not see a clear prevalence of targets corresponding to regions of negative correlations. Some target predictions support roles for previously-described stem-specific microRNAs. For example, the ES-specific mir-302 family (15, 16) is relatively devoid of predicted targets, consistent with a role in suppressing mRNAs that are not present or not regulated in these conditions. The mir-302 family, among several other significantly regulated microRNAs, is predicted to target the mRNA encoding laminin (LAMC1). Laminin expression and binding to integrin is important for cell-cell interactions during EB formation (58, 59), so expression of laminin ought to be repressed in a stem cell and should be tightly regulated. However, other than select cases such as laminin, there is no overall association of negative correlation with target prediction. Our results, therefore, are consistent with the conclusion that negative correlations of microRNA and mRNA do not directly predict functional targeting, but may identify valuable targeting predictions worth investigating further.
Discussion
Here we have used a combination of NCode™ microRNA array and qPCR to identify and validate microRNA differentially expressed between multiple cell states. The identified microRNA markers were quantitatively measured in five independent hESC lines using quantitative RTPCR.
Based on the expression pattern in ESC, differentiated cells and adult brain, three main groups of differentially expressed microRNA was identified. Group one microRNA (Figure 4, Panels A and B, similar to cluster 4 in Figure 5), are expressed at relatively high copy number in hESCs and then seem to be down regulated during differentiation. These microRNAs represent good candidates for markers of pluripotency and potentially negative regulators of gene expression that may play a role in restricting differentiation of hESCs. Indeed, they positively cross-correlate with Oct4 (POU5F1), Rex1 (ZFP42) and TDGF1, which may also be considered to be markers of pluripotency. Group two microRNAs (Figure 4, Panel C, also cluster 3 in Figure 5), are expressed at intermediate levels in hESC and significantly increase in copy number during differentiation. The expression level of these microRNAs is relatively low in the representative adult control tissue when compared to differentiating cells. This group positively correlates with two Hox family members, CDX2 and HOXA11, which may be transiently expressed during early differentiation and reduced afterwards. Finding similarities between the copy number and the intermediate differentiation state allows one to speculate that these molecules could represent a class of microRNAs that act to regulate differentiation of cells to one or more of the three embryonic lineages. Group three microRNAs (Figure 4, Panel C) decreased after differentiation but do not seem to be specifically associated with the embryonic stem cell state since they are also expressed at significant levels in adult tissue.
Regulation of gene expression by microRNA is clearly a complex process as indicated by the ability of an individual microRNA candidate to regulate several mRNA targets (60) or several microRNAs able to regulate a single mRNA (61). The mechanism of microRNA-mediated mRNA regulation itself might be either by inhibition of transcription due to chromatin modification, DNA methylation or direct translational inhibition of the mRNA (62-65). The latter can be due to repression or cleavage of target mRNA whereby the extent of base pairing between the microRNA and the mRNA determines the balance between cleavage and degradation of the transcript (66, 67). Considering this complexity, it is difficult to identify functionally relevant gene targets for candidate microRNAs. Despite the identification of several microRNAs in specific cell types and sequence based prediction of possible regulatory targets (61, 68-70), verification of specific activity has been difficult since there are many potential targets for each candidate microRNA. For example, miR200c, which is expressed at high levels in hESC, has 567 gene targets predicted by RNA22 (52). Of those present among the Illumina probe set, 33% were reliably detected in all hESC samples, suggesting the presence of both the microRNA and the predicted targets, providing the possibility of interaction. However, this analysis focused on only one microRNA and one condition. If we expand our view, we can group microRNAs through their association with many mRNAs under multiple conditions more specifically, analogous to the concept of “biclustering” (71). In our method, correlation of an mRNA to each microRNA expression level is used as a “condition” to help reveal associations between mRNAs. The inverse is applied to microRNAs as well. Therefore, cross-correlation clustering presents interpretable lists of microRNAs and associated mRNAs that may be hypothesized to interact through specific mechanisms. Such is the case in Figure 5, where mRNAs indicative of pluripotency are negatively associated with microRNAs predicted to target these mRNAs, and positively associated with microRNAs predicted to coordinately regulate other genes. This view of the data, while somewhat superficial, provides the initial impetus towards a systems analysis to recognize the role of microRNAs in stem cell differentiation.
In conclusion, microRNA analysis represents a relatively new tool for cell line profiling and discovery of putative regulatory molecules. In addition, it provides a method to study and dissect the epigenetic regulation mechanisms involved in maintenance and differentiation of hESCs. Such an insight is essential in developing methods to either maintain cells in their pluripotent undifferentiated state or to differentiate efficiently them into a desired lineage.
Supplementary Material
Gene expression data were modeled as described, and genes significantly different under at least one condition were correlated. A cross-correlation table (Supplemental Table 4) was plotted as a heatmap using R, including dendrograms showing relatedness among either the mRNAs or microRNAs. Names of microRNAs are shown but there are too many mRNAs to display.
(Excel file). Gene expression analysis was carried out using the Illumina Ref-8 bead chip and the signal intensity distribution of transcripts determined using the Illumina Beadstudio. The entire gene expression profile of undifferentiated and differentiated CyT25 and CyT203 are in worksheet 1 and expression data for ES2, ES3 and ES4 cells in worksheet 2. The signal intensity distribution obtained for each sample is tabulated in worksheet 3.
(Excel file). Raw intensity data obtained from the NCode microRNA array was statistically analyzed using the Loop model. The entire statistical analysis and raw intensities obtained on the NCode microRNA chip is given in worksheet 1. The global signal intensity distribution for each dye and channel is tabulated in worksheet 2.
(Excel File). The delta Ct values obtained by qPCR of the hESC and their corresponding differentiated cells were normalized to the house-keeping gene GAPDH. Shown in worksheet 1 is the values represented as fold change relative to the control cell type 2102Ep human EC cells.
(Excel File). Gene expression data were modeled using the groupings described, selected by F-test (p-values adjusted by Benjamini-Hochberg to a 5% FDR) and cross-correlated. The worksheet labeled “mRNA Cell Line Effect” contains linear model factors for each condition calculated from the quantile-normalized Illumina dataset for the 2,678 probes selected. The worksheet labeled “microRNA Cell Line Effect” contains linear model factors for each condition calculated from the mean Kerr-Churchill-corrected NCode™ dataset for each of the 55 probes selected. The worksheet labeled “corrmatrix R-values” is the correlation matrix, indexed by Illumina probe IDs (rows) by NCode™ probe IDs and microRNA names (columns).
Acknowledgments
We thank Dr Melissa Carpenter, Novocell, for providing us with the undifferentiated and differentiated cell pellets of CyT25 and CyT203 cells, Dr Chris Adams and Mark Landers, Invitrogen, for their technical help with NCode™ microRNA profiling tools and Drs. Bruce Davidson and Jeremy Crook, ES Cell International, for their excellent editorial advice. RPH was supported by grants from the New Jersey Commission on Spinal Cord Research, NIH, the New Jersey Commission on Science & Technology, and Invitrogen, Inc.
References
- 1.Rao S, Orkin SH. Unraveling the transcriptional network controlling ES cell pluripotency. Genome Biol. 2006;7:230. doi: 10.1186/gb-2006-7-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao L, Yuan X, Sharkis SJ. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 3.Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, Gonzalez R, Crook J, Davidson B, Schulz TC, Robins A, Khanna A, Sartipy P, Hyllner J, Vanguri P, Savant-Bhonsale S, Smith AK, Chakravarti A, Maitra A, Rao M, Barker DL, Loring JF, Fan JB. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- 5.Ciaudo C, Bourdet A, Cohen-Tannoudji M, Dietz HC, Rougeulle C, Avner P. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2006;2:e94. doi: 10.1371/journal.pgen.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Thykjaer T, Maltby E, Smith K, Dawud RA, Jones M, Matin M, Gokhale P, Draper J, Andrews PW. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 7.Brimble SN, Zeng X, Weiler DA, Luo Y, Liu Y, Lyons IG, Freed WJ, Robins AJ, Rao MS, Schulz TC. Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 2004;13:585–597. doi: 10.1089/scd.2004.13.585. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 9.Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya B, Cai J, Luo Y, Miura T, Mejido J, Brimble SN, Zeng X, Schulz TC, Rao MS, Puri RK. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and differentiating embryoid bodies. BMC Dev Biol. 2005;5:22. doi: 10.1186/1471-213X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Shin S, Zeng X, Zhan M, Gonzalez R, Mueller FJ, Schwartz CM, Xue H, Li H, Baker SC, Chudin E, Barker DL, McDaniel TK, Oeser S, Loring JF, Mattson MP, Rao MS. Genome wide profiling of human embryonic stem cells (hESCs), their derivatives and embryonal carcinoma cells to develop base profiles of U.S. Federal government approved hESC lines. BMC Dev Biol. 2006;6:20. doi: 10.1186/1471-213X-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 13.Skottman H, Mikkola M, Lundin K, Olsson C, Stromberg AM, Tuuri T, Otonkoski T, Hovatta O, Lahesmaa R. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–1356. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- 14.Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, Thies S, Wang W, Khrebtukova I, Zhou D, Liu ET, Ruan YJ, Rao M, Lim B. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 15.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 16.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, Kim VN, Kim KS. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 21.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 22.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 23.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 24.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, Pan X, Anderson TA. MicroRNA: A new player in stem cells. J Cell Physiol. 2006;209:266–269. doi: 10.1002/jcp.20713. [DOI] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell. 2005;20:3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 36.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 38.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 39.Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007 doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goff LA, Yang M, Bowers J, Getts RC, Padgett RW, Hart RP. Rational probe optimization and enhanced detection strategy for microRNAs using microarrays. RNA Biology. 2005;2:e9–e16. doi: 10.4161/rna.2.3.2059. [DOI] [PubMed] [Google Scholar]
- 45.D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 46.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 47.Costa M, Dottori M, Ng E, Hawes SM, Sourris K, Jamshidi P, Pera MF, Elefanty AG, Stanley EG. The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nat Methods. 2005;2:259–260. doi: 10.1038/nmeth748. [DOI] [PubMed] [Google Scholar]
- 48.Lakshmipathy U, Love B, Adams C, Thyagarajan B, Chesnut JD. MicroRNA Profiling: An easy and rapid method to screen and characterize stem cell populations. In: V MC, editor. Stem Cell Assays. Humana Press; New Jersey: 2006. [DOI] [PubMed] [Google Scholar]
- 49.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics (Oxford, England) 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 50.Cui X, Kerr MK, Churchill GA. Transformations for cDNA microarray data. Statistical applications in genetics and molecular biology. 2003;2 doi: 10.2202/1544-6115.1009. Article4. [DOI] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289. [Google Scholar]
- 52.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 53.Cai J, Chen J, Liu Y, Miura T, Luo Y, Loring JF, Freed WJ, Rao MS, Zeng X. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells. 2006;24:516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2006 doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 55.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007 doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 56.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9:1274. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Josephson R, Ording CJ, Liu Y, Shin S, Lakshmipathy U, Toumadje A, Love B, Chesnut JD, Andrews PW, Rao MS, Auerbach JM. Comprehensive characterization of a reference line for human embryonic stem cell research-2102Ep human embryonal carcinoma line. Stem Cells. 2006 doi: 10.1634/stemcells.2006-0236. In press. [DOI] [PubMed] [Google Scholar]
- 58.Colognato H, Yurchenco PD. Form and function: the laminin family of heterotrimers. Dev Dyn. 2000;218:213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 59.Li S, Harrison D, Carbonetto S, Fassler R, Smyth N, Edgar D, Yurchenco PD. Matrix assembly, regulation, and survival functions of laminin and its receptors in embryonic stem cell differentiation. The Journal of cell biology. 2002;157:1279–1290. doi: 10.1083/jcb.200203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 61.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 62.Hall KB. RNA-protein interactions. Curr Opin Struct Biol. 2002;12:283–288. doi: 10.1016/s0959-440x(02)00323-8. [DOI] [PubMed] [Google Scholar]
- 63.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. Embo J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 66.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 68.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smalheiser NR, Torvik VI. A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions. BMC Bioinformatics. 2004;5:139. doi: 10.1186/1471-2105-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Y, Church GM. Biclustering of expression data. Proceedings / International Conference on Intelligent Systems for Molecular Biology; ISMB. 2000;8:93–103. [PubMed] [Google Scholar]
- 72.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 73.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression data were modeled as described, and genes significantly different under at least one condition were correlated. A cross-correlation table (Supplemental Table 4) was plotted as a heatmap using R, including dendrograms showing relatedness among either the mRNAs or microRNAs. Names of microRNAs are shown but there are too many mRNAs to display.
(Excel file). Gene expression analysis was carried out using the Illumina Ref-8 bead chip and the signal intensity distribution of transcripts determined using the Illumina Beadstudio. The entire gene expression profile of undifferentiated and differentiated CyT25 and CyT203 are in worksheet 1 and expression data for ES2, ES3 and ES4 cells in worksheet 2. The signal intensity distribution obtained for each sample is tabulated in worksheet 3.
(Excel file). Raw intensity data obtained from the NCode microRNA array was statistically analyzed using the Loop model. The entire statistical analysis and raw intensities obtained on the NCode microRNA chip is given in worksheet 1. The global signal intensity distribution for each dye and channel is tabulated in worksheet 2.
(Excel File). The delta Ct values obtained by qPCR of the hESC and their corresponding differentiated cells were normalized to the house-keeping gene GAPDH. Shown in worksheet 1 is the values represented as fold change relative to the control cell type 2102Ep human EC cells.
(Excel File). Gene expression data were modeled using the groupings described, selected by F-test (p-values adjusted by Benjamini-Hochberg to a 5% FDR) and cross-correlated. The worksheet labeled “mRNA Cell Line Effect” contains linear model factors for each condition calculated from the quantile-normalized Illumina dataset for the 2,678 probes selected. The worksheet labeled “microRNA Cell Line Effect” contains linear model factors for each condition calculated from the mean Kerr-Churchill-corrected NCode™ dataset for each of the 55 probes selected. The worksheet labeled “corrmatrix R-values” is the correlation matrix, indexed by Illumina probe IDs (rows) by NCode™ probe IDs and microRNA names (columns).