Abstract
Treatment with protease inhibitors, a component of Highly Active Anti-retroviral Therapy (HAART), often results in viral resistance. Structural and biochemical characterization of a 6X mutant arising from in vitro selection with compound 1, a C2-symmetric diol protease inhibitor, has been previously described. We now show that compound 2, a copper(I)-catalyzed 1,2,3-triazole derived compound previously shown to be potently effective against wild-type protease (IC50 = 6.0 nM), has low nM activity (IC50 = 15.7 nM) against the multidrug-resistant 6X HIV-1 protease mutant. Compound 2 displays similar efficacy against wild-type and 6X HIV-1 in viral replication assays. While structural studies of compound 1 bound to wild type and mutant protease revealed a progressive change in binding mode in the mutants, the 1.3 Å resolution 6X protease–compound 2 crystal structure reveals nearly identical interactions for 2 as in the wild-type protease complex with very little change in compound 2 or protease conformation.
Keywords: HIV-1, protease, drug resistance, click chemistry
Introduction
Cumulative deaths from AIDS since its identification rank the disease as one of the deadliest in human history, and is one of the leading cause of infectious disease deaths worldwide.1 Highly Active Antiretroviral Therapy (HAART) has vastly improved the life span of HIV-infected individuals.2 However, poor patient compliance and high viral mutation rates often lead to protease inhibitor (PI)-resistant HIV strains. The emergence of specific PI–resistant viruses, as well as PI multidrug-resistant viruses, also termed cross-resistance, reduces the efficacy of treatment. The loss of PI efficacy necessitates changes in patient treatments and the need for new and more effective PIs in an ongoing battle against the viral evolution of resistance.3, 4 While PI resistant forms of protease have been extensively catalogued in HIV+ patient populations, the contribution of PI resistance mutations on protease structure and inhibitor interactions is less well understood. Many resistance mutations occur distal to the active site and do not involve amino acids contributing directly to interactions with either substrate or PIs, yet have substantial effects on inhibitor binding.5–8 Moreover, some mutations are associated with promoting cross-resistance to numerous PIs, while other mutations are associated with resistance to only one PI.9
Compound 1, a C2-symmetric diol, is a broad-based protease inhibitor that has nanomolar activity against HIV-1, SIV, FIV, and viral isolates from patients that failed treatment with indinavir, saquinavir, nelfinavir and ritonavir.10, 11 To understand the biochemical and structural contributions of mutations in PI-resistance we have previously generated a temporal series of PI-resistant HIV-1 mutants, 1X (V82A), 3X (V82A, M46I, F53L) and 6X (V82A, M46I, F53L, V77I, L24I, L63P), from wild-type virus under the selective pressure of compound 1.10 As reported, the PR mutants, 1X (PR1X), 3X (PR3X), and 6X (PR6X), exhibited 4-, 11- and 30-fold increases in IC50 to 1 respectively. Moreover, Km increased for selective substrates for all mutants and kcat decreased for PR1X, but increased for PR3X and PR6X. The net result was an increased catalytic efficiency for the PR3X and PR6X and improved viral fitness. The recovered enzymatic efficiency, kcat/Km, of the PR6X was 60% of the wild-type protease. Moreover, the PR6X variant was found to be multidrug resistant with IC50s increasing approximately 9-, 8- and 14-fold for saquinavir, nelfinavir and ritonavir, respectively.
We have previously reported on the X-ray crystallographic analyses of PR1x–6x series and showed that the dimeric PR6X–compound 1 complex assumed an asymmetric conformation relative to the wild-type dimer.12 The PI-induced mutations affected protein core packing and hydrogen bonding networks within the protease, promoting the movement of the monomer B flap region towards monomer A, resulting in a break in symmetry typically observed in dimeric HIV-1 protease structures. This asymmetric arrangement resulted in the disruption of 7 hydrogen bonds and a number of the van der Waals interactions with compound 1. The conformational change in the PR6X provides a molecular mechanism of resistance whereby PI binding affinity was considerably decreased, while efficiency of Gag-Pol substrate cleavage remained similar to the wild type enzyme.12 Therefore, the multidrug resistant PR6X variant -containing virus is useful for evaluating protease inhibitor efficacy and the cloned and expressed PR6X for determining structural mechanisms that afford drug resistance.
As a strategy to develop PIs with potential efficacy against multidrug-resistant viruses, we reported on the feasibility of rapid synthesis and selection of PIs from diverse libraries of small compounds derived from Cu(I)-catalyzed azide–alkyne [3+2] cycloaddition chemistry (click chemistry).13 The use of click chemistry can aid in synthesizing potent inhibitors by optimizing the diversity of resulting compounds with structures in the P2/P2’ positions, regions that have been shown to be critical for inhibitor binding in the protease active site.14, 15 Moreover, the rapid synthesis of potential PIs allows their use as chemical probes on our panel of PI–resistant viruses and purified PRs thereby affording biochemical and structural comparisons with reported PIs. We have previously reported that one amprenavir-like compound obtained from the synthesis, compound 2 (Figure 1), was found to have an IC50 of 6.0 nM on wild-type PR and was found to be efficacious on the protease inhibitor active single-site mutants, V82F, V82A, and G48V.13
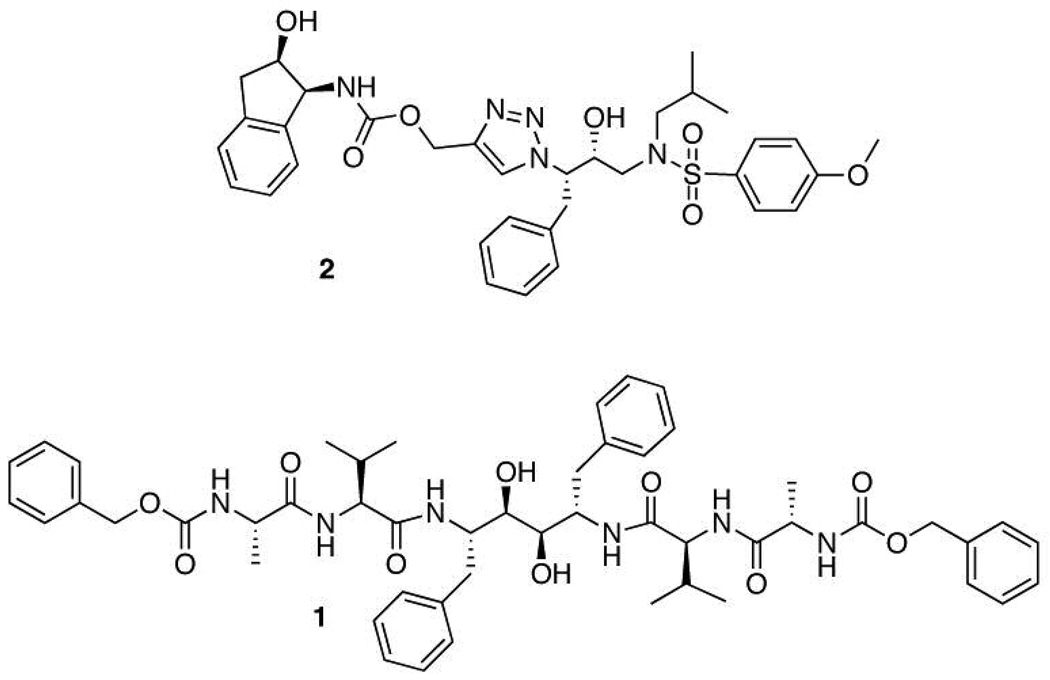
Figure 1.
Two dimensional structural representations of 2 and 1. The central triazole of 2 arises from click chemistry, while 1 is a mechanism-based inhibitor with C2 symmetry.
We report herein on our studies of the efficacy of compound 2 against the multidrug resistant PR6X containing virus and purified PR6X in biochemical assays. Compound 1 was effective against the PR6X virus in tissue culture and was found to retain low nanomolar potency against PR6X when evaluated on a fluorogenic substrate. X-ray crystallographic analysis of the PR6X–compound 2 complex revealed that the complex does not show the conformational rearrangements seen previously in the PR6X–compound 1 complex and 2 retains essentially all of the interactions with PR6X that were previously observed in the PRwt–compound 2 complex.16 Our findings provide structural insights by which compound 2, but not compound 1, maintains tight binding resulting in potent antiviral activity on a multidrug-resistant protease variant.
Results
Compound 2 Inhibits Viral Replication of the PI-cross-resistant 6X Virus
We have reported that the compound 1-evolved PR1X, PR3X, and PR6X viruses show increasing degrees of resistance to 1 with increasing resistance mutations, with PR6X virus 17-fold more resistant to 1 than PRwt virus when evaluated in tissue culture replication assays.10 In addition, PR6X is crossresistant to saquinavir, ritonavir, and nelfinavir when evaluated in cleavage assays in vitro.10 Therefore, the compound 1-resistant viral and purified PR series is useful for evaluating antiviral activity of compound 2.
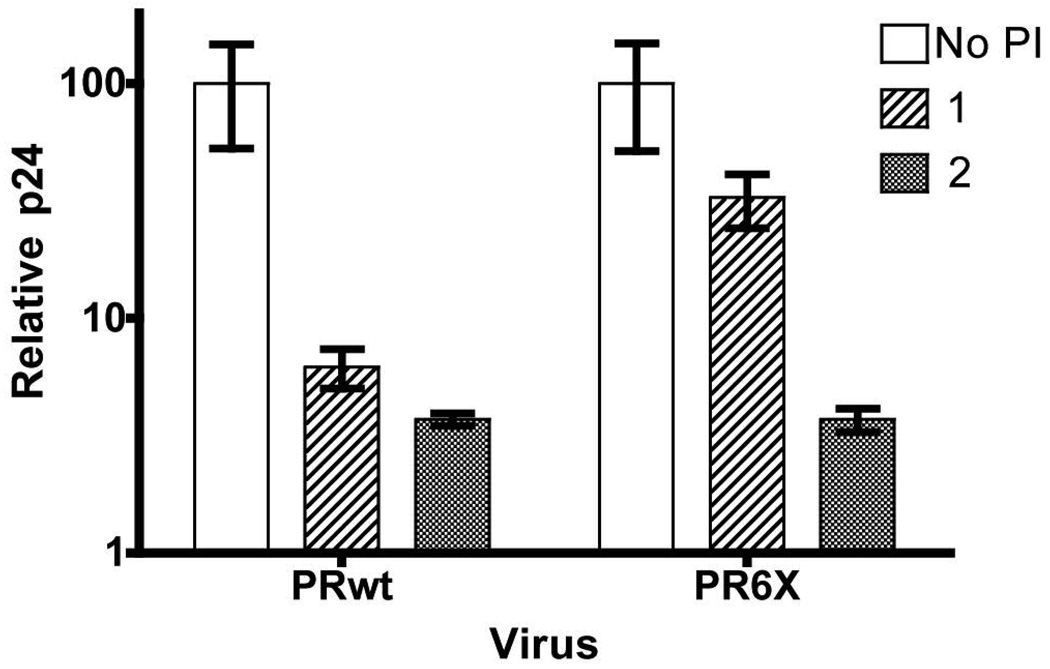
To assess the potency of compound 2 to control viral replication as compared to compound 1, SupT1 cells were infected with PRwt, and PR6X and cultured in the absence or presence of 1 or 2 at concentrations of 1–4000 nM. Replication of wild-type virus, as quantified by p24 accumulation over time and presented as relative p24 levels, was greatly reduced in the presence of 1 and 2 (Figure 2) and decreased to background levels at 250nM (data not shown). Furthermore, viral replication of the PR6X virus was inhibited by 2, with PR6X viral replication, shown in Figure 2, reduced almost 20-fold at 250 nM compound 2, relative to the infected cells in the absence of inhibitor. In contrast to inhibition of viral replication by 2, compound 1 had little effect on the compound 1-resistant PR6X virus (Figure 2).
Figure 2.
Inhibition of PRwt and PR6X viral replication by 2. SupT1 cells were infected PRwt and PR6X viruses and then cultured in the presence or absence of 100ng/mL 1 or 2. Levels of p24 were quantified 6-days post infection. Levels of p24 were detected by ELISA and are shown as a percentage of that measured from cells not treated with drug and presented as relative p24 levels.
Quantification of infectious virus production from the cell-free culture supernatants using TZM-bl cells shows EC50 values of approximately 86 nM and 93 nM for wild type virus in the presence of 2 and 1, respectively, and 260 nM for the PR6X-containing virus cultured in the presence of 2. The resulting EC50 for compound 1 against the PR6X-containing virus was greater than 1 mM. Thus, compound 2, in contrast to compound 1, is a potent inhibitor of the PR6X multidrug resistant virus. EC50 determinations based on p24 quantitation showed a similar trend, but deviated slightly in overall value (Table 1). These changes may reflect differences in the sensitivity of each assay.
Table 1.
Quantitation of viral susceptibility, EC50, to 1 and 2.
Supernatants from infected SupT1 cells were measured for HIV-1 production by p24 ELISA or HIV-1 LTR-driven luciferase activity using TZM-bl cells. Nonlinear regression results, reported in nM units, are shown.
EC50s for 1 against HIV-16X were not accurately determined, as no significant inhibition of the virus was observed, even at the highest concentration of compound tested (4µM).
Compound 2 Inhibits PRwt and PR6X in vitro
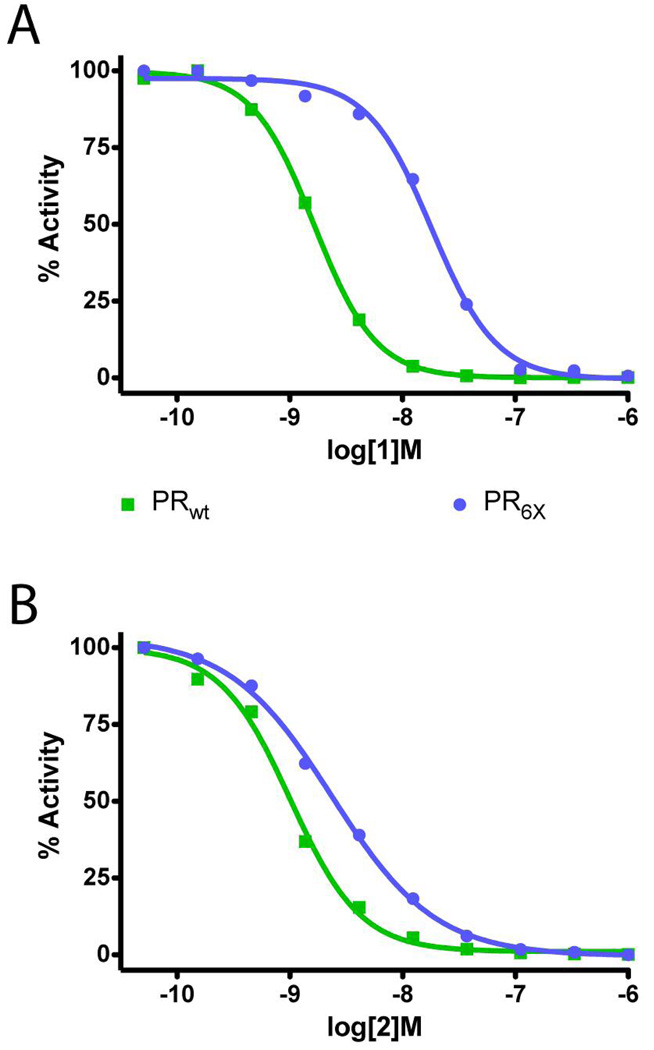
To quantify compound 2 inhibitor efficacy with respect to the PRwt and PR6X, biochemical and kinetic analyses were performed. In a fluorogenic substrate cleavage assay, 2 showed half-maximal inhibition of substrate cleavage at an inhibitor concentration of 6.0 nM for PRwt, and 15.7 nM for the PI-crossresistant PR6X (Figure 3), with Ki values of 2.7 and 7.1 nM, respectively. This finding shows that 2 has similar inhibition potency against PRwt and the PR6X proteases with IC50s differing by 2.6-fold (Figure 3). This is in contrast to our reported inhibition by compound 1, saquinavir, ritonavir, and nelfinavir, where the IC50s increased 30-fold, 9-fold, 14-fold, 8-fold, respectively, for PI activity on PR6X relative to PRwt.10 These results demonstrate that 2 has a low nanomolar inhibitory efficacy against both PRwt and PR6X, and the constellation of PI-cross-resistant mutations present in PR6X affords less than a 2.6-fold increase in the IC50.
Figure 3.
IC50 curves for 1 (A) and 2 (B) based on in vitro assay of enzyme activity. IC50 determinations are shown for each protease inhibitor against PRwt (green squares) and PR6X (blue circles) proteases. Curve fitting analysis results in an IC50 of 6.0 nM for PRwt and 15.6 nM for the PR6X. Nonlinear regression analysis in Prism (Graphpad Software) was used for curve fitting of the substrate cleavage reaction.
Crystal Structure of the PR6X–Compound 2 Complex
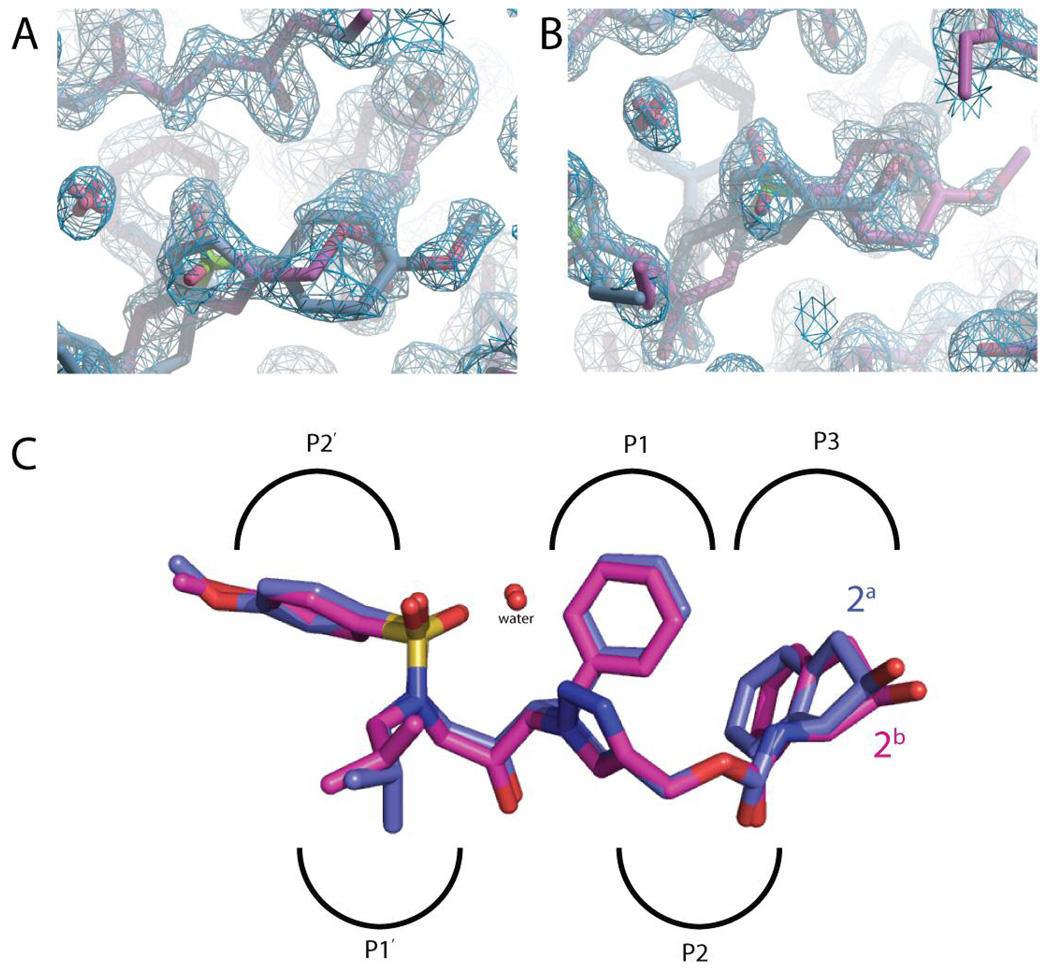
As we reported previously, the PR6X–1 crystal structure demonstrated conformational asymmetry, present in both inhibitor and protein, consistent with the decreased effectiveness of compound 1 inhibitor binding and function.12 To provide a structural basis for the differences between PI inhibitor efficacy of compound 2 relative to 1, we determined the 1.3 Å resolution crystal structure of 2 in complex with PR6X. Refinement of the structure showed that the crystals were in the P43 space group, with a dimer in the asymmetric unit (Table 2). Nevertheless, compound 2 in the PR6X–2 complex was present in two orientations within the active site related by non-crystallographic 2-fold symmetry coincident of the 2-fold axis of the PR6X dimer. Well-defined electron density was observed for both orientations of 2, which were refined with equal 50% occupancy (Figure 4A). The two orientations of 2 exhibit virtually identical conformations, the only significant differences being the torsional preferences of an isobutyl side chain and a methoxy group (Figure 4B). While the PR6X dimer was refined as monomers A and B in the asymmetric unit, the two subunits are very similar with an rms difference of 0.23 Å for all Cα atoms. Structural superposition shows that the conformations adopted by each monomer near the inhibitor are equivalent, with the protease maintaining equivalent interactions with each orientation of 2 in the active site (Figure 4B).
Table 2.
Data Collection and Refinement Statistics
| 6X HIV PR–2 | |
|---|---|
| Crystal Form | |
| Space Group | P43 |
| Unit cell parameters | |
| a (Å) | 86.61 |
| b (Å) | 86.61 |
| c (Å) | 33.66 |
| α=β=γ (°) | 90.0 |
| Solvent Content (%)/Vm (Å3/Da) | 50.0 (2.46) |
| No. Molecules per asymm. unit | 2 |
| Data Collection | |
| Resolution (Å) | 61.2-1.3 |
| No. of Unique Reflections | 57,347 |
| No. of Observations | 95,706 |
| Rmerge1 (%) | 3.9 (43.8) |
| I/σI | 10.1 (1.6) |
| Data Completeness (%) | 92.7 (74.1) |
| Mean Multiplicity | 1.67 (1.30) |
| Refinement | |
| No. of Reflections used in refinement | 57,347 |
| Rcryst ‡ (%) | 15.6 |
| Rfree ‡ (%) | 18.9 |
| % Reflections used in Rfree(|F|>0) | 5.0 |
| R.m.s. deviations, bond (Å) lengths | 0.011 |
| R.m.s. deviations, bond angles (°) | 1.24 |
| Ramachandran plot (% most favored, % additional allowed residues, % disallowed residues) | 96.8 |
| 3.2 | |
| 0.0 | |
| Estimated standard uncertainty2 (Å) | 0.070 |
| Coordinate error (Luzatti plot) (Å) | 0.237 |
| Model | |
| No. of atoms | |
| Protein (including alternate conformations) | 1518 |
| Water molecules | 286 |
| Average B-factors (Å2) | |
| Protein main chain atoms | 16.9 |
| Protein side chain atoms | 21.5 |
| Ligand | 15.2 |
| Water molecules | 29.5 |
Rsym=ΣhklΣi|Ii(hkl)- I(hkl)|/ ΣhklΣi(I(hkl)) where Ii(hkl) is the intensity of an individual measurement, and I(hkl) is the mean intensity of this reflection.
R factor = Σ hkl |Fobs| - |Fcalc | / Σ hkl | Fobs |, where |Fobs| and |Fcalc| are observed and calculated structure factor amplitudes, respectively. The values in parentheses for completeness, Rsym and I/σ correspond to the highest resolution shell.
Estimated standard uncertainty, diffraction precision index (DPI), based on Rfree 46
Figure 4.
Electron density of 2. 2|Fo|-|Fc| electron density map at 1.3Å resolution contoured at 1.0σ of 2 in the active site of the PR6X–2 complex. (A) The benzyl-methoxy group of 2a and the indene region of 2b. (B) The indene region of 2a and the benzyl-methoxy region of 2b. Some atoms of the PR6X dimer and bound H2O molecules are also shown. (C) The superposition of 2a and 2b in the PR6X-2 complex. The two orientations of 2 exhibit very similar conformations, differing only in the torsion angles of the isobutyl group in the P1’ pocket, and the methoxy group in the P2’ pocket.
Protease–Compound 2 Interactions
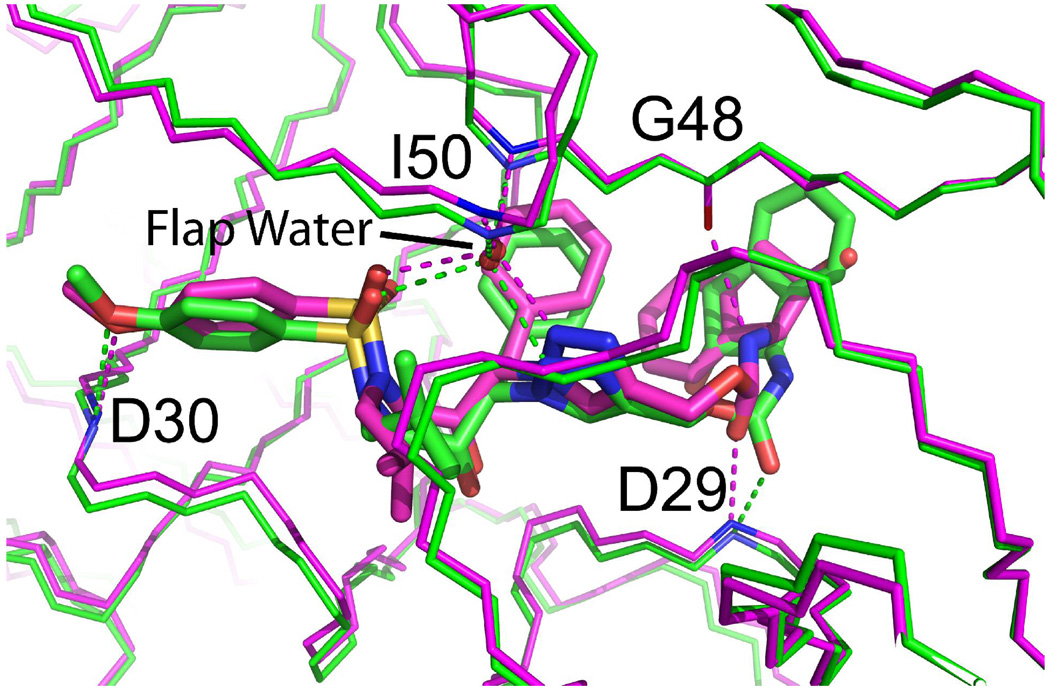
Interactions between the 2 compound and protease involve many of the same elements as those seen in protease–substrate complexes, although subtle differences exist. As previously observed in the PRwt−2 complex, the triazole linker serves to mimic of the substrate peptide bond, and provides a crucial interaction with the protease flaps via a bridging H2O molecule.13 In particular, the triazole N5 atom and a sulfonamide oxygen accept hydrogen bonds from the ‘flap water’, while this water accepts hydrogen bonds from the amides of the Ile50A and Ile50B in the protease flaps (Figure 5). This tetrahedral coordination by the flap water bridges the flaps and mimics the hydrogen bonding network seen in substrate complexes and other inhibitor complexes.17–21 The carbamate carbonyl oxygen in the P2 pocket accepts a hydrogen bond from the amide of Gly48; the hydroxyl nearest the dimer 2-fold axis donates a hydrogen bond to Asp25; and the methyoxy-benzyl oxygen accepts a hydrogen bond from the amide of Asp30 (Fig. 4B, Fig. 5). The latter hydrogen bond is important for high affinity binding in other substrate and inhibitor complexes.18–20 The hydrogen bonding interactions, present in both orientations of 2 in the active site, stabilize the closed form of the 2-fold symmetric protease dimer.
Figure 5.
The superposition of structures of PRwt and PR6X in a complex with 2. Protease is represented by main chain atoms, green for PRwt and magenta for PR6X. The conformation of 2 is represented by stick bonds with carbon atoms in green when bound to PRwt, or magenta when bound to PR6X. Hydrogen bonds are represented by dashed lines, with interacting main chain atoms colored by atom type and labeled by residue. The “flap water” is shown forming four hydrogen bonds bridging the amides of Ile50 residues with the sulfonamide and triazole of 2.
Interactions of 2 with PR6X involve several non-polar contacts. The 2a phenyl and isobutyl side chains occupy the P1 the P1′ pockets of the PR dimer, with the phenyl ring contacting Gly49A, Pro81B, Ala82B, and Ile84B, and the isobutyl group contacting Leu23A, Gly27B, Ala82A, and Ile84A (Fig. 4B, Fig. 5, Fig. 6A). The methoxy-benzyl group of 2a occupies the P2′ hydrophobic pocket formed by the side chains of residues Ala28B, Val32B, Ile47B, Ile50A, and Ile84B. At the same time, the 2a 2-hydroxy-2,3 dihydroindene group occupies the P3 specificity pocket, where it is sandwiched by π-stacking interactions with the 2a phenyl ring and the guanidinium group of Arg8B, which is salt-bridged with Asp 29A. This interaction stabilizes the dimer interface in the PR6X−2 complex, but is absent in the PRwt−2 structure where the indene group is flipped ∼180° due to rotation about the linking carbamate.
Figure 6.
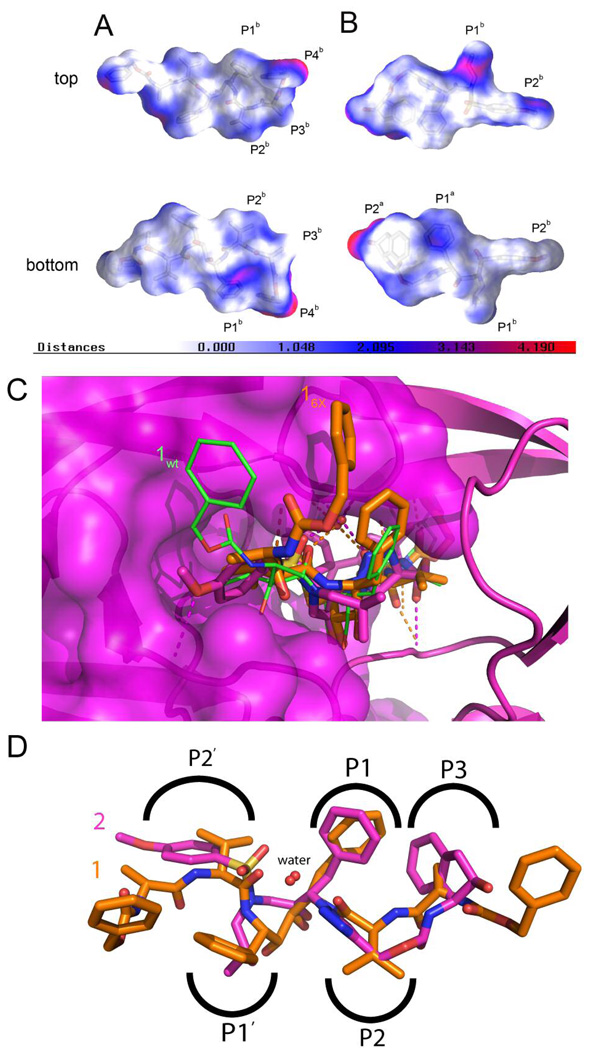
Intermolecular contact surfaces between PR6X and the two PR inhbitors, 2 (A) and 1 (B). Surfaces are colored according to intersurface distances calculated using GRASP with white indicating an intersurface distance of 0 and red indicating 4 Å. In these views the right half of each inhibitor interacts with the “B” monomer in the X-ray crystal structures, with the “top” imaging showing the flap-proximal surface and the “bottom” showing the D25-proximal surface. (C) Stick representations of 2 (magenta) and 1 (orange) in the active site of PR6x. Hydrogen bonds to the “B” PR monomer, represented as a ribbon with transparent molecular surface, are indicated by dashed lines. The conformation of 1 bound to PRwt is shown as green sticks. (D) Superposition of the PR6X-2, 1 structures (rmsd 0.70 Å for Cα atoms), as viewed from the protease flaps.
Discussion
Compound 2 Exhibits Low Nanomolar Potency Against PRwt and the Multidrug-Resistant PR6X virus and PR
We have previously reported on the biochemical and structural evolution of HIV-1 protease enzymatic function and resistance to 1,10, 12 with potency decreasing with increasing resistance-induced mutations in PR, with the PR6X virus showing only slight inhibition compared to the PRwt virus (Figure 2 and 10). The difference is most evident when comparing viral replication of the multidrug-resistant PR6X virus in the presence of 1 or 2 (Figure 2). SupT1 cells infected with PR6X virus cultured in the presence of 100 nM 1, showed approximately a 2-fold decrease in relative p24 levels when compared to the untreated control, whereas cells treated with 2 had a nearly 20-fold decrease in relative p24 levels at similar compound concentrations (Figure 2). Biochemical and kinetic analyses further underscored the potency of 2 against the PR6X with an IC50 of 15.7 nM, a modest increase relative to the 6.0 nM IC50 observed for PRwt (Figure 3). Other protease inhibitors, including 1, saquinavir, nelfinavir, and ritonavir, all show greatly increased IC50s (> log) against PR6X, consistent with the PI-cross-resistant nature of PR6X and the PR6X virus.10 Thus, 2 is effective in inhibiting a multidrug-resistant HIV-1 variant.
Compound 2 Complexes with PRwt and PR6X are Isomorphous
To gain molecular insights into the compound 2-binding site interactions responsible for its potent antiviral activity against the multidrug-resistant PR6X, we solved the crystal structure of 2 in complex with PR6X at 1.3 Å resolution (Figure 4). Compound 2 binds to the active site of the 2-fold symmetric PR6X dimer in two orientations related by a 2-fold symmetry. Clear electron density is observed for both orientations, which are 50% occupancy in the crystal (Figure 4A). Hence, 2a and 2b bind to PR6X with equivalent interactions, and superposition reveals that they align very closely (Figure 4B).
Obtaining the PR6X−2 structure allows comparison to our previously reported PRwt−2 structure.16 In the PRwt−2 complex, which crystallized in a different space group (P6122 vs. P43), the protease dimer is superposed on a crystallographic 2-fold axis; in the PR6X−2 structure the dimer represents the crystallographic asymmetric unit (Table 2).16 Despite the different symmetry restraints and packing environments, superposition of the PRwt and PR6X−2 structures reveals a high degree of similarity (Figure 5). Main chain atoms show negligible deviations and side chain atoms align well, with both proteins adopting similar side chain rotamer conformations (r.m.s. deviations 0.57 Å for 99 monomer Cα atoms, 0.60 Å for main chain and Cβ atoms). This point is further illustrated in a difference distance matrix plot showing the extent to which local 2-fold symmetry, i.e., similarity of monomer chains A and B, is retained in the PR6X−2 complex relative to the PRwt−2 complex (Figure S1, Supplement). The conformational differences that do occur are small, and confined to regions of the fold that exhibit deviations in other PR crystal structures, including the flaps, the loop involving Gly68, and the turn near Arg87.22
The protein-inhibitor interactions observed in the PRwt−2 and PR6X−2 complexes are similar, and the conformation of 2 is similar with the only significant differences being the torsion angles affecting the indene and isobutyl moieties (Figure 5). Variability in the latter group is also seen for 2a vs. 2b in the PR6X complex (Figure 4B). With respect to the indene group attached to the carbamate, analysis of the electron density and temperature factors for 2 in the PRwt structure (refined at 2.02 Å resolution; 1ZP8) indicates greater mobility in this structure. In general, the congruence of the PR6X−2 and PRwt−2 structures is in accord with the similarity of IC50 values for 2 against PR6X, IC50 = 15.7 nM, and PRwt, IC50-= 6.0 (Figure 3).
Clearly, the PRwt−2 and PR6X−2 structures differ at the sites of the six mutations. As noted, the main chain conformations of the PR dimers in each complex are very similar, so that the mutations affect only the side chains, thus limiting loss of binding affinity for compound 2(Figure 5). Moreover, four of the substitutions, L24I, M46I, L63P, V77I, are distal to the active site and do not affect compound 2 interactions or conformation. However, the remaining two mutations, F53L in the flap region and V82A, do appear to have an effect. The mutation F53L removes a packing contact with the compound 2 indene ring, which may be correlated with the flipping of the carbamate in the PR6X−2 structure, shifting the indene ring into an aromatic stack with the 2 phenyl group and Arg8 (Figure 4B, Figure 5). Similarly, mutation of Val82 to Ala in PR6X active site removes a packing contact with the 2a isobutyl group in the P1′ site, allowing it to adopt another torsion angle and be in contact with Leu23A, Gly27B, Ala82A, and Ile84A. Leu23A shifts ∼1 Å and Ile84A changes its χ1 torsion angle to favor interaction with the isobutyl group. With respect to the P1 site, the V82A mutation has a lesser effect on interactions of the 2a phenyl group with Gly49A, Pro81B, Ala82B, and Ile84B. The net effect of the V82A and F53L mutations is the loss of four contacts (isobutyl-Val82A, phenyl-Val82B, indene-Phe53 and associated carbamate-Asp30 amide hydrogen bond), however, there is a gain of three contacts (isobutyl-Gly27B, isobutyl-Leu23A, and indene stacking with Arg8). Overall, these three new contacts in PR6X stabilize compound 2 interactions and are in accordance with the modest 2.6-fold increase in the IC50 for the PR6X as compared to PRwt (Figure 3).
Different Structural Responses of PR6X to 2 and 1
Previously, we reported on the crystal structures of the PRwt–1 and PR6X–1 complexes and how the constellation of compound 1 resistance-induced mutations contributed to PR6X resistance.12 While the PRwt–1 structure is symmetric, such that a crystallographic 2-fold superposes on both the PR dimer and 1, the PR6X–1 structure is distinctly asymmetric, with the PR dimer in the asymmetric unit, and non-2-fold related changes in conformation in both the protein and 1. As discussed above, this is not the case for the PR6X–2 structure. In regards to the PR6X–1 structure, in particular, side chain packing arising from the 6 mutations in PR6X alters the dimer interface and disrupts interactions with 1 in one side of the active site, resulting in significant changes to the conformation of one flap and one-half of 1. In general terms, PR6X binds the C2-symmetric 1 inhibitor in an asymmetric fashion, but it retains the 2-fold symmetry of the PRwt complexes upon binding the distinctly asymmetric inhibitor, 2.
The structural analysis of PRwt–1 and PR6X–1 complexes, when compared to the PR6X–2 complex, reveals further insights into how PR mutations may reduce affinity for the inhibitor while maintaining enzymatic function.12 As we have reported the compound 1 IC50 for the PR6X mutant increases 30-fold as compared to PRwt.10 In contrast to the PRwt–1 and PR6X–1 complexes, the PRwt–2 and PR6X–2 structures are very similar, and 2 retains high affinity for PR6X, with the IC50 increasing 2.6-fold as compared to PRwt. Differences in the shape and volume of the intermolecular contact surfaces between the PR6X and 2 (Figure 6A) vs. PR6X and 1 (Figure 6B) further supports the notion that the active site of the compound 1 complex is expanded due to changes in one flap conformation, thereby accommodating the ‘flipped’ half of 1.12 This configuration of 1 lacks contacts with the core of the enzyme, thereby reducing PI efficacy, whereas with 2 the methoxy-benzyl with Asp30 and indene-Arg8 interactions (Figure 5) retains contacts in the 2 complex. The net result is maintenance of 2 binding in PR6X and potent antiviral activity.
It should be noted that unliganded PR6X is a symmetrical dimer, therefore the PR6X mutations do not predispose the dimer to be an asymmetric conformation.23 In particular, comparison of the apo forms of PRwt and PR6X show that the proteins adopt similar conformations with very similar hydrogen bonding networks in their core regions.23 Small differences do occur in the flaps, where the PR6X multidrug-resistant mutant exhibits more disorder, consistent with increased flexibility in this region as observed in other PI-resistant PRs.6, 24, 25 Therefore, the PR6X residues occupy the same structural context in both unliganded and 2 bound PR6X, implying that the asymmetric configuration when bound to 1 is a function of interactions with 1, and not an intrinsic conformational preference of the PI cross-resistant PR6X mutant.12
Structural Mechanisms of PI Resistance
It has been difficult to precisely define the structural mechanisms by which mutations in protease promote multidrug-resistance and loss of PI efficacy.5, 22, 26, 27 Like compound 2, saquinavir, nelfinavir, and ritonavir lack C2 symmetry, yet lose 7–14 fold affinity for PR6X relative to PRwt, whereas 2 loses only 2.6-fold affinity10. The role of conformational flexibility in the inhibitor has been noted in explaining improved binding of the inhibitor darunavir to a multidrug-resistant PR mutant.28 In a similar fashion, 2 exhibits flexibility in both the isobutyl side chain (2a vs. 2b, Figure 4B) and the carbamate-linked indene (Figure 5). These various degrees of freedom contribute to the configurational entropy of the complex, which should enhance the binding energy of the inhibitor to multidrug-resistant PRs.
Like amprenavir, darunavir, and other recently developed inhibitors,28, 29 2 is nonpeptidomimetic, underscoring an advantage for this inhibitor class against multidrug-resistant PRs. Compound 2, amprenavir, and darunavir each contain a benzyl ring with a hydrogen bonding functional group attached to the isobutyl-sulfonamide, a phenyl group in the P1/P1′ pocket, a hydroxyl group to interact with Asp25/Asp25′, and a heterocyclic ring system linked to a carbamate at the opposite end of the inhibitor (Figure 1). Compound 2 differs by having a triazole in the inhibitor core and an inverted carbamate.16 These chemical differences appear to contribute to the higher affinity of 2 for PR6X: the triazole participates in a precise hydrogen bonding network with the ‘flap water’ and the carbamate accepts a hydrogen bond from the amide of Gly48. Both of these interactions feature a conserved element of protease structure. Such interactions are less likely to be influenced by mutations, while limiting conformational change with respect to PRwt. In other words, 2 exploits structural features common to both wild-type and multidrug-resistant proteases to retain high affinity binding.
Conclusion
We have reported that 2 is effective against the single point protease inhibitor mutants, V82F, V82A, and G48V. The biochemical and structure findings reported herein demonstrate that compound 2 retains high affinity for both wild-type and the PR6X multidrug-resistant protease variant as the result of interactions with selected residues, and maintenance of hydrogen bonding to main chain atoms. These structural features of 2-protease interactions resulted in potent antiviral activity to our series of increasing PI-resistant viruses. The importance of maintaining PI interactions with the protein backbone of the active site has been found in PIs that are effective on multidrug-resistant proteases.15, 30 Furthermore, it is proposed that PIs should be developed to maintain extensive hydrogen bonding with the active site backbone to promote continued binding in the face of PI-resistance mutations.14, 15, 30 Based on our structural findings, modifications may be made to 2 to increase hydrogen bonding to backbone atoms. The use of azide–alkyne click chemistry synthesis will allow rapid and diverse chemical modification of 2. This will also allow the evaluation of whether such changes improve antiviral performance against newly appearing multidrug-resistant viruses, while maintaining efficacy to wild-type and previous versions of multidrug-resistant viruses. Lastly, insights into the contributions of multiple PI-resistance mutations and their resulting effect on protease interactions with inhibitors and substrates will continue to provide the basis for new inhibitors to target wild-type and multidrug resistant viruses.
Experimental Section
Compounds
Compound 1, phenylmethylN-[(2S)-1-[[(2S)-1-[[(2S,3R,4R,5S)-3,4-dihydroxy-5-[[(2S)-3-methyl-2-[[(2S)-2-(phenylmethoxycarbonylamino)propanoyl]amino]butanoyl]amino]-1,6-di(phenyl)hexan-2- yl]amino]-3-methyl-1-oxobutan-2-yl]amino]-1-oxopropan-2-yl]carbamate, synthesis was originally described by Lee at al as compound TL-3.11 Compound 2, (1-(3-hydroxy-4-(N-isobutyl-4-methoxyphenylsulfonamido)-1-phenylbutan-2-yl)-1H-1,2,3-triazol-4-yl)methyl 2-hydroxy-2,3-dihydro-1H-inden-1-ylcarbamate, was originally described by Brik et al as compound AB2.13, 16
Cloning, Expression and Purification of PRs
Construct design and synthesis of plasmids for the expression for all PRs as well as the detailed expression and purification protocols have been reported previously.10–12
Protease Activity Assays
HIV-1 protease enzymatic activity was assayed as described previously10, 11, using a fluorogenic substrate31. Briefly, purified HIV-1 protease was mixed with inhibitor compounds in a reaction buffer containing 25mM MES, pH 5.6, 200mM NaCl, 5% glycerol and 1mM DTT in a prewarmed 96-well plate. The reaction was started by the addition of fluorescently labeled anthranilyl protease substrate H-2992 (Bachem, CA), and the reaction progress was measured by fluorescence intensity using an FLx-800 fluoresence plate reader (BioTek, VT). Final reaction concentrations were 25nM protease, 30µM substrate, and 0.001–600µM inhibitor. Kinetics constants were determined by nonlinear regression of initial reaction velocities as a function of inhibitor concentration using Prism v4.0c (Graphpad Software, San Diego, CA). Values for Ki were calculated from the IC50 values using the Cheng-Prusoff equation32:
where S is the concentration of fluorescent substrate in the reaction.
Cells Culture Activity and Viral Expression Assays
SupT1 cells were cultured in RPMI 1640 media supplemented with 10% fetal calf serum, 2mM L-glutamine, 100U/mL penicillin, and 100µg/mL streptomycin. HIV-1 stocks were obtained from the culture supernatant of infected SupT1 cells. Cells were infected by addition of 10–100ng/mL p24 and incubated at 37°C. Unadsorbed virus was removed by washing the cells with fresh media, and resuspended in media ± protease inhibitors. Culture supernatant samples were removed for p24 quantification every second day, and then media and selected PIs were added back to the culture. Viral replication was quantified by p24 ELISA assay (Perkin Elmer).
Inhibition of infectious virion production was quantified using the TZM-bl indicator cell line, which contain a luciferase reported driven by the HIV-1 LTR.33 SupT1 cells were infected with HIV-1 expressing either PRwt or PR6X protease variants at an MOI of 0.001–0.005, and cultured with protease inhibitors 1 or 2 at concentrations from 1 – 4000 nM. On day 5–6, 50µL of cell-free supernatants were added to 104 TZM-bl cells in 96-well plates. After 48 hours, 100µL culture supernatant was removed and luciferase activity was quantified by the addition of 100µL Brite-Lite Plus reagent (Perkin Elmer) to each well, and measured with a Clarity luminometer (BioTek, VT). EC50 values were determined by nonlinear regression of normalized luciferase activity (Graphpad Software, San Diego, CA).
Crystallization and Data Collection
Crystallization
HIV-1 PR6X was mixed with a 4- to 10-fold molar excess of 2, and incubated on ice for 30–60 minutes. Insoluble material was removed by centrifugation at 14,000 × g for 15 minutes. The PR6–compound 2 complex was crystallized by mixing 1 µL complex solution with 1 µL well solution containing 0.1M sodium acetate, pH 4.5, 3.5M NaBr by sitting drop vapor diffusion at 6 °C.
Data Collection
Diffraction data for PR6X−2 crystals were collected at 100K by the rotation method (120 frames, 10 oscillation per frame) to 1.3 Å resolution on an Saturn 92 CCD system mounted on a FR-E X-ray generator equipped with VariMax optics (λ = 1.54 Å). The data were processed with d*TREK (Table 2).34
Data Collection and Structure Refinement
The structure of PR6X was solved by molecular replacement at 3 Å using a monomer of the PR6X-compound 1 complex structure (PDB I.D. 2AZC) as a search model in Molrep.35 Five percent of randomly selected reflections were designated as test reflections for use in the Free-R cross-validation method36 and used throughout the refinement. The correlation coefficient and R-factor from the molecular replacement solutions indicated that the correct space group was P43. Rigid body and restrained refinement were performed in REFMAC at 3 Å and 2.0 Å, respectively.37 Simulated annealing, Powell minimization and individual temperature factor refinements were performed using CNS.38 The model was further refined to 1.3 Å in SHELXL39 using a bulk solvent correction and isotropic B-factors, followed by several rounds of model adjustment using the SigmaA-weighted40 2|Fo|-|Fc| and |Fo|-|Fc| electron density maps using Coot41. Subsequently, Br ions from the crystallization medium and 89 water molecules were added, anisotropic temperature factors were refined (SHELXL), and 1 residue was modeled as having an alternate side chain conformation. Statistically disordered copies A and B of compound 2 were refined with 0.5 occupancy each, and two Br ions present in the active site, mutually exclusive with the alternate copies of compound 2, were refined at 0.5 occupancy. The final Rcryst/Rfree was 15.6/18.9% (Table 2). Coordinates have been deposited with RCSB, accession code 2HC0.
Structural Analysis
Structural analysis and plots of protease–inhibitor interactions were completed using LIGPLOT42. Protease–inhibitor intersurface distances and active site shape correlation were calculated using GRASP43 and sc44. Structural superpositions were performed using Cα atoms from monomers A and B of the protease dimer with CCP4. Difference-distance matrix plots were produced using the program DDMP from the Center for Structural Biology at Yale University, New Haven, CT. Structure figures were generated with PyMOL.45
Supplementary Material
Figure S1: Difference distance matrix plot showing changes in relative distances between Cα atoms in PRwt and PR6X proteases in complex with compound 2. This material is available free of charge via the Internet at http://pub.acs.org.
ACKNOWLEDGMENT
We appreciate the expert technical assistance of Jeremiah Savage, at TSRI, and Dr. Robin Rosenfeld, at Active Sight, throughout the study. We thank Drs. David Goodsell and Garrett Morris for their insightful discussions during the writing of the manuscript. The UCSD CFAR is gratefully acknowledged for p24 quantification and is funded by NIH 3 P30 AI036214-13S1. Our studies were supported by AI408820, GM48870, and GM083658 from NIH. This is publication #19205-MEM from The Scripps Research Institute.
ABBREVIATIONS
- HIV-1
human immunodeficiency virus type 1
- PI
protease inhibitor
- HAART
highly active anti-retroviral therapy
REFERENCES
- 1.del Rio C. AIDS: the second wave. Arch Med Res. 2005;36:682–688. doi: 10.1016/j.arcmed.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Piacenti FJ. An update and review of antiretroviral therapy. Pharmacotherapy. 2006;26:1111–1133. doi: 10.1592/phco.26.8.1111. [DOI] [PubMed] [Google Scholar]
- 3.Kutilek VD, Sheeter DA, Elder JH, Torbett BE. Is resistance futile? Curr Drug Targets Infect Disord. 2003;3:295–309. doi: 10.2174/1568005033481079. [DOI] [PubMed] [Google Scholar]
- 4.Machado DM, Fernandes SC, Succi RC, Freire WS, Pannuti CS, Gouveia AB, Levi JE, Diaz RS. Analysis of HIV-type 1 protease and reverse transcriptase in Brazilian children failing highly active antiretroviral therapy (HAART) Rev Inst Med Trop Sao Paulo. 2005;47:1–5. doi: 10.1590/s0036-46652005000100001. [DOI] [PubMed] [Google Scholar]
- 5.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42:631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 6.Clemente JC, Moose RE, Hemrajani R, Whitford LR, Govindasamy L, Reutzel R, McKenna R, Agbandje-McKenna M, Goodenow MM, Dunn BM. Comparing the accumulation of active-and nonactive-site mutations in the HIV-1 protease. Biochemistry. 2004;43:12141–12151. doi: 10.1021/bi049459m. [DOI] [PubMed] [Google Scholar]
- 7.Tie Y, Boross PI, Wang YF, Gaddis L, Liu F, Chen X, Tozser J, Harrison RW, Weber IT. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. Febs J. 2005;272:5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohtaka H, Schon A, Freire E. Multidrug resistance to HIV-1 protease inhibition requires cooperative coupling between distal mutations. Biochemistry. 2003;42:13659–13666. doi: 10.1021/bi0350405. [DOI] [PubMed] [Google Scholar]
- 9.Rhee SY, Taylor J, Wadhera G, Ben-Hur A, Brutlag DL, Shafer RW. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc Natl Acad Sci U S A. 2006;103:17355–17360. doi: 10.1073/pnas.0607274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buhler B, Lin YC, Morris G, Olson AJ, Wong CH, Richman DD, Elder JH, Torbett BE. Viral evolution in response to the broad-based retroviral protease inhibitor TL-3. J Virol. 2001;75:9502–9508. doi: 10.1128/JVI.75.19.9502-9508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee T, Laco GS, Torbett BE, Fox HS, Lerner DL, Elder JH, Wong CH. Analysis of the S3 and S3' subsite specificities of feline immunodeficiency virus (FIV) protease: development of a broad-based protease inhibitor efficacious against FIV, SIV, and HIV in vitro and ex vivo. Proc Natl Acad Sci U S A. 1998;95:939–944. doi: 10.1073/pnas.95.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heaslet H, Kutilek V, Morris GM, Lin YC, Elder JH, Torbett BE, Stout CD. Structural insights into the mechanisms of drug resistance in HIV-1 protease NL4-3. J Mol Biol. 2006;356:967–981. doi: 10.1016/j.jmb.2005.11.094. [DOI] [PubMed] [Google Scholar]
- 13.Brik A, Muldoon J, Lin YC, Elder JH, Goodsell DS, Olson AJ, Fokin VV, Sharpless KB, Wong CH. Rapid diversity-oriented synthesis in microtiter plates for in situ screening of HIV protease inhibitors. Chembiochem. 2003;4:1246–1248. doi: 10.1002/cbic.200300724. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh AK, Chapsal BD, Weber IT, Mitsuya H. Design of HIV Protease Inhibitors Targeting Protein Backbone: An Effective Strategy for Combating Drug Resistance. Acc Chem Res. 2008;41:78–86. doi: 10.1021/ar7001232. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh AK, Sridhar PR, Leshchenko S, Hussain AK, Li J, Kovalevsky AY, Walters DE, Wedekind JE, Grum-Tokars V, Das D, Koh Y, Maeda K, Gatanaga H, Weber IT, Mitsuya H. Structure-based design of novel HIV-1 protease inhibitors to combat drug resistance. J Med Chem. 2006;49:5252–5261. doi: 10.1021/jm060561m. [DOI] [PubMed] [Google Scholar]
- 16.Brik A, Alexandratos J, Lin YC, Elder JH, Olson AJ, Wlodawer A, Goodsell DS, Wong CH. 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. Chembiochem. 2005;6:1167–1169. doi: 10.1002/cbic.200500101. [DOI] [PubMed] [Google Scholar]
- 17.Kim EE, Baker CT, Dwyer MD, Murcko MA, Rao BG, Tung RD, Navia MA. Crystal Structure of HIV-1 Protease in Complex with VX-478, a Potent and Orally Bioavailable Inhibitor of the Enzyme. J. Am. Cltem. Soc. 1995;117:1181–1182. [Google Scholar]
- 18.Ghosh S, Rosenthal R, Zajac P, Weber WP, Oertli D, Heberer M, Martin I, Spagnoli GC, Reschner A. Culture of melanoma cells in 3-dimensional architectures results in impaired immunorecognition by cytotoxic T lymphocytes specific for Melan-A/MART-1 tumor-associated antigen. Ann Surg. 2005;242:851–857. doi: 10.1097/01.sla.0000189571.84213.b0. discussion 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King NM, Prabu-Jeyabalan M, Nalivaika EA, Wigerinck P, de Bethune MP, Schiffer CA. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J Mol Biol. 2000;301:1207–1220. doi: 10.1006/jmbi.2000.4018. [DOI] [PubMed] [Google Scholar]
- 21.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 22.Foulkes-Murzycki JE, Scott WR, Schiffer CA. Hydrophobic sliding: a possible mechanism for drug resistance in human immunodeficiency virus type 1 protease. Structure. 2007;15:225–233. doi: 10.1016/j.str.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaslet H, Rosenfeld R, Giffin M, Lin YC, Tam K, Torbett BE, Elder JH, McRee DE, Stout CD. Conformational flexibility in the flap domains of ligand-free HIV protease. Acta Crystallogr D Biol Crystallogr. 2007;63:866–875. doi: 10.1107/S0907444907029125. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Boross PI, Wang YF, Tozser J, Louis JM, Harrison RW, Weber IT. Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S. J Mol Biol. 2005;354:789–800. doi: 10.1016/j.jmb.2005.09.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Kovalevsky AY, Louis JM, Boross PI, Wang YF, Harrison RW, Weber IT. Mechanism of drug resistance revealed by the crystal structure of the unliganded HIV-1 protease with F53L mutation. J Mol Biol. 2006;358:1191–1199. doi: 10.1016/j.jmb.2006.02.076. [DOI] [PubMed] [Google Scholar]
- 26.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Logsdon BC, Vickrey JF, Martin P, Proteasa G, Koepke JI, Terlecky SR, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity. J Virol. 2004;78:3123–3132. doi: 10.1128/JVI.78.6.3123-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh AK, Ramu Sridhar P, Kumaragurubaran N, Koh Y, Weber IT, Mitsuya H. Bistetrahydrofuran: a privileged ligand for darunavir and a new generation of hiv protease inhibitors that combat drug resistance. ChemMedChem. 2006;1:939–950. doi: 10.1002/cmdc.200600103. [DOI] [PubMed] [Google Scholar]
- 29.Amano M, Koh Y, Das D, Li J, Leschenko S, Wang YF, Boross PI, Weber IT, Ghosh AK, Mitsuya H. A Novel Bis-Tetrahydrofuranylurethane-Containing Nonpeptidic Protease Inhibitor (PI), GRL-98065, Is Potent against Multiple-PI-Resistant Human Immunodeficiency Virus In Vitro. Antimicrob Agents Chemother. 2007;51:2143–2155. doi: 10.1128/AAC.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YF, Tie Y, Boross PI, Tozser J, Ghosh AK, Harrison RW, Weber IT. Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease. J Med Chem. 2007;50:4509–4515. doi: 10.1021/jm070482q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toth MV, Marshall GR. A simple, continuous fluorometric assay for HIV protease. Int J Pept Protein Res. 1990;36:544–550. doi: 10.1111/j.1399-3011.1990.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 33.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT. Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr D Biol Crystallogr. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 40.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 43.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 44.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.DeLano WL. "The PyMOL Molecular Graphics System". San Carlos, CA, USA: DeLano Scientific LLC; [Google Scholar]
- 46.Cruickshank DW. Remarks about protein structure precision. Acta Crystallogr D Biol Crystallogr. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Difference distance matrix plot showing changes in relative distances between Cα atoms in PRwt and PR6X proteases in complex with compound 2. This material is available free of charge via the Internet at http://pub.acs.org.