Abstract
Bacterial replication origins move towards opposite ends of the cell during DNA segregation. We have identified a proline-rich polar protein, PopZ, required to anchor the separated Caulobacter crescentus chromosome origins at the cell poles, a function that is essential for maintaining chromosome organization and normal cell division. PopZ interacts directly with the ParB protein bound to specific DNA sequences near the replication origin. As the origin/ParB complex is being replicated and moved across the cell, PopZ accumulates at the cell pole and tethers the origin in place upon arrival. The polar accumulation of PopZ occurs by a diffusion/capture mechanism that requires the MreB cytoskeleton. High molecular weight oligomers of PopZ assemble in vitro into a filamentous network with trimer junctions, suggesting that the PopZ network and ParB-bound DNA interact in an adhesive complex, fixing the chromosome origin at the cell pole.
Introduction
The localization of proteins and DNA/protein complexes in bacterial cells is well documented (Errington et al., 2003; Shapiro et al., 2002). Dynamic intracellular localization patterns are integral components of bacterial regulatory networks, and mediate sequential structural changes as bacteria move through their cell cycle (Goley et al., 2007; Rothfield et al., 2005) or respond to environmental challenge, as is the case in Bacillus subtilis sporulation (Barak and Wilkinson, 2005). An example of position-dependent sequential changes in cellular organization is the polar targeting of the Caulobacter crescentus replication origin/ParB complex, which, in turn, directs the selection of the mid-cell site for cytokinetic FtsZ ring formation. The C. crescentus orthologs of the plasmid partitioning proteins, ParA and ParB, are essential proteins involved in chromosome segregation and cytokinesis (Mohl et al., 2001; Figge et al. 2003). ParB binds specifically to several chromosomal parS sites near the replication origin (ori/ParB), and upon origin duplication, one copy of the ori/ParB complex travels across the long axis of the cell and becomes fixed at the cell pole (Figure 1A). A novel ATPase, MipZ, binds directly to ParB and travels with the replicated origin to the cell pole. MipZ restricts assembly of the FtsZ ring by stimulating the FtsZ GTPase activity (Thanbichler and Shapiro, 2006). Because the highest concentration of MipZ resides at the cell poles and minimal concentration is near the cell center, the cell division ring can only assemble at mid-cell. If MipZ is not concentrated at the poles, the cell division ring cannot form at the correct place in the cell and at the appropriate time in the cell cycle. Thus, in C. crescentus the molecules involved in anchoring DNA-protein complexes to a specific cellular location are critical components of an integrated temporal and spatial system controlling the cell cycle.
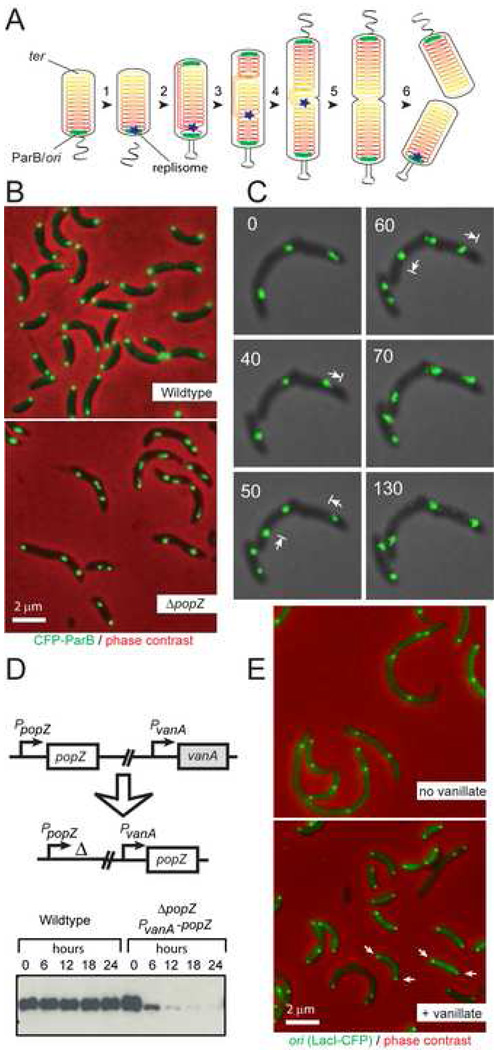
Figure 1. PopZ is required for the polar localization of chromosome origins.
(A) Chromosome segregation in C. crescentus. The chromosome (solenoid structure in diagram) is organized such that the replication origin, in complex with ParB, is found at the flagellar pole, and the chromosome arms extend the length of the cell axis, ending at the replication terminus at the opposite pole. Chromosome replication is initiated with the formation of the replisome during the swarmer to stalked cell transition (1), and a copy of the duplicated origin, in complex with ParB, is rapidly translocated to the opposite pole (2). The remaining bulk of chromosomal DNA follows this pattern of movement as it is replicated and segregated (3−5), allowing chromosome organization to be maintained in the daughter cells (6).
(B) CFP-ParB localization in the presence and absence of PopZ. Strain MT190 (cfp-parB) is compared with strain GB308 (cfp-parB; ΔpopZ). Cells were grown in minimal M2G media. Bar 2µm.
(C) Time-lapse fluorescence microscopy of living cells demonstrates movement of the CFP-ParB foci towards and away from the cell poles in ΔpopZ cells. (see Movie M1 in Supplementary Material). Strain GB308 was grown in M2G, and cells were placed on an agarose pad for microscopic analysis. Images were collected every 10 minutes for 150 minutes.
(D) Construction of the popZ depletion strain GB195 (ΔpopZ; PvanA-popZ). To demonstrate PopZ depletion, wildtype and GB195 were grown in PYE + 500µM vanillate, then transferred to vanillate free medium. At the indicated times, an aliquot of the culture was removed for immunoblot analysis with anti-PopZ antibody.
(E) Visualization of the origin region using the LacO/LacI-CFP FROS technique in a PopZ depletion strain. Strain GB196 ( popZ; PvanA-popZ; PxylX-lacI-cfp; CC0006::(lacO)n) was grown in PYE medium in the presence or absence of 500µM vanillate to control PopZ expression. Arrows point to polar foci. Bar 2µm.
Chromosomal origins are localized to and retained at the cell poles in C. crescentus (Jensen and Shapiro, 1999) and Vibrio cholerae (Fogel and Waldor, 2005), and during B. subtilis sporulation (Webb et al., 1997). In the case of both C. crescentus and V. cholerae, it is not known whether there is a mechanism that physically holds the origin region at the cell pole, or if the origin is simply transported to the pole and then maintained there within the limits of diffusion. Fogel and Waldor (2006) have provided evidence suggesting that the newly replicated origin region of chromosome I in V. cholerae traverses the cell via the pulling action of ParA polymers. The model proposed for origin movement and polar retention in V. cholerae is that the ori/ParB complex is bound to retracting ParA polymers that are attached to the pole via an unknown polar protein that serves to retain the ori/ParB/ParA complex at that pole. In C. crescentus, the actin-like protein MreB has been implicated in the moving the newly replicated origin across the cell (Gitai et al., 2005), and the polar localization of segregated origins has been shown to depend on the expression of ParB (Figge et al., 2003). During B. subtilis sporulation, the RacA protein mediates the anchoring of the duplicated origin to the cell pole (Ben-Yehuda et al., 2003). RacA binds preferentially to 25 recognition sequences spread over a large presumed centromeric region that has been postulated to interact with a polar protein complex (Ben-Yehuda et al., 2005). A protein required for the retention of the RacA/centromeric region at the cell pole is the DivIVA polymer, which resides at the pole (Ben-Yehuda et al., 2003). Because a direct interaction between RacA and DivIVA has not been demonstrated, nor has a polar binding protein been identified in V. cholerae, a protein complex that directly attaches the origin to the pole has yet to be identified in any bacterial system.
Here, we report the discovery of a polar protein, PopZ, that localizes to the cell poles and binds directly to the ParB protein positioned at the newly replicated origin region of the C. crescentus chromosome. Upon replication initiation, one duplicated origin, in complex with ParB, traverses the long axis of the cell. The concomitant assembly of PopZ at the receiving cell pole is essential for the capture and retention of the ori/ParB complex at this location. Single molecule visualization of PopZ-YFP in living cells is consistent with a diffusion/capture mechanism of PopZ accumulation at the cell pole, indicating that additional factors participate in the accumulation of PopZ at the receiving pole. One of these factors is shown to be the bacterial actin homolog, MreB.
RESULTS
PopZ is required for the polar localization of chromosomal origins
We initiated a search for a candidate polar protein that captures and retains the C. crescentus origin at the cell pole with a bioinformatic screen for factors that are associated with known polar proteins. We used a set of computational techniques that predict interactions between proteins in bacteria (Srinivasan et al., 2006) based on the observation that functionally interacting proteins tend to be expressed from genes that evolve in a correlated fashion (Pellegrini et al., 1999). Using 46 known polar localized C. crescentus proteins as “bait” (Supplementary Table S1), we identified a set of 36 conserved hypothetical proteins that are strongly predicted to interact with the members of this list.
One of the candidates was a novel 19 kDa protein encoded by the C. crescentus genetic locus CC1319 (accession number EU872488). An in-frame deletion of the CC1319 coding sequence yielded cells that lacked polar stalks and had a cell division defect (Supplementary Figure S1), yet remained viable despite poor growth. The deletion phenotype was fully complemented by expressing the CC1319 coding sequence from an inducible promoter at a distant chromosomal site, demonstrating that the defects could be attributed to the loss of CC1319. This gene, found in all alpha-proteobacteria, was named popZ for Polar Organizing Protein - Z.
Aberrant cell division is an expected phenotype of chromosome origin mislocalization (Thanbichler and Shapiro, 2006). To determine if this is the case in the popZ deletion mutant, we tracked the position of the chromosome ori/ParB complex using strains in which the parB coding sequence was replaced with cfp-parB, using fluorescence microscopy as described by Thanbichler and Shapiro (2006). In the ΔpopZ background, a high percentage of CFP-ParB foci were dissociated from the cell poles (Figure 1B). Time lapse movies (Figure 1C and Movie M1) showed that the CFP-ParB foci remain de- localized for long periods of time, and their position was not fixed within the cell. Nearly all of the foci move back-and-forth within the cytoplasm, including those that were in close proximity to the pole. By contrast, CFP-ParB foci were consistently found at the cell poles in wildtype cells, and their movement was highly constrained (Movie M2). Because ΔpopZ cells form filaments, the chromosome origin mislocalization phenotype might be caused by the inability of the nucleoid structure to compensate for the changes in cell length and volume. Accordingly, we examined origin localization in two mutants that are directly inhibited in division plane formation, and found that the CFP-ParB foci remained attached to the poles in these elongated cells (Figure S2). These results suggest that there is a specific requirement for PopZ in retaining ParB foci at the cell pole.
We confirmed that the polar retention of the chromosome origin itself is dependent on the presence or absence of PopZ using FROS (Fluorescent Repressor – Operator System) to label a DNA region adjacent to the origin of replication. To do this, we used a strain in which an array of lac operator sequences was inserted at a site 4kb away from the origin, and which carried lacI-cfp under control on an inducible promoter (Viollier et al., 2004). This chromosome labeling system was incorporated into a PopZ depletion strain (Figure 1D), in which the endogenous popZ coding sequence was deleted and a copy of popZ was placed under control of the vanillate inducible vanA promoter at the vanA chromosomal locus. When these cells were grown in the absence of vanillate, the PopZ protein was depleted (western blot shown in Figure 1D) and the LacI-CFP labeled DNA dissociated from the cell poles (Figure 1E, upper panel). When PopZ was added back to the cells by vanillate induction, the origin mislocalization phenotype was rescued (Figure 1E, arrows). Thus, the polar localization of chromosome origins can be controlled by modulating the accumulation of PopZ.
Dynamic, cell-cycle dependent localization of PopZ to the cell poles, and co-localization with ParB
PopZ protein is present at constant levels throughout the cell cycle (Figure S1). To determine if it is targeted to specific cellular locations, we replaced the chromosomal popZ coding sequence with popZ-yfp. This modification to PopZ permitted stalk formation and cell division, though some of cells were slightly elongated compared to wildtype. Time-lapse visualization of synchronized cell populations (Movie M3) showed that PopZ-YFP was located at the flagellar pole of the swarmer cell, and that it remained at this pole upon the swarmer-to-stalked cell transition. The stalked cell developed a second PopZ-YFP focus at the opposite pole, and maintained a bi-polar localization pattern of PopZ-YFP until cell division. We obtained identical localization data using a merodiploid strain in which the chromosomal popZ locus was unperturbed and popZ-yfp was transcribed at low levels from an inducible promoter at the chromosomal vanA locus (Figure 2 and not shown). In these experiments, cell morphology was indistinguishable from wildtype.
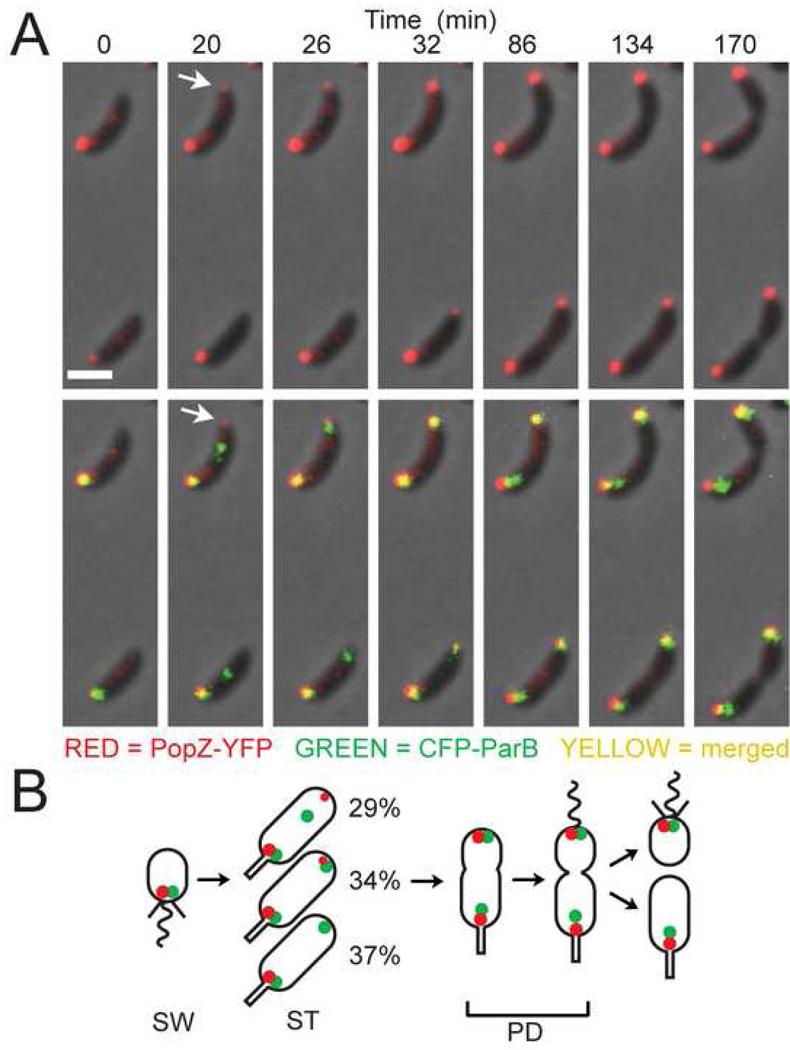
Figure 2. Upon initiation of DNA replication, PopZ co-localizes with the ori/ParB complex at the new pole.
(A) Time-lapse analysis of CFP-ParB and PopZ-YFP localization. Swarmer cells were isolated from a culture of GB301 (cfp-parB; PvanA-popZ-yfp) grown in M2G. Vanillate was added at 50µM 2h prior to synchronization of the cell culture. Swarmer cells were placed on an agar pad, and images were collected at the indicated times. Top panels: PopZ-YFP (red) over phase contrast image (grey); bottom panels: CFP-ParB (green) is added to show co-localization of the fluorescent markers (yellow). The arrow at the 20 min time point indicates a pole that accumulates PopZ-YFP before the arrival of the CFP-ParB focus. Bar 1µm.
(B) Diagram showing the localization patterns of PopZ-YFP and CFP-ParB through the cell cycle. The percentage of cells showing the different patterns is indicated (n = 406 cells, from two separate experiments that differed by less than 10% in all categories).
The cell cycle dependent localization pattern of PopZ-YFP is similar to that described for the chromosomal origin of replication (Figure 1A, (Jensen and Shapiro, 1999). To determine if the ori/ParB complex co-localizes with PopZ-YFP, we constructed a double-labeled strain, in which the chromosomal parB gene was replaced with a cfp-parB derivative and popZ-yfp was transcribed from the chromosomal vanA locus. Time-lapse visualization of a synchronized population of cells (Figure 2A) showed that PopZ-YFP and CFP-ParB co-localized at the flagellated pole of the swarmer cell. As the swarmer cell differentiated into a stalked cell, PopZ-YFP appeared at the opposite pole, close in time but not necessarily coincident with the arrival of ParB-CFP at the newly replicated origin (Figure 2A, arrow and schematic in Figure 2B). PopZ-YFP was never observed to track with CFP-ParB as it moved across the cell; in contrast, PopZ-YFP was first visible at the pole as a faint focus, increasing in intensity as the cell cycle progressed. When both markers were present at the new pole, PopZ-YFP and CFP-ParB always co-localized.
PopZ-YFP appeared before the arrival of the ori/ParB complex in approximately one-third of the observed cells (upper cell, Figure 2A), and this order was reversed at nearly the same frequency (lower cell, Figure 2A). In the remaining cases, the two markers appeared together within the same six minute time frame. When the CFP-ParB focus preceded the arrival of PopZ-YFP by more than one time frame, the CFP-ParB focus did not settle in place at the extreme end of the cell until the arrival of PopZ-YFP (Movie M4). We also observed that, while co-localization of PopZ-YFP and CFP-ParB was maintained at the new pole, the localization pattern of these two markers changed at the stalked pole: CFP-ParB became somewhat displaced from the pole, while PopZ-YFP retained its position adjacent to the base of the stalk (Figure 2A and 2B). Time-lapse movies revealed that the CFP-ParB foci at the stalked pole exhibit a slight increase in motion relative to their counterparts at the opposite pole (Movie M5). Together, these observations provide evidence that ori/ParB is fixed to the pole when it co-localizes with PopZ.
PopZ physically interacts with the chromosome origin via ParB
To investigate the possibility that there is a physical association between PopZ and the ori/ParB complex, we performed a co-immunoprecipitation experiment (Figure 3A), using a strain in which the endogenous popZ coding sequence was replaced with epitope tagged popZ-m2. A membrane permeable cross-linking agent was added to cells prior to lysis, then PopZ-M2 and associated proteins immunoprecipitated with an anti-M2 antibody. Western blots of the purified product using an anti-ParB antibody showed that ParB was part of the PopZ complex, while a control protein, FtsZ, was not.
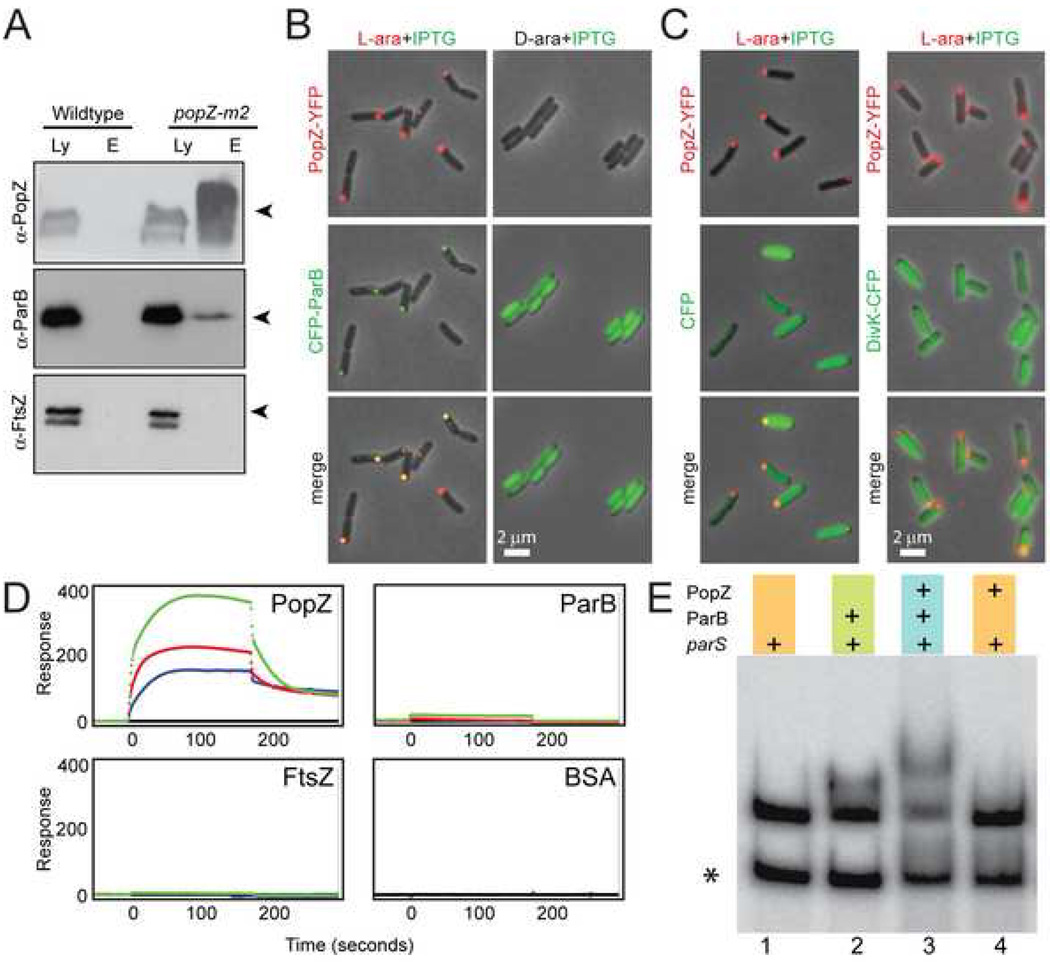
Figure 3. PopZ interacts directly with ParB.
(A) Co-immunoprecipitation assays show that PopZ forms a complex with ParB in vivo. Samples of the whole cell lysate (Ly) from strain GB135 (popZ-m2) and immunoprecipitated protein complexes (E) were probed with the indicated antibodies by western blot.
(B) CFP-ParB was expressed in the presence (left panels) or absence (right panels) of PopZ-YFP in E. coli strain GB333. Cells were grown in LB media to an optical density range of 0.3–0.5, then CFP-ParB was induced with 0.1mM IPTG and PopZ-YFP was either induced with 0.2% L-arabinose or not induced in the presence of 0.2% D-arabinose (as indicated) for 2 hours. Although PopZ-YFP alone localized to one E. coli cell pole, polar localization of CFP-ParB only occurred when PopZ-YFP was present.
(C) CFP (left panels) and DivK-CFP (right panels) were expressed in the presence of PopZ-YFP in E. coli strains GB342 and GB367, respectively. PopZ-YFP was induced with 0.2% L-arabinose, and either CFP or DivK-CFP was induced with 0.1mM IPTG or 0.01mM IPTG, respectively, to account for relative differences in expression level.
(D) Surface Plasmon Resonance analysis of the interaction between PopZ, ParB, FtsZ or BSA in solution with immobilized ParB on the chip surface. Protein was injected at concentrations of 125, 250, and 500 nM (blue, red, and green lines, respectively, at time 0), followed by injection of BC buffer only (180 seconds) to observe dissociation.
(E) Mobility shift assay. Radioactively labeled probe DNA (1.1 nM) was incubated in reaction buffer alone (lane 1); with 450 nM ParB (lane 2); with 450 nM ParB and 650 nM PopZ (lane3); or with 650 nM PopZ (lane 4). The DNA and DNA-protein complexes were resolved on a non-denaturing gel and visualized by autoradiography. The control probe (without parS) is labeled with an asterisk.
We used three independent experimental approaches to determine if PopZ interacts directly with ParB; a heterologous expression system, Surface Plasmon Resonance, and electrophoretic mobility shift assays. The heterologous expression system was designed to enable independent control of PopZ-YFP and CFP-ParB expression in Escherichia coli. Because homologs of the popZ gene are not found in E. coli, and the E. coli chromosome does not encode the ParB partitioning system, any proteins that could be promoting the indirect association of these proteins in C. crescentus are not expected to be present in this system. We constructed an E. coli strain in which PopZ-YFP and CFP-ParB are under the control of L-arabinose inducible araBAD and IPTG inducible T7 promoters, respectively. Surprisingly, when PopZ-YFP expression was induced in the absence of CFP-ParB, the protein formed foci at one pole of the E. coli cell. This effect was observed at all induction levels tested (Figure S3), indicating that this behavior is not an artifact of overexpression. When CFP-ParB was expressed in combination with PopZ-YFP, the CFP-ParB protein co-localized with the PopZ-YFP foci (Figure 3B, left panels). However, CFP-ParB was dispersed in the absence of PopZ-YFP expression (Figure 3B, right panels), indicating that CFP-ParB was recruited to the pole by interacting with the PopZ-YFP. We also found that CFP-ParB was recruited into polar foci when un-tagged PopZ protein was expressed (Figure S3), demonstrating that the native PopZ protein has the same activity. To determine if this recruitment can be explained by non-specific interactions between protein aggregates, we expressed CFP alone or the histidine kinase DivK-CFP (Jacobs et al., 2001) in the presence of PopZ-YFP (Figure 3C). Neither of these proteins was recruited to the PopZ-YFP foci, indicating that PopZ-YFP directly and specifically recruits CFP-ParB in this E. coli expression system.
The direct interaction between PopZ and ParB was demonstrated in vitro using Surface Plasmon Resonance (Figure 3D). When ParB was bound to the chip surface and probed with a buffer solution containing PopZ, we observed an increase in mass on the chip surface (representing PopZ binding) that was dependent on the concentration of PopZ protein in the buffer. Using this system, we determined the equilibrium dissociation constant for the interaction between ParB and PopZ to be 20 nanomolar, indicating a tight association. In control experiments, neither bovine serum albumin (BSA) nor C. crescentus FtsZ were capable of interacting with ParB or PopZ, and ParB exhibited minimal interaction with itself, demonstrating the specificity of the interactions we observe by this technique (Figure 3D). The reverse experiment, in which His-tagged PopZ was bound to the chip surface and probed with ParB in suspension, also indicated an interaction between these proteins, and no interaction was observed with BSA or FtsZ (not shown).
We performed electrophoretic mobility shift assays to determine if the PopZ-ParB interaction occurs in complex with parS DNA (Figure 3E). When ParB was mixed with two labeled DNA probes, a 614 base pair sequence containing two tandem parS sites and a 435 base pair sequence with no parS sites, only the parS-containing sequence was shifted to a higher molecular weight form (Figure 3E, lanes 1 and 2), demonstrating the specific association of ParB with its target site. When PopZ was added to the reaction, the band corresponding to the parS-ParB complex was shifted to a higher molecular weight form indicative of a ternary parS-ParB-PopZ complex (Figure 3E, lane3). The addition of PopZ in the absence of ParB did not cause a shift in the mobility of probe DNA (Figure 3E, lane 4), demonstrating that the association of PopZ with DNA was promoted by the presence of ParB in complex with parS. These reactions were performed in the presence of BSA and salmon sperm DNA, indicating that the complexes form in the context of competition from non-specific protein-protein and protein-DNA interactions. The specific interaction between PopZ, ParB, and parS DNA probe was also observed when the concentration of salmon sperm DNA was reduced (not shown), but some non-specific interaction between PopZ and probe DNA was also detected under these conditions.
PopZ is targeted to the cell poles
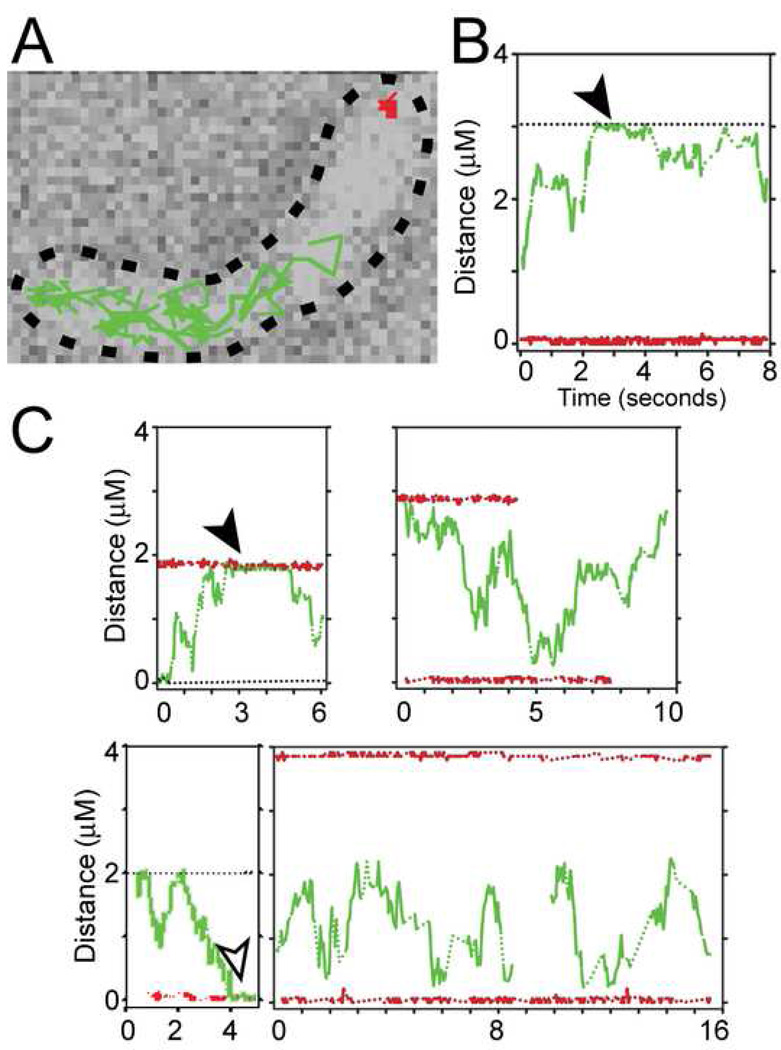
To observe the behavior of single molecules of PopZ-YFP in living cells, we used high speed video microscopy to track the motion of individual molecules with 32 millisecond time resolution (Kim et al., 2006). To resolve single molecules in these experiments, we engineered a strain that produces PopZ-YFP at roughly 1–10 PopZ-YFP molecules per cell, in addition to normal levels of untagged PopZ. Two distinct molecular behaviors were observed: pole-localized molecules stayed at the pole to the precision of the position measurement (60 nm), and mobile molecules explored a large region of the cell interior (Figure 4A and B). The mobile molecules frequently sustained brief periods of contact with the cell pole (Figure 4B and C, arrowheads), and on some occasions they appeared to become fixed at the pole, where they remained stationary until photobleaching ended the experiment (Figure 4C, open arrowhead). This dynamic behavior of single PopZ molecules is consistent with a diffusion/capture mechanism for polar localization in which PopZ exhibits diffusive behavior in the cytoplasm and can be captured and immobilized at the cell pole.
Figure 4. Visualization of single PopZ-YFP molecules in live cells.
(A) Time lapse visualization of two molecules in a cell, with colored lines tracking the distance moved between 32.2 msec frames. One molecule (red) remains localized to the pole, and the other (green) has increased mobility. The tracks are overlaid on a transmitted light image of the cell, outlined in black.
(B) A representation of the data from the experiment in A, showing the distance of the molecules from one pole as a function of time. The black horizontal dotted line marks the opposite pole; the red and green lines follow the stationary and mobilized molecules, respectively.
(C) A sampling of the time-dependent behavior of single molecules in other cells. For all graphs, the colored lines are dotted during dark (blinking-off) periods. The cells are strain GB175 (PvanA-popZ-yfp), grown without vanillate in M2G medium.
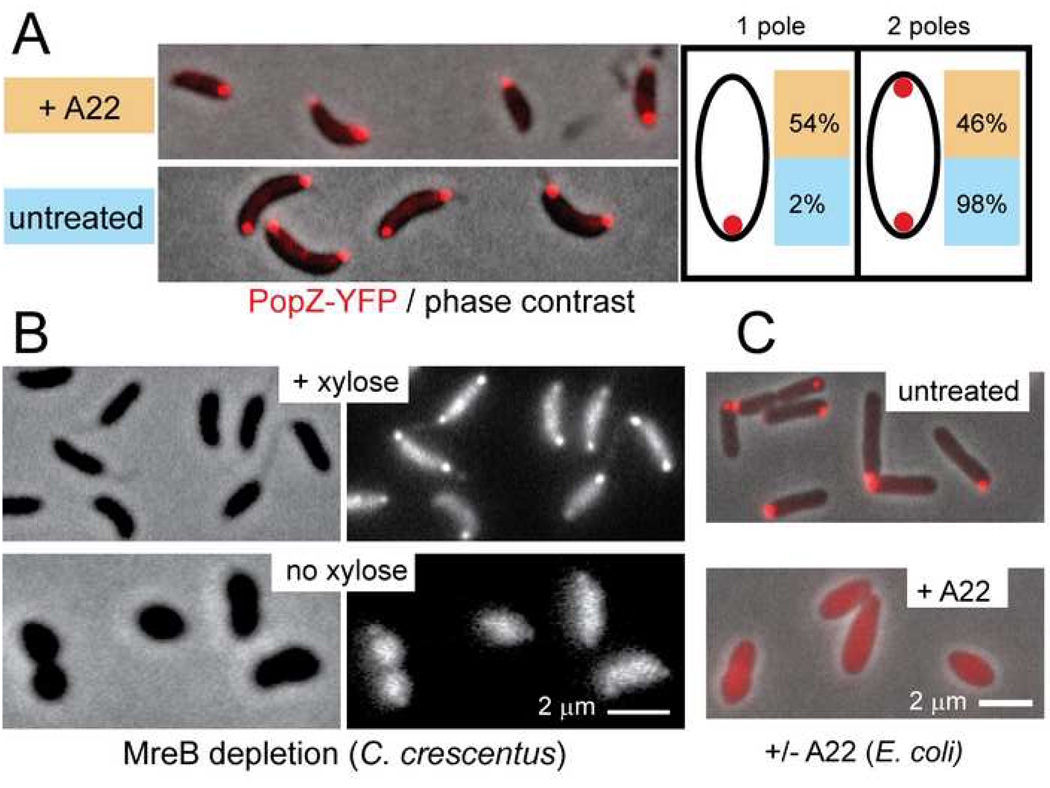
In order to identify the factors that contribute to PopZ polar localization, we disrupted the function of candidate proteins. One of these was TipN, which acts as a polar landmark by associating with the division plane, then remaining at the new pole to recruit proteins required for polar morphogenesis (Huitema et al., 2006; Lam et al., 2006). We observed normal PopZ-YFP localization in a ΔtipN deletion strain (Supplementary Figure S2). To determine if the interaction between ParB and PopZ is necessary to stabilize the accumulation of PopZ at the new pole, we also examined PopZ-YFP localization in a ParB depletion strain. Again, we found that this perturbation did not prevent the polar localization of PopZ (Supplementary Figure S2). However, we found that an actin-like protein, MreB, contributes to the localization and maintenance of PopZ at the cell pole. When we treated swarmer cells with A22, a small molecule inhibitor of MreB activity (Gitai et al., 2005), the proportion of cells at the swarmer-to-stalked-cell transition that were able to accumulate PopZ at the new pole was greatly reduced compared to controls (Figure 5A). The inhibitory effect of A22 on PopZ-YFP polar accumulation was observed on both agarose pads and in liquid media. Although long periods of growth in the absence of MreB activity causes cells to acquire a rounded morphology (Gitai et al., 2005), the cells retained their rod-shaped morphology within the short time course of these experiments.
Figure 5. MreB activity is required for the polar localization and maintenance of PopZ.
(A) A22 inhibits the formation of PopZ-YFP foci at the new pole. Strain GB301 (cfp-parB; PvanA-popZ-yfp) was grown in M2G medium, and PopZ-YFP expression was induced by the addition of 50µM vanillate for 2 hours before dividing the cells into two fractions for synchronization. One fraction was treated with 10µM A22 during synchrony. The isolated swarmer cells were allowed to grow on an agarose pad containing M2G media with or without A22 at room temperature. After 2 hours, the number of cells with one or two polar PopZ-YFP foci were quantified (+A22 n=922; no A22 n=664). A22 treatment inhibited the translocation of CFP-ParB foci to a similar degree (not shown). The data was collected from two independent experiments, which differed by less than 2%. Similar results were obtained in cultures grown in liquid media.
(B) MreB depletion causes de-localization of PopZ-YFP polar foci. Strain GB325 (ΔmreB; pMR10+PxylX-mreB; PvanA-popZ-yfp) was grown for 24 hrs in M2G medium supplemented with 0.03% xylose (top panels) or in M2G medium without xylose (bottom panels) and stimulated with 50µM vanillate for 2 hrs prior to microscopic analysis. Left panels: phase contrast images; right panels: PopZ-YFP fluorescence.
(C) A22 inhibits the polar localization of heterologously expressed PopZ-YFP. E. coli strain GB297 was grown to log phase in LB media and supplemented with either 0.2% L-arabinose (for PopZ-YFP expression, top panel) or 0.2% arabinose plus 10µM A22 (bottom panel) for 105 minutes prior to microscopic analysis. PopZ-YFP fluorescence (red) is overlaid on the phase contrast image.
To confirm the role of MreB in PopZ localization, we observed the localization of PopZ-YFP in an MreB depletion strain, in which the chromosomal mreB gene is deleted and mreB is transcribed from a xylose-inducible promoter (Figure 5B). After depletion of MreB protein in xylose-free medium, PopZ-YFP was de-localized. Because E. coli cells are able to support the polar localization of heterologously expressed C. crescentus PopZ, we asked if E. coli MreB is similarly required for PopZ localization. Treatment of PopZ-YFP expressing E. coli with A22 resulted in PopZ-YFP delocalization (Figure 5C). Thus, active MreB contributes to the maintenance of PopZ at the cell poles in two widely diverse organisms.
PopZ assembles into structured oligomers
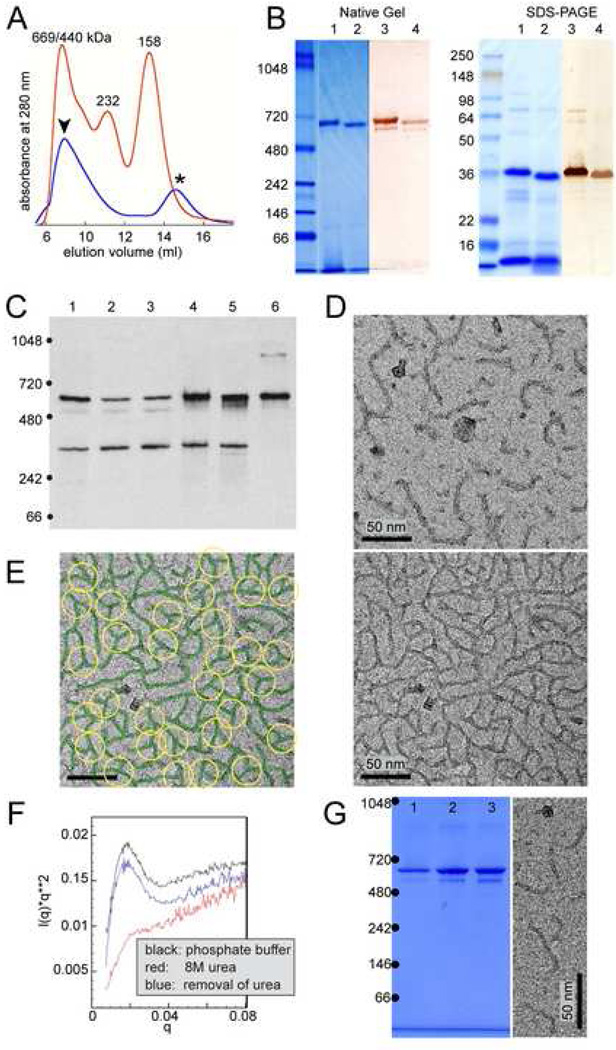
PopZ and its homologs in other α-proteobacteria are predicted to be cytoplasmic. They are rich in proline and acidic residues, but do not contain any domains that are conserved in other proteins. In the course of purifying PopZ, gel filtration chromatography revealed that His-tagged PopZ protein formed a high molecular weight complex when expressed in E. coli (Figure 6A). Analysis of the purified protein by native gel electrophoresis and Coomassie staining showed that the material was approximately 650 kDa in size (Figure 6B, left panel). The largest band, as well as a slightly smaller product, were recognized by the PopZ antibody, and persisted after removal of the His6 tag. Complete solubilization of this material for SDS-PAGE (Figure 6B, right panel) revealed that it was composed of a 37 kDa protein. There is a large disparity between the calculated molecular weight of the monomeric protein (19 kDa) and its apparent size by SDS-PAGE, perhaps because the many negatively charged residues give it an unusual mass-to-charge ratio (pI = 3.88), or because the strings of adjacent proline residues give the denatured protein a kinked conformation.
Figure 6. PopZ assembles into a structured oligomer in vitro.
(A) Size analysis of 6His-PopZ complexes purified from E. coli by gel filtration. Elution volume (X-axis) is plotted against absorbance at 280nm (Y-axis). The red trace shows the profile of molecular weight standards (size indicated above peaks), the blue trace is the profile of purified 6His-PopZ. The first peak (arrowhead) was determined to be PopZ by analyzing the corresponding fractions by SDS-PAGE and Coomassie staining; the second peak (asterisk) contained proteins that do not match the MW of PopZ.
(B) Analysis of purified PopZ by electrophoresis. His6-PopZ was purified from an E. coli lysate and separated by native gel electrophoresis (left panel) and SDS-PAGE (right panel), with molecular weight standards (in kDa) in the left-most lane. The gels were analyzed by either Coomassie staining (lanes 1 and 2) or western blotting with anti-PopZ antibody (lanes 3 and 4). In lanes 2 and 4, the 6His tag was cleaved with thrombin protease before loading.
(C) PopZ assembles into high molecular weight complexes in vivo. Proteins were isolated from whole cell lysates of wildtype cells, separated by native gel electrophoresis, and analyzed by immunoblotting with an anti-PopZ antibody. Lanes 1,4, and 5 contain lysates of C. crescentus cells (7 mg total protein) from a wildtype strain (lane 1), and MreB depletion strain LS3809 in the presence (lane 4) and absence (lane 5) of xylose-induced MreB expression; lanes 2–3 are lysates from E. coli strain GB296 (0.3 mg total protein) that were induced to express PopZ heterologously by the addition of 0.2% L-arabinose for 100 minutes prior to lysis, either left untreated (lane 2) or treated with 10 µM A22 for 3.5 hrs prior to lysis (lane 3); lane 6 contains approximately 3 ng of purified PopZ protein.
(D) Transmission electron micrographs of PopZ complexes. Purified PopZ diluted to 49 µg/ml (top panel) or 54 µg/ml (bottom panel) was placed on a carbon coated grid, negatively stained, and viewed at 86,000X magnification.
(E) PopZ filament assemblies are connected by 3-way junctions. The filaments in D were traced with a green line, and examples of 3-way junctions were circled in yellow. The field also contains a small number of 4-way junctions.
(F–G) PopZ filaments self-assemble in vitro. Purified PopZ was denatured in 1M urea or 8M urea, then allowed to renature by dialysis into buffer without urea. (F) Denaturation of the sample in 8M urea was confirmed by Small Angle X-ray Scattering (SAXS). The data is represented in a Kratky plot, in which folded proteins have parabolic features. The near elimination of this feature in the urea containing sample indicates that this sample is significantly unfolded compared to the original and renatured samples. (G) Purified PopZ (lane 1) was compared with renatured material from samples placed in 1M urea (lane 2) or 8M urea (lane 3) on a native gel stained with Coomassie (left panel), and the material that was recovered from 8M urea was analyzed by electron microscopy, as in D (right panel).
To determine if these large PopZ complexes exist in C. crescentus, we compared the mobility of PopZ in whole cell lysates of C. crescentus and E. coli cultures (expressing PopZ) with purified protein (Figure 6C). We found that the majority of PopZ in C. crescentus cells is the same size as the major band in E. coli, indicating that the high molecular weight PopZ complex is a physiologically relevant form. In the whole cell lysates, we observed an additional band (excluded from our preparations of purified PopZ during size exclusion chromatography) that is approximately half the size of the major band, suggesting that PopZ complex assembly has at least one stable alternate or intermediate form. Since we were unable to detect monomeric PopZ in whole cell lysates, we propose that PopZ is synthesized and assembled into small oligomers (approximately 325 kDa and 650 kDa) in the cytoplasm. We examined the effects of inhibiting MreB activity on the formation of PopZ oligomers, and found that the depletion of MreB from C. crescentus (Fig 6C, compare lanes 4 and 5) and the addition of A22 to E. coli (compare lanes 2 and 3) did not affect the PopZ oligomerization state in whole cell lysates, suggesting that PopZ oligomerization is independent of polar accumulation.
Electron microscopy of purified PopZ oligomers (Figure 6D) revealed filament-like structures of 5 nanometers in diameter. They were distributed in length, including some ranging from 25 to 50 nanometers, as well as some branched filaments that were over 200 nanometers long (Figure 6D, upper panel). The approximately 650 kDa PopZ oligomers in cell lysates (Figure 6A–C) may be the short filaments, which are capable of assembling into larger branched forms. At slightly higher concentrations, PopZ formed a polymeric network (Figure 6D, lower panel) in which 3-way junctions were the prominent type of intersection between filaments (Figure 6E, circles). These networks were observed directly in solution using cryogenically fixed samples (Figure S4), indicating that their assembly is not an artifact induced by drying the protein on carbon coated grids.
To determine if PopZ has an intrinsic ability to assemble into filaments, the purified protein was unfolded in the presence of 8 molar urea. The urea was then removed by dialysis, and re-folding was analyzed using a variety of techniques. Small Angle X-ray Scattering (SAXS) measurements demonstrated that PopZ was denatured in the presence of urea, and that it adapted a conformation that is similar to the original form after dialysis (Figure 6F). Using native gel electrophoresis, we found that the re-folded protein had the same high molecular weight size as the original material, and the presence of filaments was confirmed by electron microscopy (Figure 6G).
DISCUSSION
The linear order of genetic loci on bacterial chromosomes reflects their relative position in the cell (Teleman et al., 1998; Viollier et al., 2004); Figure 1A). This chromosome organization is maintained during sequential segregation of the replicating DNA (Nielsen et al., 2006; Viollier et al., 2004) such that replication initiation is followed rapidly by the segregation of origin regions to defined sub-cellular positions. The dynamic cellular organization of the replicating chromosome is analogous to the mitotic apparatus in eukaryotes, and suggests that bacterial cells may have evolved similarly complex mechanisms for DNA segregation. However, the mechanisms used by bacterial cells to separate and maintain the cellular positions of replicating chromosomes are poorly understood. In C. crescentus, as in V. cholera (chr1), the region of the chromosome bearing the origin and the nearby parS sequences reside at the cell pole in quiescent cells (Fogel and Waldor, 2005; Jensen and Shapiro, 1999). Upon initiation of replication, the newly duplicated origin region, in complex with the ParB partition protein, moves across the cell and is retained at the opposite pole (Fogel and Waldor, 2005; Thanbichler and Shapiro, 2006). The segregation of the rest of the replicating chromosome is dependent on the transport of the duplicated origin to the opposite pole (Gitai et al., 2005); Figure 1A).
We present evidence that the ori/ParB complex is specifically anchored to the pole in C. crescentus by a novel polymeric protein, PopZ. Although PopZ and ParB co-localize at the pole bearing the newly arrived ori/ParB complex, their positions are somewhat off-set at the stalked (old) pole after replication initiation (Figure 2). We propose that the anchor must be relieved to enable replication initiation and/or translocation of the replicated origin to the new pole. This likely involves the modulation of PopZ or ParB function, possibly through direct modification of the proteins, or by the activation of an inhibitor that physically disrupts the interaction between ParB and PopZ. Furthermore, the PopZ that resides at the stalked pole likely performs additional functions in polar organization, as evidenced by the inability of ΔpopZ strains to form polar stalks and the mislocalization of two integral membrane polar histidine kinases in a PopZ depletion strain (G. Bowman, unpublished). Thus, it may be that PopZ complexes perform different roles at the two poles.
Capture and assembly of a chromosomal anchoring protein at the cell pole
PopZ-YFP single molecule microscopy showed that PopZ diffuses randomly within the cytoplasm and is only immobilized when it interacts with the cell pole. This demonstrates that any non-specific interactions between PopZ and chromosomal DNA do not prevent PopZ molecules from moving rapidly within the cell, and suggests that PopZ arrives at the poles by diffusion rather than directed transport. This diffusion/capture mechanism of protein localization argues that there are polar factors enabling the capture of PopZ. Because one pole remains free of PopZ in C. crescentus swarmer cells and in E. coli that express PopZ heterologously, the mere presence of a pole is not sufficient for polar accumulation. In both of these organisms, the polar capture of PopZ is dependent on active MreB, indicating a role for the cytoskeleton in polar assembly. It has been reported previously that MreB is required for the segregation of chromosomal origins to the cell poles (Gitai et al., 2005). An unresolved question is whether the roles of MreB in origin segregation and PopZ polar accumulation are part of a single pathway, or if MreB contributes to those functions via distinct mechanisms.
MreB has been implicated in the polar localization of several proteins in C. crescentus (Gitai et al., 2004) as well as in E.coli (Nilsen et al., 2005). The underlying mechanism is not known, but may be an indirect effect of the influence of MreB on the construction of the cell wall (Carballido-Lopez et al., 2006; Divakaruni et al., 2007). Alternatively, MreB may form polar nucleation sites for cytoplasmic proteins when in the extended helical conformation at the swarmer to stalked cell transition (Gitai et al., 2004).
Purified PopZ forms a network of filaments, interconnected by trimeric junctions. We propose that a similar network forms at the poles in live cells, forming a “Velcro-like” adhesive for ParB in complex with DNA. During sporulation in Bacillus subtilis, the polar protein DivIVA is required to anchor the chromosome origin to the cell pole (Ben-Yehuda et al., 2003). Purified DivIVA assembles into protofilaments in vitro (Stahlberg et al., 2004)) though the structure (described as ribbons of “doggy bone” shaped subunits) differs from the PopZ network. The chromosome-associated target for polar DivIVA has not been established. Genetic experiments indicate that it is not the ParB homolog Spo0J, but an unrelated origin-binding protein called RacA (Ben-Yehuda et al., 2003), although a direct interaction between DivIVA and RacA has not been demonstrated. Co-immunoprecipitation experiments in the presence of a cross-linking agent have shown that DivIVA and Spo0J are in close proximity during sporulation (Perry and Edwards, 2006), yet the mechanistic relationship between these proteins is unclear, and spo0J mutants do not affect chromosome anchoring unless they are combined with a racA mutant (Wu and Errington, 2003). In this study, we establish the PopZ-ParB interaction in C. crescentus as the physical link between the cell pole and the chromosome. Despite outward similarities in structure and function, DivIVA and PopZ do not share any appreciable sequence similarity. We speculate that the common features of these two molecules have emerged through constraints associated with the convergent evolution of two different chromosome anchoring mechanisms in organisms widely separated in evolutionary time.
Making a complete origin segregation system
In order to achieve a complete mechanism for chromosome origin segregation, the anchoring function proposed in this work must be coupled with an activity that moves the newly replicated origins towards opposite ends of the cell. The best understood mechanisms for origin segregation come from studies of Type I partitioning plasmids, which encode a three component segregation mechanism that includes a ParB homolog, a plasmidic ParB binding site (parS) and a polymer forming ATPase, ParA, that provides the driving force for plasmid separation (Ebersbach and Gerdes, 2005). Comparison of these systems to other closely related plasmid segregation mechanisms (Garner et al., 2007) suggests that ParB/plasmids are propelled apart by the expansion of a ParA filament that forms between ParB foci (Ebersbach et al., 2006). In the case of Type I plasmids, a mechanism that holds the segregated plasmids at their destinations is either unnecessary for plasmid stability or has not yet been discovered. In this work, we have shown that such an anchoring mechanism exists for a chromosomally encoded ParB/parS system, through direct interaction with PopZ.
Although the factors that contribute to the movement and retention of newly replicated chromosomal origins at the cell pole have not been fully characterized, existing data implicates both the ParA/B system and the MreB cytoskeleton, two systems that are mechanistically connected to PopZ. In V. cholerae the movement of chromosome origins to the cell poles is thought to be accomplished by the retraction of a ParA filament, which is attached to an origin at one end and the cell pole at the other end (Fogel and Waldor, 2006). The proposed membrane association of V. cholerae ParA differs from the models of plasmid partitioning, and the polar anchor for the ParA filament is not known. No obvious homolog of PopZ is present in this system.
The work presented here shows that a polar anchoring protein, PopZ, directly interacts with ParB at the chromosome origin, and this interaction is required to complete the segregation of newly replicated chromosomes. The discovery of oligomeric polar proteins like PopZ, that provide critical functions for the establishment of the polar axis of the bacterial cell, is revealing the underlying principles of large-scale cellular organization.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Cell Growth, and Cell Synchronization
The growth conditions for C. crescentus and E. coli cultures and the synchronization of cells are given in detail in the Supplementary Material available on the Cell website. A list of all relevant strains and plasmids is provided in Tables S2 and S3.
Bioinformatic Screen for Proteins that are Active at the Cell poles
The probabilistic protein interaction network described by Srinivasan et al. (2006) was expanded by using publicly available sequence data from over 200 bacterial genomes. The likelihood that each annotated polypeptide was predicted to interact with an individual polar bait protein (Supplementary table 1) was tabulated in an Excel spreadsheet, and overall likelihood for polar activity was calculated as the aggregate interaction score for each polypeptide with all of the 46 bait proteins. 36 high scoring candidates were chosen for analysis.
Co-Immunoprecipitation
1 liter of cell culture in PYE medium was washed in Co-IP buffer (20mM Hepes pH 7.5, 100mM NaCl, 20% glycerol), treated with the membrane permeable cross-linking agent DSP (Peirce) at a final concentration of 2mM, then lysed in a French press. After treating the lysate with detergent (Igepal CA-630 and Sodium Deoxycholate at 1% and 0.5% final concentration, respectively, with additional 2 mM EDTA), PopZ-M2 was immunoprecipitated with the anti-FLAG affinity gel agarose beads from the FLAGIPT-1 kit (Sigma).
Preparation of PopZ protein and refolding experiments
6-His tagged PopZ was expressed at high levels in E. coli and purified from cell lysates with a cobalt affinity column. After cleavage of the 6-His tag by digestion with thrombin, PopZ was further purified by gel filtration chromatography. Measurements of protein concentration were performed using the BCA protein assay (Pierce). For refolding experiments, PopZ was purified in buffer containing 50mM NaCl, 50mM Tris pH 8.0, and urea was added to a final concentration of 1M or 8M. After 1 hr incubation at 37°C, urea was removed by four repetitions of spin dialysis and resuspension in the original buffer.
Surface Plasmon Resonance
Experiments were performed on a Biacore 3000 system. ParB was covalently immobilized on the surface of a CM5 biosensor chip (Biacore) using amine coupling chemistry, and PopZ and other proteins were diluted in BCG+M buffer (50mM Hepes/NaOH, pH 7.2, 50mM KCl, 5mM MgCl2) and injected over the ParB surface at different concentrations at a flow rate of 20µl/min. To measure the affinity of the ParB-PopZ interaction, association was monitored in Buffer 50/25PSM+BSA (50 mM Na-phosphate pH 8.0, 25 mM NaCl, 5 mM MgCl2, 0.1 mg/ml BSA), with 75µl of PopZ injected over the ParB surface at different concentrations at a flow rate of 30µl/min. Dissociation was allowed to occur at the same flow rate for 240 seconds.
Electrophoretic mobility shift assay
The probe was prepared by end-labeling linear DNA with polynucleotide kinase in the presence of [γ-32P]-ATP. The DNA probes (at 1.1nM) were then incubated with the indicated proteins for 30 min at 30° C in reaction buffer (40 mM Na-phosphate pH 8.0, 20 mM NaCl, 5 mM MgCl2, 7% glycerol containing 20 µg/ml BSA and 4.75 µg/ml sheared salmon sperm DNA), and twenty microliter samples were loaded on a non-denaturing 3% polyacrylamide gel and run in 1X TBE buffer supplemented with 5 mM MgCl2.
SAXS analysis
Solution x-ray scattering (SAXS) experiments were performed on the SIBYLS beamline at the Advanced Light Source using an energy of 8 keV (~1.55 Angstroms). The concentration of PopZ used for these experiments ranged from 1.5 to 3.3 mg/ml, with the standards in the range of 1.2 to 2.4 (no urea) or 1.6 to 5.6 mg/ml (with urea).
Electron Microscopy
The high MW PopZ peak from gel filtration chromatography (above) was diluted to the indicated concentration in buffer 50/25PS (50 mM sodium phosphate, pH 8.0, and 25 mM NaCl). 5 µl aliquots were placed on glow-discharged Formvar carbon coated grids (Ted Pella 01753-F), incubated for 2 min, washed in buffer, blotted, and stained with 2% Uranyl Acetate for 30 seconds. Images were acquired on a JEOL–3100 electron microscope at 300 kV.
Supplementary Materials
A detailed description of the Co-immunoprecipitation protocol, the preparation of PopZ protein and anti-PopZ antibodies, as well as the surface plasmon resonance and gel shift experiments is provided with this paper online.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Balaji Srinivasan and Esteban Toro for developing bioinformatics tools. We thank Martin Thanbichler for purified ParB. Mike Fero designed software used for fluorescence microscopy, Bob Glaeser and Byong-Gyoon Han provided advice on EM sample preparation, Greg Hura assisted in SAXS analysis, and James Gober and Christine Jacobs-Wagner provided antibodies and strains. This work is supported by National Institutes of Health grants GM32506, GM051426, and 5R24GM73011-3 to LS, F32GM080008 to GB, and 5 P20 HG003638-02 to MK and WEM; and Department of Energy grants DE-AC02-05CH11231 to LRC, JX, and TE, and DE-FG02-01ER63219 to LS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barak I, Wilkinson AJ. Where asymmetry in gene expression originates. Mol Microbiol. 2005;57:611–620. doi: 10.1111/j.1365-2958.2005.04687.x. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Fujita M, Liu XS, Gorbatyuk B, Skoko D, Yan J, Marko JF, Liu JS, Eichenberger P, Rudner DZ, Losick R. Defining a centromerelike element in Bacillus subtilis by Identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Ringgaard S, Moller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Easter J, Gober JW. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol Microbiol. 2003;47:1225–1237. doi: 10.1046/j.1365-2958.2003.03367.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. Distinct segregation dynamics of the two Vibrio cholerae chromosomes. Mol Microbiol. 2005;55:125–136. doi: 10.1111/j.1365-2958.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Goley ED, Iniesta AA, Shapiro L. Cell cycle regulation in Caulobacter: location, location, location. J Cell Sci. 2007;120:3501–3507. doi: 10.1242/jcs.005967. [DOI] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U S A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Easter J, Jr, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- Nilsen T, Yan AW, Gale G, Goldberg MB. Presence of multiple sites containing polar material in spherical Escherichia coli cells that lack MreB. J Bacteriol. 2005;187:6187–6196. doi: 10.1128/JB.187.17.6187-6196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Edwards DH. The Bacillus subtilis DivIVA protein has a sporulation-specific proximity to Spo0J. J Bacteriol. 2006;188:6039–6043. doi: 10.1128/JB.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL. Spatial control of bacterial division-site placement. Nat Rev Microbiol. 2005;3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- Srinivasan BS, Novak AF, Flannick JA, Batzoglou S, McAdams HH. Integrated Protein Interaction Networks for 11 Microbes. RECOMB 2006, LNBI 3909. 2006:1–14. [Google Scholar]
- Stahlberg H, Kutejova E, Muchova K, Gregorini M, Lustig A, Muller SA, Olivieri V, Engel A, Wilkinson AJ, Barak I. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol Microbiol. 2004;52:1281–1290. doi: 10.1111/j.1365-2958.2004.04074.x. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49:1463–1475. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.