Abstract
Background
Pathogens tolerate stress conditions that include low pH, oxidative stress, high salt and high temperature in order to survive inside and outside their hosts. Lipopolysaccharide (LPS), which forms the outer-leaflet of the outer membrane in Gram-negative bacteria, acts as a permeability barrier. The lipid A moiety of LPS anchors it to the outer membrane bilayer. The MsbB enzyme myristoylates the lipid A precursor and loss of this enzyme, in Salmonella, is correlated with reduced virulence and severe growth defects that can both be compensated with extragenic suppressor mutations.
Results
We report here that msbB (or msbB somA) Salmonella are highly sensitive to physiological CO2 (5%), resulting in a 3-log reduction in plating efficiency. Under these conditions, msbB Salmonella form long filaments, bulge and lyse. These bacteria are also sensitive to acidic pH and high osmolarity. Although CO2 acidifies LB broth media, buffering LB to pH 7.5 did not restore growth of msbB mutants in CO2, indicating that the CO2-induced growth defects are not due to the effect of CO2 on the pH of the media. A transposon insertion in the glucose metabolism gene zwf compensates for the CO2 sensitivity of msbB Salmonella. The msbB zwf mutants grow on agar, or in broth, in the presence of 5% CO2. In addition, msbB zwf strains show improved growth in low pH or high osmolarity media compared to the single msbB mutant.
Conclusion
These results demonstrate that msbB confers acute sensitivity to CO2, acidic pH, and high osmolarity. Disruption of zwf in msbB mutants restores growth in 5% CO2 and results in improved growth in acidic media or in media with high osmolarity. These results add to a growing list of phenotypes caused by msbB and mutations that suppress specific growth defects.
Background
Lipopolysaccharide (LPS), the most abundant molecule on the surface of Gram-negative bacteria, acts as a permeability barrier and renders the outer-leaflet of the outer membrane (OM) relatively impermeable to hydrophobic antibiotics, detergents [1], and host complement [2]. LPS consists of three major components: lipid A, core polysaccharides and O-linked polysaccharides. Lipid A, with its fatty acid anchors [lauric, myristic and sometimes palmitic acid], is an endotoxin primarily responsible for TNFα-mediated septic shock. The addition of myristic acid to the lipid A precursor is catalyzed by the enzyme MsbB [3].
It has been shown that msbB Salmonella serovar Typhimurium exhibits severe growth defects in LB and sensitivity to bile salts (MacConkey) and EGTA-containing media. However, compensatory suppressor mutants can be isolated that grow under these conditions. One of these suppressor phenotypes results from a mutation in somA, a gene of unknown function [4]. msbB Salmonella Typhimurium strains have recently been developed as potential anti-cancer agents that possess impressive anti-tumor activity in mice [5]. In a phase I clinical study msbB Salmonella were shown to be safe in humans when administered i.v. However, bacteria were rapidly cleared from the peripheral blood of humans and targeting to human tumors was only observed in few patients at the highest dose levels of 3 × 108 CFU/m2 and 1 × 109/m2 [6]. Toso et al. [6] noted that YS1646 (suppressed msbB strain, see below) grew best in air without added CO2.
The potential to grow in acidic and CO2-rich environments is a hallmark of pathogenic bacteria, enhancing persistence within phagocytes and survival inside the host. Sensitivity to CO2 and low pH of msbB Salmonella strains might explain poor colonization of tumors, which often contain high levels of CO2 and lactic acid [7,8] due to the Warburg effect, also known as aerobic glycolysis, whereby glucose uptake is elevated while oxidative phosphorylation is reduced, even in the presence of oxygen. Our previous work on suppressors of msbB Salmonella raised the possibility that secondary mutations could suppress sensitivity to 5% CO2 and acidic conditions.
Here we report that the growth of msbB Salmonella is highly inhibited (greater than 3-log reduction in plating efficiency) in a 5% CO2 atmosphere in LB media as well as under low pH conditions when compared to wild-type Salmonella. Furthermore, several CO2 resistant clones were selected from an msbB Salmonella transposon library (Tn5). Three mutations were mapped and all were shown to contain the Tn5 marker in the zwf gene, which encodes the enzyme glucose-6-phosphate-dehydrogenase and is tightly linked to the msbB gene.
Results
CO2 sensitivity of msbB Salmonella
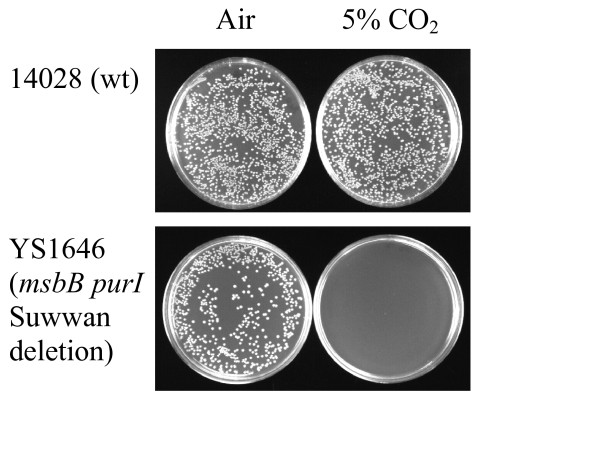
CO2 sensitivity was first observed when YS1646, an msbB purI Suwwan deletion strain of Salmonella Typhimurium, was plated on blood or LB plates and incubated in a 5% CO2 incubator (Caroline Clairmont, personal communication; Toso et al., 2002). Suwwan deletion strains lack ~100 genes in the 17.7 to 19.9 Cs region of the chromosome [9]. In our studies, plating identical amounts (e.g., 100 μl of a 10-5 dilution of a culture grown under non-selective conditions) to duplicate plates incubated at 37°C in either air or 5% CO2, few or no colonies of YS1646 were observed after 16 hours of incubation at 37°C in 5% CO2 (Figure 1). However, by plating more cells, the presence of a few resistant colonies could be detected, as we obtained 3.3 × 108 CFU/ml on plates incubated in air and 1.7 × 105 CFU/ml on plates incubated in the presence of 5% CO2, a greater than 3 log reduction. This CO2 sensitivity, first observed in YS1646, is also observed in a simple msbB mutant (see below). In contrast, wild-type Salmonella Typhimurium (ATCC 14028 and LT2), Salmonella Typhi (CS029, ATCC 33458), and Escherichia coli (MG1655, near-wild type K-12) are resistant to 5% CO2 (ATCC 14028: Figure 1; other strains: data not shown). Interestingly, msbB E. coli (KL423) was not sensitive to CO2 (not shown), consistent with there being physiologically relevant differences between the E. coli and Salmonella in regard to the loss of MsbB function, as has been previously observed [4]. These differences obscure or compensate for obvious growth defects in msbB E. coli.
Figure 1.
Sensitivity and resistance to CO2 shown by comparing colony forming units (CFUs). Each strain was grown overnight in LB broth and diluted 106 fold, and then 100 μl was spread on each plate and incubated. Upper panel: wild type Salmonella (14028) on LB media in air (left) or 5% CO2 (right). Lower panel; YS1646 on LB media in air (left) or 5% CO2 (right).
CO2 sensitivity was found in all msbB Salmonella strains tested so far, indicating that CO2 sensitivity is a direct result of the lack of lipid A myristoylation (data not shown, see list of strains in Table 1). Consistent with these results, normal growth in CO2 was completely restored when msbB was expressed from a plasmid (pSM21(msbB+)) (see Table 1).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Parental strain | Genotype | Derivation or source |
|---|---|---|---|
| S. enterica serovar Typhimurium | |||
| 14028 | 14028 | Wild type | ATCC 14028 |
| 14028Δzwf | 14028 | Δzwf82 | Replacement of zwf gene with Δzwf82 by homologous recombination |
| 14028 gnd | 14028 | gnd-189::MudJ (KanR) | P22·DM4483 × 14028 → Kan40R |
| YS1646(=VNP20009) | 14028 | ΔmsbB2 ΔpurI ΔSuwwan | [9,31] |
| VNP20057 | YS1646 | ΔmsbB2 ΔpurI zwf81::Tn5 (KanR) ΔSuwwan | YS1646 × P22 Tn5 pool (on 14028) → selection on LB plates in 5% CO2 |
| YS1 | 14028 | msbB1::Ωtet | [4] |
| YS1 msbB+ | YS1 | msbB1::Ωtet/pSM21msbB+ (AmpR) | Plasmid pSM21 [4] into YS1 |
| YS1 zwf(=YS11) | YS1 | msbB1::Ωtet zwf:Tn5 (KanR) | P22·VNP20057 × YS1 → Kan20R |
| YS873 | 14028 | msbB1::Ωtet somA1 zbj10:Tn10 | [4] |
| YS873 msbB+ (=YS8731) | YS873 | msbB1::Ωtet somA1 zbj10:Tn10/pSM21msbB+ (AmpR) | Plasmid pSM21 [4] into YS873 |
| YS873 zwf(=YS8732) | YS873 | msbB1::Ωtet somA1 zbj10:Tn10 zwf81:Tn5 (KanR) | P22·VNP20057 × YS873 → Kan20R |
| YS873 Δzwf (=YS8733) | YS873 | msbB1::Ωtet somA1 zbj10:Tn10 Δzwf82 | Replacement of zwf81::Tn5 gene in YS873zwf with Δzwf82 by homologous recombination |
| YS873 gnd (=YS8734) | YS873 | msbB1::Ωtet somA1 zbj10:Tn10 gnd-189::MudJ (KanR) | P22·DM4483 × YS873 → Kan10R |
| LT2 | LT2 | Wild type | ATCC 15277 |
| DM4483 | LT2 | gnd-189::MudJ (KanR) | Gift of Diana Downs and Eugene I. Vivas, U. of Wisconsin |
| YS501 | LT2 | recD541::Tn10dCm hsdSA29 hsdSB121 hsdL6 metA22 metE551 trpC2 ilv-452 H1-b H2-e,n,x fla-66 nml(-) rpsL120 xyl-404 galE719 | [5] |
| Salmonella enterica serovar Typhi CS029 | |||
| Salmonella enterica serovar Typhi ATCC 33458 | |||
| E. coli K-12 MG1655 | MG1655 | F- l- rph-1 | [32] |
| KL423 | MG1655 | F- l- rph-1 msbB1:: ΩCm | [4] |
| pCVD442 | AmpR | [10] | |
| pCVD442Δzwf82 | AmpR | This study | |
| pSP72 | AmpR | Promega Corporation | |
| pSP72lacZ | lacZ, AmpR | This study | |
| pSM21 | msbB, AmpR | [4] | |
The somA (for EGTA and salt resistance) and Suwwan deletion (for EGTA, salt, and galactose-MacConkey resistance) msbB suppressors do NOT suppress sensitivity to 5% CO2
Two msbB Salmonella strains with secondary mutations that allow faster growth are YS873 and YS1646. YS873 has a loss-of-function mutation in somA [4] and YS1646 has a large deletion, referred to as the Suwwan deletion [9], that includes somA plus ~100 other genes. The somA mutation in YS873 suppresses growth defects on EGTA and salt-containing media [4] and the Suwwan deletion in YS1646 suppresses sensitivity to EGTA, salt, and galactose MacConkey media [9]. However, neither the somA mutation nor the Suwwan deletion suppresses MsbB-mediated sensitivity to 5% CO2 (Suwwan deletion in YS1646, Figure 1; somA in YS873, see below). As shown in Figure 1, when plating identical dilutions containing greater than 100 CFU onto LB agar from an MSB broth culture of YS1646 and wild type Salmonella, no YS1646 colonies are detected after 24 hours of incubation in 5% CO2 at 37°C. Since we have not yet identified all of the genes within the Suwwan deletion that are responsible for the suppressor phenotype, we focused our study on YS873, which has clearly defined mutations in msbB and somA.
CO2 resistant mutations are detected at high frequency in msbB somA Salmonella
Subsequent experiments revealed that spontaneous CO2 resistant mutants are detected when higher numbers of YS873 bacteria are plated and incubated under 5% CO2 conditions. The mutation frequency of spontaneous CO2 mutants from an MSB broth culture was determined to be ~3 out of 104 (not shown), which is similar to the frequency that EGTA and galactose MacConkey suppressor mutations arise in msbB Salmonella [4].
A loss-of-function mutation in zwf suppresses CO2 sensitivity
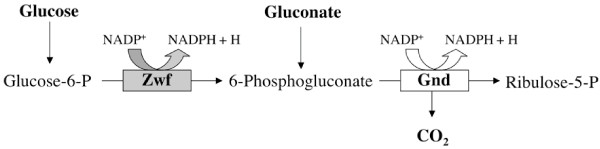
In our preliminary studies, several spontaneous CO2 resistant mutants were isolated that showed a high degree of instability. Therefore, we subsequently focused on the use of Tn5 mutagenesis, which is known to generate stable insertions primarily associated with null mutations. To screen for a mutation that would compensate for CO2 sensitivity, a random Tn5 insertion library of YS1646 was created and selected on LB agar in 5% CO2. 9 clones were isolated, of which we determined the insertion sites in three of the clones using a genome-walking method. All of the Tn5 insertions identified were located in the monocistronic zwf gene. Two of the insertions (clones 14.2 and 32.2) were identical (possible siblings), located after open reading frame nucleotide 1019, and the third (clone 37.2) was located at after base pair 1349. Because we focused our screening on Tn5 insertions, we do not know if other mutagenesis methods would have isolated clones with mutations in other genes. zwf encodes glucose-6-phosphate-dehydrogenase, an enzyme of the pentose-phosphate-pathway (PPP). In this pathway, Zwf converts glucose-6-phosphate, from glycolysis, to 6-phosphogluconate, generating NADPH + H. The subsequent reaction, catalyzed by Gnd, converts 6-phosphogluconate to ribulose-5-phosphate, generating NADPH + H and CO2 (Figure 2). A non-polar deletion (see materials and methods) was created in zwf (Δzwf82) using the pCVD442 vector [10] to test if the phenotypes arise from loss of the zwf gene or a polarity effect. The zwf non-polar deletion was found to exhibit the same CO2 growth phenotypes as the zwf Tn5 insertions. Subsequent experiments use the non-polar deletion in zwf in 14028 and YS873. A loss-of-function mutation in zwf results in smaller colony size than zwf+ strains on agar media in both wild type and msbB genetic backgrounds.
Figure 2.
Steps of the Pentose Phosphate Pathway (PPP) highlighting the relationship of the Zwf enzyme, gluconate, and Gnd-based production of CO2.
Gluconate prevents suppression of CO2 sensitivity by zwf
Zwf catalyzes the first step of the pentose phosphate pathway (PPP). PPP produces NADPH for anabolic pathways and the molecules generated by this pathway serve as building blocks for nucleotides, sugars, amino acids, and vitamins [11]. As shown in Figure 2, Zwf catalyzes the conversion of glucose-6-phosphate to 6-phosphogluconate. 6-phosphogluconate can also be formed from gluconate by gluconate kinase [12], which bypasses the PPP's requirement for Zwf (Figure 2). The addition of gluconate to media thereby allows for the production of 6-phosphogluconate in the absence of Zwf. The enzyme gluconate-6-phosphate dehydrogenase (Gnd) then decarboxylates 6-phosphogluconate, converting it from a 6-carbon to a 5-carbon (ribulose-5-phosphate) sugar and releasing CO2 gas. Perhaps a threshold of CO2 must be passed to inhibit the growth of msbB Salmonella and a loss-of-function mutation in zwf allows for the CO2 level to remain below this threshold. Previous reports of zwf E. coli show reduced CO2 production when grown in minimal media with acetate or pyruvate as a carbon source. However, zwf E. coli produced more CO2 than wild type when grown in minimal media with glucose [13,14]. Further studies will be required to clarify the production of CO2 by Salmonella grown in Luria-Bertani-based media and its contribution to CO2 sensitivity.
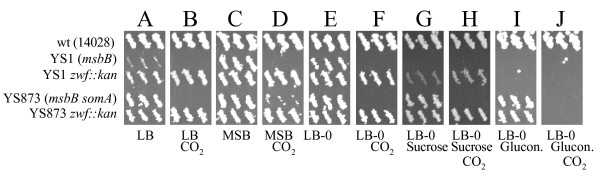
To test whether zwf's suppressive effects result from its role in PPP pathway products and not from some unknown function, we observed the effect of gluconate on CO2 sensitivity in our mutants. Growth of YS873 zwf was tested on LB-0 plates containing 0.33% gluconate in ambient air and 5% CO2 (Figures 3I and 3J). As we hypothesized, YS873 zwf was not able to grow on LB-0 gluconate in 5% CO2. Thus, we confirmed that the zwf's suppression of CO2 sensitivity results from its known enzymatic step in the PPP pathway. We also found a new phenotype for unsuppressed msbB Salmonella: YS1 does not grow on LB-0 agar in the presence of 0.33% gluconate (Figure 3I). To test if the production of 6-phosphogluconate or a downstream PPP metabolite is responsible for mediating CO2 resistance, we tested for CO2 resistance in a YS873 gnd-189::MudJ mutant (Gnd catalyzes the second step of the PPP pathway, Figure 2) and found that the strain remained CO2 sensitive (data not shown). Therefore, we conclude that the production of 6-phosphogluconate, by either Zwf or gluconate kinase, contributes to CO2 sensitivity in an msbB genetic background.
Figure 3.
zwf mutation suppresses both msbB-induced CO2 sensitivity and osmotic defects. Double velvet replica plates with different media were used to indicate the ability of small patches of bacteria (3 each) to grow. The strains used are listed on the left. Growth conditions (all at 37°C) included: A, LB media in air; B, LB media in 5% CO2; C, MSB media in air; D, MSB media in 5% CO2; E, LB-0 media in air; F, LB-O media in 5% CO2; G, LB-0 media containing sucrose (total 455 miliosmoles) in air; H, LB-0 media containing sucrose in 5% CO2; I, LB-0 + gluconate (glucon.) in air; J, LB-0 + gluconate in 5% CO2.
zwf mutation suppresses both msbB-induced CO2 sensitivity and osmotic defects
For further analysis of the msbB zwf phenotype, the zwf (zwf81::Tn5) mutation was transduced into msbB (YS1) and msbB somA (YS873) genetic backgrounds to generate strains YS1 zwf and YS873 zwf respectively. As shown in the replica plate series of Figure 3, growth of unsuppressed YS1 is inhibited on LB (Figure 3A) and LB-0 gluconate (Figure 3I) but it grew well on MSB and LB-0 agar (Figures 3C and 3E), confirming the results of Murray et al. [4]. In contrast, growth of YS1 on MSB and LB-0 agar is completely inhibited when the plates are incubated in the presence of 5% CO2. The introduction of the zwf mutation completely compensates for the phenotype and allows the bacteria to grow under 5% CO2 on all three media (Figures 3B, 3D and 3F). However, it does not rescue YS1 from gluconate sensitivity (Figure 3I).
When NaCl in LB plates is substituted with sucrose at iso-osmotic concentrations (Figures 3G), growth of YS1 is also inhibited, indicating osmosensitivity of YS1. Interestingly, introduction of the zwf mutation improves growth of YS1 on LB and on LB-0 5% sucrose agar, indicating that the zwf mutation can partially compensate for the msbB-induced osmotic growth defect.
MSB media contains high levels of divalent cations, which have been proposed to increase lateral interactions between the phosphate groups of neighboring lipid A molecules [15]. Based on Murray et al.'s finding [16] that a decrease in electrostatic repulsion between the phosphates of lipid A can help to compensate for the lack of the myristic acid residue, we investigated whether Mg2+ and Ca2+ would protect against the detrimental effects of 5% CO2. On agar plates, Mg2+ and Ca2+showed partial protection in YS873 (Figure 3D).
YS873, which contains the EGTA and salt resistance suppressor mutation somA [4], grows well on LB (Figure 3A), MSB (Figure 3C), LB-0 (Figure 3E) and LB-0 sucrose (Figure 3G) agar plates in air, but not when the plates are incubated in 5% CO2 (Figures 3B, 3D, 3F, and 3H). In contrast, the strain YS873 zwf is able to grow on all of these media in CO2, indicating that the zwf mutation can compensate for the growth defect of msbB strains in CO2 (Figure 3). Subsequent experiments were performed using the YS873 (msbB somA) genetic background because unsuppressed msbB Salmonella can not grow under mammalian physiological salt conditions [4].
msbB somA Salmonella are sensitive to CO2 in LB and LB-0 broth
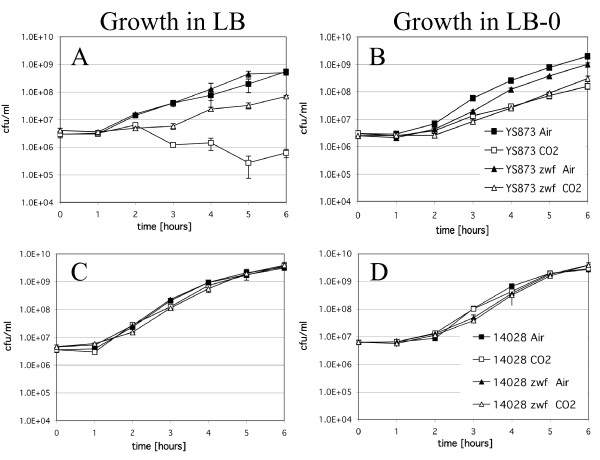
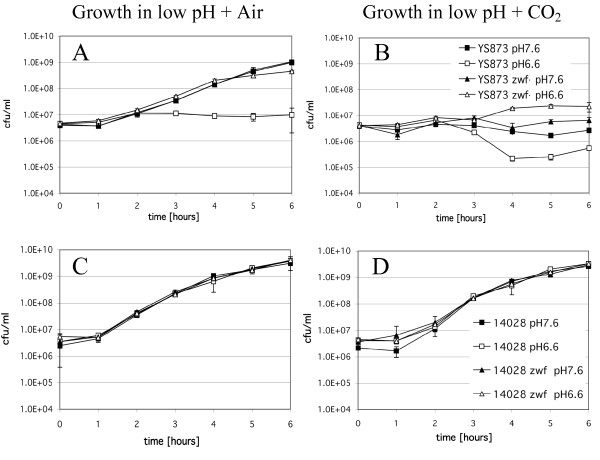
Figure 4 shows the growth of wild type ATCC 14028, 14028 zwf, YS873, and YS873 zwf in LB and LB-0 broth, incubated in the presence or absence of 5% CO2. As shown in Figure 4, the growth of YS873 (Figure 4A), but not ATCC 14028 (Figure 4C) is greatly impaired in LB broth in the presence of 5% CO2. A significant decrease in CFU is observed (Figure 4A), indicating that YS873 cells lose viability in the presence of 5% CO2 in LB broth. When a loss-of-function mutation in zwf is incorporated into YS873, no loss in viability is observed under identical conditions, although there is a longer lag phase of growth (Figure 4A). In LB-0 broth, there are no growth defects in 14028 or 14028 zwf (Figure 4D). For YS873 and YS873 zwf, the growth defects in LB-0 in the presence of 5% CO2 are attenuated in comparison to those observed in LB broth. There is no decrease in viability in YS873 in LB-0 in 5% CO2, although there is impaired growth in both YS873 and YS873 zwf in LB-0 in the presence of CO2 compared to growth in the absence of CO2 (Figure 4B).
Figure 4.
msbB confers growth sensitivity in liquid media under CO2 conditions containing physiological amounts of salt and this is suppressed by zwf. Two sets of Salmonella strains (YS873 and YS873 zwf; 14028 and 14028 zwf) were grown on either LB (A and C) or LB-0 (B and D) in either air or 5% CO2.
YS873 has severe morphological defects in LB broth under 5% CO2 conditions that are suppressed by a loss-of-function mutation in zwf
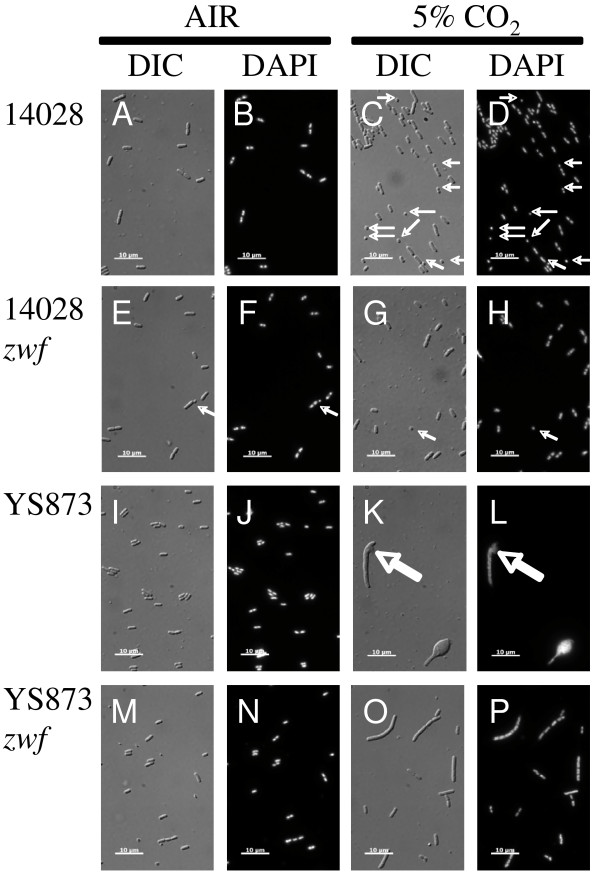
Since our results show that msbB Salmonella lose viability in the presence of 5% CO2 (Figure 4), we examined msbB mutants grown in the presence of 5% CO2 to determine if there are any defects in cell morphology or chromosome segregation. Differential interference contrast (DIC) microscopy shows striking morphological defects under CO2 conditions (Figure 5K), with long, bulging filamentous YS873 cells. DAPI staining shows no apparent chromosomal segregation defects, as no cells lacking DNA were observed (Figure 5L). However, the cell directly under the "K" and "L" labels appears to be lysing (see thick arrow).
Figure 5.
YS873 has severe morphological defects in LB broth under 5% CO2 conditions that are suppressed by a loss-of-function mutation in zwf. DIC, Differential Interference Contrast; DAPI, 4'6-diamidino-2-phenylindole (DNA stain); Thick arrows point to lysis; Thin arrows point to mini-cells.
As shown in Figures 5O and 5P, zwf suppresses the severe morphological defects in YS873 grown in LB in the presence of 5% CO2. Many cells are elongated but lack gross morphological defects. Growth in LB in a 5% CO2 environment caused wild type ATCC 14028 Salmonella to form minicells, with minicells (see thin arrows) accounting for ~15% of the cells (21/144) (Figure 5C and 5D as compared to Figures 5A and 5B). As seen in Figure 5E and 5F, 14028 zwf exhibits ~21% minicell formation in LB broth, even without CO2 (20/95 cells). Thus, we conclude that both CO2 and Zwf can, either directly or indirectly, affect cell division.
β-galactosidase assays confirm cell lysis in LB in the presence of 5% CO2
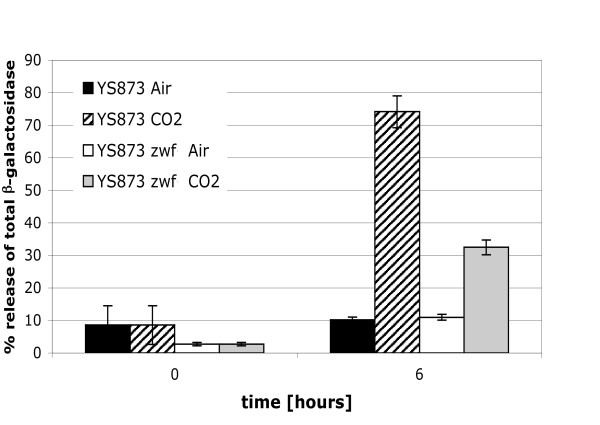
Microscopy (Figure 5K and 5L) suggested that some YS873 cells were lysing in LB in the presence of 5% CO2. To test if the decrease in CFU observed in YS873 in LB in the presence of 5% CO2 resulted from cell lysis, a plasmid expressing β-galactosidase was electroporated into YS873 and YS873 zwf and the cells were grown in LB in the presence or absence of CO2. As shown in Figure 6, after 6 hours of growth, significant cell lysis is observed in YS873 grown in the presence of 5% CO2 as measured by the release of the cytoplasmic enzyme β-galactosidase. Furthermore, a loss-of-function mutation in zwf significantly reduces cell lysis in YS873. No significant cell lysis is observed in the absence of CO2.
Figure 6.
β-galactosidase release assays confirm cell lysis in LB in the presence of 5% CO2 and that zwf confers resistance. Release of β-galactosidase from the cytosol of the bacteria was used to test if the decrease in CFU observed in YS873, in LB in the presence of 5% CO2, resulted from cell lysis. The strains were grown under either ambient air or 5% CO2 conditions.
CO2 sensitivity does not result from increased acidification of LB media and zwf suppresses sensitivity to acidic pH in LB broth
During this study, we observed that the pH of LB broth dropped from pH 7.0 to pH 6.6 after equilibration in 5% CO2. Since CO2 can acidify bicarbonate buffered media, we tested whether part of the CO2 sensitivity was due to acidification of the media. Thus, to test if increased or decreased pH would alter sensitivity to CO2 in LB broth, we buffered LB broth to pH 7.6, or 6.6, and cultures were grown in the presence or absence of 5% CO2. As shown in Figure 7, wild type ATCC 14028 and ATCC 14028 zwf grow normally under all conditions in LB broth in the absence (Figure 7C) or presence (Figure 7D) of 5% CO2. In contrast, the growth of YS873 is significantly impaired when the pH of LB is 6.6, with no significant increase in CFU after 6 hours (Figure 7A), whereas when the pH of LB is 7.6, YS873 grows well (Figure 7A). A loss-of-function mutation in zwf allows for YS873 to grow well in LB broth at a pH of 6.6 (Figure 7A). 5% CO2 inhibited the growth of YS873 and YS873 zwf in LB pH 6.6 and 7.6 (Figure 7B). Although zwf protects against 5% CO2 in LB broth pH 6.6 (Fig 7B), it does not significantly improve survival in the presence of 5% CO2 in LB broth pH 7.6 (Figure 7B), suggesting that an acidic pH is a component for zwf to suppress msbB-mediated sensitivity to 5% CO2.
Figure 7.
zwf suppresses sensitivity to acidic pH in LB broth in air, and to 5% CO2 in LB broth pH 6.6, but not pH 7.6. Strains were grown in LB broth buffered to pH 6.6, or pH 7.6, in either air (A and C) or 5% CO2 (B and D).
β-galactosidase assays confirm cell lysis in LB broth, pH 6.6, in air
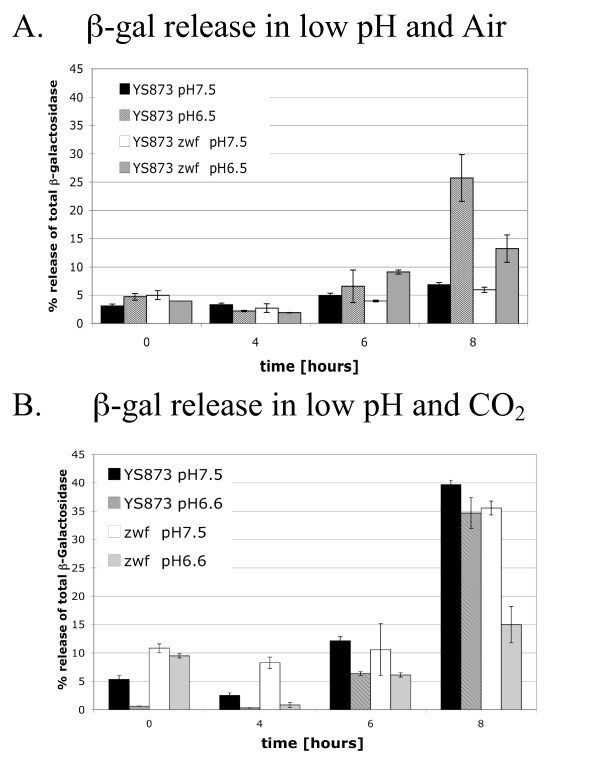
To test if the loss of growth of YS873 in LB broth pH 6.6 was the result of cell death or simply the result of inhibition or delay of cell division, β-galactosidase release was measured. As shown in Figure 8A, significant cell lysis occurs after growth of YS873 for 8 hours in LB broth, pH 6.5 but not pH 7.5 (pH shifted slightly [+/-0.1 units] during autoclaving). Furthermore, a loss-of-function mutation in zwf significantly reduces cell lysis of YS873 grown in LB broth pH 6.5. This reduction in cell lysis, as measured by release of the cytoplasmic enzyme β-galactosidase, correlates with increased CFU/ml numbers observed in YS873 zwf (as compared to YS873) grown in LB broth, pH 6.6 (Figure 7A).
Figure 8.
β-galactosidase release assays confirm cell lysis in LB broth, pH 6.6, in air; zwf inhibits cell lysis in LB broth, pH 6.6, in air and in LB broth, pH 6.6, but not pH 7.6, in the presence of 5% CO2. Release of β-galactosidase from the cytosol of the bacteria was used to test if the growth defects observed in YS873 and YS873 zwf resulted from cell lysis. Strains grown in LB broth at either pH 6.5, or pH 7.5, under either ambient air (A) or 5% CO2 (B) conditions.
zwf reduces YS873 cell lysis in the presence of 5% CO2 in LB broth pH 6.6, but not pH 7.6
Since we observed that YS873 lysed when there was no net growth in LB broth pH 6.5 while maintaining a relatively constant CFU/ml, we investigated if cell lysis occurs in YS873 zwf, which also exhibits little net growth with a relatively constant CFU/ml in the presence of 5% CO2 in LB broth pH 6.6 or 7.5 (Figure 7B). Growth curves for these strains indicated that there was a decrease in CFU/ml when YS873 was grown in LB broth pH 6.6 in the presence of 5% CO2, but that CFU/ml remained relatively constant if a loss-of-function mutation in zwf was present or if the pH of LB broth was 7.5 (Figure 7B). Figure 8 (8 hours) shows that significant cell lysis, as indicated by release of the cytoplasmic enzyme β-galactosidase, occurs when YS873 is grown in the presence of 5% CO2 at pH 6.6 or 7.6, and in YS873 zwf grown in the presence of 5% CO2 in LB pH 7.5. YS873 zwf exhibited significantly less lysis in the presence of 5% CO2 in LB broth pH 6.6, showing that a loss-of-function mutation in zwf significantly suppresses sensitivity to CO2 at neutral (as shown in Figure 6) or slightly acidic pH (Figure 8B). Again, we found that significant cell lysis can occur with a relatively constant CFU/ml (Figure 8B: YS873 zwf in LB pH 7.6).
Discussion
msbB Salmonella pleiotropy
The msbB gene was mutated to reduce the toxicity of Salmonella in mice and humans [5,6]. In order for these strains to function within mammalian systems they must be able to persist under normal mammalian physiological conditions. In contrast to other reports [17-20], we found msbB Salmonella to have striking growth defects, demonstrating sensitivity to salt, EGTA, MacConkey media, and polymyxin B sulfate [4,9,16]. Here we report additional sensitivity to osmolarity, gluconate, acidic pH and 5% CO2 growth conditions. Significantly, msbB Salmonella are sensitive to the conditions found within mammals, where blood has significant levels of salt and CO2; we therefore we screened for a suppressor of msbB-associated CO2 sensitivity.
zwf supresses CO2 sensitivity in msbB Salmonella
Glucose-6-phosphate-dehdrogenase (encoded by zwf) catalyzes the first enzymatic step in the pentose phosphate pathway (PPP), which converts glucose-6-phosphate to 6-phosphogluconate and NADPH + H. In E. coli, zwf is regulated by several mechanisms including anaerobic growth [21], growth rate [22], weak acids as well as superoxide [23]. Weak acids appear to regulate zwf through the multiple antibiotic resistance (mar) regulon, whereas superoxide exposure induces zwf through the Sox R/S regulon and contributes to DNA repair [24]. zwf mutants of Pseudomonas are hypersensitive to superoxide generating agents such as methyl viologen [25].
Salmonella Typhimurium zwf might be regulated by a different set of environmental signals than E. coli. Superoxide, while clearly activating other SoxR/S regulated genes like sodA and fumC, does not induce zwf transcription [26]. S. Typhimurium zwf mutants have been shown to be less virulent in mice and more sensitive to reactive oxygen and nitrogen intermediates [27]. In general, it is thought that the expression of zwf and subsequent generation of NADPH helps cells to combat oxidative stress. Interestingly, SoxS mutants of Salmonella are not attenuated in mice [28], suggesting that even though zwf expression is important for survival, superoxide generated responses might not be required. In the case of msbB mutants, the zwf mutation restores wild type growth under 5% CO2 and pH 6.5 conditions, suggesting that the expression of zwf is detrimental for growth of msbB mutants in an acidic or increased CO2 atmosphere. Furthermore, our data showing that a loss-of-function mutation in gnd (which produces the second enzyme of the PPP pathway, Figure 2) does not suppress sensitivity to CO2 suggests that the production of 6-phosphogluconate, by either Zwf or gluconate kinase, contributes to CO2 sensitivity in msbB Salmonella.
MsbB as a virulence factor?
Several publications cite MsbB as a virulence factor that is necessary for both septic shock and the ability to invade and persist in mammalian cells [5,17,29]. However, owing to the fact that msbB Salmonella were tested under 5% CO2 conditions, the lack of virulence may be partially or fully due to the inability of msbB Salmonella to grow in the presence of the 5% CO2. Further experimentation with msbB zwf Salmonella will be necessary to determine which virulence defects are attributable to msbB lipid A and those that arise from sensitivity to 5% CO2. Based upon this study and earlier studies on the sensitivity of zwf mutant to superoxides, zwf may both reduce virulence on one hand, yet potentiate growth under CO2 conditions on the other, further complicating virulence analyses.
Conclusion
Here, we report new growth defects in msbB Salmonella: sensitivity to gluconate and growth in hypertonic, acidic or 5% CO2 conditions. These characteristics are in addition to the previously reported growth defects in the presence of salt, EGTA, polymyxin, or MacConkey media. Previous studies showing that MsbB is a virulence factor require further evaluation of the role that CO2 sensitivity plays. The potential for cryptic, spontaneous mutations remains a possibility that should be addressed by re-transduction under non-selective conditions followed by plating independently under CO2 and ambient air. We have created an msbB somA zwf Salmonella strain that is resistant to growth under acidic or 5% CO2 conditions. This strain contains a loss-of-function mutation in zwf, an enzyme in the pentose phosphate pathway that produces CO2 as it converts a 6 carbon sugar to a 5 carbon sugar. The study of the virulence of msbB zwf Salmonella will allow the determination of what types of virulence are attributable to cells having an MsbB lipid A independent of sensitivity to 5% CO2, which is required for in vitro and in vivo virulence assays.
Methods
Bacterial strains, plasmids, phage and media
The bacterial strains and plasmids used in this study are listed in Table 1. The Salmonella msbB insertion/deletion for tetracycline resistance was described by Low et al. [5]. P22 mutant HT105/1int201 (obtained from the Salmonella Genetic Stock Center, Calgary, Canada) was used for Salmonella transductions. Salmonella enterica serovar Typhimurium strains were grown on LB-0 or MSB agar or in LB, LB-0, buffered LB or MSB broth. MSB media consists of LB (Luria-Bertani media, [30]) with no NaCl and supplemented with 2 mM MgSO4 and 2 mM CaCl2. LB-0 is LB media with no NaCl. Buffered LB pH 7.5 and pH 6.5 consisted of LB-0 with 100 mM NaPO4 adjusted to 455 mOsmol by adding NaCl. MSB broth and agar were used for the growth of strains under non-selective conditions. LB-0 agar was used when using selective antibiotics in transductions and transformations. Plates were solidified with 1.5% agar. LB-0 agar or MSB broth were supplemented as needed with ampicillin (100 μg/ml) or kanamycin (20 μg/ml). Antibiotics were added to LB-0 agar after cooling to 45 degrees Celsius.
Restoring msbB+ genotype
In order to confirm that the observed CO2 sensitivity results simply from knocking out MsbB function, wild type msbB was expressed from the msbB promoter using plasmid pSM21 [4]. Purified plasmids were transformed into electroporation-competent cells of strains YS1 and YS873.
Growth Analysis
Phenotypes of strains were determined by replica plating. Master plates were made on either MSB or LB-0 agar. Replica plating was performed using a double velvet technique [4]. Replica plates were incubated for 16 hours at 37°C. To generate growth curves, 3 ml broth tubes were inoculated with single colonies and grown on a shaker overnight at 37°C in air. Cells were diluted 1:1000 or 1:500 (β-gal strains) in LB broth. Cells were held on ice until all inoculations were completed. Triplicate cultures were then placed in a 37°C shaker with 250 rpm in air or 5% CO2. O.D.600 was measured every 60 minutes and dilutions of bacteria were plated onto MSB or LB agar plates to calculate the number of colony forming units (CFU) per ml.
Microscopic Observation
Strains 14028, 14028 zwf, YS873 and YS873 zwf were grown for 6 hours, as described above for growth curves, at 200 RPM. The cells were then fixed for microscopy using a solution of 30 mM sodium phosphate buffer (pH 7.5) and 2.5% formaldehyde. Cell morphology was observed with a Zeiss Axiovision microscope using differential interference contrast settings and DNA was detected via DAPI fluorescence. Fixed cells were incubated with 2 μg/ml DAPI for 10 minutes in the dark and aliquoted onto a 1% agarose pad.
Mutation Frequency Determination
A frozen stock of YS873 was streaked on MSB media and incubated overnight at 37°C to isolate individual clones. Triplicate 3 ml of LB broth were inoculated with independent YS873 colonies. They were grown at 37°C in a shaker over night. The tubes were then placed on ice and diluted in 0.9% saline. 10-6 and 10-4 dilutions were plated in duplicates onto LB agar and incubated in air and CO2 incubators respectively overnight at 37°C to calculate the number of CFU per ml.
Transduction and Transformation
Salmonella P22 transductions were performed by the method of Davis et al. [30], except that LB-0 plates supplemented with the appropriate antibiotic were used. EGTA was not added to the antibiotic plates for transductions. A BioRad Gene Pulser was used for electroporation with the following settings: 2.5 kV, 1000 ohms and 25 μFD for transformation of YS1 and 1.7 kV, 186 ohms and 25 μFD were used for YS873, YS1646, and ATCC 14028 [4].
Tn5 mutagenesis and mapping
A library of transposons in YS1646 was made using the EZ::TN <Kan-2> insertion kit from Epicentre (Madison, WI). Over 56,000 kanamycin resistant (KanR) clones of YS1646 were pooled. The pool was screened for mutation rate and auxotrophy for different biosynthetic pathways by replica plating onto minimal media and media containing various pools of amino acids and bases [30]. Following selection for CO2 resistance by plating dilutions to LB-Kan and incubating in 5% CO2, the colonies were again pooled and a P22 lysate was generated and transduced to a non-suppressed strain and purified for kanamycin resistance under non-CO2 conditions in order to separate spontaneous mutants from Tn5-based suppressors. Transposon-associated Tn5 insertions were identified by replica plating in air and CO2. Mapping of the insertion sites was performed by using the GenomeWalker™ kit (Clonetech, Mountain View, CA) according to the manufacture's instructions.
Construction of non-polar deletion in zwf
A non-polar deletion in zwf was generated by constructing a pCVD442 vector capable of deleting the entire zwf coding region by homologous recombination with the Salmonella chromosome [10]. Primers for PCR were designed that would generate one product immediately upstream of the 5' ATG start codon and a separate product immediately downstream of the 3' stop codon of the zwf coding region. The two separate products could then be ligated sequentially into the pCVD442 vector. The primers were: zwf-5'-reverse: 5'-GTGTGAGCTCGTGGCTTCGCGCGCCAGCGG CGTTCCAGC-3' (with added SacI), zwf-5'-forward: 5'-GTGTGCATGCGGGGGG CCATATAGGCCGGGGATTTAAATGTCATTCTCCTTAGTTAATCTCCTGG-3' (with added SphI), zwf-3' reverse: 5'-GTGTGCATGCGGGGTTAATTAA GGGGGCGGCCGCATTTGCCACTCACTCTTAGGTGG-3', and zwf-3'-forward: 5'-GTGTGTCGACCCTCGCGCAGCGGCGCATCCGGATGC-3'). The primers also generate internal NotI, PacI, SphI, SfiI, and SwaI in order to facilitate cloning of DNA fragments into the Δzwf for stable chromosomal integration without antibiotic resistance. This vector is referred to as pCVD442-Δzwf. The presence of the deletion, in AmpS SucR colonies, was detected by PCR using the following primers:zwf-FL-forward: 5'-ATATTACTCCTGGCGACTGC-3' and zwf-FL-reverse: 5'-CGACAATACGCTGTGTTACG-3'. Wild type produces a 2,026 base pair product whereas the mutant produces a 608 base pair (bp) product, a difference of 1418 bp, which corresponds to the size of the zwf gene (1475 bp minus a 57 bp multiple cloning site that replaces the open reading frame).
β-galactosidase Assay
For β-galactosidase expression, lacZ was cloned into the high copy vector pSP72 (Promega) in E. coli, transformed into Salmonella strains (via restriction defective Salmonella strain YS501 [31], and screened for bright blue colonies on LB agar containing 40 μg/ml X-gal. lacZ was cloned from E. coli K-12 MG1655 [32] obtained from the Yale E. coli Genetic Stock Center (New Haven, CT) by PCR using the primers BGF1 5'-GATCGGATCCATGACCATGATTACGGATTCACTGGC-3' and BGR1 5'-GATCAAGCTTTTATTTTTGACACCAGACCAACTGG-3'. The PCR product was cut with BamHI and HindIII and cloned into the plasmid pSP72 (Promega, Madison, WI) which had been cut with the same enzymes, transformed into DH5α, and selected for bright blue colonies on LB-amp plates containing 40 μg/ml X-gal. The plasmid was subsequently transformed to the restriction minus methylation plus strain YS501 before transforming other Salmonella strains. β-gal assays were performed according to the instructions for the Galacto-Star™ chemiluminescent reporter gene assay system (Applied Biosystems, Bedford, Massachusetts). Briefly, 1 ml of bacterial culture expressing β-gal from pSP72lacZ was pelleted at 13,000 × g for 5 min. Supernatants were filtered through a 0.2 μm syringe filter and then assayed immediately or frozen at -80°C until assayed with no further processing. Cell pellets were quickly freeze-thawed and suspended in 50 μl or 200 μl B-PER™ bacterial cell lysis reagent (Pierce Chemical) containing 10 mg/ml lysozyme (Sigma). Bacteria were allowed to lyse for 10–20 min. at room temperature and were then placed on ice. All reagents and samples were allowed to adjust to room temperature before use. Filtered supernatants and bacterial lysates were diluted as needed in Galacto-Star™ Lysis Solution or assayed directly. β-gal standard curves were made by preparing recombinant β-gal (Sigma, 600 units/mg) to 4.3 mg/ml stock concentration in 1× PBS. The stock was diluted in Lysis Solution to prepare a standard curve of 100 ng/ml- 0.05 ng/ml in doubling dilutions. 20 μl of standard or sample was added to each well of a 96-well tissue culture plate. 100 μl of Galacto-Star™ Subtrate, diluted 1:50 in Reaction Buffer Diluent, was added to each well and the plate rotated gently to mix. The plate was incubated for 90 minutes at 25°C in the dark and then read for 1 second/well in an L-max™ plate luminometer (Molecular Devices). Sample light units/ml were compared to the standard curve and values converted to units β-gal/ml. Percent release of β-gal was determined by dividing units/ml supernatant by total units/ml (units/ml supernatant + units/ml pellet). All samples were assayed in triplicate.
Authors' contributions
DB was responsible for the overall project concept and design. VK, SRM and DB designed and planned the experiments. VK, SRM, JP, KT, MI, MK, KBL and DB performed the experiments and analyzed the results. VK, SRM, KBL and DB wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Verena Karsten, Email: vkarsten@vionpharm.com.
Sean R Murray, Email: sean.murray@csun.edu.
Jeremy Pike, Email: jeremypike@sbcglobal.net.
Kimberly Troy, Email: ktroydesign@aol.com.
Martina Ittensohn, Email: mittens@vionpharm.com.
Manvel Kondradzhyan, Email: manvel.kondradzhyan@csun.edu.
K Brooks Low, Email: brooks.low@yale.edu.
David Bermudes, Email: dbermudes@gmail.com.
Acknowledgements
We wish to thank the reviewers for helpful suggestions, and Diana Downs and Eugenio I. Vivas (University of Wisconsin, Madison) for expeditiously providing gnd mutants. This work was supported by Vion Pharmaceuticals, New Haven, CT. SRM was supported by NIH Grant 1SC2 GM084860-01. DB thanks Caroline Clairmont for informing him of the plating results at the NCI.
References
- Nikaido H. In: Escherichia coli and Salmonella: Cellular and molecular biology. Neidhardt F, Curtiss R, Ingraham J, Lin ECC, Low KB, Magasanik B, Reznikoff M, Riley M, Schaechter M, Umbager HE, editor. Washington D.C.: ASM Press; 1996. Outer membrane; pp. 29–47. [Google Scholar]
- Grossman N, Schmetz MA, Foulds J, Klima EN, Jimenez-Lucho VE, Leive LL, Joiner KA. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol. 1987;169(2):856–863. doi: 10.1128/jb.169.2.856-863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz T, Zhou Z, Raetz CR. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J Biol Chem. 1997;272(16):10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- Murray SR, Bermudes D, de Felipe KS, Low KB. Extragenic suppressors of growth defects in msbB Salmonella. J Bacteriol. 2001;183(19):5554–5561. doi: 10.1128/JB.183.19.5554-5561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KB, Ittensohn M, Le T, Platt J, Sodi S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fischer J. Lipid A mutant Salmonella with suppressed virulence and TNFalpha induction retain tumor-targeting in vivo. Nat Biotechnol. 1999;17(1):37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20(1):142–152. doi: 10.1200/JCO.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullino PM, Grantham FH, Smith SH, Haggerty AC. Modifications of the acid-base status of the internal milieu of tumors. J Natl Cancer Inst. 1965;34(6):857–869. [PubMed] [Google Scholar]
- Helmlinger G, Sckell A, Dellian M, Forbes NS, Jain RK. Acid production in glycolysis-impaired tumors provides new insights into tumor metabolism. Clin Cancer Res. 2002;8(4):1284–1291. [PubMed] [Google Scholar]
- Murray SR, de Felipe KS, Obuchowski PL, Pike J, Bermudes D, Low KB. Hot spot for a large deletion in the 18- to 19-centisome region confers a multiple phenotype in Salmonella enterica serovar Typhimurium strain ATCC 14028. J Bacteriol. 2004;186(24):8516–8523. doi: 10.1128/JB.186.24.8516-8523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59(12):4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger GA. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch Microbiol. 1995;164(5):324–330. doi: 10.1007/BF02529978. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita T. Effect of mutations causing gluconate kinase or gluconate permease deficiency on expression of the Bacillus subtilis gnt operon. J Bacteriol. 1989;171(3):1751–1754. doi: 10.1128/jb.171.3.1751-1754.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab Eng. 2004;6(2):164–174. doi: 10.1016/j.ymben.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K. Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities. Appl Microbiol Biotechnol. 2004;64(1):91–98. doi: 10.1007/s00253-003-1458-5. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SR, Ernst RK, Bermudes D, Miller SI, Low KB. pmrA(Con) confers pmrHFIJKL-dependent EGTA and polymyxin resistance on msbB Salmonella by decorating lipid A with phosphoethanolamine. J Bacteriol. 2007;189(14):5161–5169. doi: 10.1128/JB.01969-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Everest P, Servos S, Foxwell N, Zahringer U, Brade H, Rietschel ET, Dougan G, Charles IG, Maskell DJ. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29(2):571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- Everest P, Ketley J, Hardy S, Douce G, Khan S, Shea J, Holden D, Maskell D, Dougan G. Evaluation of Salmonella typhimurium mutants in a model of experimental gastroenteritis. Infect Immun. 1999;67(6):2815–2821. doi: 10.1128/iai.67.6.2815-2821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PR, Benmore A, Khan SA, Jones PW, Maskell DJ, Wallis TS. Mutation of waaN reduces Salmonella enterica serovar Typhimurium-induced enteritis and net secretion of type III secretion system 1-dependent proteins. Infect Immun. 2000;68(6):3768–3771. doi: 10.1128/IAI.68.6.3768-3771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKelvie ND, Khan SA, Karavolos MH, Bulmer DM, Lee JJ, DeMarco R, Maskell DJ, Zavala F, Hormaeche CE, Khan CM. Genetic detoxification of an aroA Salmonella enterica serovar Typhimurium vaccine strain does not compromise protection against virulent Salmonella and enhances the immune responses towards a protective malarial antigen. FEMS Immunol Med Microbiol. 2008;52(2):237–246. doi: 10.1111/j.1574-695X.2007.00368.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87(16):6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf RE Jr, Prather DM, Shea FM. Growth-rate-dependent alteration of 6-phosphogluconate dehydrogenase and glucose 6-phosphate dehydrogenase levels in Escherichia coli K-12. J Bacteriol. 1979;139(3):1093–1096. doi: 10.1128/jb.139.3.1093-1096.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett WP, Wolf RE Jr. Genetic definition of the Escherichia coli zwf "soxbox," the DNA binding site for SoxS-mediated induction of glucose 6-phosphate dehydrogenase in response to superoxide. J Bacteriol. 1995;177(7):1742–1750. doi: 10.1128/jb.177.7.1742-1750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giro M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152(Pt 4):1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- Ma JF, Hager PW, Howell ML, Phibbs PV, Hassett DJ. Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat) J Bacteriol. 1998;180(7):1741–1749. doi: 10.1128/jb.180.7.1741-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello PJ, Demple B. Identification of SoxS-regulated genes in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182(1):23–29. doi: 10.1128/JB.182.1.23-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg BE, Wolf RE Jr, Dinauer MC, Xu Y, Fang FC. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect Immun. 1999;67(1):436–438. doi: 10.1128/iai.67.1.436-438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Vazquez-Torres A, Xu Y. The transcriptional regulator SoxS is required for resistance of Salmonella typhimurium to paraquat but not for virulence in mice. Infect Immun. 1997;65(12):5371–5375. doi: 10.1128/iai.65.12.5371-5375.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville JE Jr, Cassiano L, Darveau RP. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect Immun. 1999;67(12):6583–6590. doi: 10.1128/iai.67.12.6583-6590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor; 1980. [Google Scholar]
- Low KB, Ittensohn M, Luo X, Zheng LM, King I, Pawelek JM, Bermudes D. Construction of VNP20009: a novel, genetically stable antibiotic-sensitive strain of tumor-targeting Salmonella for parenteral administration in humans. Methods Mol Med. 2004;90:47–60. [PubMed] [Google Scholar]
- Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]