Abstract
We screened an existing collection of zebrafish insertional mutants for cancer susceptibility by histological examination of heterozygotes at two years of age. As most mutants had no altered cancer predisposition, this provided the first comprehensive description of spontaneous tumor spectrum and frequency in adult zebrafish. Moreover, the screen identified four lines, each carrying a different dominant mutant allele of Hagoromo previously linked to adult pigmentation defects, which develop tumors with high penetrance that histologically resemble neuroblastoma. These tumors are clearly neural in origin, although they do not express catecholaminergic neuronal markers characteristic of human neuroblastoma. The zebrafish tumors result from inappropriate maintenance of a cell population within the cranial ganglia that are likely neural precursors. These neoplasias typically remain small but they can become highly aggressive, initially traveling along cranial nerves, and ultimately filling the head. The developmental origin of these tumors is highly reminiscent of human neuroblastoma. The four mutant Hagoromo alleles all contain viral insertions in the fbxw4 gene, which encodes an F-box WD40 domain containing protein. However, while one allele clearly reduced the levels of fbxw4 mRNA, the other three insertions have no detectable effect on fbw4 expression. Instead, we show that all four mutations result in the post-embryonic upregulation of the neighboring gene, fibroblast growth factor 8 (fgf8). Moreover, fgf8 is highly expressed in the tumorigenic lesions. While fgf8 overexpression is known to be associated with breast and prostate cancer in mammals, this study provides the first evidence that fgf8 misregulation can lead to neural tumors.
Keywords: neural precursors, retrovirus, insertional mutagenesis, cancer
INTRODUCTION
The zebrafish is an increasingly popular model organism in which to study cancer (1). Mutation of known tumor suppressors such as nf2, p53, apc, and mlh1 (2-5) result in cancer susceptibility, and tumorigenesis can also be driven by the expression of a number of oncogenes such as c-myc (6), mutant Kras (7) or mutant braf (8). Additionally, forward genetic screens in zebrafish can identify novel mutations that lead to cancer susceptibility. For example, by screening initially for mutations that affect the cell cycle in homozygous embryos, mutations in bmyb and separase were found to modestly increase the rate of carcinogen-induced cancer in heterozygous adults (9,10). In another study, by conducting a pilot screen of an existing collection of heterozygous carriers of embryonic lethal insertional mutations for the development of externally visible spontaneous tumors, we established that haploinsufficiency of 17 different ribosomal protein genes is highly tumorigenic (2,11). This validates the use of zebrafish to identify novel cancer genes.
The mutant collection used for this pilot screen includes over 500 mutations in over 370 genes (12). These mutations were made by retroviral insertion, allowing rapid identification of the mutated gene in almost all cases by cloning the genomic DNA flanking the mutagenic insertion (13). These mutants were recovered in a large-scale screen designed to find recessive mutations with embryonic phenotypes (14) and nearly all of the mutants are embryonic or larval lethal in their homozygous state. In the course of the screen we also recovered a few dominant mutations with viable adult phenotypes affecting either pigmentation or fin growth. None of these dominant mutations had any visible phenotypes during embryogenesis as either heterozygotes or homozygotes.
We maintain all of the zebrafish mutant lines as stocks of 15−25 heterozygotes until two years of age, thereby allowing for an assessment of adult phenotypes in the heterozygous state. In this study, we have extended our analysis of the full mutant collection by screening for spontaneous tumor development by histological examination as opposed to just externally visible tumors. This revealed that all four alleles of the dominant mutation Hagoromo (14,15) were highly predisposed to the development of neuroblastoma-like tumors arising in the cranial ganglia.
RESULTS
Cancer screen of insertional mutants identifies Hagoromo
We had previously discovered that heterozygous mutation of many ribosomal protein (rp) genes, as well as the nf2a gene (one of two paralogs of the mammalian tumor suppressor NF2) predisposed zebrafish to the development of cancer, primarily malignant peripheral nerve sheath tumors (2). Notably, these mutants develop tumors spontaneously, in the absence of chemical carcinogens. While the ribosomal protein mutants generally developed large, externally visible tumors, the nf2a heterozygotes usually developed much smaller tumors that were only evident upon histological examination of the fish. Thus it is unlikely that we would have discovered that these fish were tumor-prone had we not examined sections of a cohort of apparently healthy fish. Reasoning that there might be other mutations which similarly predisposed fish to tumors that might not be externally visible, we set about to systematically examine heterozygotes from all of the insertional mutant lines in our collection by histology. Initially we examined at least fifteen fish for each line at approximately two years of age. In most cases two longitudinal sections near either side of the midline were examined, although in some cases six sections were screened. For any families that had a tumor frequency in the first cohort greater than background (see below), we collected additional two-year-old fish to determine if this was a true tumor predisposition or just a sampling artifact.
All of the analyzed fish, with the exception of those from lines that we determined to be tumor-prone, served as a control set. Analysis of these 10,000 fish from 437 lines (including mutations in 341 different genes) indicated that the “background” frequency of tumorigenic lesions, including pre-adenoma bile-duct hyperplasia, was about 5% at two years of age (473/9988). The most common tumor types were seminomas and bile duct lesions (hyperplasia, adenoma, and adenocarcinoma), followed by pancreatic islet cell adenoma and leukemia/lymphoma (Figure 1). None of these tumor types appeared in greater than 2% of the fish and we did not find any families in which these tumors were significantly overrepresented. As far as we are aware, this is the first comprehensive analysis of spontaneous tumor incidence and spectrum in zebrafish.
Figure 1.
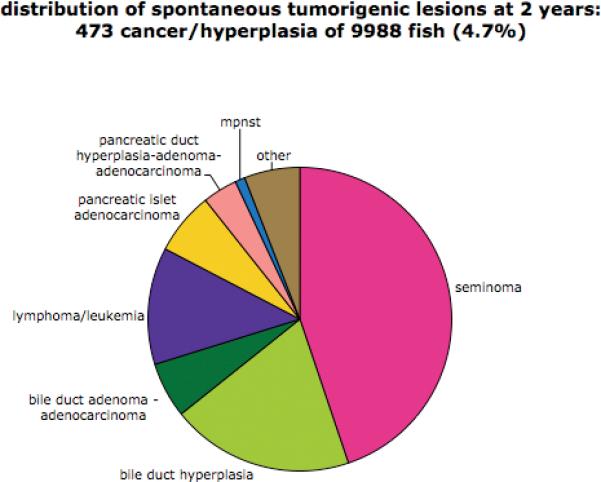
Distibution of types of tumorigenic lesions in the background population.
Of the 342 loci screened in this manner, 339 were recessive embryonic lethal mutations. Beyond the previously noted rp and nf2a genes, none of the heterozygous carriers of recessive lethal mutations had a spontaneous cancer frequency significantly above background. This included mutations in two genes, c-myb and separase, whose heterozygous mutation has been shown to increase cancer frequency in carcinogen-treated fish (9,10). In addition to these recessive mutations, the collection contains dominant mutations in three loci that have no embryonic phenotype (as homozygotes or heterozygotes) but rather have viable visible adult phenotypes either in fin growth or stripe patterning. One of these loci, for which we have four mutant alleles, is Hagoromo (Hag). Hag mutants, named for “the dress of a goddess”, are characterized by disorganized stripes in the adult pigment pattern that are first apparent during the reorganization of iridophores and melanophores during metamorphosis at 3−4 weeks of age (15). Neither heterozygotes nor homozygotes have any embryonic phenotype; homozygotes are viable and have, on average, more severe stripe defects than heterozygotes, sometimes resulting in a spotty appearance in the anterior flank. All four Hag alleles carry insertions in the fbxw4 gene, which encodes an F-box containing WD-40 repeat protein (15). Our screen revealed a significant tumor predisposition in these four Hag alleles. In each of these lines, 25−50% of two-year-old heterozygotes had tumors that histologically resembled neuroblastoma (Table 1, Figure 2). Importantly, these neuroblastoma-like tumors were not observed in any of the ∼10,000 screened fish that lacked insertions in the fbxw4 gene, including 150 non-carrier siblings from the Hag families. Thus, we conclude that fish without insertions in the Hag locus spontaneously develop this tumor type with a frequency less than 1/10,000. This, coupled with the observation that four different insertions in this locus lead to this tumor type, indicates that the insertions at the Hag locus are unquestionably responsible for these cancers.
Table 1.
Incidence of neuroblastoma in 2 year old Hag heterozygotes and their wild type siblings
| Allele | Wild type | Heterozygote | |
|---|---|---|---|
| hiD1 | Total tumors | 0/34 | 16/34 (47%) |
| Small neoplasias | 0/34 | 7/34 (21%) | |
| Advanced tumors | 0/34 | 9/34 (26%) | |
| hiD2 | Total tumors | 0/56 | 19/77 (25%) |
| Small neoplasias | 0/56 | 19/77 (25%) | |
| Advanced tumors | 0/56 | 0/77 | |
| hiD2058 | Total tumors | 0/39 | 17/38 (45%) |
| Small neoplasias | 0/39 | 13/38 (34%) | |
| Advanced tumors | 0/39 | 4/38 (11%) | |
| hiD4000 | Total tumors | 0/24 | 18/59 (30%) |
| Small neoplasias | 0/24 | 9/59 (15%) | |
| Advanced tumors | 0/24 | 9/59 (15%) |
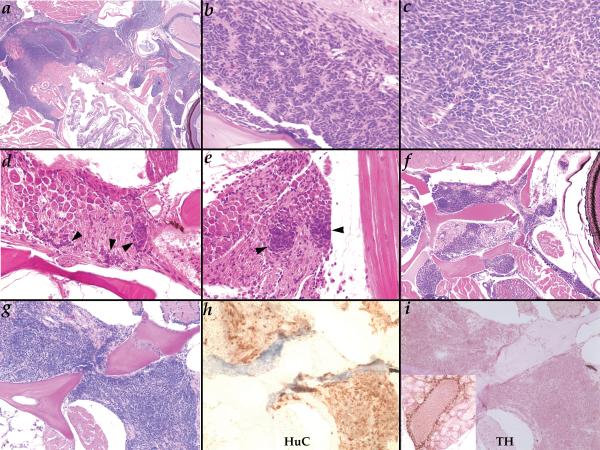
Figure 2.
Neuroblastoma-like tumors in Hag heterozygotes at two years of age. A. (40X) a very advanced tumor fills most of the head space between the esophagus and the skull and invades the body wall musculature; B-C. (400X) rosette arrangement of cells is occasionally observed in tumors (B) but more commonly cellular arrangement is more variable (C); D-E. (400X) very small neoplasias (black arrowheads) observed in the ganglia within the skull below the midbrain (D) or posterior of the ear (E); F. (100X) this tumor can be seen growing along the nerve running behind the eye as well as another below the skull; G-I. (200X) H&E (G), anti-HuC (H), and anti-TH (I) of the same tumor; inset in I shows TH staining of adrenal cells in the kidney on the same slide as a positive control for the antibody.
Hagoromo tumors resemble neuroblastoma and are preceded by the inappropriate maintenance of a putative neural precursor cell population
To better understand the nature and origin of these tumors, we conducted a careful analysis of the Hag fish. Hematoxylin and eosin staining indicated that the tumors in Hag fish indicated that the tumors were made up of small, densely packed, oval cells with very little cytoplasm (Figure 2). While in some tumors, cells were arranged in “rosettes” (Figure 2B) commonly seen in partly differentiated human neuroblastoma (16), more frequently the cells were simply present as sheets with little organization (Figure 2C) as is seen in undifferentiated human neuroblastoma (16). The tumors were found in a wide range of sizes, from small neoplasias (20−30 cells per planar section) to large tumors that occupied nearly the entire head, pushing aside (though never invading) the brain and pushing into musculature of the cheeks and dorsal body wall. The highly aggressive tumors, such as that shown in Figure 2A, were least common, and we rarely observed fish with any external signs of tumor growth, such as a bump on the dorsal surface or bulging of an eye. Small neoplasias were most common (Table 1) and always arose in cranial ganglia, either just below the midbrain but still within the skull (Figure 2D) or in ganglia projecting posteriorly from behind the ear (Figure 2E). As these tumors grew larger, they clearly overtook the entire ganglia and tracked along nerves, for example those leaving the skull and projecting towards the ear or behind the eye (Figure 2F). These tumors did not appear to be ganglioneuromas as they did not produce ganglion cells as they grew. Thus, based upon the origin of these tumors in ganglia as well as their histological appearance, we suspected that they were neuroblastomas. By staining with the pan-neural marker HuC we confirmed that these tumors were of neural origin (Figure 2G-H). However, the tumors failed to stain for tyrosine hydroxylase (Figure 2I). This marker is expressed in the catecholaminergic neurons of the peripheral sympathetic nervous system that are most often the cell of origin for human neuroblastoma (17,18). So while these tumors were unquestionably neural and histologically resemble human neuroblastoma, they did not appear to arise from the same population of neurons that contributes to most human neuroblastoma. However, they may share other characteristics with the human disease.
Neuroblastoma is usually a neonatal or childhood cancer in humans. Tumors of this class typically orginate from a precursor/progenitor population that persists inappropriately because of a failure to instigate the normal differentiation and/or apoptotic program (18,19). In order to determine the origin of the tumors in Hag zebrafish, we conducted a time course analysis of the progeny of Hag heterozygous crosses (including wild type, heterozygous, and homozygous sibs) by analyzing a subset of each cohort at one-month intervals. The cranial ganglia of one-month-old fish contained clusters of cells that appeared very similar in size, shape, and organization to the tumor cells (Figure 3A-B). These were found in both wild type and mutant fish, and thus we conclude that this is a normal developmental stage. However, by two months of age, these cells appeared less frequently and are fewer in number in wild type fish than in either heterozygous or homozygous Hag fish (data not shown). This was even more apparent at three months, by which time these cells were completely absent in the wild type fish, but they were maintained within almost all of the heterozygous and homozygous Hag mutants (Figure 3C-D, Table 2). These cells were neural as they strongly express HuC (Figure 3E-H). Taken together, these data suggest that the presence of this cell population in the wild type zebrafish denotes a stage of neuronal development that is normally completed before 3 months of age. The simplest explanation of this finding is that these cells represent a neural precursor population. We believe that the persistence of these cells in 3 month old Hag mutants is an early event in the tumorigenic process. Notably, these small neoplasias occurred at a similar frequency in both heterozygous and homozygous Hag mutants.
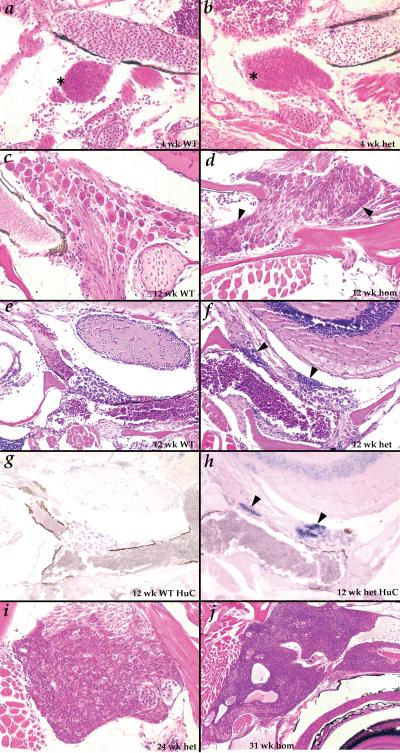
Figure 3.
Tumors begin as inappropriate maintenance of a normal developmental stage of putative neural precursors. A-B. (400X) at 4 weeks small cells resembling the tumor cells (asterisks) are seen in cranial ganglia of both wild type (A) and Hag (B) fish; C-D. (400X) at 12 weeks, only fully developed ganglion cells are observed in wild type fish (C), but the small precursor-like cells (black arrowheads) are still observed in most Hag mutants (D). E-H. (200X) the small cells seen uniquely in Hag mutants (black arrowheads) at 12 weeks (F) stain strongly for HuC mRNA by in situ hybridization (H); ganglion cells in wild type (E) and Hag mutants (F) stain weakly for HuC (G,H); I. (400X) tumor beginning to spread over entire ganglia in 24 week Hag heterozygote; J. (100X) tumor in 31 week homozygote growing within the skull and along nerves behind and above the eye.
Table 2.
Incidence of neural neoplasias and advanced neuroblastoma in Hag heterozygotes, homozygotes and wild type siblings over time
| Allele | Age (weeks) | Wild types | Heterozygotes | Homozygotes | |
|---|---|---|---|---|---|
| hiD4000 | 12 | Small neoplasias | 0/17 | 28/30 (93%) | 17/20 (85%) |
| Advanced tumors | 0/17 | 0/30 | 0/20 | ||
| Dead* | 0/17 | 0/30 | 0/20 | ||
| 24 | Small neoplasias | 0/10 | 20/28 (71%) | 7/9 (78%) | |
| Advanced tumors | 0/10 | 1/28 (4%) | 0/9 | ||
| Dead* | 0/10 | 0/28 | 4/13 (31%) | ||
| 31 | Small neoplasias | 0/16 | 18/28 (64%) | 0/2 | |
| Advanced tumors | 0/16 | 0/28 | 1/2 (50%) | ||
| Dead* | 0/16 | 0/28 | 6/8 (75%) | ||
| 39 | Small neoplasias | N.D. | 6/14 (43%) | 3/10 (30%) | |
| Advanced tumors | N.D. | 2/14 (14%) | 4/10 (40%) | ||
| Dead* | N.D. | 0/14 | 7/17 (41%) | ||
| hiD1 | 17 | Small neoplasias | 0/10 | 22/26 (85%) | 9/11 (82%) |
| Advanced tumors | 0/10 | 0/26 | 0/11 | ||
| Dead* | N.D. | N.D. | N.D. | ||
| 24 | Small neoplasias | 0/21 | 37/52 (71%) | 3/13 (38%) | |
| Advanced tumors | 0/21 | 6/52 (11%) | 4/13 (31%) | ||
| Dead* | N.D. | N.D. | N.D. | ||
| 34 | Small neoplasias | N.D. | 3/10 (30%) | 3/7 (43%) | |
| Advanced tumors | N.D. | 3/10 (30%) | 2/7 (29%) | ||
| Dead* | N.D. | 0/10 | 2/7 (29%) | ||
| hiD2058 | 17 | Small neoplasias | 0/16 | 14/17 (82%) | 9/11 (82%) |
| Advanced tumors | 0/16 | 0/17 | 0/11 | ||
| Dead* | N.D. | N.D. | N.D. | ||
| 24 | Small neoplasias | 0/15 | 29/37 (78%) | 8/11 (73%) | |
| Advanced tumors | 0/15 | 0/37 | 2/11 (18%) | ||
| Dead* | N.D. | N.D. | N.D. | ||
| 34 | Small neoplasias | 0/6 | 5/11 (45%) | 4/14 (29%) | |
| Advanced tumors | 0/6 | 4/11 (36%) | 6/14 (43%) | ||
| Dead* | 0/6 | 0/11 | 4/18 (22%) | ||
| hiD2 | 24 | Small neoplasias | 0/12 | 33/38 (87%) | 11/17 (64%) |
| Advanced tumors | 0/12 | 0/38 | 1/17 (6%) | ||
| Dead* | N.D. | N.D. | N.D. |
these fish died from wasting, presumably subsequent to severe esophageal/gut inflammation, not from tumors. N.D., not determined.
By as early as 5 months these neoplasias began to grow into advanced tumors in a small proportion of Hag fish, spreading throughout entire ganglia and following nerve bundles away from their site of origin (Fig 3I-J). These advanced tumors emerged at a slightly higher rate in homozygotes than in heterozygotes (Table 2). However, our ability to track the tumor development in homozygotes was compromised by an additional phenotype of the homozygous fish; with variable penetrance they develop severe inflammation in the wall of the posterior esophagus and anterior gut, leading to wasting and eventual mortality, possibly due to starvation (Table 2, Figure S1). Taken as a whole, this time course experiment indicates that heterozygotes and homozygotes have a similar propensity to inappropriately maintain this presumed precursor cell population but these neoplasias develop into more advanced tumors at a somewhat faster rate in homozygous versus heterozygous Hag mutants.
Hag mutations upregulate fgf8 expression rather than affect fbxw4
We wished to determine how the Hag mutant alleles contribute to the tumorigenic state. The insertional mutants used in this screen were made with Moloney Murine Leukemia Virus (MoMLV)-based vectors. In mammals, MoMLV insertions can cause either gene activation through enhancer activity of the viral LTRs or inactivation by disruption of splicing or coding sequences (20,21). Most of the mutants we recovered in our screen were recessive (491/497), and we isolated only six dominant mutations that represent insertions in three different loci (12,14, A.A. and N.H. unpublished data). Molecular analysis demonstrates that in all of the recessive mutations that we have examined, expression of the gene in which the insertion lies is reduced or abrogated (11,13,22, A.A. and N.H. unpublished data). In contrast, the cause of the dominant mutations is less clear; they could be loss-of-function alleles of haploinsufficient loci or gain-of-function alleles.
To understand why the Hag mutants are tumor-prone, we first examined if expression of the fbxw4 gene was affected, as all of the insertions lie within this gene, hiD2058 at the splice donor of the first exon and hiD1, hiD2 and hiD4000 all in the fifth intron (Figure 4A). We chose to analyze gene expression in homozygotes as this would facilitate the detection of gene expression changes in cis to the insertion, especially if the effect was down-regulation. We used real time reverse transcriptase polymerase chain reaction (RTPCR) on RNA isolated from whole fish. We examined expression of fbxw4 in wild type fish and homozygotes for three of the lines at two developmental stages: the end of embryogenesis (5 days) and after juvenile morphogenesis (6 weeks), when the pigment phenotype that is the defining characteristic of the Hag mutants is clearly visible (Figure 4B). For hiD2058, the mutant with an insertion at the splice donor of the first exon, expression was reduced to approximately 1% of wild type levels (Figure 4B). Additional analysis indicated that this insertion interferes with the splicing of the message, utilizing a cryptic splice site in the first exon upstream of the initiation codon (data not shown). Thus for hiD2058, the level of fbxw4 is greatly reduced and also incapable of producing full-length protein, if any protein at all. However, for two of the lines with insertions in the fifth intron (hiD1 and hiD4000), there was no change in level of fbxw4 mRNA at either of these time points (Figure 4B). Since all of the alleles have the same phenotype (both pigment and tumor phenotypes) but at least two of the three alleles with insertions in the large intron do not affect fbxw4 expression (expression in hiD2 was not examined), it is unlikely that the loss of fbxw4, even in hiD2058, is responsible for any of the observed phenotypes.
Figure 4.
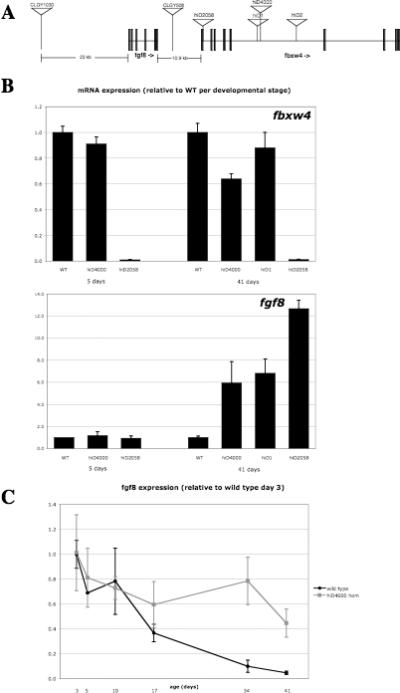
Hag mutations are insertions in the fbxw4 gene but affect expression of the neighboring fgf8 gene. A. Location of the mutagenic insertions in Hag mutants. hiD2058 is precisely at the splice donor of exon 1 of fbxw4; the other three insertions are in the 24 kb fifth intron. Enhancer trap insertions CLGY1030 and CLGY508 are in intergenic sequence upstream and downstream of fgf8, respectively. B. fbxw4 and fgf8 mRNA measured in whole fish by real-time RT-PCR in wild type and homozygotes of multiple Hag alleles at 5 and 41 days. Units are normalized to wild type sample of each developmental age. C. fgf8 mRNA measured in whole fish by real-time RT-PCR in wild type and hiD4000 homozygotes from 3 to 41 days. Units are normalized to the 3 day wild type sample. For B and C, error bars show the standard deviation of 2−3 technical replicates.
The gene immediately upstream of fbxw4 is fgf8 (Figure 4A), and this juxtaposition is conserved between fish and mammals. Fgf8 is a common site for mouse mammary tumor virus (MMTV) insertions in murine mammary cancer, where viral insertions lead to fgf8 overexpression (23-25). There are even some reported cases where MMTV had inserted in the murine fbxw4 gene to cause activation of fgf8 (25). Although we have not previously seen the MoMLV virus activate genes in zebrafish, given the mouse results we thought it would be prudent to examine the effects of these insertions upon fgf8 expression in a manner identical to that used for fbxw4. During embryogenesis, fgf8 expression is very dynamic and its function is especially important, as both loss-of-function mutants and mutations that lead to fgf8 overexpression have early embryonic phenotypes (26,27). In wild type fish, fgf8 expression declines after embryogenesis (Figure 4C) though it is known to remain expressed in some tissues (28,29). During embryogenesis, we did not see any difference in fgf8 mRNA levels between whole wild type fish and Hag homozygotes (Figure 4C). However, at six weeks of age, fgf8 mRNA levels were 6−12 times higher in Hag homozygotes than in wild type controls for all three lines tested (Figure 4B). To further explore this deregulation, we examined fgf8 mRNA levels in wild type and homozygous fish over a number of intervening time points for one allele (hiD4000, Figure 4C). In wild type fish, the overall level of fgf8 mRNA began to decline 1−2 weeks after embryogenesis was complete and fgf8 mRNA dropped to very low levels by 5−6 weeks of age. In contrast, the amount of fgf8 mRNA was maintained at near-embryonic levels in Hag homozygotes throughout the time course. Notably, there is an excellent correlation between the loss or maintenance of fgf8 mRNA in the wildtype and Hag mutant fish and the loss or maintenance of the presumed neural precursor population. Importantly, the timeframe of fgf8 downregulation precedes the loss of the putative precursor cells in wild type fish suggesting that high fgf8 somehow contributes to the maintenance of this population in the Hag mutants. As expression was measured from RNA prepared from whole fish, it is not clear if this difference is the result of higher levels of fgf8 in cells that normally express it or ectopic expression of fgf8. Nonetheless, it is clear that fgf8 is dramatically upregulated in Hag mutants and that it is likely fgf8, not fbxw4, which is the gene responsible for the Hagoromo phenotypes. Consistent with this hypothesis, we note that an enhancer trap screen using another MoMLV-based retrovirus (30) has yielded two insertions, CLGY1030 and CLGY508 (31), that lie in intergenic sequences upstream and downstream of fgf8, respectively (fig 4A). Analysis of fgf8 expression in these mutants revealed both increased fgf8 expression in the midbrain-hindbrain boundary at 3 dpf and ectopic fgf8 expression in the spinal region of 35 dpf fish, and these two mutants develop a stripe phenotype that is highly reminiscent of our Hagoromo mutants (A.Z.K and T.S.B., unpublished data). Histological analysis of these lines show that they also develop neuroblastoma tumors (Figure S2). Taken together, these data strongly suggest that the tumor predisposition in these six lines result from a failure to downregulate fgf8 subsequent to embryogenesis and the consequent persistence of a putative neuronal precursor population.
As a further test of this hypothesis, we wished to determine if increased fgf8 expression occurs in the region of the cranial ganglia where the tumors originate. Thus we analyzed fgf8 expression by both semi-quantitative RT-PCR and real-time RT-PCR on mRNA from several parts of the head of wild type and both heterozygous and homozygous Hag fish at 3 months of age, the time at which the clusters of neural precursors are first completely absent in wild type fish but maintained in the Hag mutants. The tissues analyzed included the brain, the skull, the soft tissue ventral to the skull (which included the cranial ganglia), and the jaws and skin. Of these tissues, the brain had the highest overall fgf8 expression in wild type fish, and fgf8 expression appeared slightly higher in the brain of Hag mutants. The other three tissues, where fgf8 was expressed at a low level in wild type fish, showed dramatically higher levels of fgf8 in Hag mutants (Figure 5A). Thus, we conclude that elevated fgf8 expression is not restricted to the cranial ganglia but is clearly present in this region of the Hag mutants. Most importantly, in situ hybridization with antisense fgf8 probe on sections cut from fish with tumors shows robust expression of fgf8 in most, although not all, cells in the tumor (Figure 5B-C). Thus, we conclude that the neuroblastomas themselves express fgf8 at a very high level.
Figure 5.
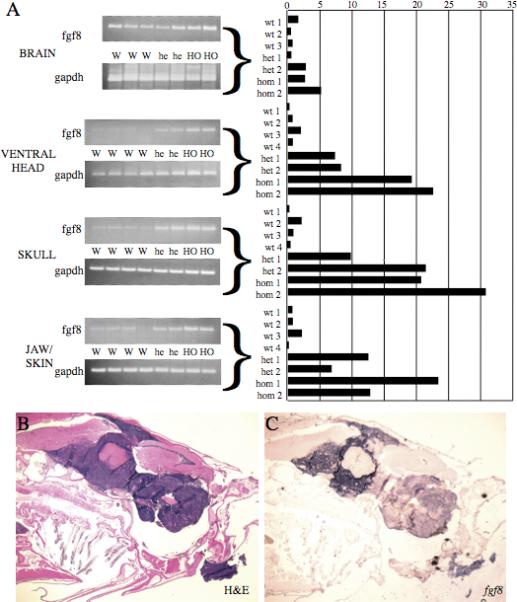
Hag mutants overexpress fgf8 in the head region of young adults and in tumors A. fgf8 mRNA in multiple parts of the head of 3 month old wild type, hiD2058 heterozygote and hiD2058 homozygote fish were measured by real-time and semi-quantitative RT-PCR; gapdh was used as a normalization control. Samples were isolated from three (brain) or four wild type fish and two each Haghi2058 heterozygotes and homozygotes. Ethidium bromide stained gels from semi-quantitative RT-PCR are shown on the left; quantification of the real-time PCR is shown on the right. Quantification is normalized within the dataset for each tissue to the average wild type sample of that tissue; error bars show the standard deviation of three technical replicates. B-C. (20X) H&E (B) and fgf8 in situ hybridization (C) on a very advanced tumor in a 2 year old hiD2058 homozygote.
DISCUSSION
Through the unbiased histological screening of a collection of zebrafish insertional mutants, we determined that four independent lines carrying mutations in the Hagoromo locus develop neuroblastoma-like tumors at high penetrance. In some regards, these tumors are quite different than human neuroblastoma, which arises from neural crest-derived neural precursors in the sympathetic nervous system and grow predominantly in the abdomen or neck (17,18). Tumors in Hagoromo fish arise in cranial ganglia and grow in the head. While they are clearly of neural origin, as judged by their expression of HuC, these zebrafish tumors do not express markers of catecholaminergic neurons indicating that they are derived from a different neural lineage than human neuroblastoma. Despite this difference, the Hagoromo tumors seem to arise through a similar mechanism as the human disease. Specifically, human neuroblastoma typically arises in infants and is thought to be a tumor of embryonic origin, i.e. derived from neural precursors present during fetal development (18,19). Our analysis shows that wildtype zebrafish have a population of cells which exists in the cranial ganglia of juvenile fish but is lost as the fish grow to adulthood. We hypothesize that these cells represent neural precursors and their disappearance, presumably through differentiation or apoptosis, reflects the normal developmental program. These cells are inappropriately maintained in Hagoromo mutants and they are the originating cell of the Hagoromo mutant tumors. While in many cases these clusters of cells fail to grow past the size of small neoplasias, in 10−20% of the mutants they eventually grow into advanced tumors beginning as early as five months. This stochastic event likely reflects the need to acquire additional mutations within these cells. The Hagoromo mutation seems to act by prolonging the existence of, and possibly driving the expansion of, this target cell population.
The four Hagoromo mutant lines each carry viral insertions in the fbxw4 gene. However, our data show that they do not have a shared effect on fbxw4 expression: one allele, in which the insertion lies at the splice donor of the first exon, does diminish fbxw4 mRNA levels, but the other insertions, which lie in the fifth intron, have no detectable effect on fbxw4. Instead, our data suggest that the key consequence of the Hagoromo insertions is to maintain high levels of fgf8 expression after embryogenesis. Specifically, we find that fgf8 mRNA is dramatically upregulated in Hagoromo mutants after the completion of embryonic development, including expression in the head region where the inappropriately maintained precursors are first observed. In addition, very high fgf8 expression is seen in the tumors themselves. Finally, insertional mutants have been recovered with a related virus from a different screen with phenotypes including both the stripe phenotype and the neuroblastoma phenotype in which the insertions lay outside of fbxw4, either between fgf8 and fbxw4 or upstream of fgf8, distal to fbxw4.
The mechanism by which these insertions activate fgf8 expression is unclear. That the gene containing the viral insertions is not truly the affected gene should serve as a cautionary example for the analysis of other insertional mutants. In this case, although we cannot rule out direct viral activation, our previous studies suggest that the MoMLV LTR does not activate transcription in zebrafish cells; viruses relying upon the LTR to drive expression of the lacZ gene produced no ß-galactosidase activity while viruses with an internal promoter driving lacZ were able to do so (32). Additionally, in all other cases where the expression of genes at the site of MoMLV insertions in zebrafish has been examined, gene expression has either decreased or been unaffected (11,13,22,33). Thus, we instead favor the notion that the virus disrupts the transcriptional control of fgf8 by distal cis-acting elements. This hypothesis is supported by several lines of evidence showing that fgf8 expression is controlled by a number of enhancers upstream, within, and downstream of the gene, including elements within the fbxw4 gene (31,34,35). Additionally, retroviral insertions in the Fgf8/Fbxw4 locus in the mouse (36-37) or genomic duplications near FGF8 in humans (38-39) cause a limb outgrowth phenotype (dactylaplasia or split hand foot malformation). The mechanism of these mouse and human mutations is not understood. However, the mouse mutants have been shown to have a defect in the maintenance of the apical ectodermal ridge (AER) of the developing limb, and Fgf8 expression is normal at the beginning of limb development but is quickly lost (36). While it has been suggested that this altered Fgf8 expression is a secondary consequence of AER loss, it also seems possible that the insertions in the mouse dactylaplasia mutants affect fgf8 expression in a direct, but complex, way that somehow accounts for the observed downregulation after limb development begins. Although the molecular and phenotypic consequences of insertions and chromosomal rearrangements of the Fgf8/Fbxw4 locus clearly differ between mammals and fish, it seems plausible that the resulting phenotypes in each organism (limb outgrowth defects in mammals, maintenance of neural precursor cells and stripe disruption in zebrafish) can be accounted for by differential effects upon fgf8 expression.
Regardless of the mechanism by which the zebrafish insertions cause continued high levels of fgf8 expression after embryogenesis, it clearly leads to the inappropriate maintenance and expansion of a putative neural precursor population that ultimately become tumors. We note that the increase in fgf8 expression in the head region of Hagoromo fish occurs at a time when this cell population diminishes in wild type fish. This suggests that fgf8 is capable of contributing to the maintenance of these cells, either by blocking differentiation or promoting survival. It would seem paradoxical that fgf8 should prevent differentiation of these precursor cells as many studies have associated fgf8 with neural differentiation. A requirement for fgf8 in the development of neurons in various areas of both the central and peripheral nervous system, including the epibranchial placodes that give rise to the cranial ganglia, has been demonstrated both in zebrafish, (40-42) and in mice (43). Furthermore, Fgf8 cooperates with retinoic acid to force mouse embryonic carcinoma P19 cells to differentiate into neurons while inhibition of fgf signaling in this cell culture system attenuates neural differentiation (44). However, this pro-differentiation role for fgf8 might be restricted to specific developmental stages such as embryogenesis. For example, in some settings fgf8 signaling can promote proliferation of neural precursors at the expense of differentiation (45). The other possibility, that fgf8 acts as a pro-survival signal for these cells, is supported by the observation that several fgfs, including fgf8, have anti-apoptotic functions in both neural crest and ectoderm-derived neurons (43,46). The effects of fgf8 upon differentiation and survival may depend upon timing and context, such that early in development neural differentiation is the favored response, but sustained fgf8 expression past the time when such differentiation normally occurs may result in inappropriate survival and/or proliferation of the few remaining neural precursor cells.
Thus fgf8 appears to be an oncogene, in that its overexpression predisposes zebrafish to tumorigenesis. However, the tumor phenotype in Hagoromo mutants is not fully penetrant. Notably, it appears that fgf8 overexpression in Hagoromo mutants is always sufficient to disrupt the orderly alignment of pigment cells (which are moving into their stripe pattern at the time when we first see fgf8 overexpression in the mutants), as well as the inappropriate maintenance of putative neural precursor cells in cranial ganglia. However, these only progress to tumors in a subset of cases, suggesting that additional mutations are required. It is possible that there is only a limited window of opportunity for the precursor cell population to acquire these changes. This model is supported by our finding that small neoplasias exist in nearly all of the mutants at 3−6 months of age (and we might miss some due to sampling only three sections per fish) but are detected in fewer than half of the fish at two years of age. Thus, we speculate that fgf8 overexpression allows for the persistence of a cell population that is then a target for additional mutations, and there may be a limited time in which such mutations can contribute to tumorigenesis. We do not currently know what these additional mutagenic events are, and this is an important question for future study. However, we believe it is unlikely that mutation of the tumor suppressor gene p53 is one such key event, as fish mutant for both Hagoromo and p53 (3) do not have a higher rate of neuroblastoma formation by one year of age than fish mutant for Hagoromo alone (data not shown).
Fgf8 has also been implicated in tumorigenesis in other tumor types in mammals. Overexpression of FGF8 has been observed in a number of human tumors, especially prostate and breast cancer (47-49). Furthermore, whole genome association studies indicate that specific polymorphisms in the fgf receptor FGFR2 correlate with an increased frequency of breast cancer (50,51). Direct evidence for the oncogenic properties of Fgf8 in these tissues is even clearer in mice. The Fgf8 locus is a common insertion site for MMTV in retrovirus-induced mammary tumors (23-25), implying that its activation in mammary tissue can lead to tumorigenesis. Additionally, transgenic tissue-specific overexpression of Fgf8 cooperates with Pten loss in mouse models of prostate cancer (52). Thus, FGF8 appears to function as oncogene in numerous settings including prostate and breast cancer in mammals, and now neuroblastoma in zebrafish.
MATERIALS AND METHODS
Fixation and Histology
Adult fish were euthanized in 500mg/L Tricaine and fixed in either Bouin's fixative or 10% neutral buffered formalin (in cases where we wished to retain the option of immunohistochemistry or in situ hybridization). Embedding in paraffin and sectioning were performed as previously described (2,53). Prior to fixation a piece of tail tissue was retained for the isolation of genomic DNA and either Southern analysis or PCR was conducted to determine the genotype of fish from the Hag heterozygous crosses (54).
Antibody staining
Slides from fish fixed in 4% paraformaldehyde/PBS and paraffin embedded were stained as previously described (55). Primary antibodies used were anti-TH (P40101−0, 1:100, Pel-Freez, Rogers AR) and anti-HuC (16A11, 1:250, Molecular Probes, Eugene OR)
RNA In Situ Hybridization
Dig-labeled riboprobes were produced for both sense and antisense strands of the coding regions of the zebrafish fgf8 and HuC genes using a dig-labeling kit (Roche 11175033910) Slides from fish fixed in 4% paraformaldehyde/PBS and paraffin embedded were dewaxed (55), rinsed with PBS, treated with Proteinase K (40 μg/ml) for 7 min, rinsed with PBS, refixed in 4% paraformaldehyde/PBS for 20 min, rinsed with PBS, andrinsed twice in 2X SSC. They were then prehybridized in hybridization solution (50% formamide, 5X SSC, 0.1% Tween, 1 mg/ml tRNA, 50 μg/ml heparin) for 3 hours at 70°C, and hybridized overnight at 70°C in a humid box with 100 μl hybridization solution plus probe (1 μg/ml) under a cover slip. Slides were washed at 70°C for 10 min with 75% hyb/25% 2X SSC, 50% hyb/50% 2X SSC, 25% hyb/75% 2X SSC, 2X SSC, and for 30 min with 0.2X SSC. Slides were equilibrated in MAB + 0.1% tween for 30 min, blocked MAB + 0.1% tween + 10% lamb serum + 2% blocking reagent (Roche 11096176001) then AP-conjugated anti-DIG Fab fragments were applied (1:1000, Roche 11093274910) overnight at 4°C. Antibody was washed 5 × 30 min in MAB + 0.1% Tween, and AP activity was detected with NBT/BCIP as recommended by the maufacturer (Roche, 11383213001 and 11383221001). Figures show results with antisense probes; no blue stain was observed with sense-strand probes of simultaneously processed adjacent-level slides.
Quantitative RNA analysis
RNA was prepared from embryos, adult fish tissues, or whole juvenile fish using Trizol reagent (Invirtrogen, Carlsbad CA); in the case of whole fish 17dpf or older, fish were flash-frozen in liquid nitrogen and mashed in a mortar and pestle prior to homogeniztion in Trizol. First strand cDNA was prepared using Superscript III reverse transcriptase (Invirtrogen, Carlsbad CA) and gene expression levels were determined using real time PCR with Sybr Green Master Mix (ABI). Standard curves were established for each primer set with cDNA dilutions and all samples were run in triplicate and normalized with primers for gapdh. All primer sequences are available upon request.
Supplementary Material
Figure S1: Hagoromo homozygotes often develop severe inflammation in the wall of the posterior esophagus and anterior gut. A-B. (200X) A typical homozygote (A) compared to a wild type sibling (B) at 8 weeks of age; arrowheads in (A) indicate inflammatory cells. C-D. (100X) A typical homozygote (C) compared to a heterozygous sibling (D) at 12 weeks of age. The fish shown in (C), as with many of the homozygotes, was much smaller than its heterozygous and wild type siblings, possibly due to decreased food uptake resulting from blockage of the food tract or scarring of the smooth muscle around the esophagus and foregut; many of these fish die after a period of wasting. g, gut; e, esophagus.
Figure S2: Neuroblastomas develop in mutants with intergenic insertions upstream and downstream of fgf8. A. (20X) a very advanced tumor in a 16.5 month old CLGY508 homozygote fills most of the skull and invades the dorsal body wall musculature; B. the same tumor at 400X shows that the cellular morphology is identical to those seen in the Hagoromo mutant tumors, C. (100X) typical medium sized tumor in a 15 month CLGY1030 homozyogte, beginning in the skull and following nerves ventrally out of the skull D. (400X) very small neoplasia (black arrowheads) observed in the ganglia posterior of the ear of a 15 month CLGY1030 heterozygote.
ACKNOWLEDGEMENTS
We thank Sarah Farrington and Kate Anderson for maintenance of the mutant lines of fish and assistance in sample collection, and Tim Angelini and Sam Farrington for maintenance of the zebrafish colony. We thank the Histology Facility of the MIT Koch Institute for Integrative Cancer Research, especially Alicia Caron and Weijia Zhang, for sample processing and sectioning. We thank Drs. A. Thomas Look and Jeffery Kutok for helpful discussions and consultation on histopathology. N.H. and J.A.L. were funded by an NIH-NCI grant (CA106416). T.S.B. was supported by grants from the Sars Centre, the National Programme in Functional Genomics (FUGE) in Norway, and by the European Commission as part of the ZF-Models Integrated Project in the 6th Framework Programme (Contract No. LSHG-CT-2003−503496). J.A.L. is a Ludwig Scholar at MIT.
REFERENCES
- 1.Feitsma H, Cuppen E. Zebrafish as a Cancer Model. Mol Cancer Res. 2008;6:685–94. doi: 10.1158/1541-7786.MCR-07-2167. [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam A, Sadler KC, Lai K, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berghmans S, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102:407–12. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haramis AP, Hurlstone A, van der Velden Y, et al. Adenomatous polyposis coli-deficient zebrafish are susceptible to digestive tract neoplasia. EMBO Rep. 2006;7:444–9. doi: 10.1038/sj.embor.7400638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feitsma H, Kuiper RV, Korving J, Nijman IJ, Cuppen E. Zebrafish with mutations in mismatch repair genes develop neurofibromas and other tumors. Cancer Res. 2008;68:5059–66. doi: 10.1158/0008-5472.CAN-08-0019. [DOI] [PubMed] [Google Scholar]
- 6.Langenau DM, Traver D, Ferrando AA, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–90. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 7.Langenau DM, Keefe MD, Storer NY, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–95. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patton EE, Widlund HR, Kutok JL, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–54. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 9.Shepard JL, Amatruda JF, Stern HM, et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci U S A. 2005;102:13194–9. doi: 10.1073/pnas.0506583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepard JL, Amatruda JF, Finkelstein D, et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007;21:55–9. doi: 10.1101/gad.1470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai K, Amsterdam A, Farrington S, et al. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn. 2008 doi: 10.1002/dvdy.21815. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amsterdam A, Nissen RM, Sun Z, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–7. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaiano N, Amsterdam A, Kawakami K, et al. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–32. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 14.Amsterdam A, Burgess S, Golling G, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–24. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami K, Amsterdam A, Shimoda N, et al. Proviral insertions in the zebrafish hagoromo gene, encoding an F-box/WD40-repeat protein, cause stripe pattern anomalies. Curr Biol. 2000;10:463–6. doi: 10.1016/s0960-9822(00)00444-9. [DOI] [PubMed] [Google Scholar]
- 16.Machin GA. Histogenesis and histopathology of neuroblastoma. In: Pochedly C, editor. Neuroblastoma: Clinical and biological manifestations. Elsevier Biomedical; New York: 1982. pp. 195–231. [Google Scholar]
- 17.McConville CM, Forsyth J. Neuroblastoma – a developmental perspective. Cancer Lett. 2003;197:3–9. doi: 10.1016/s0304-3835(03)00075-2. [DOI] [PubMed] [Google Scholar]
- 18.Brodeur GM, Maris JM. Neuroblastoma. In: Pizzo PA, Poplack DG, editors. Principles and practice of pediatric oncology. Lippincott Williams & Wilkins; Philadelphia: 2002. pp. 895–937. [Google Scholar]
- 19.Dyer MA. Mouse models of childhood cancer of the nervous system. J Clin Pathol. 2004;57:561–76. doi: 10.1136/jcp.2003.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaenisch R, Soriano P. Retroviruses as tools for mammalian development. Symp Fundam Cancer Res. 1986;39:59–65. [PubMed] [Google Scholar]
- 21.Mikkers H, Berns A. Retroviral insertional mutagenesis: tagging cancer pathways. Adv Cancer Res. 2003;88:53–99. doi: 10.1016/s0065-230x(03)88304-5. [DOI] [PubMed] [Google Scholar]
- 22.Golling G, Amsterdam A, Sun Z, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–40. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 23.MacArthur CA, Shankar DB, Shackleford GM. Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol. 1995;69:2501–07. doi: 10.1128/jvi.69.4.2501-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valve EM, Tasanen MJ, Ruohola JK, Harkonen PL. Activation of Fgf8 in S115 mouse mammary tumor cells is associated with genomic integration of mouse mammary tumor virus. Biochem Biophys Res Commun. 1998;250:805–8. doi: 10.1006/bbrc.1998.9397. [DOI] [PubMed] [Google Scholar]
- 25.Theodorou V, Kimm MA, Boer M, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–69. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 26.Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrainhindbrain boundary development and somitogenesis. Development. 1998;125:2381–95. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- 27.Heisenberg C-P, Brennan C, Wilson SW. Zebrafish aussichtmutant embryos exhibit widespread overexpression of ace (fgf8) and coincident defects in CNS development. Development. 1999;126:2129–2140. doi: 10.1242/dev.126.10.2129. [DOI] [PubMed] [Google Scholar]
- 28.Albertson RC, Yelick PC. Fgf8 haploinsufficiency results in distinct craniofacial defects in adult zebrafish. Dev Biol. 2007;306:505–15. doi: 10.1016/j.ydbio.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Topp S, Stigloher C, Komisarczuk AZ, Adolf B, Becker TS, Bally-Cuif L. Fgf signaling in the zebrafish adult brain: association of Fgf activity with ventricular zones but not cell proliferation. J Comp Neurol. 2008;510:422–39. doi: 10.1002/cne.21802. [DOI] [PubMed] [Google Scholar]
- 30.Ellingsen S, Laplante MA, Konig M, et al. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- 31.Kikuta H, Laplante M, Navratilova P, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–55. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaiano N, Allende M, Amsterdam A, Kawakami K, Hopkins N. Highly efficient germ-line transmission of proviral insertions in zebrafish. Proc Natl Acad Sci U S A. 1996;93:7777–82. doi: 10.1073/pnas.93.15.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Jao LE, Zheng N, et al. Efficient genome-wide mutagenesis of zebrafish genes by retroviral insertions. Proc Natl Acad Sci USA. 2007;104:12428–33. doi: 10.1073/pnas.0705502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue F, Nagayoshi S, Ota S, et al. Genomic organization, alternative splicing, and multiple regulatory regions of the zebrafish fgf8 gene. Dev Growth Differ. 2006;48:447–62. doi: 10.1111/j.1440-169X.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 35.Beermann F, Kaloulis K, Hofmann D, Murisier F, Bucher P, Trumpp A. Identification of evolutionarily conserved regulatory elements in the mouse Fgf8 locus. Genesis. 2006;44:1–6. doi: 10.1002/gene.20177. [DOI] [PubMed] [Google Scholar]
- 36.Sidow A, Bulotsky MS, Kerrebrock AW, et al. A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet. 1999;23:104–7. doi: 10.1038/12709. [DOI] [PubMed] [Google Scholar]
- 37.Kano H, Kurahashi H, Toda T. Genetically regulated epigenetic transcriptional activation of retrotransposon insertion confers mouse dactylaplasia phenotype. Proc Natl Acad Sci U S A. 2007;104:19034–19039. doi: 10.1073/pnas.0705483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Mollerat XJ, Gurrieri F, Morgan CT, et al. A genomic rearrangement resulting in a tandem duplication is associated with split hand-split foot malformation 3 (SHFM3) at 10q24. Hum Mol Genet. 2003;12:1959–1971. doi: 10.1093/hmg/ddg212. [DOI] [PubMed] [Google Scholar]
- 39.Lyle R, Radhakrishna U, Blouin JL, et al. Split-hand/split-foot malformation 3 (SHFM3) at 10q24, development of rapid diagnostic methods and gene expression from the region. Am J Med Genet A. 2006;140:1384–95. doi: 10.1002/ajmg.a.31247. [DOI] [PubMed] [Google Scholar]
- 40.Ye W, Shimamura K, Rubenstein JLR, Hynes MA, Rosenthal A. FGF and Shh Signals Control Dopaminergic and Serotonergic Cell Fate in the Anterior Neural Plate. Cell. 1998;93:755–66. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- 41.Guo S, Brush J, Teraoka H, et al. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–66. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- 42.Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134:611–23. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- 43.Kawauchi S, Shou J, Santos R, et al. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–23. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- 44.Wang C, Xia C, Bian W, et al. Cell aggregation-induced FGF8 elevation is essential for P19 cell neural differentiation. Mol Biol Cell. 2006;17:3075–84. doi: 10.1091/mbc.E05-11-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimers D, López-Toledano MA, Mason I, et al. Developmental expression of fibroblast growth factor (FGF) receptors in neural stem cell progeny. Modulation of neuronal and glial lineages by basic FGF treatment. Neurol Res. 2001;23:612–21. doi: 10.1179/016164101101199090. [DOI] [PubMed] [Google Scholar]
- 46.Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess S. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–2554. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- 47.Marsh SK, Bansal GS, Zammit C, et al. Increased expression of fibroblast growth factor 8 in human breast cancer. Oncogene. 1999;18:1053–60. doi: 10.1038/sj.onc.1202392. [DOI] [PubMed] [Google Scholar]
- 48.Dorkin TJ, Robinson MC, Marsh C, Bjartell A, Neal DE, Leung HY. FGF8 over-expression in prostate cancer is associated with decreased patient survival and persists in androgen independent disease. Oncogene. 1999;18:2755–61. doi: 10.1038/sj.onc.1202624. [DOI] [PubMed] [Google Scholar]
- 49.Mattila MM, Harkonen PL. Role of fibroblast growth factor 8 in growth and progression of hormonal cancer. Cytokine Growth Factor Rev. 2007;18:257–266. doi: 10.1016/j.cytogfr.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, et al. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9:R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong C, Saribekyan G, Liao CP, Cohen MB, Roy-Burman P. Cooperation between FGF8b overexpression and PTEN deficiency in prostate tumorigenesis. Cancer Res. 2006;66:2188–94. doi: 10.1158/0008-5472.CAN-05-3440. [DOI] [PubMed] [Google Scholar]
- 53.Moore JL, Aros M, Steudel KG, Cheng KC. Fixation and decalcification of adult zebrafish for histological, immunocytochemical, and genotypic analysis. BioTechniques. 2002;32:300–4. doi: 10.2144/02322st03. [DOI] [PubMed] [Google Scholar]
- 54.Amsterdam A, Hopkins N. Retroviral-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 2004;77:3–30. doi: 10.1016/s0091-679x(04)77001-6. [DOI] [PubMed] [Google Scholar]
- 55.Danielian PS, Bender Kim CF, Caron AM, Vasile E, Bronson RT, Lees JA. E2f4 is required for normal development of the airway epithelium. Dev Biol. 2007;305:564–76. doi: 10.1016/j.ydbio.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Hagoromo homozygotes often develop severe inflammation in the wall of the posterior esophagus and anterior gut. A-B. (200X) A typical homozygote (A) compared to a wild type sibling (B) at 8 weeks of age; arrowheads in (A) indicate inflammatory cells. C-D. (100X) A typical homozygote (C) compared to a heterozygous sibling (D) at 12 weeks of age. The fish shown in (C), as with many of the homozygotes, was much smaller than its heterozygous and wild type siblings, possibly due to decreased food uptake resulting from blockage of the food tract or scarring of the smooth muscle around the esophagus and foregut; many of these fish die after a period of wasting. g, gut; e, esophagus.
Figure S2: Neuroblastomas develop in mutants with intergenic insertions upstream and downstream of fgf8. A. (20X) a very advanced tumor in a 16.5 month old CLGY508 homozygote fills most of the skull and invades the dorsal body wall musculature; B. the same tumor at 400X shows that the cellular morphology is identical to those seen in the Hagoromo mutant tumors, C. (100X) typical medium sized tumor in a 15 month CLGY1030 homozyogte, beginning in the skull and following nerves ventrally out of the skull D. (400X) very small neoplasia (black arrowheads) observed in the ganglia posterior of the ear of a 15 month CLGY1030 heterozygote.