Abstract
The heat shock proteins (HSPs), originally identified as heat-inducible gene products, are a highly conserved family of proteins that respond to a wide variety of stress. Although HSPs are among the most abundant intracellular proteins, they are expressed at low levels under normal physiological conditions, and show marked induction in response to various stressors. HSPs function primarily as molecular chaperones, facilitating the folding of other cellular proteins, preventing protein aggregation, or targeting improperly folded proteins to specific pathways for degradation. By modulating inflammation, wound debris clearance, cell proliferation, migration and collagen synthesis, HSPs are essential for normal wound healing of the skin. In this review, our goal is to discuss the role and clinical implications of HSP with respect to skin wound healing and diabetes. The numerous defects in the function of HSPs associated with diabetes could contribute to the commonly observed complications and delayed wound healing in diabetics. Several physical, pharmacological and genetic approaches may be considered to address HSP-directed therapies both in the laboratory and in the clinics.
Keywords: Heat shock proteins, Wound healing, Tissue protection, Diabetes
Introduction
The heat shock proteins (HSPs), originally identified as heat-inducible gene products, are a highly conserved family of proteins (Table 1) that respond to a wide variety of stress. HSPs protect against tissue injury by maintaining synthesis and proper conformation of proteins, repairing damaged proteins, and promoting the healing of injured tissue. The synthesis of HSPs can be induced by either physical or pharmacological inducers, resulting in cytoprotection against a subsequent potentially irreversible injury [1].
Table 1. Current mouse HSP nomenclature.
HSP names based on Mouse Genome Informatics database (MGI 3.54, updated January 05, 2008), The Jackson Laboratory.
| Current Name | Gene Symbols | Additional information |
|---|---|---|
| HSP110 | Hsp110 | Aka: HSP-E7I, HSP105, HSP110. Cytoplasm, nucleus. Protein domain: HSP70 |
| HSP90kDaβ, member1 | Hsp90b1 | Aka: endoplasmin, ERp99, gp96, GRP94, Targ2, Tra-1, Tra1, tumor rejection antigen (gp96) 1. Endoplasmic reticulum, extracellular space. Protein domains: HSP90, endoplasmin. |
| HSP90kDaα class B, member 1 | Hsp90ab1 | Aka: 90kDa, C81438, HSP84, HSP84-1, HSP90, HSPcb. Cytoplasm, mitochondria. Protein domain: HSP90. |
| HSP90kDaα class A, member 1 | Hsp90aa1 | Aka: 86kDa, 89kDa, hSP4, HSP86-1, HSP89, HSP90, HSPca. Cytoplasm. Protein domain: HSP90. |
| HSP70, ps1 | Hsp70-ps1 | Aka: 70kDa. |
| HSP34 | Hsp34 | Aka: 34kDa, p34. |
| HSP25, ps1 | Hsp25-ps1 | Aka: 25kDa, ENSMUSG00000007852. |
| HSP14 | Hspa14 | Aka: 70kDa, HSP70-4, HSP70L1, NST-1. Protein domain: HSP70 |
| HSP12B | Hspa12b | Aka: 2700081N06Rik. Protein domain: HSP70 |
| HSP12A | Hspa12a | Aka: 1700063D12Rik, HSPa12a, mKIAA0417. Protein domain: HSP70 |
| HSP 86 ps1-4 | Hsp86-ps1-4 | ps1 aka: 86kDa, Hsp86-2, Hsp90. ps2 aka: 86kDa, Hsp86-3, Hsp90. ps3 & ps4 aka: 86kDa. |
| HSP9 | Hspa9 | Aka: C3H-specific antigen, CSA, GRP75, Hsc74, HSP74, HSP74a, Hspa9a, mortalin, mot-2, mthsp70, PBP74. Cytoplasmic, mitochondrial. Protein Domains: HSP70, chaperone DnaK, calcium-binding EF-hand |
| HSP9, ps1-2 | Hspa9-ps1-2 | ps1 aka 70kDa, Hsc74-ps1, HSP74-ps1, HSP74b, HSPa9b; ps2 aka: Hsc74-ps2 |
| HSP, α-crystallin related B9 | Hspb9 | Protein domain: HSP20 |
| HSP8 | Hspa8 | Aka: 2410008N15Rik, 70kDa, Hsc70, Hsc71, Hsc73, HSP73, Hspa10. Cytoplasmic, Nuclear. Protein domain: HSP70 |
| HSP8 | Hspb8 | Aka: Cryac, D5Ucla4, E2IG1, H11, H11K, HSP20-like, HSP22 |
| HSP7 (CV) | Hspb7 | Aka: 27kDa, cvHsp, HSP25-2. Cytoskeletal |
| HSP, α-crystallin related B6 | Hspb6 | Aka: HSP20. Eye lens |
| HSP5 | Hspa5 | Aka: 78kDa, Bip, D2Wsu141e, D2Wsu17e, Grp78, Hsce70, mBiP, Sez7. Endoplasmic reticulum. |
| HSP4-like | Hspa4l | Aka: 94kDa, APG-1, OSP94. Cytoplasm, nucleus. Protein domain: HSP70 |
| HSP4 | Hspa4 | Aka: 70kDa, APG-2, Hsp110, Hsp70RY. Cytoplasm. Protein domain: HSP70 |
| HSP,3 | Hsp84-3 | Aka: 84kDa, HSP3, HSP90. |
| HSP,2 | Hsp84-2 | Aka: 84kDa, hsp2, Hsp90. |
| HSP3 | Hspb3 | Aka: 2310035K17Rik, 27kDa, spb3. Protein domain: HSP20 |
| HSP,2 ostu | Hsp70-2os | Chromosome 12 |
| HSP2 | Hspa2 | Aka: 70kDa, Hsp70-2. Mitochondria. Protein domain: HSP70 |
| HSP2 | Hspb2 | Aka: 27kDa, 2810021G24Rik, HSP27, MKBP. Soluble fraction. Protein domain: HSP20 |
| HSP1A | Hspa1a | Aka: Hsp70-3, Hsp70.3. Cell survival, DNA repair |
| HSP1B | Hspa1b | Aka: HSP68, HSP70, HSP70-1, HSP70.1, HSP70A1. Cell survival, DNA repair |
| HSP1-like | Hspa1l | Aka: 70kDa, Hsc70t, Msh5. Protein domain: HSP70 |
| HSP1 | Hspb1 | Aka: HSP25. Cytoplasm, contractile fiber. Protein domain: HSP20 |
| HSP1 (Cpn) | Hspd1 (HSP60) | Mitochondria |
| Hspe1 (Mt Cpn 10) | Mitochondria | |
| Hspe1-ps1-6 | HSP1 pseudogenes | |
| Hspe1-rs1 | 10kDa, Cpn10-rs1, EPF | |
| DnaJ (HSP40) homolog, subfamily A, member 1 | Dnaja1 | Old name: HSP DNAJ-like 2. Membrane. Protein domain: HSP DnaJ |
| DnaJ (Hsp40) homolog, subfamily A, member 1, pseudogene | Dnaja1-ps | Old name: HSP DNAJ-like 2, pseudogene. Aka: Hsj2-ps. |
| DnaJ (Hsp40) homolog, subfamily A, member 4 | Dnaja4 | Old name: HSP DNAJ-like 4. Membrane. |
| DnaJ (Hsp40) homolog, subfamily B, member 3 | Dnajb3 | Old name: HSP DNAJ-like 3. Aka: Hsj3, MSJ-1, Msj1.Protein domain: HSP40. |
| DnaJ (Hsp40) homolog, subfamily C, member 4 | Dnajc4 | Old name: heat shock 40kD protein 2. Aka: 2010301J22Rik, Hspf2, Mcg18. Membrane, extracellular space. |
| Serine (or cysteine) peptidase inhibitor, clade H, member 1 | Serpinh1 | Old name: heat shock protein 47 kDa. Endoplasmic reticulum, extracellular space. |
| Transmembrane protein 132A | Tmem132a | Old name: heat shock protein 5 binding protein 1. Aka: 6720481D13Rik, Hspa5bp1. Endoplasmic reticulum, golgi apparatus. |
| AHA1, activator of heat shock protein ATPase homolog 1 (yeast) | Ahsa1 | Aka: MGC:36589, MGC:36618, p38. Cytoplasm, endoplasmic reticulum. |
| AHA1, activator of heat shock protein ATPase homolog 2 (yeast) | Ahsa2 | Aka: 1110064P04Rik. |
| human ortholog name: HSPB (heat shock 27kDa) associated protein 1 | Hspbap1 | Aka: 3830421G21Rik. Protein domains: Protein Associated with Small Stress protein 1 (PASS1); Transcription factor jumonji/aspartyl β-hydroxylase. |
Cpn, Chaperonin; EPF, early pregnancy factor; Aka, also known as; CV, cardiovascular; ps, pseudogene; ostu, opposite strand transcription unit.
Barrier function represents a major role of skin. Wounding induces HSPs, particularly in the epidermis [2]. In the initial phase of wound healing there is an inflammatory response followed by organization of the fibrin-rich exudates and subsequent re-epithelialization and formation of granulation tissue. Complicated by ischemia, infection and related factors the wound site represents a hostile environment for the cells involved in the repair process. The wound bed contains abundant inducible HSP70 which contributes to protein homeostasis and cell survival within the healing wound [3]. HSP functions are compromised under conditions of diabetes. Here, our goal is to discuss the role and clinical implications of HSP with respect to skin wound healing and diabetes. In addition, we posit mechanisms which may lead to impaired HSP defenses and wound healing in diabetics.
1. HSPS: functions and implications in wound healing
HSPs are among the most abundant intracellular proteins. Although expressed at low levels under normal physiological conditions, HSPs show dramatically increased expression in response to cellular stress. HSPs function primarily as molecular chaperones, facilitating the folding of other cellular proteins, preventing protein aggregation, or targeting improperly folded proteins to specific pathways for degradation [4–6]. HSPs improve cellular survival by repairing denatured proteins, dissociating initial loose protein aggregates and ensuring correct folding and translocation of proteins [7]. This function is also needed in physiological conditions during de novo protein synthesis, folding of nascent polypeptides and transport [8]. In case of severe damage, HSPs direct damaged proteins for degradation within the proteasome system [4–6]. HSPs play a key role in facilitating immune responses, because they can bind antigenic peptides and transport them to antigen-presenting cells and T lymphocytes [9]. Initially, HSPs were thought to be only functional in the cytoplasm and nucleus. However, recently they have been implicated in intercellular signaling and transport after release to extracellular space and to bloodstream [10–12] (Table 1). HSPs are also capable of binding to adjacent cells, initiating signal transduction [11,13]. The heat shock response attenuates pro-inflammatory mechanisms and inducible nitric oxide synthase activity. It also stabilizes IκBα, which inhibits NF-κB-dependent transcription [14]. Recently, it has been recognized that HSPs play a dual role in regulating immune responses [15]. Indeed, while intracellular HSP induction in response to pro-inflammatory stimuli can exert anti-inflammatory effects, extracellular HSPs may signal danger activating immune cells [15]. Because of the overlap in their functions, HSPs have been classified into families according to their rough molecular weight and homology, i.e. HSP100, HSP90, HSP70, HSP60, HSP40 and small HSPs.
a. HSP90
The HSP90 family comprises of HSP90α and HSP90β, which form an inactive complex with the steroid hormone receptor before agonist binding and therefore participate in steroid hormone signaling [16]. HSP90 is responsible for catalyzing the interaction with several substrate proteins and co-chaperones involved in cell regulation and intracellular signaling [17]. HSP90 is a potent autoantigen and thought to have a role in various inflammatory diseases including arteriosclerosis [18]. HSP90 also plays an important role in the activation of endothelial nitric oxide synthase (eNOS) resulting in increased synthesis of vasoregulatory NO and concomitant reduction of the eNOS-derived radical, the superoxide anione [19]. Despite the fact that HSP90 is one of the most abundant proteins in other tissues, it is not present in large amounts in the normal skin [20]. Nevertheless, HSP90 is constitutively expressed in the superficial epidermis in the suprabasal keratinocytes [2,21] and has been shown to be induced by heat stress [22] and skin wounding in the superficial thickened epidermis [21]. The presence of HSP90 in the suprabasal layers during wound healing suggests a role in keratinocyte differentiation [21]. Wound hypoxia, a consequence of alterations in systemic hemodynamics and vascular disruption associated with wounding, promotes fibroblast migration especially in the absence of growth factors via a mechanism dependent on hypoxia-inducible factor-1alpha (HIF-1α) -mediated secretion of HSP90α into the extracellular space [23]. This HIF-1α-mediated pathway also controls the secretion of multiple growth factors complementing the another pathway dependent on signaling via reactive oxygen species [24,25]. Interestingly, application of purified recombinant HSP90α protein within carboxymethylcellulose cream enhances skin wound healing during days 5–13, with an overall improvement of 30% in the time needed for wound healing [23].
b. HSP70 family
The HSP70 family comprises eight isoforms, namely three different forms of HSP72, HSPA2, Grp78, HSP70B, HSP73 and Grp75 [26,27]. The isoforms of HSP are located mainly in the cytosol and nucleus, but they have also been detected in the lysosomes and endoplasmatic reticulum [26]. Grp75 has also been detected in the mitochondria [27]. HSP72 is the major inducible HSP found in the nucleus and cytosol [28]. HSP72 requires ATP for its chaperone activity [8] and minimizes aggregation of newly synthesized proteins. Stress-induced HSP72 is effective against aggregation of denatured proteins. Moreover, HSP72 is also capable of inhibiting stress induced apoptosis [29], even after the activation of effector caspases [30]. Interestingly, HSP72 is located on the luminal side of the lysosomes where it stabilizes the lysosomal membrane and inhibits the release of hydrolases and subsequent mediators of cell death [31]. Both excessive or impaired synthesis of HSP72 may result in disturbances of cell proliferation [32]. Overexpression of HSP72 also increases the survival and reduces oxidative damage of murine fibroblasts against ultraviolet light and suppresses the inflammatory response [33]. Interestingly, in these experiments, the effect of HSP72 seemed not to be mediated by antagonism of ultraviolet light-induced reactive oxygen radicals [33]. The role of HSP73 is controversial. It seems to act as a chaperone in non-stressed cells [1] and it is also likely to have a role in the formation of clathrin and several other factors involved in intracellular transportation [34].
HSPs may be viewed as survival proteins possessing an intrinsic ability to confer protection against most apoptotic stimuli [35–37]. Both HSP70 (HSP72) and the heat-shock cognate protein70 (HSC70, HSP73) are capable of inhibiting apoptosis by inhibiting release of cytochrome c, processing of pro-caspase-9 and activation of initiator caspases [29]. Heat stress results in inhibition of effector caspases upon subsequent heat shock [29], and thereby confers protection against heat-induced apoptosis. HSP70 is capable of rendering cells resistant to cell death induced by TNF-α [36] as well as by caspase-3 overexpression [30].
HSP72 is constitutively expressed in keratinocytes, especially in the upper layer of the epidermis [38–40] and also in fibroblasts, macrophages and endothelium [41]. HSP72 has been proposed to have a role in the wound healing process, which is supported by the finding that its expression is rapidly induced after skin wounding in animal models [42]. Polymer implants (polypropylene, polyester, polypropylene polyglactin) intended to be used in the repair of large abdominal wounds induce expression of HSP70 predominantly in macrophages and local inflammatory response dependent on the material [43]. Interestingly HSP70 expression shows an inverse correlation with the local inflammatory response [43]. The anti-inflammatory and pro-proliferative effects of HSP72 support its potential role in wound healing [21,42,44].
In chronic wounds, such as decubitus ulcers and in post-traumatic wounds, the expression of HSP70 protein has been noted to be low. In contrast, in healing wounds with rich granulation tissue the expression of HSP70 is high [44]. In the granulation tissue of healing wounds, only the endothelial cells are rich in HSP72 [44]. Of note, there is an inverse correlation between HSP72 levels and the latency of the healing process, and HSP72 and essential wound healing-related growth factors are expressed in a co-ordinated manner during the initial phase of healing at 7–14 days [42]. HSP72 is passively released after cell death or plasma membrane wounding but also actively secreted via the exosome system [10,45,46]. Subcutaneous HSP72 delivery into murine wounds accelerates the wound closure in a dose-dependent manner [47]. This effect was attributed to improved clearance of wound debris by enhanced macrophage-dependent phagocytosis [47]. After skin wounding there are significant differences in the expression pattern of different HSPs [48]. Whereas HSP72 and HSP32 are expressed only in the epidermis, HSP47 is expressed in both the epidermis and dermis, and only after skin wounding has occurred [48]. Whether exogenously applied HSP72 may stimulate wound re-epithelialization remains to be proven.
c. HSP70 and HSF-1
The regulation of HSP synthesis is controlled mainly by a major transcription factor heat shock factor-1 (HSF-1) which binds to the heat shock elements (HSE) present in promoter region of specific genes. Posttranscriptional mechanisms are also implicated in the regulation of HSP synthesis [49]. Under physiological conditions, HSF-1 monomers are co-localized with HSP70 in the nucleus. HSF-1 is activated by cellular stress [49]. The activation process involves trimerization of HSF-1 monomers, translocation of the trimers, hyperphosphorylation and binding to the promoter of heat shock genes [50–52]. The end-products of this process, such as HSP70, exert negative feedback regulation [53]. The posttranscriptional mechanism involves stabilization of HSP70 mRNA [54]. HSF-1 and nuclear factor-κB (NF-κB) signaling are tightly linked because in certain conditions, HSF-1 inhibits NF-κB activation [55], Fig. 1(1). In addition to protein denaturation, stress signals may also originate from cell membranes. Recently it has been proposed that the lipid composition and the architecture of membranes act as membrane sensors and modulate HSP response through the activation of HSF-1 [56].
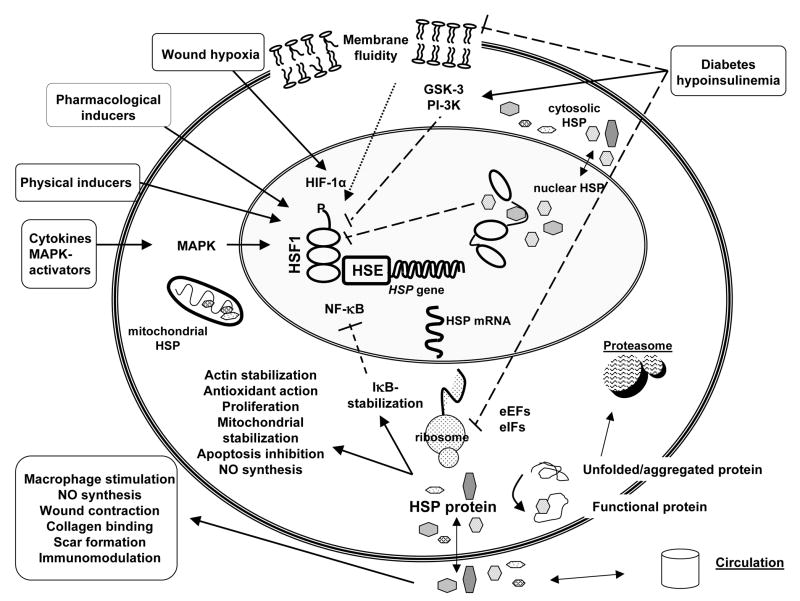
Fig. 1.
Schematic presentation of the regulation, mechanisms, inducers and inhibitors of heat shock protein (HSP) synthesis and the effect of HSPs on key factors in wound healing. Physical inducers include heat shock, ischemia-reperfusion, physical exercise, heavy metals, toxins, radiation, UV-light, laser, decreased ATP levels, pH and osmolarity changes. Pharmacological inducers include bimoclomol, geranylgeranylacetone (GGA), α-lipoic acid, ansamycins, butyrate, prostaglandins, celastrol, terrecyclin-A, BRX-220, PLA2 and nitric oxide (NO). Solid lines represent stimulation and dashed lines inhibition. Dot lines represent both stimulation and inhibition. HSF, heat shock factor; HSE, heat shock element; NF-κB, nuclear factor-κB; eEFs, eukaryotic elongation factors; eIFs, eukaryotic translation initiation factors.
d. HSP60
The HSP60 family is made up of only HSP60 itself which is also stress inducible [28]. In contrast to HSP70, HSP60 is present in both mitochondria and the cytosol. HSP60 has a complicated role in innate immune response [57] and in mitochondrial protein biogenesis [58,59]. HSP60 may inhibit caspase-3 [60] or facilitate the maturation of pro-caspase-3 to its active form [61]. The capacity of HSP60 to stabilize mitochondrial proteins, promote mitochondrial protein biosynthesis and prevent induction of mitochondrial apoptosis seems to be crucial for its cytoprotective function [62]. HSP60 is constitutively expressed in skin mainly in the epidermis [2, 21] and is induced by skin wounding in the deep neoepidermis in the basal and suprabasal keratinocytes [21]. It is likely that HSP60 is associated with proliferation and migration of keratinocytes in close proximity to the wound microenvironment [21].
e. HSP40 family and hemeoxygenase (HSP32)
The HSP40 family is made up of HSP47, which is involved in the procollagen synthesis [63]. HSP47 closely interacts with HSP70 [64]. HSP47 also has the capability to bind to collagen types I-III [65]. It is expressed in the epidermal basal cell layer of rat fetal and neonatal skin [66], and is induced only in the neonatal skin for up to 7 days after wounding in the dermis and subcutaneous tissue. Induction of HSP47 is paralleled by increased synthesis of type I collagen and cellular proliferation in subcutaneous tissue [66]. The rat neonatal skin is characterized by wound healing without scar formation in the absence of the HSP47 response [66]. Daily injections of antisense oligonucleotide preventing HSP47 expression in the rat neonatal skin suppressed the accumulation of type I collagen in the skin wound, resulting in minimized scar formation [67].
The heme oxygenases, i.e. HSP32 (HO-1), although not functioning as molecular chaperones, catalyze the conversion of heme to iron, biliverdin and carbon monoxide and regulates inflammatory and immune responses [68,69]. HO-1, biliverdin and its catalytic product bilirubin have well-characterized antioxidant properties. Iron regulates the expression of genes such as nitric oxide synthase. HO-1 induction has been recognized as a sensitive consequence of oxidative stress, and overexpression of HO-1 protects against oxidative damage in several cell types [1,70].
f. HSP27 and small HSPs
HSP27 and αB crystalline are members of the small heat shock proteins that stabilize actin microfilaments [71] and inhibit stress-induced apoptosis [72]. HSP27 function is modulated by phosphorylation [73]. The overexpression of HSP27 results in promotion of endothelial cell migration [74]. HSP27 is constitutively expressed in the epidermis in the superficial layers [21]. During wound healing, HSP27 becomes more phosphorylated at the wound edge one day after skin incision [73]. The expression is also increased in the deep epidermis, in the suprabasal keratinocytes. Suprabasal keratinocytes are characterized by low proliferation and high differentiation, suggesting a role in migration [21]. Overexpression of HSP27 in dermal fibroblasts improves wound contraction. Consistently, wound contraction is compromised if fibroblasts possess low levels of HSP27 [75]. Increased rapid elongation and formation of pseudopods, elongated stress fibers, enhanced attachment of cells to the dishes and enhanced migration has been reported in fibroblasts overexpressing HSP27 [74]. The cellular proliferation rate did not correlate with HSP27 expression, which indicates that the effects on wound contraction and migration was independent of cell growth [75]. The effect of HSP27 on wound contraction in the same model was shown to be mediated by mitogen-activated protein kinase (MAPK)- activated protein kinase 2/3 (MAPKAPK 2/3) -mediated HSP27 phosphorylation [73]. HSP27 phosphorylation correlated with both fibroblast-populated collagen lattice model contraction and wound contraction in rats in vivo [73]. These findings appear to be clinically relevant since wound contraction represents a significant factor in closing an open wound. In addition to acting as a chaperone, HSP22 i.e. α-crystallin, regulates cell proliferation, apoptosis, neoplasia, cell motility and cellular redox state [76]. However, its plausible role in cutaneous wound healing remains unknown.
2. Diabetes and HSP
Both type 1 and type 2 diabetes are characterized by an increased risk for the development of microvascular and macrovascular complications. In diabetes, endogenous defense systems are overwhelmed causing various types of stress. Uncontrolled oxidative stress represents a characteristic feature of diabetes [77–81]. Among the other important conditions related to diabetes are dyslipidemia, modification of proteins and lipids, and perturbations in the tissue antioxidant defense network [77–80,82,83]. These disturbances are exacerbated in diabetes with microvascular complications such as nephropathy, retinopathy and neuropathy. The antioxidant functions of HSP should therefore prove to be helpful in fighting diabetic complications. Indeed, the crucial role of HSPs in diabetes is highlighted by their ability to counteract denaturation of tissue proteins and facilitate cellular repair and defense mechanisms. Of note in this context is the observation that the diabetic state may interfere with the synthesis of HSPs [84], utilizing either a transcriptional or post-translational mechanism [85]. In turn, compromised HSP expression may contribute to diabetic complications [86,87] resulting in a vicious cycle.
a, Experimental and in vitro studies
The effect of diabetes on HSPs is tissue-specific. Under conditions of experimental diabetes, reduced expression of HSP72 has been associated with impaired cytoprotective function and impaired tissue regeneration [41,88]. On the other hand, although heat stress induces HSP72 in diabetic rats, it failed to protect the isolated heart against ischemia-reperfusion injury despite insulin treatment [89]. Our group has previously noted that experimental diabetes limits the expression of HSP72 in heart, liver and vastus lateralis skeletal muscle [77], and also in the kidney [Atalay et al., unpublished observation]. Furthermore, the impaired HSP response in the diabetic heart was associated with tissue inflammation and overt oxidative stress [77].
Our group has also utilized endurance type of exercise training as a therapeutic tool to restore HSP synthesis in tissues. Although training induced the activation and expression of HSF-1 in the skeletal muscle and increased HSP72 levels in every tissue examined, such favorable induction was shown to be blunted in diabetic rats [77]. It has been shown that HSP60 levels are lowered in the heart [77,90,91], and increased in the kidney and liver of diabetic rats [91,92]. Expression of HSPs is altered in diabetes in a tissue specific manner because of altered susceptibility of the tissues to injury [77]. In line with decreased myocardial HSP60 expression, another mitochondrial chaperone, namely glucose regulated protein (GRP75), has recently been shown to be decreased in the mitochondria of heart tissue of diabetic rats [93].
Of interest in the context of diabetes, the synthesis of HSP60 is closely linked to insulin as the deficiency of insulin down-regulates HSP60 [90,94], insulin-like growth factor-1 (IGF-1) and its receptor (IGF-1R) in the diabetic heart [95]. In this context it should be acknowledged that IGF-1 is known to be cardioprotective [96,97]. HSP60 increases the amount of functional IGF-1R by inhibiting its ubiquitination in cardiomyocytes [90,98] thereby averting proteosome-dependent degradation of the protein. This is important because lower levels of IGF-1R can potentially limit myocardial defense systems leading to diabetic cardiomyopathy. In contrast, increased expression of IGF-1 and increased IGF-1 signaling in the diabetic kidney causes diabetic nephropathy [99]. Thus, the mechanism through which diabetes modulates HSP60 levels seems to be organ-specific. Despite the various protective actions of the HSPs, some of these proteins have been associated with disease progression in diabetes. For example, HSP47, a collagen-binding stress protein that has a specific role in the intracellular processing of pro-collagen molecules during collagen synthesis, plays a pathological role in the later stages of diabetic nephropathy and has been associated with glomerulosclerosis and tubulointerstitial fibrosis in rats with experimental diabetes [100].
The mechanisms by which diabetes may impair HSP responses remain sketchy at best. Impaired or deficient insulin secretion in diabetes may be one mechanism leading to attenuated protein synthesis, especially related to the acute induction of stress proteins, including HSPs. Insulin regulates both the initiation and elongation phases of translation by altering the phosphorylation of eukaryotic translation initiation factors (eIF2, eIF2B, eIF3, eIF4B, eIF4E, and eIF4G) and eukaryotic elongation factors (eEF1 and eEF2) [101]. Generally, EF-1alpha plays a major role in the regulation of mRNA translation. In rats, experimental diabetes decreased the rate of peptide-chain elongation which was associated with a marked reduction in the amount of elongation factor 2 (EF-2). Insulin therapy reversed the effects of diabetes on protein synthesis and increased the total EF-2 content of diabetic rats to control values [102]. Recently, it has been noted that experimental diabetes increases the level of EF-1alpha in rats, which was almost completely reversed after a ten-day vitamin E supplementation. [103]. EF-1alpha mRNA expression was found to be rapidly affected by oxidative stress [104]. Interestingly, in rat cardiac myocytes elevated glucose alone significantly reduced EF-2 phosphorylation [105] which is an essential step involved in extension of the polypeptide chain in protein translation.
Impaired HSF-1 activation via up-regulation of glycogen synthase kinase 3 (GSK-3) represents another important pathway in diabetes. GSK-3, an enzyme initially described as a key regulator of glycogen metabolism, is currently known to be involved in a diverse array of cell functions, including suppression of HSF-1 activity [106]. Limited membrane fluidity has been proposed to represent another factor limiting HSP synthesis in diabetics [84]. The physical state and lipid composition of cellular membranes, subtle alterations of membrane fluidity, phase state, and microheterogeneity may function as membrane sensors to regulate HSP response [56]. Various pathological states, including diabetes are associated with membrane defects, which can be restored with insulin treatment [107].
b. Human diabetes
In human studies, HSP72 levels in peripheral blood leukocytes are considerably lower in type 1 diabetic patients with polyneuropathy than in healthy volunteers [108]. Similarly, intramuscular HSP72 and HO-1 mRNA levels are lower in type 2 diabetic patients [109]. A study performed in twins showed that lower levels of HSP72 mRNA in skeletal muscle is associated with some markers of insulin resistance in patients with type 2 diabetes [87]. However, HSP72 mRNA levels did not correlate with either fasting plasma glucose or insulin level in any of the subgroups investigated [87] suggesting that alterations in HSP mRNA is not simply the consequence of chronically elevated plasma glucose or insulin levels. Nonetheless, it has been shown in diabetic rodents that pharmacological induction of HSP72 expression improves insulin sensitivity [110], providing evidence that HSP72 is directly involved in the pathogenesis of insulin resistance. It was later demonstrated that skeletal muscle HSP72 mRNA levels indeed tightly correlate with glucose disposal rate and oxidative capacity of tissues [109]. On the other hand, using a quantitative proteome analysis, Hojlund et al. reported increased levels of HSP90 and GRP78 in the skeletal muscle of diabetic subjects, but no relationship between fasting plasma glucose and HSP90 levels were noted [111]. Of interest is the observation that HSP90 plays an important role in maintaining the activity of protein kinase B (PK-B) [112], which is involved in the regulation of insulin-mediated glucose transport and glycogen synthesis.
Abnormal HSP and EF expression may also contribute to the altered protein synthesis in diabetes, in addition to the direct effect of insulin on protein synthesis. A marked increase in expression of EF-1alpha and an imbalance with other subunits of the EF-1 complex has been reported in skeletal muscle of type 1 type 2 diabetic subjects [113].
Both type 1 and type 2 diabetes are well-established risk factor for cardiovascular disease. Firm evidence lend support to the contention that immunity to HSP60 and its derived peptides is involved in type 1 diabetes [114,115]. Moreover, it has been recently suggested that circulating HSP60 may contribute to the cardiovascular pathology associated with diabetes [116], supporting earlier observations on the association of HSP60 and atherosclerosis [117]. Interestingly, HSP60 shifts the cytokine secretion profile towards Th2 type via toll-like receptor-2 (TLR2)-dependent signaling in human T cells [118], which could have a modifying effect on the immunological response in the pathogenesis of type 1 diabetes
3. Impaired wound healing in diabetes and the possible role of HSPs
Diabetes affects approximately 170 million people worldwide, including 20.8 million in the USA. By the year 2030, these numbers are expected to double [119]. The diabetic foot ulcer is a major diabetic complication, representing a serious medical, social and economic problem all over the world [119]. The cumulative lifetime incidence of diabetic foot ulcers may be as high as 25%. Foot ulcers precede 84% of all diabetes-related lower leg amputations [120].
A number of factors contribute to wound healing deficiencies in individuals with diabetes. Hyperglycemia leads to increased glycosylation of immune cells, such as neutrophils and macrophages by inhibiting their normal function and predisposing both to chronic inflammation and increased susceptibility to infection. Glycosylation of erythrocytes increases their rigidity, which may predispose to sludging and local ischemia in the microvasculature. Susceptibility to ulceration and impairment of wound healing in diabetes increases dramatically when accompanied by diabetic peripheral neuropathy and peripheral vascular disease. These complications predispose to microtrauma, foot deformities and ischemia [120].
Wound healing occurs as a cellular response to injury and involves a number of coordinated events including clot formation, inflammation, re-epithelialization, angiogenesis, granulation tissue formation, wound contraction, scar formation and tissue remodeling [121,122]. Pathophysiological processes that impair wound healing in diabetes include decreased or impaired growth factor production [123,124], angiogenesis [125], leukocyte function and chemotaxis [126], keratinocyte and fibroblast migration and proliferation, function of epidermal nerves [127], collagen accumulation, re-epithelialization, granulation tissue formation [124], and tissue remodeling by matrix-metalloproteinases (MMPs) [128]. In addition, narrowing or occlusion of the blood vessels within the edge of the wound leads to tissue hypoxia and abnormal wound healing. Oxidative stress, abnormal nitric oxide production and disturbed redox status may also play a role [121].
Little is known about the role of HSPs in wound healing or in the abnormal wound healing in diabetes. The importance of HSPs in several of the processes involved in would healing and their expression as a protective mechanism suggests that they may be important in the response to wounds. The diabetic state results in delayed expression of HSP72 at the protein level, despite mRNA upregulation in the epithelial cells and inflammatory cells during the wound healing process [40]. On the other hand, HSP70 expression in the wound bed in diabetic mice increased after a delay, suggesting that the poorly healing, chronically open wound may result in a more potent HSP induction than a normally healing wound. Similarly, in skin fibroblasts isolated from a patient with diabetic nephropathy, the levels of HSP70, HSP60 and HSP27 were increased [129]. In diabetic streptozotocin-induced rats, during wound healing, the levels of HSP72 and HSP73 were roughly 60% lower than in normal control rats 7–14 days after the incision [41]. This effect of diabetes could be prevented by administration of insulin and normalization of blood glucose levels [41]. According to these results, it seems clear that the diabetic state interferes with the expression of HSP72 and HSP73, but the mechanism remains unclear [41]
4. Strategies to induce HSPs in diabetes
a. Physical inducers of HSPs
An increased intracellular pool of aggregating denatured proteins is the major inducer of HSP70 accumulation [1]. HSP expression is upregulated by various physical stresses including heat, ischemia, reperfusion, physical exercise, heavy metals and toxins [4,77,130]. Transient exposure of the tissue to heat results in release of arachidonic acid from plasma membrane resulting in leukotriene and prostaglandin synthesis [11,72,131]. Changes in interstitial fluid osmolarity [132], pH and cellular ATP content can also induce the heat shock response [133]. Heat shock response and synthesis of growth factors are interrelated, because transforming growth factor-β (TGF-β) regulates synthesis of HSP70 and HSP90 in cultured chicken embryo cells, whereas fibroblast growth factor (FGF) and epidermal growth factor (EGF) are ineffective [134]. In cultured fibroblasts exposed to ultraviolet B irradiation, TGF-β mediates the upregulation of HSP70 synthesis [135]. Hyperthermia induces expression of HSP70 in the keratinocytes in human skin [38].
Treatment of skin incisions with 815 nm laser light accelerates wound closure without prominent scar formation, along with increased expression of HSP70 in the epidermal and dermal layers [136]. In addition, exposure of rat skin to laser irradiation resulted in increased expression of HSP72 a day after the exposure in the stratum spinosum and paralleled with reduced thickness of the skin layer [39]. The elevated expression of HSP72 was sustained for up to 7 days [39].
b. Pharmacological inducers of HSPs
Anti-inflammatory drugs such as indomethacin induce HSF-1 DNA-binding activity by lowering the temperature threshold for HSF-1 activation to physiologic temperature range without activation of HSP gene itself, resulting in increased protection against lethal conditions [72]. Exposure to sodium salicylate has similar effects [131]. In tissue injury and various inflammatory states, the release of arachidonic acid has been shown to hyperphosphorylate HSF-1 in a dose-dependent manner, and to induce DNA binding activity [131] resulting in HSP gene activation and HSP protein synthesis. HSF-1 is rapidly activated by synthesis of HSF-1-activating prostaglandins, especially cyclopentenone prostaglandins [137] and some cytokines [138]. Exposure of the cell to exogenous phospholipase A2 results in HSF-1 DNA-binding and modification of HSF-1 similar to that observed after heat shock [131]. Exposure to hyperthermia results in activation of membrane bound A2 [11]. Similar to indomethacin and arachidonic acid, phospholipase A2 lowers the thermal threshold for HSF-1 activation [131].
Geranylgeranylacetone (GGA) is an antiulcer drug which induces gastroprotection, HSF-1 activation and HSP70 gene activation [139]. Short chain fatty acids namely butyrate induce dose dependent increase of HSP25 in rat IEC-18 cells and tolerance against monochloramine induced oxidative stress [140]. Alpha lipoic acid (α-LA) is a naturally occurring antioxidant precursor which induces HSP60 in rat heart [91]. At high doses α-LA has been suggested to function as a HSP inducer by activating HSF-1 via increasing disulfide formation in certain target proteins [141]. Indeed, Strokov et al. [108] observed that α-LA administration restored HSP levels and NO production and improved neuropathic symptoms in a study of type 1 diabetic patients with neuropathy.
Proteasome inhibitors, including MG-132, lactacystin and bortezomib, are potent inducers of the heat shock response [142–144]. Proteasome inhibitors are proposed to increase the amount of misfolded proteins, leading to HSF-1-dependent transcription [143,145]. Certain proteasome inhibitors induce HSP expression by directly inducing HSPs [146]. Another HSF-1 activator, celastrol results in hyperphosphorylation and transcriptional activation of HSF-1 possibly via proteasome inhibition [147]. Terrecyclic acid A is a novel HSP inducer leading to HSF-1 activation, the mechanism of which seems to be related to perturbation of the redox state and increase of reactive oxygen species [148]. Interestingly, based on interaction of HSP90 with HSF-1, several inhibitors of HSP90, namely radicicol and the benzoquinone ansamycins [149], have been found to stimulate the HSF-1 activity and subsequently induce HSP expression.
Specific drugs designed to co-induce HSP expression, namely bimoclomol [150] and BRX-220 [151], improve diabetic retinopathy, neuropathy, nephropathy, wound healing, cardiac ischemia and insulin resistance in animal models [110,152,153]. Bimoclomol interacts with HSF-1 prolonging the binding to DNA, and it also upregulates HSP72 in the diabetic wound [152, 154]. Altered lipid composition and physical state (fluidity) of biological membranes are decisive factors in the processes of perception and transduction of stress into a signal that triggers the transcriptional activation of genes encoding stress protein. Bimoclomol and its derivatives specifically interact with and increase the fluidity of negatively charged membrane lipids [155]. Accordingly, the HSP-related activity of bimoclomol is highly susceptible to the fatty acid composition and fluidity of membrane of target cells [155]. Because the plasma membrane also acts as an important regulatory interface, it has therefore been recently speculated that the lower HSF-1 and HSP levels in diabetes could be the result of compromised membrane fluidity [84].
Metformin, a dimethylbiguanide drug that is widely used for the treatment of hyperglycemia in type 2 diabetes also has effects on vascular endothelial function, presumably via activation of eNOS [156]. HSP90 has an important role in the activation of eNOS [157]. Interestingly, metformin has been shown to enhance endothelial function by AMPK-dependent and HSP90-mediated eNOS activation [156]. Because mitogen-activated protein kinases modulate phosphorylation of HSP27, and therefore its activity in actin microfilament function, MAPK modulators are also potentially useful for HSP27 function [73].
Nitric oxide (NO) synthesis induces HSP expression. Inhibition of NO synthesis limits the expression of HSPs [158]. Medications that have been associated with improved outcome in diabetes, including HMG CoA reductase inhibitors [159], angiotensin-converting enzyme (ACE) inhibitors [160], and thiazolidinediones [161] all can restore endothelial NO synthase function which in turn may induce HSP expression. Physical exercise provides a physiological stimulus for increased NO production [162] and increased HSP and HSF-1 expression [77,163], thus contributing to the improved outcomes associated with exercise in diabetes.
c. Gene delivery agents
Transfection of cells with appropriate vectors provide a good experimental approach to understand the protective effects of HSPs. Fibroblasts overexpressing HSP70 demonstrate improved survival against ultraviolet radiation [33]. Similarly, HSP70 overexpressing endothelial cells and astrocytes are more resistant to metabolic stressors including hypoxia and hypoglycemia [164]. Several HSP72 overexpressing mice strains have higher protection against hyperthermia, circulatory shock, and cardiac and cerebral ischemia [165–167]. Systemic gene delivery in the clinic continues to pose serious challenges. Whether topical approaches to deliver HSP-related genes may benefit wound healing is a hypothesis testable not only in the laboratory but also in clinical practice.
5. Conclusions
Members of the extensive HSP family play a key role in cellular protein homeostasis and cytoprotection. By modulating inflammation, cell proliferation, migration and collagen synthesis, HSPs play important functions in normal skin wound healing. Diabetes is associated with defects in HSP function. Such impairments could contribute to complications in wound healing commonly observed in diabetics. Several physical, pharmacological and genetic approaches may be considered to address HSP-directed therapies in the laboratory as well as in the clinics.
Acknowledgments
Supported in part by grants from the Finnish Ministry of Education, COST Actions B35, BM0602, Juho Vainio Foundation and Yrjo Jahnsson Foundations, Helsinki, Finland. Also supported by NIH awards GM077185, GM069589 and DK 76566.
References
- 1.Benjamin IJ, McMillan DR. Circ Res. 1998;83:117–132. doi: 10.1161/01.res.83.2.117. [DOI] [PubMed] [Google Scholar]
- 2.Charveron M, Calvo M, Gall Y. Cell Biol Toxicol. 1995;11:161–165. doi: 10.1007/BF00756518. [DOI] [PubMed] [Google Scholar]
- 3.Wagstaff MJ, Shah M, McGrouther DA, Latchman DS. J Plast Reconstr Aesthet Surg. 2007;60:974–982. doi: 10.1016/j.bjps.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 4.Morimoto RI. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 5.Li GC, Werb Z. Proc Natl Acad Sci U S A. 1982;79:3218–3222. doi: 10.1073/pnas.79.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riabowol KT, Mizzen LA, Welch WJ. Science. 1988;242:433–436. doi: 10.1126/science.3175665. [DOI] [PubMed] [Google Scholar]
- 7.Buchner J. Faseb J. 1996;10:10–19. [PubMed] [Google Scholar]
- 8.McKay DB. Adv Protein Chem. 1993;44:67–98. doi: 10.1016/s0065-3233(08)60564-1. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava P. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 10.Asea A. J Biosci. 2007;32:579–584. doi: 10.1007/s12038-007-0057-5. [DOI] [PubMed] [Google Scholar]
- 11.Calderwood SK, Mambula SS, Gray PJ., Jr Ann N Y Acad Sci. 2007;1113:28–39. doi: 10.1196/annals.1391.019. [DOI] [PubMed] [Google Scholar]
- 12.Campisi J, Fleshner M. J Appl Physiol. 2003;94:43–52. doi: 10.1152/japplphysiol.00681.2002. [DOI] [PubMed] [Google Scholar]
- 13.Javid B, MacAry PA, Lehner PJ. J Immunol. 2007;179:2035–2040. doi: 10.4049/jimmunol.179.4.2035. [DOI] [PubMed] [Google Scholar]
- 14.Wong HR. New Horiz. 1998;6:194–200. [PubMed] [Google Scholar]
- 15.Chen Y, Voegeli TS, Liu PP, Noble EG, Currie RW. Inflamm Allergy Drug Targets. 2007;6:91–100. doi: 10.2174/187152807780832274. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan G, Post JF, Thompson EB. J Steroid Biochem Mol Biol. 1997;60:1–9. doi: 10.1016/s0960-0760(96)00182-3. [DOI] [PubMed] [Google Scholar]
- 17.Whitesell L, Lindquist SL. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 18.Rigano R, Profumo E, Buttari B, Tagliani A, Petrone L, D’Amati G, Ippoliti F, Capoano R, Fumagalli L, Salvati B, Businaro R. Ann N Y Acad Sci. 2007;1107:1–10. doi: 10.1196/annals.1381.001. [DOI] [PubMed] [Google Scholar]
- 19.Pritchard KA, Jr, Ackerman AW, Gross ER, Stepp DW, Shi Y, Fontana JT, Baker JE, Sessa WC. J Biol Chem. 2001;276:17621–17624. doi: 10.1074/jbc.C100084200. [DOI] [PubMed] [Google Scholar]
- 20.Morris SD. Clin Exp Dermatol. 2002;27:220–224. doi: 10.1046/j.1365-2230.2002.01012.x. [DOI] [PubMed] [Google Scholar]
- 21.Laplante AF, Moulin V, Auger FA, Landry J, Li H, Morrow G, Tanguay RM, Germain L. J Histochem Cytochem. 1998;46:1291–1301. doi: 10.1177/002215549804601109. [DOI] [PubMed] [Google Scholar]
- 22.Wilson N, McArdle A, Guerin D, Tasker H, Wareing P, Foster CS, Jackson MJ, Rhodes LE. J Cutan Pathol. 2000;27:176–182. doi: 10.1034/j.1600-0560.2000.027004176.x. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Li Y, Guan S, Fan J, Cheng CF, Bright AM, Chinn C, Chen M, Woodley DT. Embo J. 2007;26:1221–1233. doi: 10.1038/sj.emboj.7601579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen CK, Khanna S, Babior BM, Hunt TK, Ellison EC, Roy S. J Biol Chem. 2002;277:33284–33290. doi: 10.1074/jbc.M203391200. [DOI] [PubMed] [Google Scholar]
- 25.Sen CK, Khanna S, Gordillo G, Bagchi D, Bagchi M, Roy S. Ann N Y Acad Sci. 2002;957:239–249. doi: 10.1111/j.1749-6632.2002.tb02920.x. [DOI] [PubMed] [Google Scholar]
- 26.Daugaard M, Rohde M, Jaattela M. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 27.Tavaria M, Gabriele T, Kola I, Anderson RL. Cell Stress Chaperones. 1996;1:23–28. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink AL. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 29.Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, Massie B. Mol Cell Biol. 2000;20:7146–7159. doi: 10.1128/mcb.20.19.7146-7159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaattela M, Wissing D, Kokholm K, Kallunki T, Egeblad M. Embo J. 1998;17:6124–6134. doi: 10.1093/emboj/17.21.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nylandsted J, Gyrd-Hansen M, Danielewicz A, Fehrenbacher N, Lademann U, Hoyer-Hansen M, Weber E, Multhoff G, Rohde M, Jaattela M. J Exp Med. 2004;200:425–435. doi: 10.1084/jem.20040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nollen EA, Morimoto RI. J Cell Sci. 2002;115:2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- 33.Simon MM, Reikerstorfer A, Schwarz A, Krone C, Luger TA, Jaattela M, Schwarz T. J Clin Invest. 1995;95:926–933. doi: 10.1172/JCI117800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapoport I, Boll W, Yu A, Bocking T, Kirchhausen T. Mol Biol Cell. 2007;19:405–413. doi: 10.1091/mbc.E07-09-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buzzard KA, Giaccia AJ, Killender M, Anderson RL. J Biol Chem. 1998;273:17147–17153. doi: 10.1074/jbc.273.27.17147. [DOI] [PubMed] [Google Scholar]
- 36.Jaattela M, Wissing D. J Exp Med. 1993;177:231–236. doi: 10.1084/jem.177.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Awasthi N, Wagner BJ. Invest Ophthalmol Vis Sci. 2005;46:2082–2091. doi: 10.1167/iovs.05-0002. [DOI] [PubMed] [Google Scholar]
- 38.Trautinger F, Trautinger I, Kindas-Mugge I, Metze D, Luger TA. J Invest Dermatol. 1993;101:334–338. doi: 10.1111/1523-1747.ep12365491. [DOI] [PubMed] [Google Scholar]
- 39.Souil E, Capon A, Mordon S, Dinh-Xuan AT, Polla BS, Bachelet M. Br J Dermatol. 2001;144:260–266. doi: 10.1046/j.1365-2133.2001.04010.x. [DOI] [PubMed] [Google Scholar]
- 40.McMurtry AL, Cho K, Young LJ, Nelson CF, Greenhalgh DG. J Surg Res. 1999;86:36–41. doi: 10.1006/jsre.1999.5700. [DOI] [PubMed] [Google Scholar]
- 41.Bitar MS, Farook T, John B, Francis IM. Surgery. 1999;125:594–601. [PubMed] [Google Scholar]
- 42.Shukla A, Dubey MP, Srivastava R, Srivastava BS. Biochem Biophys Res Commun. 1998;244:434–439. doi: 10.1006/bbrc.1998.8286. [DOI] [PubMed] [Google Scholar]
- 43.Klosterhalfen B, Klinge U, Tietze L, Henze U, Muys L, Mittermayer C, Bhardwaj RS. J Mater Sci Mater Med. 2000;11:175–181. doi: 10.1023/a:1008931725401. [DOI] [PubMed] [Google Scholar]
- 44.Oberringer M, Baum HP, Jung V, Welter C, Frank J, Kuhlmann M, Mutschler W, Hanselmann RG. Biochem Biophys Res Commun. 1995;214:1009–1014. doi: 10.1006/bbrc.1995.2386. [DOI] [PubMed] [Google Scholar]
- 45.Gallucci S, Lolkema M, Matzinger P. Nat Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- 46.Hightower LE, Guidon PT., Jr J Cell Physiol. 1989;138:257–266. doi: 10.1002/jcp.1041380206. [DOI] [PubMed] [Google Scholar]
- 47.Kovalchin JT, Wang R, Wagh MS, Azoulay J, Sanders M, Chandawarkar RY. Wound Repair Regen. 2006;14:129–137. doi: 10.1111/j.1743-6109.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 48.Keagle JN, Welch WJ, Young DM. Wound Repair Regen. 2001;9:378–385. doi: 10.1046/j.1524-475x.2001.00378.x. [DOI] [PubMed] [Google Scholar]
- 49.Anckar J, Sistonen L. Adv Exp Med Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- 50.Baler R, Dahl G, Voellmy R. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarge KD, Murphy SP, Morimoto RI. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarge KD. Ann N Y Acad Sci. 1998;851:112–116. doi: 10.1111/j.1749-6632.1998.tb08983.x. [DOI] [PubMed] [Google Scholar]
- 53.Abravaya K, Myers MP, Murphy SP, Morimoto RI. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 54.Kaarniranta K, Elo M, Sironen R, Lammi MJ, Goldring MB, Eriksson JE, Sistonen L, Helminen HJ. Proc Natl Acad Sci U S A. 1998;95:2319–2324. doi: 10.1073/pnas.95.5.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wirth D, Bureau F, Melotte D, Christians E, Gustin P. Am J Physiol Lung Cell Mol Physiol. 2004;287:L953–961. doi: 10.1152/ajplung.00184.2003. [DOI] [PubMed] [Google Scholar]
- 56.Vigh L, Horvath I, Maresca B, Harwood JL. Trends Biochem Sci. 2007;32:357–363. doi: 10.1016/j.tibs.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Vabulas RM, Wagner H, Schild H. Curr Top Microbiol Immunol. 2002;270:169–184. doi: 10.1007/978-3-642-59430-4_11. [DOI] [PubMed] [Google Scholar]
- 58.Moseley P. Immunopharmacology. 2000;48:299–302. doi: 10.1016/s0162-3109(00)00227-7. [DOI] [PubMed] [Google Scholar]
- 59.Voos W, Rottgers K. Biochim Biophys Acta. 2002;1592:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- 60.Gupta S, Knowlton AA. Circulation. 2002;106:2727–2733. doi: 10.1161/01.cir.0000038112.64503.6e. [DOI] [PubMed] [Google Scholar]
- 61.Xanthoudakis S, Roy S, Rasper D, Hennessey T, Aubin Y, Cassady R, Tawa P, Ruel R, Rosen A, Nicholson DW. Embo J. 1999;18:2049–2056. doi: 10.1093/emboj/18.8.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta S, Knowlton AA. J Cell Mol Med. 2005;9:51–58. doi: 10.1111/j.1582-4934.2005.tb00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohba S, Wang ZL, Baba TT, Nemoto TK, Inokuchi T. Arch Oral Biol. 2003;48:627–633. doi: 10.1016/s0003-9969(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 64.Fan CY, Lee S, Cyr DM. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macdonald JR, Bachinger HP. J Biol Chem. 2001;276:25399–25403. doi: 10.1074/jbc.M102471200. [DOI] [PubMed] [Google Scholar]
- 66.Wang ZL, Inokuchi T, Ikeda H, Baba TT, Uehara M, Kamasaki N, Sano K, Nemoto TK, Taguchi T. Int J Oral Maxillofac Surg. 2002;31:179–184. doi: 10.1054/ijom.2001.0191. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z, Inokuchi T, Nemoto TK, Uehara M, Baba TT. Plast Reconstr Surg. 2003;111:1980–1987. doi: 10.1097/01.PRS.0000054844.41243.F2. [DOI] [PubMed] [Google Scholar]
- 68.Srisook K, Kim C, Cha YN. Antioxid Redox Signal. 2005;7:1674–1687. doi: 10.1089/ars.2005.7.1674. [DOI] [PubMed] [Google Scholar]
- 69.Camara NO, Soares MP. Free Radic Biol Med. 2005;38:426–435. doi: 10.1016/j.freeradbiomed.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 70.Motterlini R, Foresti R, Intaglietta M, Winslow RM. Am J Physiol. 1996;270:H107–114. doi: 10.1152/ajpheart.1996.270.1.H107. [DOI] [PubMed] [Google Scholar]
- 71.Lavoie JN, Gingras-Breton G, Tanguay RM, Landry J. J Biol Chem. 1993;268:3420–3429. [PubMed] [Google Scholar]
- 72.Welsh N, Margulis B, Borg LA, Wiklund HJ, Saldeen J, Flodstrom M, Mello MA, Andersson A, Pipeleers DG, Hellerstrom C, et al. Mol Med. 1995;1:806–820. [PMC free article] [PubMed] [Google Scholar]
- 73.Hirano S, Rees RS, Gilmont RR. J Surg Res. 2002;102:77–84. doi: 10.1006/jsre.2001.6315. [DOI] [PubMed] [Google Scholar]
- 74.Rousseau S, Houle F, Landry J, Huot J. Oncogene. 1997;15:2169–2177. doi: 10.1038/sj.onc.1201380. [DOI] [PubMed] [Google Scholar]
- 75.Hirano S, Shelden EA, Gilmont RR. Cell Stress Chaperones. 2004;9:29–37. doi: 10.1379/471.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shemetov AA, Seit-Nebi AS, Gusev NB. J Neurosci Res. 2008;86:264–269. doi: 10.1002/jnr.21441. [DOI] [PubMed] [Google Scholar]
- 77.Atalay M, Oksala NK, Laaksonen DE, Khanna S, Nakao C, Lappalainen J, Roy S, Hanninen O, Sen CK. J Appl Physiol. 2004;97:605–611. doi: 10.1152/japplphysiol.01183.2003. [DOI] [PubMed] [Google Scholar]
- 78.Ceriello A, Bortolotti N, Falleti E, Taboga C, Tonutti L, Crescentini A, Motz E, Lizzio S, Russo A, Bartoli E. Diabetes Care. 1997;20:194–197. doi: 10.2337/diacare.20.2.194. [DOI] [PubMed] [Google Scholar]
- 79.Dandona P, Thusu K, Cook S, Snyder B, Makowski J, Armstrong D, Nicotera T. Lancet. 1996;347:444–445. doi: 10.1016/s0140-6736(96)90013-6. [DOI] [PubMed] [Google Scholar]
- 80.Kennedy AL, Lyons TJ. Metabolism. 1997;46:14–21. doi: 10.1016/s0026-0495(97)90311-5. [DOI] [PubMed] [Google Scholar]
- 81.Atalay M, Laaksonen DE. Journal of Sports Science and Medicine. 2002;1:1–14. [PMC free article] [PubMed] [Google Scholar]
- 82.Atalay M, Laaksonen DE, Niskanen L, Uusitupa M, Hanninen O, Sen CK. Acta Physiol Scand. 1997;161:195–201. doi: 10.1046/j.1365-201X.1997.00200.x. [DOI] [PubMed] [Google Scholar]
- 83.Gul M, Laaksonen DE, Atalay M, Vider L, Hanninen O. Scand J Med Sci Sports. 2002;12:163–170. doi: 10.1034/j.1600-0838.2002.120307.x. [DOI] [PubMed] [Google Scholar]
- 84.Hooper PL, Hooper JJ. Diabetes Technol Ther. 2005;7:204–208. doi: 10.1089/dia.2005.7.204. [DOI] [PubMed] [Google Scholar]
- 85.Li M, Guo D, Isales CM, Eizirik DL, Atkinson M, She JX, Wang CY. J Mol Med. 2005;83:504–513. doi: 10.1007/s00109-005-0645-5. [DOI] [PubMed] [Google Scholar]
- 86.Hooper PL. Diabetes Care. 2003;26:951–952. doi: 10.2337/diacare.26.3.951. [DOI] [PubMed] [Google Scholar]
- 87.Kurucz I, Morva A, Vaag A, Eriksson KF, Huang X, Groop L, Koranyi L. Diabetes. 2002;51:1102–1109. doi: 10.2337/diabetes.51.4.1102. [DOI] [PubMed] [Google Scholar]
- 88.Yamagishi N, Nakayama K, Wakatsuki T, Hatayama T. Life Sci. 2001;69:2603–2609. doi: 10.1016/s0024-3205(01)01337-6. [DOI] [PubMed] [Google Scholar]
- 89.Joyeux M, Faure P, Godin-Ribuot D, Halimi S, Patel A, Yellon DM, Demenge P, Ribuot C. Cardiovasc Res. 1999;43:939–946. doi: 10.1016/s0008-6363(99)00185-6. [DOI] [PubMed] [Google Scholar]
- 90.Shan YX, Yang TL, Mestril R, Wang PH. J Biol Chem. 2003;278:45492–45498. doi: 10.1074/jbc.M304498200. [DOI] [PubMed] [Google Scholar]
- 91.Oksala NK, Laaksonen DE, Lappalainen J, Khanna S, Nakao C, Hanninen O, Sen CK, Atalay M. J Diabetes Complications. 2006;20:257–261. doi: 10.1016/j.jdiacomp.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 92.Oksala NK, Lappalainen J, Laaksonen DE, Khanna S, Kaarniranta K, Sen CK, Atalay M. Antioxid Redox Signal. 2007;9:497–506. doi: 10.1089/ars.2006.1450. [DOI] [PubMed] [Google Scholar]
- 93.Turko IV, Murad F. J Biol Chem. 2003;278:35844–35849. doi: 10.1074/jbc.M303139200. [DOI] [PubMed] [Google Scholar]
- 94.Chen HS, Shan YX, Yang TL, Lin HD, Chen JW, Lin SJ, Wang PH. Diabetes. 2005;54:175–181. doi: 10.2337/diabetes.54.1.175. [DOI] [PubMed] [Google Scholar]
- 95.Chen HS, Jia J, Su HF, Lin HD, Chen JW, Lin SJ, Yang JY, Lai HC, Mestril R, Wang PH. J Endocrinol. 2006;190:433–440. doi: 10.1677/joe.1.06692. [DOI] [PubMed] [Google Scholar]
- 96.Chae HJ, Kim HR, Bae J, Chae SU, Ha KC, Chae SW. Arch Pharm Res. 2004;27:324–333. doi: 10.1007/BF02980068. [DOI] [PubMed] [Google Scholar]
- 97.Lee WL, Chen JW, Ting CT, Ishiwata T, Lin SJ, Korc M, Wang PH. Endocrinology. 1999;140:4831–4840. doi: 10.1210/endo.140.10.7082. [DOI] [PubMed] [Google Scholar]
- 98.Lai HC, Liu TJ, Ting CT, Yang JY, Huang L, Wallace D, Kaiser P, Wang PH. Am J Physiol Endocrinol Metab. 2007;292:292–297. doi: 10.1152/ajpendo.00189.2006. [DOI] [PubMed] [Google Scholar]
- 99.Lupia E, Elliot SJ, Lenz O, Zheng F, Hattori M, Striker GE, Striker LJ. Diabetes. 1999;48:1638–1644. doi: 10.2337/diabetes.48.8.1638. [DOI] [PubMed] [Google Scholar]
- 100.Liu D, Razzaque MS, Cheng M, Taguchi T. Histochem J. 2001;33:621–628. doi: 10.1023/a:1016398200087. [DOI] [PubMed] [Google Scholar]
- 101.Kimball SR, Vary TC, Jefferson LS. Annu Rev Physiol. 1994;56:321–348. doi: 10.1146/annurev.ph.56.030194.001541. [DOI] [PubMed] [Google Scholar]
- 102.Vary TC, Nairn A, Lynch CJ. Am J Physiol. 1994;266:E628–634. doi: 10.1152/ajpendo.1994.266.4.E628. [DOI] [PubMed] [Google Scholar]
- 103.Al-Maghrebi M, Cojocel C, Thompson MS. Mol Cell Biochem. 2005;273:177–183. doi: 10.1007/s11010-005-0552-7. [DOI] [PubMed] [Google Scholar]
- 104.Chen E, Proestou G, Bourbeau D, Wang E. Exp Cell Res. 2000;259:140–148. doi: 10.1006/excr.2000.4952. [DOI] [PubMed] [Google Scholar]
- 105.Yeshao W, Gu J, Peng X, Nairn AC, Nadler JL. Metabolism. 2005;54:1453–1460. doi: 10.1016/j.metabol.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 106.Bijur GN, Jope RS. J Neurochem. 2000;75:2401–2408. doi: 10.1046/j.1471-4159.2000.0752401.x. [DOI] [PubMed] [Google Scholar]
- 107.Patel SP, Katyare SS. Lipids. 2006;41:819–825. doi: 10.1007/s11745-006-5036-3. [DOI] [PubMed] [Google Scholar]
- 108.Strokov IA, Manukhina EB, Bakhtina LY, Malyshev IY, Zoloev GK, Kazikhanova SI, Ametov AS. Bull Exp Biol Med. 2000;130:986–990. [PubMed] [Google Scholar]
- 109.Bruce CR, Carey AL, Hawley JA, Febbraio MA. Diabetes. 2003;52:2338–2345. doi: 10.2337/diabetes.52.9.2338. [DOI] [PubMed] [Google Scholar]
- 110.Kurthy M, Mogyorosi T, Nagy K, Kukorelli T, Jednakovits A, Talosi L, Biro K. Ann N Y Acad Sci. 2002;967:482–489. doi: 10.1111/j.1749-6632.2002.tb04306.x. [DOI] [PubMed] [Google Scholar]
- 111.Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. J Biol Chem. 2003;278:10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- 112.Sato S, Fujita N, Tsuruo T. Proc Natl Acad Sci U S A. 2000;97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reynet C, Kahn CR. Proc Natl Acad Sci U S A. 2001;98:3422–3427. doi: 10.1073/pnas.051630398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Abulafia-Lapid R, Elias D, Raz I, Keren-Zur Y, Atlan H, Cohen IR. J Autoimmun. 1999;12:121–129. doi: 10.1006/jaut.1998.0262. [DOI] [PubMed] [Google Scholar]
- 115.Elias D, Markovits D, Reshef T, van der Zee R, Cohen IR. Proc Natl Acad Sci U S A. 1990;87:1576–1580. doi: 10.1073/pnas.87.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shamaei-Tousi A, Stephens JW, Bin R, Cooper JA, Steptoe A, Coates AR, Henderson B, Humphries SE. Eur Heart J. 2006;27:1565–1570. doi: 10.1093/eurheartj/ehl081. [DOI] [PubMed] [Google Scholar]
- 117.Xu Q, Schett G, Perschinka H, Mayr M, Egger G, Oberhollenzer F, Willeit J, Kiechl S, Wick G. Circulation. 2000;102:14–20. doi: 10.1161/01.cir.102.1.14. [DOI] [PubMed] [Google Scholar]
- 118.Zanin-Zhorov A, Bruck R, Tal G, Oren S, Aeed H, Hershkoviz R, Cohen IR, Lider O. J Immunol. 2005;174:3227–3236. doi: 10.4049/jimmunol.174.6.3227. [DOI] [PubMed] [Google Scholar]
- 119.Wild S, Roglic G, Green A, Sicree R, King H. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 120.Rathur HM, Boulton AJ. Clin Dermatol. 2007;25:109–120. doi: 10.1016/j.clindermatol.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 121.Blakytny R, Jude E. Diabet Med. 2006;23:594–608. doi: 10.1111/j.1464-5491.2006.01773.x. [DOI] [PubMed] [Google Scholar]
- 122.Brem H, Tomic-Canic M. J Clin Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Galkowska H, Wojewodzka U, Olszewski WL. Wound Repair Regen. 2006;14:558–565. doi: 10.1111/j.1743-6109.2006.00155.x. [DOI] [PubMed] [Google Scholar]
- 124.Falanga V. Lancet. 2005;366:1736–1743. doi: 10.1016/S0140-6736(05)67700-8. [DOI] [PubMed] [Google Scholar]
- 125.Galiano RD, Tepper OM, Pelo CR, Bhatt KA, Callaghan M, Bastidas N, Bunting S, Steinmetz HG, Gurtner GC. Am J Pathol. 2004;164:1935–1947. doi: 10.1016/S0002-9440(10)63754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maruyama K, Asai J, Ii M, Thorne T, Losordo DW, D’Amore PA. Am J Pathol. 2007;170:1178–1191. doi: 10.2353/ajpath.2007.060018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gibran NS, Jang YC, Isik FF, Greenhalgh DG, Muffley LA, Underwood RA, Usui ML, Larsen J, Smith DG, Bunnett N, Ansel JC, Olerud JE. J Surg Res. 2002;108:122–128. doi: 10.1006/jsre.2002.6525. [DOI] [PubMed] [Google Scholar]
- 128.Lobmann R, Ambrosch A, Schultz G, Waldmann K, Schiweck S, Lehnert H. Diabetologia. 2002;45:1011–1016. doi: 10.1007/s00125-002-0868-8. [DOI] [PubMed] [Google Scholar]
- 129.Tessari P, Puricelli L, Iori E, Arrigoni G, Vedovato M, James P, Coracina A, Millioni R. J Proteome Res. 2007;6:976–986. doi: 10.1021/pr060443n. [DOI] [PubMed] [Google Scholar]
- 130.Powers SK, Locke, Demirel HA. Med Sci Sports Exerc. 2001;33:386–392. doi: 10.1097/00005768-200103000-00009. [DOI] [PubMed] [Google Scholar]
- 131.Jurivich DA, Sistonen L, Kroes RA, Morimoto RI. Science. 1992;255:1243–1245. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- 132.Sheikh-Hamad D, Di Mari J, Suki WN, Safirstein R, Watts BA, 3rd, Rouse D. J Biol Chem. 1998;273:1832–1837. doi: 10.1074/jbc.273.3.1832. [DOI] [PubMed] [Google Scholar]
- 133.Riordan M, Sreedharan R, Wang S, Thulin G, Mann A, Stankewich M, Van Why S, Kashgarian M, Siegel NJ. Am J Physiol Renal Physiol. 2005;288:F1236–1242. doi: 10.1152/ajprenal.00438.2004. [DOI] [PubMed] [Google Scholar]
- 134.Takenaka IM, Hightower LE. J Cell Physiol. 1992;152:568–577. doi: 10.1002/jcp.1041520317. [DOI] [PubMed] [Google Scholar]
- 135.Cao Y, Ohwatari N, Matsumoto T, Kosaka M, Ohtsuru A, Yamashita S. Pflugers Arch. 1999;438:239–244. doi: 10.1007/s004240050905. [DOI] [PubMed] [Google Scholar]
- 136.Capon A, Souil E, Gauthier B, Sumian C, Bachelet M, Buys B, Polla BS, Mordon S. Lasers Surg Med. 2001;28:168–175. doi: 10.1002/lsm.1035. [DOI] [PubMed] [Google Scholar]
- 137.Amici C, Giorgi C, Rossi A, Santoro MG. J Virol. 1994;68:6890–6899. doi: 10.1128/jvi.68.11.6890-6899.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schett G, Redlich K, Xu Q, Bizan P, Groger M, Tohidast-Akrad M, Kiener H, Smolen J, Steiner G. J Clin Invest. 1998;102:302–311. doi: 10.1172/JCI2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hirakawa T, Rokutan K, Nikawa T, Kishi K. Gastroenterology. 1996;111:345–357. doi: 10.1053/gast.1996.v111.pm8690199. [DOI] [PubMed] [Google Scholar]
- 140.Ren H, Musch MW, Kojima K, Boone D, Ma A, Chang EB. Gastroenterology. 2001;121:631–639. doi: 10.1053/gast.2001.27028. [DOI] [PubMed] [Google Scholar]
- 141.McCarty MF. Med Hypotheses. 2001;57:313–317. doi: 10.1054/mehy.2001.1320. [DOI] [PubMed] [Google Scholar]
- 142.Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, Bradley S, Gobburu JV, Goheer A, Lee SL, Leighton J, Liang CY, Lostritto RT, McGuinn WD, Morse DE, Rahman A, Rosario LA, Verbois SL, Williams G, Wang YC, Pazdur R. Clin Cancer Res. 2004;10:3954–3964. doi: 10.1158/1078-0432.CCR-03-0781. [DOI] [PubMed] [Google Scholar]
- 143.Kim D, Kim SH, Li GC. Biochem Biophys Res Commun. 1999;254:264–268. doi: 10.1006/bbrc.1998.9840. [DOI] [PubMed] [Google Scholar]
- 144.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M, Libermann TA, Treon SP, Munshi NC, Richardson PG, Hideshima T, Anderson KC. Proc Natl Acad Sci U S A. 2002;99:14374–14379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Westerheide SD, Morimoto RI. J Biol Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 146.Kawamura H, Otsuka T, Matsuno H, Niwa M, Matsui N, Kato K, Uematsu T, Kozawa O. Am J Physiol. 1999;277:E1046–1054. doi: 10.1152/ajpendo.1999.277.6.E1046. [DOI] [PubMed] [Google Scholar]
- 147.Westerheide SD, Bosman JD, Mbadugha BN, Kawahara TL, Matsumoto G, Kim S, Gu W, Devlin JP, Silverman RB, Morimoto RI. J Biol Chem. 2004;279:56053–56060. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- 148.Turbyville TJ, Wijeratne EM, Whitesell L, Gunatilaka AA. Mol Cancer Ther. 2005;4:1569–1576. doi: 10.1158/1535-7163.MCT-05-0050. [DOI] [PubMed] [Google Scholar]
- 149.Whitesell L, Bagatell R, Falsey R. Curr Cancer Drug Targets. 2003;3:349–358. doi: 10.2174/1568009033481787. [DOI] [PubMed] [Google Scholar]
- 150.Biro K, Jednakovits A, Kukorelli T, Hegedus E, Koranyi L. Brain Res Bull. 1997;44:259–263. doi: 10.1016/s0361-9230(97)00118-4. [DOI] [PubMed] [Google Scholar]
- 151.Rakonczay Z, Jr, Ivanyi B, Varga I, Boros I, Jednakovits A, Nemeth I, Lonovics J, Takacs T. Free Radic Biol Med. 2002;32:1283–1292. doi: 10.1016/s0891-5849(02)00833-x. [DOI] [PubMed] [Google Scholar]
- 152.Vigh L, Literati PN, Horvath I, Torok Z, Balogh G, Glatz A, Kovacs E, Boros I, Ferdinandy P, Farkas B, Jaszlits L, Jednakovits A, Koranyi L, Maresca B. Nat Med. 1997;3:1150–1154. doi: 10.1038/nm1097-1150. [DOI] [PubMed] [Google Scholar]
- 153.Nanasi PP, Jednakovits A. Cardiovasc Drug Rev. 2001;19:133–151. doi: 10.1111/j.1527-3466.2001.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 154.Hargitai J, Lewis H, Boros I, Racz T, Fiser A, Kurucz I, Benjamin I, Vigh L, Penzes Z, Csermely P, Latchman DS. Biochem Biophys Res Commun. 2003;307:689–695. doi: 10.1016/s0006-291x(03)01254-3. [DOI] [PubMed] [Google Scholar]
- 155.Torok Z, Tsvetkova NM, Balogh G, Horvath I, Nagy E, Penzes Z, Hargitai J, Bensaude O, Csermely P, Crowe JH, Maresca B, Vigh L. Proc Natl Acad Sci U S A. 2003;100:3131–3136. doi: 10.1073/pnas.0438003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Davis BJ, Xie Z, Viollet B, Zou MH. Diabetes. 2006;55:496–505. doi: 10.2337/diabetes.55.02.06.db05-1064. [DOI] [PubMed] [Google Scholar]
- 157.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 158.Malyshev I, Manukhina EB, Mikoyan VD, Kubrina LN, Vanin AF. FEBS Letters. 1995;370:159–162. doi: 10.1016/0014-5793(95)00801-f. [DOI] [PubMed] [Google Scholar]
- 159.Hattori Y, Nakanishi N, Kasai K. Cardiovascular Research. 2002;54:649–658. doi: 10.1016/s0008-6363(02)00266-3. [DOI] [PubMed] [Google Scholar]
- 160.Nikolaidis LA, Doverspike A, Huerbin R, Hentosz T, Shannon RP. Circulation. 2002;105:2785–2790. doi: 10.1161/01.cir.0000017433.90061.2e. [DOI] [PubMed] [Google Scholar]
- 161.Fujishima S, Ohya Y, Nakamura Y, Onaka U, Abe I, Fujishima M. Am J Hyperten. 1998;11:1134–1137. doi: 10.1016/s0895-7061(98)00130-7. [DOI] [PubMed] [Google Scholar]
- 162.Roberts CK, Barnard RJ, Jasman A, Balon TW. Am J Physiol. 1999;277:E390–394. doi: 10.1152/ajpendo.1999.277.2.E390. [DOI] [PubMed] [Google Scholar]
- 163.Walsh RC, Koukoulas I, Garnham A, Moseley PL, Hargreaves M, Febbraio MA. Cell Stress Chaperones. 2001;6:386–393. doi: 10.1379/1466-1268(2001)006<0386:eishih>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Suzuki K, Sawa Y, Kaneda Y, Ichikawa H, Shirakura R, Matsuda H. J Mol Cell Cardiol. 1998;30:1129–1136. doi: 10.1006/jmcc.1998.0678. [DOI] [PubMed] [Google Scholar]
- 165.Lee WC, Wen HC, Chang CP, Chen MY, Lin MT. J Appl Physiol. 2006;100:2073–2082. doi: 10.1152/japplphysiol.01433.2005. [DOI] [PubMed] [Google Scholar]
- 166.Hutter JJ, Mestril R, Tam EK, Sievers RE, Dillmann WH, Wolfe CL. Circulation. 1996;94:1408–1411. doi: 10.1161/01.cir.94.6.1408. [DOI] [PubMed] [Google Scholar]
- 167.Radford NB, Fina M, Benjamin IJ, Moreadith RW, Graves KH, Zhao P, Gavva S, Wiethoff A, Sherry AD, Malloy CR, Williams RS. Proc Natl Acad Sci U S A. 1996;93:2339–2342. doi: 10.1073/pnas.93.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]