Abstract
Androgens are an important output of the hypothalamic-pituitary-gonadal (HPG) axis that controls reproduction in all vertebrates. In male teleosts two androgens, testosterone and 11-ketotestosterone, control sexual differentiation and development in juveniles and reproductive behavior in adults. Androgenic signals provide feedback at many levels of the HPG axis, including the hypothalamic neurons that synthesize and release gonadotropin-releasing hormone 1 (GnRH1), but the precise cellular site of androgen action in the brain is not known. Here we describe two androgen receptor subtypes, ARα and ARβ, in the cichlid Astatotilapia burtoni and show that these subtypes are differentially located throughout the adult brain in nuclei known to function in the control of reproduction. ARα was expressed in the ventral part of the ventral telencephalon, the preoptic area (POA) of the hypothalamus and the ventral hypothalamus, whereas ARβ was more widely expressed in the dorsal and ventral telencephalon, the POA, and the ventral and dorsal hypothalamus. We provide the first evidence in any vertebrate that the GnRH1-releasing neurons, which serve as the central control point of the HPG axis, express both subtypes of AR. Using quantitative real-time PCR, we show that A. burtoni AR subtypes have different expression levels in adult tissue, with ARα showing significantly higher expression than ARβ in the pituitary, and ARβ expressed at a higher level than ARα in the anterior and middle brain. These data provide important insight into the role of androgens in regulating the vertebrate reproductive axis.
Keywords: in situ hybridization, gonadotropin-releasing hormone, androgen receptor subtypes, teleost
Androgens are one endocrine output of the hypothalamic-pituitary-gonadal (HPG) axis that controls reproduction in all vertebrates. In male fish, the major androgens testosterone and 11-ketotestosterone (11-KT) are important for regulating sexual differentiation and functional development at all levels of the HPG axis (Borg, 1994). For example, exogenous androgens accelerate gonadal differentiation, spermatogenesis, and maturation of the hypothalamic and pituitary cells that synthesize and release the signaling peptides to mediate reproductive function (Amano et al., 1994; Goos et al., 1986; Miura et al., 1991a; Montero et al., 1995; Piferrer et al., 1993; Schreibman et al., 1986). If administered early enough in development in some teleost species, sex steroids can induce complete gonadal sex inversion in either direction (Hunter and Donaldson, 1983). In addition androgens also influence the adult HPG axis, mediating important social and reproductive behaviors, including courtship, territoriality, and aggression (Borg, 1994; Breton and Sambroni, 1996; Trudeau et al., 1991).
Many physiological actions of androgens are mediated by the androgen receptor (AR) family. As with other steroid hormone receptors, ARs are ligand-dependent transcription factors, containing a highly conserved DNA-binding domain (DBD) and a moderately well conserved ligand-binding domain (LBD). After ligand binding, AR monomers undergo conformational change, dissociate from chaperone proteins (mostly of the heat-shock protein family), dimerize, and bind coactivator proteins (Bohen et al., 1995; Pratt and Toft, 1997). Steroid receptor dimerization, which induces activation of the receptor, is followed by translocation of the complex to the nucleus, where ARs recognize and bind to hormone response elements (HRE) in androgen-sensitive genes (Gelmann, 2002; Whitfield et al., 1999).
Having multiple nuclear steroid receptor isoforms within one species is a common theme among mammals and teleosts alike (Hawkins et al., 2000). For example, the cichlid fish Astatotilapia burtoni has three isoforms of glucocorticoid receptor, GR1, GR2a, and GR2b (Greenwood et al., 2003) as well as three subtypes of estrogen receptor (ERα, ERβa, and ERβb; Burmeister et al., 2006). To date, multiple isoforms of the AR have been described in several fish species, including rainbow trout and Japanese eel, which each have two isoforms, ARα and ARβ (Ikeuchi et al., 1999; Takeo and Yamashita, 1999, 2000; Todo et al., 1999). Biochemical characterization has also identified two forms of AR (AR1 and AR2) in the Atlantic croaker and Kelp bass (Sperry and Thomas, 1999a,b). Most teleost ARs display high testosterone affinity and significantly lower 11-KT affinity, as shown by binding studies (Pasmanik and Callard, 1988; Pottinger, 1987; Slater et al., 1995), and the ARα/AR1 subtypes show a similar ligand-binding profile (Ikeuchi et al., 1999; Sperry and Thomas, 1999a,b, 2000; Takeo and Yamashita, 2000; Todo et al., 1999). In contrast, ARβ/AR2 subtypes in the Japanese eel, Atlantic croaker, and kelp bass demonstrate high binding affinity for a broader range of natural androgens (Ikeuchi et al., 1999; Sperry and Thomas, 1999a,b, 2000). In males of many fish species, the circulating levels of 11-KT are higher than levels of other androgens, and 11-KT, rather than testosterone, appears to be the major influence on many androgen-dependent changes in male reproductive state (Borg, 1994). Recently an AR subtype (ARβ2) that is preferentially activated by 11-KT compared with testosterone has been identified in the male stickle-back (Olsson et al., 2005).
The number and functional diversity of teleost AR subtypes is particularly intriguing given the number of fish species that show androgen-induced sex-changing ability and multiple sexual or social phenotypes correlated with circulating androgen levels (Hirschenhauser et al., 2004; Oliveira et al., 1996, 2002). For example, reproductively competent, territorial (T) male A. burtoni have high circulating levels of both testosterone and 11-KT, whereas non-territorial (NT) males, which are not reproductively competent, have lower circulating levels of both androgens compared with T males (Burmeister et al., 2005; Fernald, 1977; Fernald and Hirata, 1977; Francis et al., 1993; Parikh et al., 2005; White et al., 2002) so that the social system of A. burtoni provides a unique opportunity to investigate the role of androgens in the regulation of reproductive capacity.
Androgen production is controlled by the hypothalamic neurons that synthesize and release gonadotropin-releasing hormone 1 (GnRH1) and serve as the final control point of the HPG axis to regulate reproduction in all vertebrates. GnRH1 induces the pituitary to release gonadotropins (GtH), which in turn act on the gonads to regulate gametogenesis and gonadal steroidogenesis and hence reproductive capacity (Kalra and Kalra, 1994). In A. burtoni males, the soma size of GnRH1-releasing neurons, as well as the amount of GnRH1 synthesized by these neurons, changes as a function of social status (Davis and Fernald, 1990; White et al., 2002). Increased cell size and GnRH1 production in T males induced by social ascent causes up-regulation of the entire HPG axis, and consequent increased reproductive capacity (Francis et al., 1993; Parikh et al., 2005). Although it has long been known that androgenic signals provide feedback to the pituitary and brain, the precise effect of androgens on the HPG axis at the level of GnRH1-releasing neurons, both in mammals and in fish, appears to depend on life stage. In juvenile fish of many species, exogenous androgens increase GnRH1 expression in the hypothalamus (Amano et al., 1994; Dubois et al., 1998; Montero et al., 1995). In adults, however, castration of adult T A. burtoni males, which dramatically reduces the levels of circulating testosterone and 11-KT (Francis et al., 1993), results in a further increase in the size of the GnRH1-releasing neurons in these animals, suggesting that androgens are normally a limiting factor in GnRH1 neuron size (Soma et al., 1996). The hypertrophy of the GnRH1 neuron soma size in castrated T A. burtoni, can be reversed by exogenous treatment with either testosterone or 11-KT, which demonstrates that T males have large GnRH1 neurons despite high circulating androgens and must therefore be under the influence of a socially induced trophic signal (see the discussion of the “social set point” by Soma et al., 1996). Similarly, exogenous testosterone decreases GnRH1 mRNA levels in castrated males of the closely related Nile tilapia (Soga et al., 1998). These data have led to the hypothesis of a socially regulated negative feedback control of gonadal steroid levels on GnRH1-releasing neurons in adult A. burtoni.
However, the precise sites of androgen action in the teleost brain are not known. One hypothesis is that androgen effects on GnRH1-releasing neurons are indirect, via androgen-sensitive interneurons, or following aromatization of testosterone to estrogen (Belsham and Lovejoy, 2005). Estrogen alone has no effect on GnRH1-neuron soma size in A. burtoni, whereas the nonaromatizable androgen 11-KT is as effective as testosterone in reversing the hypertrophy induced by castration (Soma et al., 1996). Because GnRH1 release from dispersed teleost brain cultures can be modulated by both testosterone and 11-KT (Lee et al., 2004), and A. burtoni GnRH1 contains several putative hormone response elements (HREs; White and Fernald, 1998), it is possible that endogenous androgen action on the HPG axis is mediated by AR expressed on the GnRH1-releasing neurons themselves. The precise cellular and tissue distributions of AR subtypes within one species are unknown, so we have localized multiple AR subtypes in A. burtoni. Because social regulation of maturation and reproduction occurs in many species, discovering the cellular circuits responsible for androgen action in A. burtoni will be an important step in understanding how social information is translated into physiological change.
MATERIALS AND METHODS
Animals
A. burtoni were taken from a laboratory breeding population derived from wild-caught stock originating in Lake Tanganyika, Africa (Fernald and Hirata, 1977). The animals were kept under conditions matching their natural environment and were maintained according to the animal care guidelines of Stanford University. Aquaria were filled with treated water (Cichlid Lake Salt and Tanganyika Buffer; Seachem Laboratories, Stone Mountain, GA) to maintain pH at 8.6 and carbonate hardness of 170 mg/liter CaCO3. Temperature was 29°C with a 12-hour light/12-hour dark cycle with full-spectrum illumination (Vita-Lite; Duro-Test, Fairfield, NJ). Fish were fed once daily at 9:00–9:30 AM with cichlid formula pellets and flakes (Aquadine, Healdsburg, CA). Gravel and terra cotta pot shards were provided to allow the males to establish and maintain territories, which is an integral component of their reproductive and social behavior (Fernald, 1977). Focal observations were performed to determine the dominance status of individual males over a period of >3 weeks using previously established criteria (Fernald, 1977).
AR sequence
To identify the ARα sequence, total RNA from the whole brain of an adult male A. burtoni was prepared (Ultraspec-II RNA; Biotecx Laboratories, Houston, TX). The RNA was treated (DNAse I, proteinase K), and 2 μg per reaction was reverse transcribed (Superscript II RT; Life Technologies, Gaithersburg, MD) as recommended by the manufacturer, except that oligo-(dT18) priming was used. Polymerase chain reaction (PCR) was used to amplify the A. burtoni ARα with degenerate primers. Degenerate primer sequences were identified from previously cloned fish androgen receptors (rainbow trout ARα and red sea bream AR; see Table 1) using the web-based COnsensus-DEgenerate Hybrid Oligonucleotide Primers program (CODEHOP; http://blocks.fhcrc.org/blocks/codehop.html). The 5′ degenerate primer sequence was GGYAGCTGCAARGTNTTYTTYAA and the 3′ degenerate primer sequence was MACAAGRTCYGGRGCRAARTANA. PCR was done in 20-μl glass microcapillary tubes (Rapidcycler; Idaho Technologies, Idaho Falls, ID) with a touchdown cycling protocol as follows: an initial 15-second denaturation at 94°C, followed by 40 cycles of a 15-second hold at 94°C, a 15-second primer annealing step at 55°C (every five cycles the annealing temperature was lowered 1°C), and a 15-second extension step at 72°C. Klentaq1 DNA polymerase was used with supplied buffer (AB Peptides, St. Louis, MO), prebound with TaqStart Antibody (Clontech, Palo Alto, CA) for an automatic hot-start to reduce nonspecific priming. Fluorescent DNA sequencing was performed by dideoxy termination methods with automated base calling. To obtain the complete cDNA, 5′- and 3′-RACE was performed on brain total RNA (Marathon cDNA Amplification Kit; Clontech). Based on the cDNA sequence amplified by the degenerate primers, unique A. burtoni androgen receptor primers (3′-RACE TGTCATGGATGGGGGTGATGGTGTTTGC and 5′-RACE AGGTCCCGAAACCCTGGTATTGCCTTGG) were designed (Oligo; Molecular Biology Insights, Cascade, CO). The RACE products from this cDNA were sequenced and were found to contain the start and end codons. New primers, specific for the 5′- and 3′-UTRs of the A. burtoni ARα, were then used to obtain the full-length cDNA.
TABLE 1.
Species and Genbank Accession Number for AR Genes Used in Phylogenetic Comparison
| Scientific name | Common name | Gene | Genbank accession No. |
|---|---|---|---|
| Anolis carolinensis | Green anole | Androgen receptor | AAF28356 |
| Pimephales promelas | Fathead minnow | Androgen receptor | AAF88138 |
| Xenopus laevis | African claw frog | Androgen receptor | CAA41726 |
| Carassius auratus | Goldfish | Androgen receptor | AAM09278 |
| Homo sapiens | Human | Androgen receptor | AAD45921 |
| Cotournix japonica | Japanese quail | Androgen receptor | BAD38679 |
| Gambusia affinis | Mosquitofish | Androgen receptor alpha | BAD81045 |
| Gambusia affinis | Mosquitofish | Androgen receptor beta | BAD81046 |
| Mus musculus | House mouse | Androgen receptor | NP 038504 |
| Rattus norvegicus | Norweigen rat | Androgen receptor | AAA40759 |
| Pagrus major | Red seabream | Androgen receptor | BAA33451 |
| Halichoeres trimaculatus | Threespot wrasse | Androgen receptor | AAG48340 |
| Danio rerio | Zebrafish | Androgen receptor | AAS80170 |
| Oryzias latipes | Japanese medaka | Androgen receptor alpha | BAC98301 |
| Gasterosteus aculeatus | Three spined stickleback | Androgen receptor beta 2 | AAO83572 |
| Anguilla japonica | Japanese eel | Androgen receptor alpha | BAA75464 |
| Anguilla japonica | Japanese eel | Androgen receptor beta | BAA83805 |
| Oreochromis niloticus | Nile tilapia | Androgen receptor alpha | BAB20081 |
| Oreochromis niloticus | Nile tilapia | Androgen receptor beta | BAB20082 |
| Oncorhynchus mykiss | Rainbow trout | Androgen receptor alpha | BAA32784 |
| Oncorhynchus mykiss | Rainbow trout | Androgen receptor beta | BAA32785 |
| Salmo salar | Atlantic Salmon | Androgen receptor beta | AAL29928 |
| Astatotilapia burtoni | Burton’s mouthbrooder | Androgen receptor alpha | AAD25074 |
| Astatotilapia burtoni | Burton’s mouthbrooder | Androgen receptor beta | AAL92878 |
| Homo sapiens | Human | Progesterone receptor | AAA60081 |
To identify ARβ, we designed primers to recognize O. niloticus ARβ (3′ CCAGCCATTCGCCGACCTC, 5′ CCTTTTCTGCCTGCGCCACAA). Based on the sequence amplified, specific A. burtoni primers (5′-RACE TGAACAGTGGTACCC TGGCGGAATGCTG, 3′-RACE CTCGTGTCA CCGGACTCCATGGTCAGAC) were used to generate RACE products, and these in turn were subsequently used to design primers to amplify a full-length transcript of 2,290 bp.
Phylogenetic analysis
Translated protein sequences based on A. burtoni ARα and ARβ gene sequences were compared with AR sequences from 23 additional androgen receptors and one progesterone receptor using ClustalW (ClustalW 1.8; http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html; BCM Search Launcher). Full species names and GenBank accession numbers for the receptor cDNAs are shown in Table 1. Human progesterone receptor was included as an outgroup. Phylogenetic trees were generated by using full protein sequences of the multiple species (Mega http://www.megasoftware.net/m_con_select.html) to identify relationships between the alpha and beta androgen receptors of A. burtoni and among other known sequences. Bootstrap values are shown.
Localizing AR receptors in the brain by using in situ hybridization
Gene-specific primers for amplification of ARα and ARβ were designed (Oligo; Molecular Biology Insights, Cascade, CO). The oligonucleotide sequences used are given in Table 2. Adult brain cDNA from A. burtoni was generated as described above and used as template for touchdown PCR (PCR Express, Hybaid, Basingstoke, United Kingdom) under the following conditions; an initial 3-minute 95°C denaturation step, 17 cycles of 30-second 95°C denaturation, 65°C annealing (this step reduced by 0.5°C each cycle), and 1.5-minute 72°C elongation step, 25 cycles of 30 seconds at 95°C, 30 seconds at 57°C and 1.5min at 72°C, followed by a final elongation step of 15 minutes at 72°C. The amplified AR subsequences were a 1,052-bp fragment containing parts of the hinge region and the LBD (ARα) and a 1,020-bp fragment within the hypervariable transactivation domain (ARβ). The AR fragments were cloned into the pcRTOPO-II plasmid (Invitrogen) for subsequent in vitro transcription. Radioisotopically labeled AR sense and antisense probes (see Table 2) were generated by using 35S-UTP (Amersham) and Maxiscript in vitro transcription kit (Ambion) with either T7 or SP6 polymerase. Digoxigenin (DIG)-labeled GnRH1 probe was generated as described by Chen and Fernald (2006).
TABLE 2.
A. burtoni Primers and Probes Used in This Study
| Gene | Genbank No. | Forward primer | Reverse primer | Amplicon length |
Amplicon purpose |
|---|---|---|---|---|---|
| ARα | AF121257 | GCC CAC CAG CCT GAA CGA GTT | AAG CTA CCG GGA TGT GC | 1,052 | To generate in situ hybridization probe |
| ARβ | AY082342 | TGC GGC TCA CTT AGG ACC TTT | TTG ACT TTG ACG CCG TAC C | 1,020 | To generate in situ hybridization probe |
| ARα | AF121257 | CGC TGT ATC TGG TAC GGT AG | TGA GGA ATC GCA CTT GG | 104 | To amplify gene of interest by QRT-PCR |
| ARβ | AY082342 | TTC GGC GAC AAG TAC AAC TC | ACT GTT CAC GGC GCA TTA | 124 | To amplify gene of interest by QRT-PCR |
| GnRH1 | U31865 | CAG ACA CAC TGG GCA ATA TG | GGC CAC ACT CGC AAG A | 128 | To amplify gene of interest by QRT-PCR |
| 18S | U67333 | CCC TTC AAA CCC TCT TAC CC | CCA CCG CTA AGA GTC GTA TT | 82 | To amplify gene of interest by QRT-PCR |
| Gene | Genbank No. | Antisense probe sequence | Sense probe sequence | Probe length | Probe purpose |
|---|---|---|---|---|---|
| ARα | AF121257 | CAAGCTACCGGGATGTGCTGCCTTCA AACTTGGCCCCCTGCACCGCACGCCC CAGAAGAGTTTGTGATTATCCACATG AAGAGCCGAAGAAGTTCACATGCCAT GAGGTTCACCATTAGTATGTGGACAG AAATGCATGCAAGTATTTTAATCAGC CCCCGAAAAGCATTTGTGTATGTGCG GGATACATTCAAACAGGCAATAAGAA GAAGAAGTGAGAAGATGAAATCAAAC TACATTAGACTGGAAGAATGTGCTGT CTGCCCTTCTGTTAACATCTGCTTAC ACCTTGACTGCACCCTACTCTCAGGA CAGAAGTGTAAAAATCATTGAGAACT CTTTCCGGTCGGTGTGGCTGATGAAA AGACAGTTTTGGAGACTCTCACTCAC TAGGCCGCGTTGTGGAAAAGGATGGG CTTGACCATCCCAGAGAGGATTTTGG GCACGTGGACGCTCACAATCTCCGAG ATCATCTCTGGGAAGCTGACGTGCAT TTGCATGGACTGAGCTTGGATGAACA GATCGTAGGTGAACTGGTGTAATTTC CTCACAACCGACTGGAGGTAGTCCAG CAGCTGCGTGAGCTGAAACAGTCTCT GCGTGCGGGTGGTCTCTCCGTGGTGG CTGGCCAAACGGTCTAGCTCCTTGAT GTAGGAGGTCCGCAGTTTGTCAAAAC AACTCTGGCTCTTCAGGCCCTGCACT GGCATGATGCTGAGGAGGACCAGGGC CTTCATGCAGAGGAACTCCTCCTGGG TAACCTTGAGCATGCACAGCCTTTGG GATAGTAGCTTCATCCTCACACAGTG TTCATACATGCTGGACACTTGCATCC GCTGGTCATTGAAAACCAGGTCTGGA GCAAAGTACAGCATAGAGCTGTTAGT CAGCGTGTAGGATCTCCAGCCTAAGG CAAACACCATCACCCCCATCCATGAC AGCTGAATCAGTGACATCTGGTCATC CACATACAGGTCCCGAAACCCTGGTA TTGCCTTGGCCCAGTGGACCACGGTT ACCAGCTGCCGTTCTCCCAACTCGTT CAGGCTGGTGGGC |
GCCCACCAGCCTGAACGAGTTGGGAG AACGGCAGCTGGTAACCGTGGTCCAC TGGGCCAAGGCAATACCAGGGTTTCG GGACCTGTATGTGGATGACCAGATGT CACTGATTCAGCTGTCATGGATGGGG GTGATGGTGTTTGCCTTAGGCTGGAG ATCCTACACGCTGACTAACAGCTCTA TGCTGTACTTTGCTCCAGACCTGGTT TTCAATGACCAGCGGATGCAAGTGTC CAGCATGTATGAACACTGTGTGAGGA TGAAGCTACTATCCCAAAGGCTGTGC ATGCTCAAGGTTACCCAGGAGGAGTT CCTCTGCATGAAGGCCCTGGTCCTCC TCAGCATCATGCCAGTGCAGGGCCTG AAGAGCCAGAGTTGTTTTGACAAACT GCGGACCTCCTACATCAAGGAGCTAG ACCGTTTGGCCAGCCACCACGGAGAG ACCACCCGCACGCAGAGACTGTTTCA GCTCACGCAGCTGCTGGACTACCTCC AGTCGGTTGTGAGGAAATTACACCAG TTCACCTACGATCTGTTCATCCAAGC TCAGTCCATGCAAATGCACGTCAGCT TCCCAGAGATGATCTCGGAGATTGTG AGCGTCCACGTGCCCAAAATCCTCTC TGGGATGGTCAAGCCCATCCTTTTCC ACAACGCGGCCTAGTGAGTGAGAGTC TCCAAAACTGTCTTTTCATCAGCCAC ACCGACCGGAAAGAGTTCTCAATGAT TTTTACACTTCTGTCCTGAGAGTAGG GTGCAGTCAAGGTGTAAGCAGATGTT AACAGAAGGGCAGACAGCACATTCTT CCAGTCTAATGTAGTTTGATTTCATC TTCTCACTTCTTCTTCTTATTGCCTG TTTGAATGTATCCCGCACATACACAA ATGCTTTTCGGGGGCTGATTAAAATA CTTGCATGCATTTCTGTCCACATACT AATGGTGAACCTCATGGCATGTGAAC TTCTTCGGCTCTTCATGTGGATAATC ACAAACTCTTCTGGGGCGTGCGGTGC AGGGGGCCAAGTTTGAAGGCAGCACA TCCCGGTAGCTTG |
1,052 | To localize mRNA of interest by in situ hybridization |
| ARβ | AY082342 | TTTGACTTTGACGCCGTACCGTTCTG CCTGAAACCCCGTGGACCTATTCTCC ACGGCATCCACGACGTCTCCGGCTTC CTCCACGGAGTTGTAAGCTCCGCATG GCCTCTGTGCAGCCGCGGGGCCGAAC TGTGCCAAAGGCTCCGGCGCAGCTGG GGCGTAAGGGCAAGAGGCAGCGCACG CTAAGTCCAGATGATCTATCTCTTTC GGAGTTATGTCATCAGCCAGGCACAG CGTGAAGTTTTGCGCGTTCACAGATG TCGCCGTGGAAGTGAGGTGATGAAAA GCCGCCGCGGGCTCTGAACTTTTGAA CATTTTCACCTGTTGCTTGTGGTCGT GTGGCGGGCGCTCATCTCGCTCGGGA CACCTGTAATCGGAGAGAGAAGCCGG GGCTCCGGGACAGTTCGCAGGCACGG CTTCCACCCCGAATAAATACTCTGCG CTCATTTGGTCATTTGCGGCACACGG GGGGAGAGAAACGTGCGCCTCGCTCG TGTCACCGGACTCCATGGTCAGACCC AGAGACACGGACACAGCTTTGCAGAG CTCCCTGGCTGTTTCTGAGATTGTGG CGCAGGCAGAAAAGGAGCGAATGTCG CCCACTCGGCACTCAGAGCTTAACTC CTCCTGAGGAGCAGCAGCTGTCTGGC AGCAGCGTTTCTCCATGTCACAGGCT AGCGGAATAATGTTTCCGGATCCGTA CAGGTTTGCATTCTCGGATTCTGCCA AACTCCCGGCACTGCTTCCAGTCGGG GTTTTGGTGAAATAGACGCGACTTTC TTCAGTTTTTTGCGCCATACTGGCAG CGGTCACGACGTTGCCTGTGTATGAT TTCCTATCCTCTGGCCAGATTTTGGC ACAAAATAACTGTCTGCTCGTTTGGC TCATGTCCCAGAAAGTGTTTGCTTTG CCAGGATACTGGACTTTGCACGTCAA AAAGTCTTCGCAGTTCTCAGCAGCAC AACAGCCCAACTCTCACGTTGTGAAA AGCGTTTTGGTAAAAGGTCCTAAGTG AGCCGCA |
TGCGGCTCACTTAGGACCTTTTACCA AAACGCTTTTCACAACGTGAGAGTTG GGCTGTTGTGCTGCTGAGAACTGCGA AGACTTTTTGACGTGCAAAGTCCAGT ATCCTGGCAAAGCAAACACTTTCTGG GACATGAGCCAAACGAGCAGACAGTT ATTTTGTGCCAAAATCTGGCCAGAGG ATAGGAAATCATACACAGGCAACGTC GTGACCGCTGCCAGTATGGCGCAAAA AACTGAAGAAAGTCGCGTCTATTTCA CCAAAACCCCGACTGGAAGCAGTGCC GGGAGTTTGGCAGAATCCGAGAATGC AAACCTGTACGGATCCGGAAACATTA TTCCGCTAGCCTGTGACATGGAGAAA CGCTGCTGCCAGACAGCTGCTGCTCC TCAGGAGGAGTTAAGCTCTGAGTGCC GAGTGGGCGACATTCGCTCCTTTTCT GCCTGCGCCACAATCTCAGAAACAGC CAGGGAGCTCTGCAAAGCTGTGTCCG TGTCTCTGGGTCTGACCATGGAGTCC GGTGACACGAGCGAGGCGCACGTTTC TCTCCCCCCGTGTGCCGCAAATGACC AAATGAGCGCAGAGTATTTATTCGGG GTGGAAGCCGTGCCTGCGAACTGTCC CGGAGCCCCGGCTTCTCTCTCCGATT ACAGGTGTCCCGAGCGAGATGAGCGC CCGCCACACGACCACAAGCAACAGGT GAAAATGTTCAAAAGTTCAGAGCCCG CGGCGGCTTTTCATCACCTCACTTCC ACGGCGACATCTGTGAACGCGCAAAA CTTCACGCTGTGCCTGGCTGATGACA TAACTCCGAAAGAGATAGATCATCTG GACTTAGCGTGCGCTGCCTCTTGCCC TTACGCCCCAGCTGCGCCGGAGCCTT TGGCACAGTTCGGCCCCGCGGCTGCA CAGAGGCCATGCGGAGCTTACAACTC CGTGGAGGAAGCCGGAGACGTCGTGG ATGCCGTGGAGAATAGGTCCACGGGG TTTCAGGCAGAACGGTACGGCGTCAA AGTCAAA |
1,020 | To localize mRNA of interest by in situ hybridization |
By following standard in situ hybridization procedures as modified in our laboratory (Burmeister et al., 2005; Grens et al., 2005), stable T adult males were killed by rapid decapitation, and the brain was removed and snap frozen in OCT embedding medium (Ted Pella Inc, Redding, CA). Brains were sectioned at 14 μm in the transverse plane and stored overnight at -80°C before being fixed in 4% formaldehyde in PBS (pH 7.4) for 10 minutes. Slides were washed twice in PBS, washed once in 0.1 M triethanolamine (TEA; pH 8.0), and then incubated in 0.25% acetic anhydride in TEA. After washing twice in 2× SSC (pH 7.0) buffer, slides were dehydrated in a series of ethanol dilutions and left to air dry. The labeled probes were diluted in hybridization buffer (Sigma, St. Louis, MO) to a final concentration equivalent to 5 × 105 CPM per slide (AR probes) or 2 ng/μl (GnRH1 probe). The slides were coverslipped and incubated overnight in a mineral oil bath at 60°C. After two chloroform washes to remove the mineral oil, labeled slides were washed twice in 4× SSC, washed twice in 2× SSC containing 1 mM dithiothreitol (DTT), and then incubated in 10 μg/ml RNAseA in 2× SSC-DTT at 37°C for 30 minutes. After washing twice in 2× SSC-DTT at room temperature, slides were washed in 50% formamide-2× SSC-DTT at 60°C, followed by two washes in 0.1× SSC containing 1 mM DTT at 60°C and one further wash in 0.1× SSC-DTT at RT. For double in situ hybridization studies, slides were rinsed in PBS, quenched with 3% H2O2 in PBS for 10 minutes, rinsed once in PBS and once in PBS-0.05% Tween-20 (PBS-T), blocked in PBSB (PBS containing 0.5% blocking reagent from TSA biotin kit; Perkin Elmer Life Sciences, Boston, MA) for 30 minutes, and incubated in primary antibody (anti-Digoxigenin-POD; Roche Diagnostics, Mannheim, Germany) diluted 1:250 in PBSB for 1 hour at RT. After two 5-minute washes in PBS-T, slides were incubated in biotinyl tyramide diluted 1:50 in amplification diluent (both TSA biotin kit; Perkin Elmer Life Sciences) for 10 minutes, washed twice in PBS-T, and incubated for 30 minutes in streptavidin-HRP diluted 1:100 in PBSB. After two further washes in PBS-T, slides were incubated in diaminobenzidine (DAB) solution (0.05%) in PBS with 0.012% H2O2 for 8–20 minutes and washed in PBS.
After dehydration in a series of ethanol dilutions, slides were coated with NTB-2 emulsion (Kodak), exposed at 4°C for 4 weeks, developed, and counterstained with 0.5% cresyl violet acetate. Sense probes for A. burtoni ARα and ARβ were used to test the specificity of the in situ hybridization data, and in no case was signal above background levels observed with the sense probes. A. burtoni midbrain nuclei were identified with nomenclature from Fernald and Shelton (1985), and telencephalic nuclei were identified by using zebrafish, tilapia, rainbow trout, and sea bream brain atlases (Ando et al., 1999; Munoz-Cueto et al., 2001; Pepels et al., 2002; Wullimann et al., 1996) as used by Burmeister and Fernald (2005). Brain sections were viewed (Axioscope; Carl Zeiss Inc.) and digital images captured (Spot camera; Diagnostic Instruments, Sterling Heights, MI) for subsequent analysis. The silver grains in the darkfield and merged images were pseudocolored green (Adobe Photoshop 7; Adobe Systems Inc., San Jose, CA) to facilitate demonstration of colocalization with the DAB product. The contrast of darkfield images was adjusted for clarity, and the illumination of bright-field images was equalized (Adobe Photoshop 7).
Measuring AR mRNA abundance in A. burtoni brains
Three dominant males from three different community tanks (total n = 9) were chosen for quantitative real-time polymerase chain reaction (QRT-PCR; see below) based on maintaining a consistent social status over time. Subjects were weighed and then killed by rapid decapitation, and their brains and pituitaries were collected. For QRT-PCR the brain was divided into anterior, middle, and posterior parts by sectioning the tissue, at an angle of 45° to horizontal, at the anterior commissure and between the optic tectum and cerebellum with a scalpel. As a result of the trisection, the anterior portion contained the entire telencephalon and a portion of the anterior parvocellular preoptic nucleus (aPPn). The middle portion contained the diencephalon, including the remainder of the aPPn, the pretectum, and the midbrain; the posterior portion contained the cerebellum and brainstem. We were able to determine that the anterior portion of the brain included part, but not all, of the aPPn, because both the anterior and middle portions show GnRH1 expression (see below) and GnRH1-expressing neurons are located in the most anterior part of the aPPn (Davis and Fernald, 1990; Fernald and Shelton, 1985).
After dissection, tissue was immersed in Trizol, frozen in an ethanol-dry ice bath, and stored at – 80°C until RNA extraction. For QRT-PCR, tissue was homogenized in Trizol (Invitrogen, Carlsbad, CA) and the RNA was extracted according to the standard Trizol protocol. Because of the large number of samples, we extracted RNA in multiple groups organized by tissue and tank (e.g., we extracted RNA from all the testes of males from each tank simultaneously). RNA integrity and concentration were estimated spectrophotometrically, and the RNA was treated with DNase (Turbo DNA-free; Ambion, Austin, TX) to remove contaminating genomic DNA before synthesizing cDNA with random hexamer primers and Transcriptor reverse transcription (Roche Applied Science, In-dianapolis, IN). The primers for QRT-PCR were designed by using a strategy described by Greenwood et al. (2003) that maximizes reaction efficiency, and the sequences are given in Table 2. During development of the QRT-RCR primers, QRT-PCRs were analyzed by gel electrophoresis to verify the presence of a single amplicon of the predicted size. Melt-curve analysis was also performed on a subset of PCR plates to verify that the reaction produced a single amplicon. The absence of multiple amplicons indicated that the primers bound only to single sites in the template DNA, along with the absence of contaminating genomic DNA. Negative controls (no DNA template) were performed for all QRT-PCR. The level of 18S mRNA in each sample was measured to control for variation between total RNA levels between samples. For the QRT-PCR, iQ Sybr Green Supermix (Bio-Rad Laboratories, Hercules, CA) was used with 0.5 μM of each primer. The PCR parameters were 3 minutes at 95°C, followed by 40 cycles of 95°C, 60°C, and 72°C for 30 seconds each, and PCR Miner (Zhao and Fernald, 2005) was used to calculate the reaction efficiencies and cycle thresholds from the fluorescence readings of individual wells during the reaction. The reaction efficiencies were 1.9 –2.1. For each sample, the mean cycle threshold of three PCRs was determined. The expression of each target gene of interest relative to 18S mRNA levels was calculated using the equation: relative target gene expression = 100 × (E18SCT18S)/ (EtargetCTtarget), where E was reaction efficiency and CT was cycle threshold (Pfaffl, 2001). Thus, expression of the target genes is expressed as a percentage of 18S mRNA expression. Sample sizes were 9 for each tissue and gene. In addition to the primary genes of interest, GnRH1 mRNA expression levels were measured in the anterior and middle portions of the brain to determine the location of the GnRH1-expressing neurons in the dissected brain tissue. GnRH1 mRNA expression levels were similar in the anterior and middle portions of the brain (data not shown), suggesting that the aPPn was divided between these two brain regions.
Statistical analysis
To compare the level of AR mRNA expressed in each tissue, multiple general linear models with AR subtype as a within-subject variable and with tank/RNA-extraction group as between-subject variables were used. Error attributable to the confounding variables of community tank and RNA extraction group were accounted for by including the between-subject factor “tank/group” in the analyses. The effects of status or tank/group are not reported or discussed, and in all ANOVA models the type III sums of squares was used. When comparing expression levels of the two AR subtypes in our graphs, the estimated marginal means and standard errors (which represent the effect tested by the model in which is the receptor subtype with effects of group/tank removed) are reported. Sample sizes were 9 for each tissue and gene.
RESULTS
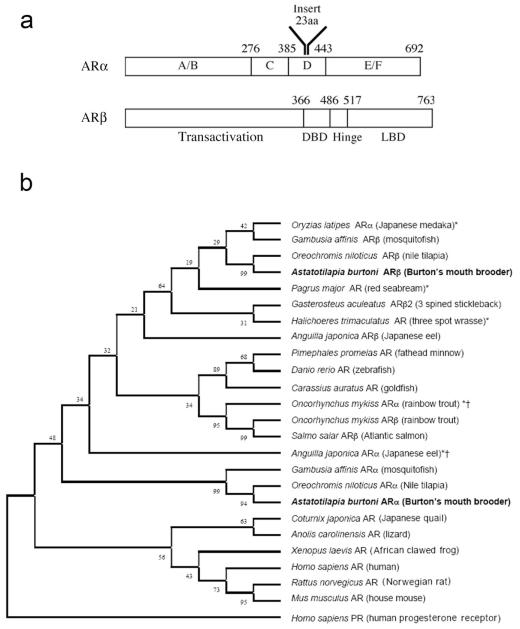
We found two transcripts in A. burtoni encoding putative androgen receptors. Both identified sequences have canonical steroid receptor domains [Krust et al., 1986; transactivation, DNA binding domain (DBD), hinge, ligand binding domain (LBD); see Fig. 1B] and, based on comparison with other androgen receptors, are designated ARα and ARβ following the convention of Ikeuchi et al. (1999). ARα encoded a predicted 692-aa protein, with a 23-aa insert in the hinge region compared with ARβ encoding a 763-aa predicted protein. Alignment of the two A. burtoni AR predicted protein sequences revealed only moderate overall homology between the transcripts and 39% identity overall (Table 3). Similar to other steroid receptors, the A. burtoni ARα and ARβ had the highest homology within the putative DBD (75% identical; Table 3). In the DBD, eight cysteine residues in two zinc-finger motifs and the P-box important for HRE recognition were completely conserved between both A. burtoni AR sub-types and human AR (hAR). Within the A. burtoni AR D-box, which plays a role in half-site spacing, the arginine residue at position 597 in hAR was substituted by a lysine. This substitution is found in some, but not all, known teleost AR subtypes. For example, sea bream AR and Nile tilapia ARβ each contain R597K, but neither Japanese eel AR subtypes have the substitution (Ikeuchi et al., 1999, 2001; Todo et al., 1999). A. burtoni ARα and ARβ each had a putative nuclear localization sequence (ARα aa 367–383 and ARβ aa 467– 483) and shared high homology within the LBD (62% identical; Table 3). The four LBD amino acids that are thought to interact directly with ligand in hAR (N705, Q711, R52, and T877) were all conserved in A. burtoni ARα and ARβ. In contrast, the A/B domain, which contains the cell-type-and promoter-specific transactivation function and is commonly the region of lowest homology between steroid receptors, was only 18% identical between ARα and ARβ (Table 3). The previously described rainbow trout isoforms (ARα and ARβ) are likely the result of salmonid tetraploidy, since they show very high overall identity (85%; Takeo and Yamashita, 1999).
Fig. 1.
a: Comparison of A. burtoni ARα and ARβ predicted protein sequences. The canonical functional steroid receptor domains (A—F) are represented schematically, and their roles are given accordingly. Numbers indicate amino acid position from the start of the coding sequence. ARα contains a 23-aa insert in the hinge region (domain C) absent in ARβ. b: Phylogenetic comparison of A. burtoni ARα and ARβ protein sequences with AR in other species. As shown, A. burtoni ARβ clusters with the other ARβ/AR2 family members identified to date in teleosts, and this AR subtype is more similar to mammalian ARs than is A. burtoni ARα. A. burtoni ARα clusters with other ARα/AR1-type receptors previously described in teleosts. ARs that are marked with an asterisk have nomenclature that does not correspond to their position in the ARβ/AR2 family, defined by their position in the phylogenetic tree. Some of these species (marked with a dagger) likely have two forms of the ARβ/AR2 family as a result of tetraploidy. Bootstrap values are shown.
TABLE 3.
Percentage Identity Between A. burtoni ARα and ARβ Predicted Protein Sequence
| Domain | Amino acid identity (%) |
|---|---|
| Entire sequence | 39 |
| A/B [Transactivation] | 18 |
| C [DNA binding] | 75 |
| D [Hinge] | 20 |
| E/F [Ligand binding] | 62 |
Sequence comparison analysis showed that each of the A. burtoni AR subtypes clustered with the appropriate teleost ARα or ARβ isotypes (Fig. 1B). Generally, teleost ARβ/2-like receptors have greater homology with mam-malian ARs than do ARα/1-like receptors (Ikeuchi et al., 1999; Sperry and Thomas, 1999a,b; Takeo and Yamashita, 1999, 2000; Todo et al., 1999), and this trend was maintained by the A. burtoni AR subtypes, illustrated by the clustering of A. burtoni ARβ closer to the mammalian ARs (Fig. 1B). The high similarity between ARβ and mammalian ARs is particularly pronounced in the ARβ DBD, which was 92% identical to the DBD of the hAR, compared with the ARα DBD, which shared only 72% identity with the equivalent region in hAR.
Expression of ARα and ARβ in the brain and pituitary of A. burtoni
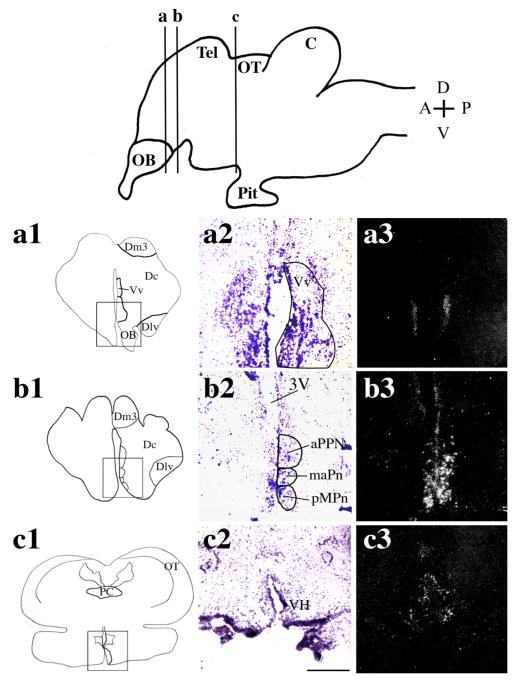
By using in situ hybridization, we found that the two AR subtypes were expressed in the A. burtoni telencephalon and diencephalon but with little detectable expression in the hindbrain. In particular ARα and ARβ expression patterns had considerable overlap within the preoptic area (POA) of the hypothalamus. ARα mRNA was expressed in the ventral portion of the ventral telencephalon (Vv; Fig. 2a2,a3). In the diencephalon, a high density of ARα-expressing neurons was located in the POA, specifically the anterior portion of the parvocellular preoptic nucleus (aPPn), and dorsally along the edge of the third ventricle (3V; Fig. 2b2,b3, arrow). ARα-expressing cells were also located throughout the ventral hypothalamus (VH; Fig. 2c2,c3), distributed over a total rostrocaudal distance of more than 400 μm.
Fig. 2.
In situ hybridization showing the distribution of ARα expression in A. burtoni brain. Cartoon of a sagittal section through the A. burtoni brain (top) shows the positions of the coronal sections shown in a1—c3. OB, olfactory bulb; Tel, telencephalon; OT, optic tectum; C, cerebellum; Pit, pituitary. Cartoon of coronal sections through the A. burtoni brain (a1,b1,c1) show the region magnified in a2,3, b2,3, and c2,3, respectively. Brightfield images (a2,b2,c2) show cresyl violet counterstain (purple) of coronal brain sections, with the brain nuclei depicted schematically. Darkfield images (a3,b3,c3) show silver grains specific for the antisense ARα probe in the corresponding nuclei. Specific expression of A. burtoni ARα can be seen in the ventral portion of the ventral telencephalon (Vv; a2,a3), anterior portion of the parvocellular preoptic nucleus (aPPn) and extending dorsally along the boundary of the third ventricle (3V; b2,b3, arrow) and scattered diffusely in the ventral hypothalamus (VH; c2,c3). Scale bar = 250 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
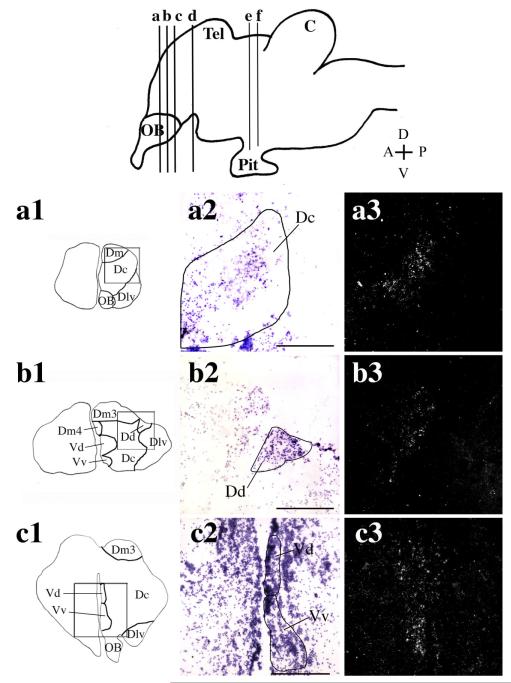
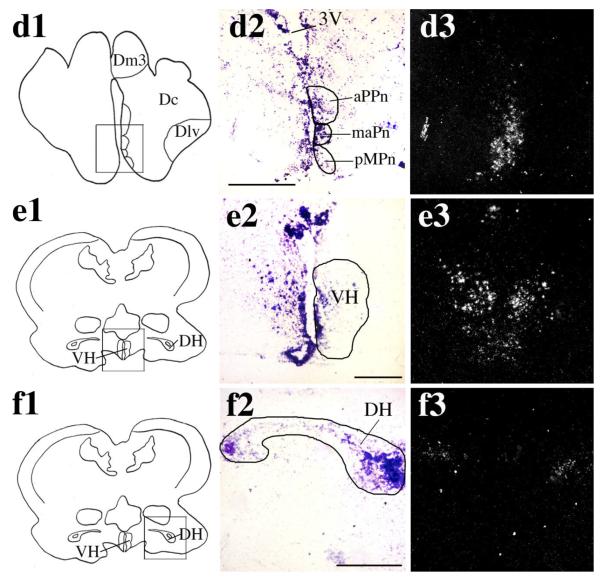
Overall, ARβ shows a more widespread distribution than ARα in the A. burtoni brain. There was dense ARβ expression in the central nucleus of the dorsal telencephalon (Dc; Fig. 3a2,a3), and more diffuse labeling at the lateral edge of part 3 of the medial zone of the dorsal telencephalon (Dm3; Fig. 3b2,b3) bordering the dorsal nucleus of the dorsal telencephalon (Dd). In both of these nuclei, ARβ-expressing neurons were distributed over a similar rostrocaudal distance (approximately 400 μm in total). ARβ-expressing neurons were also localized to the dorsal (Vd) and ventral (Vv) portions of the ventral telencephalon (Fig. 3c2,c3). In the POA, a very dense pattern of ARβ mRNA was detected in the anterior portion of the parvocellular preoptic nucleus (aPPn), the parvocellular portion of the magnocellular preoptic nucleus (pMPn), a nucleus not evident in the zebrafish brain, but with a direct anatomical equivalent in the goldfish (Bradford and Northcutt, 1983; Wullimann et al., 1996) and the magnocellular portion of the anterior preoptic nucleus (maPn Fig. 3d2,d3). Unlike ARα expression, ARβ expression does not extend dorsally from the aPPn along the edge of 3V (compare Fig. 2b2,b3 with Fig. 3d2,d3). ARβ-expressing neurons were also detected in the ventral hypothalamus (VH; Fig. 3e2,e3) and, to a lesser extent, in the dorsal hypothalamus (DH; Fig. 3f2,f3).
Fig. 3.
In situ hybridization showing the distribution of ARβ in A. burtoni brain. Cartoon of a sagittal section through the A. burtoni brain (top) shows the positions of the coronal sections shown in panels a1—h3. OB, olfactory bulb; Tel, telencephalon; OT, optic tectum; C, cerebellum; Pit, pituitary. Cartoon of coronal sections through the A. burtoni brain (a1,b1,c1,d1,e1,f1,g1,h1) show the region magnified in a2,3, b2,3, c2,3, d2,3, e2,3, f2,3, g2,3, and h2,3, respectively. Brightfield images (a2,b2,c2,d2,e2,f2,g2,h2) show cresyl violet counterstain (purple) of coronal sections, with the corresponding brain nuclei depicted schematically. Darkfield images (a3,b3,c3,d3,e3,f3,g3,h3) show silver grains specific for the antisense ARβ probe in the corresponding nuclei. Specific A. burtoni ARβ expression can be seen in the central nucleus of the dorsal telencephalon (Dc; a2,a3), at the lateral edge of part 3 of the medial zone of the dorsal telencephalon (Dm3; b2,b3), dorsal (Vd) and ventral (Vv) portion of the ventral telencephalon (c2,c3), in the anterior portion of the parvocellular preoptic nucleus (aPPn), the magnocellular portion of the anterior preoptic nucleus (maPn) and the parvocellular portion of the magnocellular preoptic nucleus (pMPn; d2,d3). More posterior sections through the A. burtoni brain reveal ARβ expression in the ventral (VH; e2,e3) and dorsal (DH; f2,f3) hypothalamus. Scale bars = 250 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Both ARα and ARβ mRNA were detected in the A. burtoni pituitary, with a similarly striking expression pattern. Expression of ARα (Fig. 4a1,a2) and ARβ (Fig. 4b1,b2) is limited to the area of the ventral pituitary that corresponds to the pars distali, with no expression in the dorsal portion. Coronal sections shown in Figure 4 are through the posterior pituitary.
Fig. 4.
In situ hybridization showing the distribution of ARα (a1,a2) and ARβ (b1,b2) in the A. burtoni pituitary. Brightfield images (a1,b1) show cresyl violet counterstain (purple) and silver grains (black spots) of coronal sections through the posterior pituitary, and darkfield images. b1 and b2 show silver grains specific for antisense ARα (a2) and ARβ (b2). Expression of both A. burtoni ARα (a1,a2) and ARβ (b1,b2) is seen restricted to the ventral pituitary. Scale bars = 250 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Colocalization of AR mRNA with GnRH1-releasing neurons
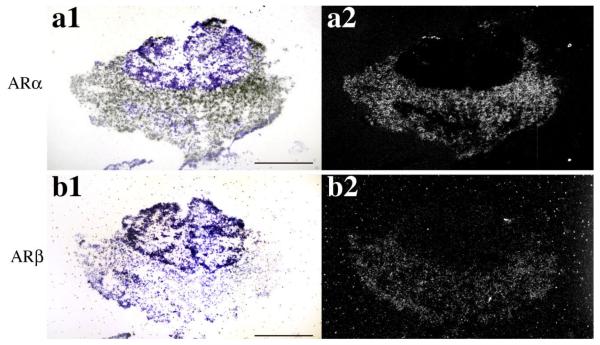
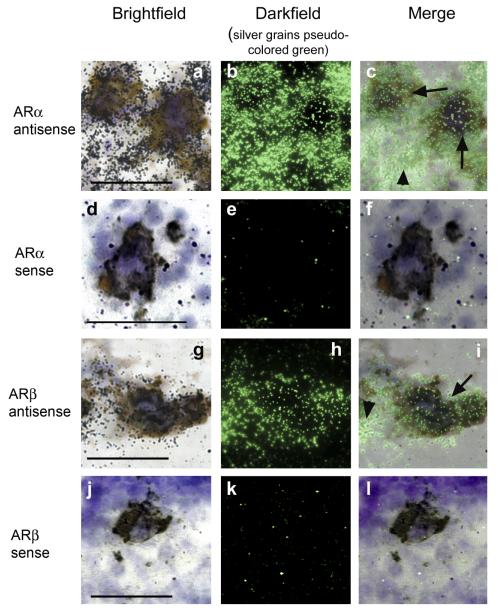
As described above, androgen levels influence reproductive competence via feedback on GnRH1 release, and, as shown, both ARα and ARβ expression levels are high in aPPn of A. burtoni, where GnRH1-releasing neurons are located (Chen and Fernald, 2006; White and Fernald, 1998). Consequently, we mapped the distribution of AR mRNA in relation to the GnRH1-releasing neurons. A double in situ hybridization for GnRH1 and ARα or ARβ revealed that GnRH1-releasing neurons in the aPPn of A. burtoni expressed both ARα (Fig. 5c, arrows) and ARβ (Fig. 5i, arrows; note that the silver grains in Fig. 5 have been psuedocolored green in Fig. 5 to facilitate demonstration of colocalization). Expression of either AR mRNA subtype in the aPPn was not limited to the GnRH1-releasing neurons; surrounding cells also expressed both (Fig. 5, arrowheads). It should be noted that the non-GRH1-releasing neurons were more densely labeled with probes for both AR subtypes, suggesting that the GnRH1-releasing neurons express ARα and ARβ at lower levels than the surrounding cells. Double in situ hybridization with the equivalent AR sense probes (Fig. 5d-f,j-l) demonstrates that the ARα antisense signal (Fig. 5a-c) and the ARβ antisense signal (Fig. 5g-i) that is colocalized with GnRH1 are well above background levels. To our knowledge, this is the first description of coexpression of ARs and GnRH1 in a teleost brain.
Fig. 5.
Double in situ hybridization showing coexpression pattern of GnRH1 with ARα (a—c) and ARβ (g—i) in the POA of A. burtoni. Brightfield images (a,d,g,j) show DAB-labeled GnRH1 mRNA (brown), cresyl violet counterstain (purple) and silver grains (black spots). Darkfield (b,e,h,k) and merged (c,f,i,l) images show silver grains pseuocolored in green to facilitate demonstration of colocalization. Both A. burtoni ARα (c) and ARβ (i) are expressed in GnRH1-releasing neurons (c, i, arrows) and surrounding neurons (c, i, arrowheads). Double in situ hybridization with the equivalent sense probes for ARα (d—f) and ARβ (j—l) demonstrate the specificity of the antisense probe above background. Scale bars = 50 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Relative expression levels of A. burtoni ARα and ARβ mRNA in the brain and pituitary
In situ hybridization data suggested that the level of ARα expression in the A. burtoni pituitary was higher than that of ARβ but that ARβ, which was more widespread throughout the telencephalon and diencephalon, had higher expression levels in these regions of the brain. By using QRT-PCR, we found that expression of ARβ mRNA was significantly higher than expression of ARα in the anterior (F1,6 = 95.1, P < 0.001) and middle (F1,6 = 162.9, P < 0.001) A. burtoni brain (Fig. 6a,b), which is consistent with the observation that ARβ is expressed in more nuclei throughout the telencephalon and diencephalon by in situ hybridization. In contrast, ARα expression in the A. burtoni pituitary (Fig. 6d) was significantly higher than ARβ (F1,6 = 45.2, P = 0.001). Our in situ hybridization studies did not reveal AR signal above background in the posterior A. burtoni brain for either transcript (data not shown). The more sensitive technique of QRT-PCR, however, detected very low expression of AR subtypes in the posterior brain compared with other brain regions (Fig. 6c), with ARα mRNA expressed at higher levels in the posterior brain than ARβ (F1,6 = 24.5, P = 0.003). This is the first report of differential expression levels of AR subtypes in one species.
Fig. 6.
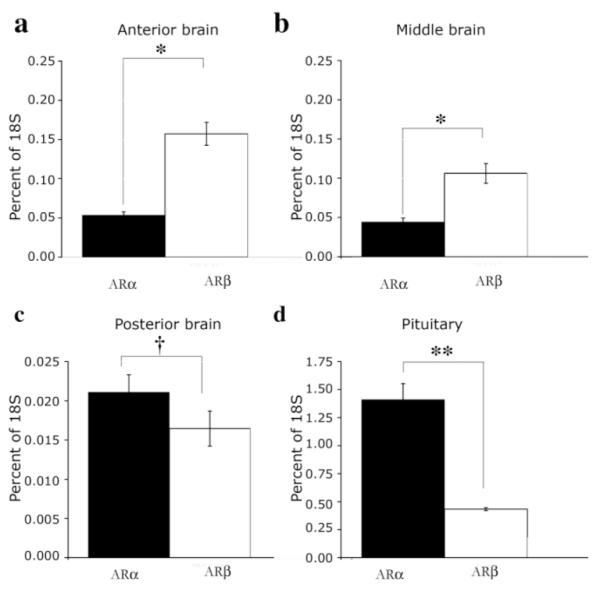
Relative expression levels of ARα and ARβ in brain and pituitary of A. burtoni. QRT-PCR reveals that ARα and ARβ are differentially expressed in regions of the brain (see Materials and Methods) and in the pituitary of adult A. burtoni males. ARβ mRNA is expressed at significantly higher levels than ARα in the anterior (a) and middle (b) brain regions (*P < 0.001), but ARα expression is significantly higher than ARβ expression in the pituitary (d, **P = 0.001). Although AR expression levels in the posterior brain are low compared with those in other brain regions (c), expression of ARα is significantly higher than ARβ in this area (†P = 0.003). Data are plotted as mean percentage of 18s mRNA expression as a control for variation in total mRNA levels between samples, with standard errors designated by error bars.
DISCUSSION
Here we identify two AR subtypes expressed by A. burtoni and demonstrate that phylogenetic analysis classifies the predicted A. burtoni AR proteins as ARα and ARβ. We provide a detailed description of the expression patterns of the two AR subtypes in the brain and pituitary of A. burtoni. We show for the first time in a teleost that GnRH1 and both AR types are coexpressed by neurons in the POA and that AR subtypes within one species are differentially expressed in the brain and pituitary.
A. burtoni express two AR subtypes
The two predicted A. burtoni AR proteins likely arise from different genes. Both the comparatively low homology between ARα and ARβ and the observation that these amino acid differences are distributed throughout the protein suggest that these receptors are unlikely to be the result of tetraploidy or alternative splicing of a single gene product. The presence of two subtypes of AR in A. burtoni was not unexpected given the number of teleost species that express multiple AR isoforms. In general, teleost ARs of the ARα/AR1 family show high ligand binding affinity for testosterone and its metabolite DHT, but lower affinity for 11-KT, whereas ARβ/AR2-like receptors tend to have a high affinity for a broad range of androgens (Ikeuchi et al., 1999; Sperry and Thomas, 1999a,b, 2000; Takeo and Yamashita, 2000; Todo et al., 1999). In cases of gene duplication, one or both of the subtypes can assume one of three fates; become nonfunctional, assume a related but different function (subfunctionalization), or assume a new, less related function (neofunctionalization; Postlethwait et al., 2004). The expression of A. burtoni ARα and ARβ in overlapping regions of the brain, particularly their coexpression on GnRH1-releasing neurons, suggests that the two subtypes are subfunctionalized. Few studies have directly compared the ligand-induced transactivation profiles of AR subtypes in any one species, but it has recently been shown that the stickleback ARβ2 is preferentially activated by 11-KT compared with testosterone (Olsson et al., 2005). A. burtoni ARβ is very closely related to the stickleback ARβ2 (for example 89.9% identical in the LBD) as evidenced by their proximity in the phylogenetic tree presented in Figure 1a. This raises the intriguing possibility that A. burtoni ARβ shows preferential activity with 11-KT, but ARα exhibits ARα/AR1-like characteristics in its preferential activation by testosterone. For teleosts, data suggest that 11-KT is the major physiological androgen in the control of male reproduction (Borg, 1994). For example 11-KT is far more potent than testosterone at inducing male sexual differentiation in female salmonids (Piferrer et al., 1993), and, of all androgens tested, only 11-KT can induce all stages of spermatogenesis in the Japanese eel (Miura et al., 1991a,b). In A. burtoni males of either social status, the levels of circulating 11-KT are an order of magnitude lower than those of testosterone (Parikh et al., 2005). Teleost sex steroid binding protein (SBP), identified in numerous teleost species to date, reduces the effectiveness of sex steroids and is known to have a higher affinity for testosterone than for 11-KT (Borg, 1994). It is therefore possible that the circulating level of available 11-KT in male teleosts may be effectively greater relative to that of testosterone. Preliminary data on A. burtoni suggest that a dominance hierarchy within the territorial male population may correlate with 11-KT, but not testosterone (W.J. Korzan and R.D. Fernald, unpublished results).
Expression of two AR subtypes throughout the brain and pituitary of A. burtoni
We found ARα and ARβ mRNA in distinct nuclei throughout the A. burtoni telencephalon and diencephalon, with ARβ expressed more widely and at higher levels than ARα. Many of the brain areas for which we report AR expression, including Dc, Vv, POA, and Vd, maintain strong reciprocal interactions in a variety of fish species (Folgueira et al., 2004a,b; Rink and Wullimann, 2004), suggesting multiple levels of potential androgen influence within the teleost brain.
In most cases, the regions of A. burtoni brain that express AR are known to play a role in reproductive function and/or behavior in fish. Of particular note is the expression of AR in regions of the brain closely associated with the olfactory system. We found ARα and ARβ in Vv and ARβ in Dc, both of which receive olfactory projections in cod (Rooney et al., 1992), zebrafish (Rink and Wullimann, 2004), and rainbow trout (Folgueira et al., 2004a,b). AR immunoreactivity has been described for these regions of the goldfish brain, as well as the goldfish olfactory bulb itself (Gelinas and Callard, 1997). We do not see AR transcripts in the A. burtoni olfactory bulb, which we believe is more likely to reflect a discrepancy in the specificities of the techniques, perhaps arising from the use of an antibody raised against mammalian AR (Gelinas and Callard, 1997), rather than differential expression between teleost species. The influence of sex steroids in the behavioral response to olfactory cues in mammals is well described (Bakker, 2003; Kelliher and Baum, 2002; Woodley and Baum, 2003), and there is increasing evidence that androgens exert a stimulatory effect on the olfactory-driven mating responses of male fish (Bhatt et al., 2002; Cardwell et al., 1995; Murphy and Stacey, 2002). In addition to its role in olfactory responses, Dc receives a visual input in fish (Lee and Bullock, 1990; Saidel et al., 2001). The expression of ARβ mRNA in this region in A. burtoni, in addition to the demonstration of AR protein in the goldfish retina (Gelinas and Callard, 1997), suggests that androgens may also influence visual processing in teleosts.
A. burtoni AR subtypes are segregated in some regions of the brain, whereas other regions contain mRNA for both AR isoforms. Without double labeling of both subtypes of AR, we cannot conclude that the two forms are coexpressed on the same cells, but this is a tempting hypothesis, in that both ARα and ARβ are expressed by GnRH1-releasing neurons (see below) and are localized in the same brain nuclei. In the Japanese eel, ARα and ARβ are coexpressed in cells in the testes (Ikeuchi et al., 2001). Both subtypes of A. burtoni AR contain the expected leucine-zipper, which is essential for steroid receptor dimerization. Colocalization would suggest the possibility that the two AR receptor types could form heterodimers, as shown in vitro the Japanese eel AR subtypes (Ikeuchi et al., 2001), facilitating a more complex interaction between them.
The main brain areas where expression of ARα and ARβ overlapped were the preoptic area, the ventral hypothalamus, and the ventral pituitary. These areas participate in regulating gonadotropin release, control of sexual differentiation and regulation of reproductive behavior in fish, all of which are androgen-sensitive processes. In the pituitary, ARα and ARβ were expressed in similar patterns, with ARα mRNA at a higher level. The cells in the ventral region of the pituitary, to which AR expression was limited, are the gonadotropin (GtH)-releasing cells, whereas AR expression was excluded from the dorsal cells, which are growth hormone (GH)-releasing cell (Chen and Fernald, 2006; Parhar et al., 2002). There are conflicting reports regarding the effect of androgen on GH release in fish, likely resulting from methodological differences in vitro and seasonal variations in vivo (Degani et al., 1998; Huggard et al., 1996; Larson et al., 2003; Onuma et al., 2005). There is some evidence that the effect of testosterone on plasma GH levels is dependent on testosterone’s conversion to E2 (Holloway and Leatherland, 1997). The absence of AR on presumptive GH-releasing cells in the A. burtoni pituitary is consistent with the idea that ER, rather than AR, mediates androgenic effects on GH-releasing neurons.
In teleosts, both T and 11-KT can potentiate GnRH-induced GtH release (Antonopoulou et al., 1999; Borg, 1994; Breton and Sambroni, 1996; Lo and Chang, 1998a,b; Weil and Marcuzzi, 1990). The results presented here, together with a recent report that describes GnRH-R1 but not GnRH-R2 expression in the ventral pituitary of A. burtoni (Chen and Fernald, 2006), suggest that the effect of androgen on GtH release at the level of the pituitary is potentially sensitive to influence by the GnRH-R1 pathway in LH/FSH-releasing gonadotropes.
Previous research has not examined the relative expression levels of AR mRNA between isoforms (Sperry and Thomas, 1999a,b; Takeo and Yamashita, 1999; Todo et al., 1999). Our data showing differential expression levels of AR subtypes in the brain and pituitary suggest differential influence of these two forms of receptor.
Expression of two AR subtypes by GnRH1-releasing neurons in A. burtoni
GnRH1-releasing neurons in A. burtoni express both ARα and ARβ, which, as discussed above, may have different ligand-induced transactivation profiles. Although mammalian neurons in the preoptic area near GnRH1-releasing cells express AR, expression has not been detected in mammalian GnRH1-releasing cells (Huang and Harlan, 1993). Functional ARs are expressed at low levels by the immortalized mouse GnRH-releasing cell line GT1 (Belsham et al., 1998; Poletti et al., 1994, 2001). If the GT1 AR expression levels are representative of those in vivo, it is possible that mammalian AR expression in GnRH neurons is below levels detectable by the techniques used to date (Belsham and Lovejoy, 2005). In the teleost brain, AR expression levels are 100–1,000-fold higher than in mammals (Pasmanik and Callard, 1988), which may reflect fundamental differences in androgen regulation of reproduction in teleosts and mammals.
The prediction that A. burtoni ARα and ARβ will show different activation profiles in response to 11-KT and testosterone based on comparative data makes this report of the expression of both subtypes by GnRH1-releasing neurons particularly important. Both testosterone and 11-KT, but not E2, restore the soma size of the GnRH1-releasing neurons in the aPPn of A. burtoni following castration (Soma et al., 1996). AR expression by GnRH1-releasing neurons themselves supports the idea of direct effects (i.e., not via aromatization) of testosterone and 11-KT on these GnRH1 neurons. In isolated black porgy neurons, there was no significant difference between 11-KT- and testosterone-induced GnRH1 release over a short time (1 hour; Lee at al., 2004). However, extended 11-KT treatment stimulated a greater total amount of GnRH1 release over 6 hours (Lee et al., 2004), suggesting potential differential regulation of GnRH1-releasing neurons by 11-KT and testosterone.
The expression of both ARα and ARβ in the aPPn is extremely dense. The results presented here are a representative example of AR expression in the brain of a stable T male, in which the entire HPG axis is up-regulated relative to NT males (Davis and Fernald, 1990; Francis et al., 1993; Parikh et al., 2005; White et al., 2002). ARα and ARβ mRNA levels are significantly lower in the telencephalon of NT males compared with T males (Burmeister et al., 2006). Since GnRH1-releasing neurons are the central point of control for the up-regulation of the reproductive axis in A. burtoni, future studies revealing whether social regulation of AR mRNA in the brain occurs at the level of the GnRH1 neurons themselves, the surrounding neurons in the aPPn, or both, will be important.
AR mRNA expression and possible interaction with GnRHR
ARα and ARβ mRNA levels were detected in nuclei throughout the brain that express both GnRH-R1 and GnRH-R2 (Chen and Fernald, 2006). In the absence of double in situ hybridization studies, we cannot conclude definitively that AR and GnRHR are coexpressed in the ventral telencephalon, ventral and dorsal hypothalamus, and ventral pituitary. However, given the evidence that all GnRH1-releasing neurons in the POA of A. burtoni express both ARα and ARβ mRNA (this study) and GnRH-R2 (Chen and Fernald, 2006), this suggests that there is coexpression of GnRHR and AR on hypothalamic neurons in teleosts. This raises the intriguing possibility that androgens not only regulate basal GnRH release but might also have a modulatory role in the presumptive GnRH feedback loop.
ACKNOWLEDGMENTS
The authors thank Dr. S. Zhou, Dr. A. Greenwood, Mr. S. Singh, and Ms. C-C. Chen for technical advice and Mr. J. Durand for assistance with images.
Abbreviations
- 3V
third ventricle
- aPPn
anterior part of the parvocellular preoptic nucleus
- C
cerebellum
- Dc
central nucleus of the dorsal telencephalon
- Dd
dorsal nucleus of the dorsal telencephalon
- DH
dorsal hypothalamus
- Dlv
ventral nucleus of the lateral part of the dorsal telencephalon
- Dm3
part 3 of the medial zone of the dorsal telencephalon
- Dm4
part 4 of the medial zone of the dorsal telencephalon
- icOB
internal cell layer of OB
- maPn
magnocellular portion of the anterior preoptic nucleus
- OB
olfactory bulb
- OT
optic tectum
- PC
posterior commisure
- Pit
pituitary
- pMPn
parvocellular portion of the magnocellular preoptic nucleus
- Tel
telencephalon
- Vd
dorsal nucleus of the ventral telencephalon
- VH
ventral hypothalamus
- Vv
ventral nucleus of the ventral telencephalon
LITERATURE CITED
- Amano M, Hyodo S, Urano A, Okumoto N, Kitamura S, Ikuta K, Suzuki Y, Aida K. Activation of salmon gonadotropin-releasing hormone synthesis by 17 alpha-methyltestosterone administration in yearling masu salmon, Oncorhynchus masou. Gen Comp Endocrinol. 1994;95:374–380. doi: 10.1006/gcen.1994.1136. [DOI] [PubMed] [Google Scholar]
- Ando H, Hasegawa M, Ando J, Urano A. Expression of salmon corticotropin-releasing hormone precursor gene in the preoptic nucleus in stressed rainbow trout. Gen Comp Endocrinol. 1999;113:87–95. doi: 10.1006/gcen.1998.7182. [DOI] [PubMed] [Google Scholar]
- Antonopoulou E, Swanson P, Mayer I, Borg B. Feedback control of gonadotropins in Atlantic salmon, Salmo salar, male. Part II. Aromatase inhibitor and androgen effects. Gen Comp Endocrinol. 1999;114:142–150. doi: 10.1006/gcen.1998.7248. [DOI] [PubMed] [Google Scholar]
- Bakker J. Sexual differentiation of the neuroendocrine mechanisms regulating mate recognition in mammals. J Neuroendocrinol. 2003;15:615–621. doi: 10.1046/j.1365-2826.2003.01036.x. [DOI] [PubMed] [Google Scholar]
- Belsham DD, Lovejoy DA. Gonadotropin-releasing hormone: gene evolution, expression, and regulation. Vitam Horm. 2005;71:59–94. doi: 10.1016/S0083-6729(05)71003-7. [DOI] [PubMed] [Google Scholar]
- Belsham DD, Evangelou A, Roy D, Duc VL, Brown TJ. Regulation of gonadotropin-releasing hormone (GnRH) gene expression by 5alpha-dihydrotestosterone in GnRH-secreting GT1–7 hypothalamic neurons. Endocrinology. 1998;139:1108–1114. doi: 10.1210/endo.139.3.5846. [DOI] [PubMed] [Google Scholar]
- Bhatt JP, Kandwal JS, Nautiyal R. Water temperature and pH influence olfactory sensitivity to pre-ovulatory and post-ovulatory ovarian pheromones in male Barilius bendelisis. J Biosci. 2002;27:273–281. doi: 10.1007/BF02704916. [DOI] [PubMed] [Google Scholar]
- Bohen SP, Kralli A, Yamamoto KR. Hold ’em and fold ’em: chaperones and signal transduction. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. [DOI] [PubMed] [Google Scholar]
- Borg B. Androgens in teleost fish. Comp Biochem Physiol. 1994;109C:219–245. [Google Scholar]
- Bradford MR, Northcutt RG. Organization of the diencephalon and pretectum of the ray-finned fishes. In: Davis RE, Northcutt RG, editors. Fish neurobiology, higher brain areas and functions. vol 2. University of Michigan Press; Ann Arbor, MI: 1983. pp. 117–140. [Google Scholar]
- Breton B, Sambroni E. Steroid activation of the brain-pituitary complex gonadotropic function in the triploid rainbow trout Oncorhynchus mykiss. Gen Comp Endocrinol. 1996;101:155–164. doi: 10.1006/gcen.1996.0017. [DOI] [PubMed] [Google Scholar]
- Burmeister SS, Fernald RD. Evolutionary conservation of the egr-1 immediate-early gene response in a teleost. J Comp Neurol. 2005;481:220–232. doi: 10.1002/cne.20380. [DOI] [PubMed] [Google Scholar]
- Burmeister SS, Jarvis ED, Fernald RD. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 2005;3:e363. doi: 10.1371/journal.pbio.0030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister SS, Kailasanath V, Fernald RD. Social dominance regulates androgen and estrogen receptor gene expression. Horm Behav. 2006 doi: 10.1016/j.yhbeh.2006.09.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell JR, Stacey NE, Tan ESP, McAdam DS, Lang SLC. Androgen increases olfactory receptor response to a vertebrate sex pheromone. J Comp Physiol. 1995;A176:55–61. [Google Scholar]
- Chen CC, Fernald RD. Distributions of two gonadotropin-releasing hormone receptor types in a cichlid fish suggest functional specialization. J Comp Neurol. 2006;495:314–323. doi: 10.1002/cne.20877. [DOI] [PubMed] [Google Scholar]
- Davis MR, Fernald RD. Social control of neuronal soma size. J Neurobiol. 1990;21:1180–1188. doi: 10.1002/neu.480210804. [DOI] [PubMed] [Google Scholar]
- Degani G, Boker R, Jackson K. Growth hormone, sexual maturity and steroids in male carp (Cyprinus carpio) Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;120:433–440. doi: 10.1016/s0742-8413(98)10020-8. [DOI] [PubMed] [Google Scholar]
- Dubois EA, Florijn MA, Zandbergen MA, Peute J, Goos HJ. Testosterone accelerates the development of the catfish GnRH system in the brain of immature African catfish (Clarias gariepinus) Gen Comp Endocrinol. 1998;112:383–393. doi: 10.1006/gcen.1998.7141. [DOI] [PubMed] [Google Scholar]
- Fernald RD. Quantitative behavioural observations of Haplochromis burtoni under semi-natural conditions. Anim Behav. 1977;25:643. [Google Scholar]
- Fernald RD, Hirata NR. Field study of Haplochromis burtoni: habitats and co-habitant. Environ Biol Fish. 1977;2:299–308. [Google Scholar]
- Fernald RD, Shelton LC. The organization of the diencephalon and the pretectum in the cichlid fish, Haplochromis burtoni. J Comp Neurol. 1985;238:202–217. doi: 10.1002/cne.902380207. [DOI] [PubMed] [Google Scholar]
- Folgueira M, Anadon R, Yanez J. An experimental study of the connections of the telencephalon in the rainbow trout (Oncorhynchus mykiss). I: olfactory bulb and ventral area. J Comp Neurol. 2004a;480:180–203. doi: 10.1002/cne.20340. [DOI] [PubMed] [Google Scholar]
- Folgueira M, Anadon R, Yanez J. Experimental study of the connections of the telencephalon in the rainbow trout (Oncorhynchus mykiss). II: dorsal area and preoptic region. J Comp Neurol. 2004b;480:204–233. doi: 10.1002/cne.20341. [DOI] [PubMed] [Google Scholar]
- Francis RC, Soma K, Fernald RD. Social regulation of the brain-pituitary-gonadal axis. Proc Natl Acad Sci U S A. 1993;90:7794–7798. doi: 10.1073/pnas.90.16.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas D, Callard GV. Immunolocalization of aromatase- and androgen receptor-positive neurons in the goldfish brain. Gen Comp Endocrinol. 1997;106:155–168. doi: 10.1006/gcen.1997.6891. [DOI] [PubMed] [Google Scholar]
- Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- Goos HJ, de Leeuw R, Cook H, van Oordt PG. Gonadotropic hormone-releasing hormone (GnRH) bioactivity in the brain of the immature rainbow trout, Salmo gairdneri: the effect of testosterone. Gen Comp Endocrinol. 1986;64:80–84. doi: 10.1016/0016-6480(86)90031-6. [DOI] [PubMed] [Google Scholar]
- Greenwood AK, Butler PC, White RB, DeMarco U, Pearce D, Fernald RD. Multiple corticosteroid receptors in a teleost fish: distinct sequences, expression patterns, and transcriptional activities. Endocrinology. 2003;144:4226–4236. doi: 10.1210/en.2003-0566. [DOI] [PubMed] [Google Scholar]
- Grens KE, Greenwood AK, Fernald RD. Two visual processing pathways are targeted by gonadotropin-releasing hormone in the retina. Brain Behav Evol. 2005;66:1–9. doi: 10.1159/000085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins MB, Thornton JW, Crews D, Skipper JK, Dotte A, Thomas P. Identification of a third distinct estrogen receptor and reclassification of estrogen receptors in teleosts. Proc Natl Acad Sci U S A. 2000;97:10751–10756. doi: 10.1073/pnas.97.20.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschenhauser K, Taborsky M, Oliveira T, Canario AVM, Oliveira RF. A test of the “challenge hypothesis” in cichlid fish: simulated partner and territory intruder experiments. Anim Behav. 2004;68:741–750. [Google Scholar]
- Holloway AC, Leatherland JF. Effect of gonadal steroid hormones on plasma growth hormone concentrations in sexually immature rainbow trout, Oncorhynchus mykiss. Gen Comp Endocrinol. 1997;105:246–254. doi: 10.1006/gcen.1996.6826. [DOI] [PubMed] [Google Scholar]
- Huang X, Harlan RE. Absence of androgen receptors in LHRH immunoreactive neurons. Brain Res. 1993;624:309–311. doi: 10.1016/0006-8993(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Huggard D, Khakoo Z, Kassam G, Habibi HR. Effect of testosterone on growth hormone gene expression in the goldfish pituitary. Can J Physiol Pharmacol. 1996;74:1039–1046. [PubMed] [Google Scholar]
- Hunter GA, Donaldson EM. Hormonal sex control and its application to fish culture. In: Hoar WS, Randall DJ, Donaldson EM, editors. Fish physiology. volume 9. Academic Press; New York: 1983. pp. 223–303. part B. [Google Scholar]
- Ikeuchi T, Todo T, Kobayashi T, Nagahama Y. cDNA cloning of a novel androgen receptor subtype. J Biol Chem. 1999;274:25205–25209. doi: 10.1074/jbc.274.36.25205. [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, Todo T, Kobayashi T, Nagahama Y. Two subtypes of androgen and progestogen receptors in fish testes. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:449–455. doi: 10.1016/s1096-4959(01)00375-x. [DOI] [PubMed] [Google Scholar]
- Kalra SP, Kalra PS. Regulation of gonadotropin secretion. In: Imura H, editor. The pituitary gland. Raven Press Ltd.; New York: 1994. pp. 285–307. [Google Scholar]
- Kelliher K, Baum M. Effect of sex steroids and coital experience on ferrets’ preference for the smell, sight and sound of conspecifics. Physiol Behav. 2002;76:1–7. doi: 10.1016/s0031-9384(02)00691-1. [DOI] [PubMed] [Google Scholar]
- Krust A, Green S, Argos P, Kumar V, Walter P, Bornert JM, Chambon P. The chicken oestrogen receptor sequence: homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 1986;5(5):891–897. doi: 10.1002/j.1460-2075.1986.tb04300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson ET, Norris DO, Summers CH. Monoaminergic changes associated with socially induced sex reversal in the saddleback wrasse. Neuroscience. 2003;119:251–263. doi: 10.1016/s0306-4522(03)00119-2. [DOI] [PubMed] [Google Scholar]
- Lee LT, Bullock TH. Responses of the optic tectum to telencephalic stimulation in catfish. Brain Behav Evol. 1990;35:313–324. doi: 10.1159/000115877. [DOI] [PubMed] [Google Scholar]
- Lee YH, Du JL, Shih YS, Jeng SR, Sun LT, Chang CF. In vivo and in vitro sex steroids stimulate seabream gonadotropin-releasing hormone content and release in the protandrous black porgy, Acanthopagrus schlegeli. Gen Comp Endocrinol. 2004;139:12–19. doi: 10.1016/j.ygcen.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Lo A, Chang JP. In vitro action of testosterone in potentiating gonadotropin-releasing hormone-stimulated gonadotropin-II secretion in goldfish pituitary cells: involvement of protein kinase C, calcium, and testosterone metabolites. Gen Comp Endocrinol. 1998a;111:318–333. doi: 10.1006/gcen.1998.7116. [DOI] [PubMed] [Google Scholar]
- Lo A, Chang JP. In vitro application of testosterone potentiates gonadotropin-releasing hormone-stimulated gonadotropin-II secretion from cultured goldfish pituitary cells. Gen Comp Endocrinol. 1998b;111:334–346. doi: 10.1006/gcen.1998.7117. [DOI] [PubMed] [Google Scholar]
- Miura T, Yamauchi K, Takahashi H, Nagahama Y. Hormonal induction of all stages of spermatogenesis in vitro in the male Japanese eel (Anguilla japonica) Proc Natl Acad Sci U S A. 1991a;88:5774–5778. doi: 10.1073/pnas.88.13.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Yamauchi K, Takahashi H, Nagahama Y. Human chorionic gonadotropin induces all stages of spermatogenesis in vitro in the male Japanese eel (Anguilla japonica) Dev Biol. 1991b;146:258–262. doi: 10.1016/0012-1606(91)90468-i. [DOI] [PubMed] [Google Scholar]
- Montero M, Le Belle N, King JA, Millar RP, Dufour S. Differential regulation of the two forms of gonadotropin-releasing hormone (mGnRH and cGnRH-II) by sex steroids in the European female silver eel (Anguilla anguilla) Neuroendocrinology. 1995;61:525–535. doi: 10.1159/000126876. [DOI] [PubMed] [Google Scholar]
- Munoz-Cueto JA, Sarasquete C, Zohar Y, Kah O. An atlas of the brain of the gilthead seabream (Sparus aurata) Maryland Sea Grant; College Park, MD: 2001. [Google Scholar]
- Murphy CA, Stacey NE. Methyl-testosterone induces male-typical ventilatory behavior in response to putative steroidal pheromones in female round gobies (Neogobius melanostomus) Horm Behav. 2002;42:109–115. doi: 10.1006/hbeh.2002.1810. [DOI] [PubMed] [Google Scholar]
- Oliveira RF, Almada VC, Canario AV. Social modulation of sex steroid concentrations in the urine of male cichlid fish Oreochromis mossambicus. Horm Behav. 1996;30:2–12. doi: 10.1006/hbeh.1996.0002. [DOI] [PubMed] [Google Scholar]
- Oliveira RF, Hirschenhauser K, Carneiro LA, Canario AV. Social modulation of androgen levels in male teleost fish. Comp Biochem Physiol B Biochem Mol Biol. 2002;132:203–215. doi: 10.1016/s1096-4959(01)00523-1. [DOI] [PubMed] [Google Scholar]
- Olsson PE, Berg AH, von Hofsten J, Grahn B, Hellqvist A, Larsson A, Karlsson J, Modig C, Borg B, Thomas P. Molecular cloning and characterization of a nuclear androgen receptor activated by 11-ketotestosterone. Reprod Biol Endocrinol. 2005;3:37. doi: 10.1186/1477-7827-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma T, Ando H, Koide N, Okada H, Urano A. Effects of salmon GnRH and sex steroid hormones on expression of genes encoding growth hormone/prolactin/somatolactin family hormones and a pituitary-specific transcription factor in masu salmon pituitary cells in vitro. Gen Comp Endocrinol. 2005;143:129–141. doi: 10.1016/j.ygcen.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Parhar IS, Soga T, Sakuma Y, Millar RP. Spatio-temporal expression of gonadotropin-releasing hormone receptor subtypes in gonadotropes, somatotropes and lactotropes in the cichlid fish. J Neuroendocrinol. 2002;14:657–665. doi: 10.1046/j.1365-2826.2002.00817.x. [DOI] [PubMed] [Google Scholar]
- Parikh VN, Clement TS, Fernald RD. Androgen level and male social status in the African cichlid, Astatotilapia burtoni. Behav Brain Res. 2005 doi: 10.1016/j.bbr.2005.07.011. (in press) [DOI] [PubMed] [Google Scholar]
- Pasmanik M, Callard GV. A high abundance androgen receptor in goldfish brain: characteristics and seasonal changes. Endocrinology. 1988;123:1162–1171. doi: 10.1210/endo-123-2-1162. [DOI] [PubMed] [Google Scholar]
- Pepels PP, Meek J, Bonga SE Wendelaar, Balm PH. Distribution and quantification of corticotropin-releasing hormone (CRH) in the brain of the teleost fish Oreochromis mossambicus (tilapia) J Comp Neurol. 2002;453:247–268. doi: 10.1002/cne.10377. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piferrer F, Baker IJ, Donaldson EM. Effects of natural, synthetic, aromatizable, and nonaromatizable androgens in inducing male sex differentiation in genotypic female chinook salmon (Oncorhynchus tshawytscha) Gen Comp Endocrinol. 1993;91:59–65. doi: 10.1006/gcen.1993.1104. [DOI] [PubMed] [Google Scholar]
- Poletti A, Melcangi RC, Negri-Cesi P, Maggi R, Martini L. Steroid binding and metabolism in the luteinizing hormone-releasing hormone-producing neuronal cell line GT1–1. Endocrinology. 1994;135:2623–2628. doi: 10.1210/endo.135.6.7988451. [DOI] [PubMed] [Google Scholar]
- Poletti A, Rampoldi A, Piccioni F, Volpi S, Simeoni S, Zanisi M, Martini L. 5Alpha-reductase type 2 and androgen receptor expression in gonadotropin releasing hormone GT1–1 cells. J Neuroendocrinol. 2001;13:353–357. doi: 10.1046/j.1365-2826.2001.00635.x. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Pottinger TG. Androgen binding in the skin of mature male brown trout, Salmo trutta L. Gen Comp Endocrinol. 1987;66:224–232. doi: 10.1016/0016-6480(87)90271-1. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Connections of the ventral telencephalon (subpallium) in the zebrafish (Danio rerio) Brain Res. 2004;1011:206–220. doi: 10.1016/j.brainres.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Rooney D, Doving KB, Ravaille-Veron M, Szabo T. The central connections of the olfactory bulbs in cod, Gadus morhua L. J Hirnforsch. 1992;33:63–75. [PubMed] [Google Scholar]
- Saidel WM, Marquez-Houston K, Butler AB. Identification of visual pallial telencephalon in the goldfish, Carassius auratus: a combined cytochrome oxidase and electrophysiological study. Brain Res. 2001;919:82–93. doi: 10.1016/s0006-8993(01)03001-3. [DOI] [PubMed] [Google Scholar]
- Schreibman MP, Margolis-Nunno H, Halpern-Sebold LR, Goos HJ, Perlman PW. The influence of androgen administration on the structure and function of the brain-pituitary-gonad axis of sexually immature platyfish, Xiphophorus maculatus. Cell Tissue Res. 1986;245:519–524. doi: 10.1007/BF00218552. [DOI] [PubMed] [Google Scholar]
- Slater CH, Fitzpatrick MS, Schreck CB. Characterization of an androgen receptor in salmonid lymphocytes: possible link to androgen-induced immunosuppression. Gen Comp Endocrinol. 1995;100:218–225. doi: 10.1006/gcen.1995.1151. [DOI] [PubMed] [Google Scholar]
- Soga T, Sakuma Y, Parhar IS. Testosterone differentially regulates expression of GnRH messenger RNAs in the terminal nerve, preoptic and midbrain of male tilapia. Brain Res Mol Brain Res. 1998;60:13–20. doi: 10.1016/s0169-328x(98)00153-3. [DOI] [PubMed] [Google Scholar]
- Soma KK, Francis RC, Wingfield JC, Fernald RD. Androgen regulation of hypothalamic neurons containing gonadotropin-releasing hormone in a cichlid fish: integration with social cues. Horm Behav. 1996;30:216–226. doi: 10.1006/hbeh.1996.0026. [DOI] [PubMed] [Google Scholar]
- Sperry TS, Thomas P. Characterization of two nuclear androgen receptors in Atlantic croaker: comparison of their biochemical properties and binding specificities. Endocrinology. 1999a;140:1602–1611. doi: 10.1210/endo.140.4.6631. [DOI] [PubMed] [Google Scholar]
- Sperry TS, Thomas P. Identification of two nuclear androgen receptors in kelp bass (Paralabrax clathratus) and their binding affinities for xenobiotics: comparison with Atlantic croaker (Micropogonias undulatus) androgen receptors. Biol Reprod. 1999b;61:1152–1161. doi: 10.1095/biolreprod61.4.1152. [DOI] [PubMed] [Google Scholar]
- Sperry TS, Thomas P. Androgen binding profiles of two distinct nuclear androgen receptors in Atlantic croaker (Micropogonias undulatus) J Steroid Biochem Mol Biol. 2000;73:93–103. doi: 10.1016/s0960-0760(00)00069-8. [DOI] [PubMed] [Google Scholar]
- Takeo J, Yamashita S. Two distinct isoforms of cDNA encoding rainbow trout androgen receptors. J Biol Chem. 1999;274:5674–5680. doi: 10.1074/jbc.274.9.5674. [DOI] [PubMed] [Google Scholar]
- Takeo J, Yamashita S. Rainbow trout androgen receptor-alpha fails to distinguish between any of the natural androgens tested in transactivation assay, not just 11-ketotestosterone and testosterone. Gen Comp Endocrinol. 2000;117:200–206. doi: 10.1006/gcen.1999.7398. [DOI] [PubMed] [Google Scholar]
- Todo T, Ikeuchi T, Kobayashi T, Nagahama Y. Fish androgen receptor: cDNA cloning, steroid activation of transcription in transfected mammalian cells, and tissue mRNA levels. Biochem Biophys Res Commun. 1999;254:378–383. doi: 10.1006/bbrc.1998.9919. [DOI] [PubMed] [Google Scholar]
- Trudeau VL, Peter RE, Sloley BD. Testosterone and estradiol potentiate the serum gonadotropin response to gonadotropin-releasing hormone in goldfish. Biol Reprod. 1991;44:951–960. doi: 10.1095/biolreprod44.6.951. [DOI] [PubMed] [Google Scholar]
- Weil C, Marcuzzi O. Cultured pituitary cell GtH response to GnRH at different stages of rainbow trout spermatogenesis and influence of steroid hormones. Gen Comp Endocrinol. 1990;79:492–498. doi: 10.1016/0016-6480(90)90080-6. [DOI] [PubMed] [Google Scholar]
- White RB, Fernald RD. Genomic structure and expression sites of three gonadotropin-releasing hormone genes in one species. Gen Comp Endocrinol. 1998;112:17–25. doi: 10.1006/gcen.1998.7125. [DOI] [PubMed] [Google Scholar]
- White SA, Nguyen T, Fernald RD. Social regulation of gonadotropin-releasing hormone. J Exp Biol. 2002;205:2567–2581. doi: 10.1242/jeb.205.17.2567. [DOI] [PubMed] [Google Scholar]
- Whitfield GK, Jurutka PW, Haussler CA, Haussler MR. Steroid hormone receptors: evolution, ligands, and molecular basis of biologic function. J Cell Bio. 1999;75:110–122. doi: 10.1002/(sici)1097-4644(1999)75:32+<110::aid-jcb14>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Woodley SK, Baum MJ. Effects of sex hormones and gender on attraction thresholds for volatile anal scent gland odors in ferrets. Horm Behav. 2003;44:110–118. doi: 10.1016/s0018-506x(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Rupp B, Reichert H. A topological atlas. Birkauser Verlag; Basel: 1996. Neuroanatomy of the zebrafish brain. [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]