Abstract
Tumor hypoxia is one of the features of tumor microenvironment that contributes to chemoresistance in particular by cellular adaptations that modulate the apoptotic process. However, the mechanisms involved in this resistance still need deeper understanding. In this study, we investigated the involvement of four transcription factors, c-Myc, nuclear factor κB (NF-κB), p53, and c-jun/activator protein 1 (AP-1) in the hypoxia-induced resistance to etoposide in HepG2 cells. Whereas the profile of c-Myc and NF-κB activity did not fit the effect of hypoxia on caspase 3 activity, hypoxia decreased basal p53 abundance and DNA binding activity as well as p53 etoposide-induced activation. Short interfering RNA (siRNA) silencing evidenced that p53 was required for etoposide-induced apoptosis under normoxia. An inhibition of its activity under hypoxia could thus be responsible at least in part for the protection observed under hypoxic conditions. Moreover, p53 was found to induce the expression of Bak1. We showed that Bak1 was involved in the etoposide-induced apoptosis because Bak1 siRNA decreased it. Conversely, hypoxia increased c-jun DNA binding activity in the presence of etoposide. siRNA-mediated silencing of c-jun increased the responsiveness of cells to etoposide under hypoxia, as shown by an increase in caspase 3 activity and lactate dehydrogenase release. These effects occurred in a p53-independent manner. These data evidenced that hypoxia decreased the responsiveness of HepG2 cells to etoposide at least by two independent pathways involving p53 inhibition and c-jun activation.
Introduction
Tumor hypoxia is present in most solid tumors because of the abnormal vascular network and the high cancer cell proliferation rate. Two types of hypoxia are described: transient hypoxia and chronic hypoxia. Transient hypoxia is a consequence of abnormal vasculature network and high interstitial pressure that cause inadequate blood flow. In contrast, chronic hypoxia is caused by increased oxygen diffusion distance due to tumor expansion. Chronic hypoxia affects tumor area at a distance greater than 70 to 100 µm from the nearest capillary [1]. Besides its influence on the delivery and the uptake of chemotherapeutic drugs, hypoxia also affects the responsiveness of cancer cells to chemotherapy by inducing cellular adaptations through posttranslational modifications and gene expression regulation [2]. Hypoxia promotes angiogenesis, glucose transport, and anaerobic metabolism, invasion, resistance, and survival particularly by modulating the apoptotic process. The main transcription factor involved in the adaptation to hypoxia is the hypoxia-inducible factor 1 (HIF-1). HIF-1 plays a role in the hypoxia-induced metabolic switch, in the regulation of tumor pH, in triggering angiogenesis, and in the regulation of apoptotic process. Besides HIF-1, hypoxia can modulate the activity of other transcription factors, among them are nuclear factor κB (NF-κB), p53, c-myc, c-jun, ETS-1, Egr-1, or SP-1 [3,4]. Some of them have been described to be involved in the down-regulation of apoptosis. Indeed, NF-κB regulates the expression of antiapoptotic effectors such as Bcl-2, Bcl-xL, c-IAP2, or A1/Bfl1. In contrast, p53 is able to induce apoptosis in response to various stresses through its transcriptional activity and by direct interaction with Bcl-2 family members [5]. Some transcription factors can also be proapoptotic or antiapoptotic according to the stress or the cell line. It is the case of c-jun, a component of the transcription factor activator protein 1 (AP-1). c-jun can regulate pathways leading either to apoptosis or to cell survival and proliferation. For example, c-jun -/- fibroblasts have been shown to be more resistant to UV irradiation [6]. In contrast, a protective role of c-jun has been observed in T98G glioblastoma cells in response to DNA-damaging agents such as cisplatin or etoposide but not in response to thapsigargin or taxol [7].
Our previous studies have shown that hypoxia protects human hepatoma cell line HepG2 from DNA damage-induced apoptosis [8–10]. Etoposide, a topoisomerase II inhibitor, was used to induce double-strand breaks in DNA. In this study, we investigated the involvement of different hypoxia- or DNA damage-responsive transcription factors, c-myc, NF-κB, p53, and c-jun in the hypoxia-induced resistance to etoposide. We observed that hypoxia decreased the responsiveness of HepG2 to etoposide by regulating p53 abundance and c-jun DNA binding activity.
Materials and Methods
Cell Culture and Hypoxia Incubation
Human hepatoma cells HepG2 were maintained in culture in 75-cm2 polystyrene flasks (Costar, New York, NY) with 15 ml of Dulbecco's modified Eagle medium liquid and 10% of fetal calf serum and incubated under an atmosphere containing 5% CO2. For hypoxia experiments (1% O2), cells were incubated in serum-free CO2-independent medium (Invitrogen, Carlsbad, CA) supplemented with 1 mM l-glutamine (Sigma, St. Louis, MO) with or without etoposide (Sigma) at 50 µM. Normoxic control cells were incubated in the same conditions but in normal atmosphere (21% O2).
Lactate Dehydrogenase Release
Lactate dehydrogenase (LDH) release was measured with the cytotoxicity detection kit from Roche Molecular Biochemical (Indianapolis, IN) according to the manufacturer's protocol.
Caspase 3 Activity
The fluorogenic substrate Ac-DEVD-AFC was used to measure caspase 3 activity according to Lozano et al. [11]. Cell extracts were prepared as described by Wellington et al. [12]. Cells were seeded in six-well plates (250,000 cells per well). The assay for caspase 3 activity was performed according to Cosse et al. [8].
Western Blot Analysis
Cells, seeded in 25-cm2 flasks, were scrapped in 200 µl of lysis buffer (40 mM Tris pH 7.5, 150 mM KCl, 1 mM EDTA, 1% Triton X-100) containing a protease inhibitor mixture (« Complete » from Roche Molecular Biochemicals; 1 tablet in 2 ml of H2O, added at a 1: 25 dilution) and phosphatase inhibitors (25 mM NaVO3, 250 mM PNPP, 250 mM α-glycerophosphate, and 125 mM NaF, at a 1: 25 dilution). Western blot analysis was performed as described in Piret et al. [9] using mouse anti-poly (ADP-ribose) polymerase 1 (PARP-1) monoclonal antibody (no. 556493; PharMingen, San Diego, CA), rabbit anti-c-jun antibody (SC-1694; Santa Cruz, Santa Cruz, CA), mouse anti-PTEN antibody (SC-7974; Santa Cruz), caspase 3 (nos. 9662 and 9664; Cell Signaling, Beverly, MA), and anti-Bak1 antibody (no. 556382; PharMingen) were used at 1:5000 dilutions. Mouse antip53 (8–224; Upstate, Temecula, CA) was used at a 1:10,000 dilution. Membranes were reprobed with α-tubulin antibody (final dilution, 1:50,000; Sigma) or with β-actin antibody (final dilution, 1:50,000; Sigma) for normalization.
DNA Binding Assay
Nuclear protein extractions in high-salt buffer were prepared as previously described [8]. DNA binding assays using TransAM ELISA kit (Active Motif, Carlsbad, CA) for detecting transcription factor DNA binding activity was performed according to the manufacturer's recommendations.
Gene Expression Analysis on DNA Microarray
We used a low-density DNA array allowing the gene expression analysis for 123 genes related to apoptosis (DualChip Human Apoptosis; Eppendorf, Hamburg, Germany). HepG2 cells cultured in 75-cm2 flasks (Corning, New York, NY) were incubated for 16 hours with or without etoposide under normoxic and hypoxic conditions. At the end of the incubation, total RNA was extracted with the TRIzol Extraction Kit (Invitrogen), quality was checked with a bioanalyzer (Agilent Technologies, Waldbronn, Germany), and 25 µg was used for retrotranscription in the presence of biotin-11-dCTP (Perkin-Elmer, Boston, MA) and SuperScript II Reverse Transcriptase (Invitrogen), as described previously [13]. Hybridizations on the arrays were carried out as described by the manufacturer. Detection was performed with a cyanin 3-conjugated immunoglobulin antibiotin (Jackson ImmunoResearch Laboratories, West Grove, PA). Fluorescence of hybridized arrays was scanned using a Packard ScanArray (Perkin-Elmer) at a resolution of 10 µm.
Real-time Reverse Transcription-Polymerase Chain Reaction
After the incubation, total RNA was extracted using the TRIzol Extraction Kit (Invitrogen). Messenger RNA (mRNA) contained in 2 µg of total RNA was reverse-transcribed using SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Forward and reverse primers for Bak1, PUMA, CDKN1A, c-jun, p53, and RPL13A were designed using the Primer Express 1.5 software (Applied Biosystems, Cheschire, UK). Amplification reaction assays contained 1x SYBR Green PCR Mastermix (Applied Biosystems) and primers (Eurogentec, Seraing, Belgium) at the optimal concentrations. RPL13A was used as the reference gene for normalization, and mRNA expression level was quantified using the threshold cycle method.
Short Interfering RNA Transfection
The siGENOME smartpool of short interfering RNA (siRNA) anti-p53 (M-003329-01), anti-Bak1 (M-003305-01), and anti-c-jun (M-003268-02) were obtained from Dharmacon (Lafayette, CO). The siGENOME smartpool of siRNA was transfected with the DharmaFECT1 transfection reagent (T-2001; Dharmacon) according to the manufacturer's recommendations. Cells seeded at 40,000 cells/cm2 were incubated with transfection medium for 24 hours. After this incubation, transfection medium was replaced by fresh medium with 10% of fetal calf serum. DharmaFECT1 was used at a final dilution equal to 1:500, and siRNA were used at 50 nM. siCONTROL Nontargeting no. 1 (D-001210-01) and siCONTROL RISC free (D-001220-01) siRNA were used as negative controls.
Results
Effect of Hypoxia and/or Etoposide on the DNA Binding Activity of p53, c-jun, c-myc, and NF-κB
To determine the involvement of c-myc, NF-κB, p53, and c-jun in the protection of HepG2 cells by hypoxia, we compared the etoposide-induced caspase 3 activity with the DNA binding activity of these transcription factors for increasing periods from 2 to 16 hours of incubation with or without etoposide under normoxia or under hypoxia. What we called “normoxia” in standard culture conditions is not the physiological oxygen pressure that cells encounter within organs and tissues. Relevant physiological oxygen pressure ranges from 10% to 3% oxygen. In these conditions, both in vivo and in vitro, HIF-1 is not active, and there is no detectable HIF-1α protein. That is the case for most normal human tissues. However, in tumors, including the ones in liver, a true hypoxia can occur due to the oxygen diffusion limit as well as to the dysfunctioning tumor blood vessels. In this case, the oxygen pressure falls to values less than 1%, and HIF-1α is stabilized. In this work, we used in vitro conditions that mimic the “true” hypoxia that occurs in tumors. Indeed, HIF-1 has been shown to be activated in these conditions [10].
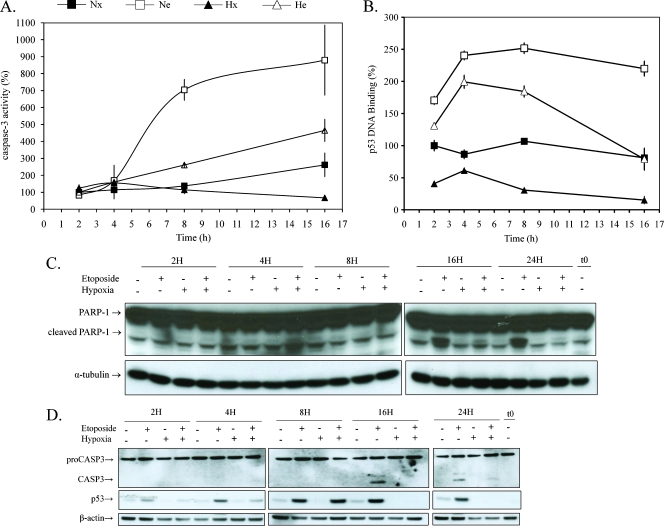
As a first step, the effect of hypoxia on the etoposide-induced apoptosis was studied by following the abundance of active caspase 3 and its activity. For an incubation time longer than 8 hours with etoposide, we observed an accumulation of the active caspase 3 and the cleavage of PARP, an endogenous substrate of caspase 3 by Western blot (Figure 1, C and D). Neither the active caspase 3 nor the cleaved form of PARP is observed during the first 8 hours of incubation with etoposide. However, the etoposide-induced activity of caspase 3 was detected from 8 hours of incubation using a more sensitive method based on the cleavage of a specific fluorogenic substrate Ac-DEVD-AFC (Figure 1A). Hypoxia per se had no effect on caspase 3 activity but inhibited the effect of etoposide on the caspase 3 activity as soon as its activity was detected.
Figure 1.
Effect of hypoxia and/or etoposide on caspase 3 activity and p53 regulation. HepG2 cells were incubated under normoxic (Nx; 21% O2) or hypoxic (Hx; 1% O2) conditions with (Ne-He; 50 µM) or without etoposide (Nx-Hx) for increasing periods from 2 to 16 hours or from 2 to 24 hours. (A) Caspase 3 activity was assayed by measuring fluorescence intensity associated to free AFC released from the cleavage of caspase 3 substrate Ac-DEVD-AFC. Data are expressed in percentages of caspase 3 activity in control cells (Nx) as means ± 1 SD (n = 3). (B) p53 DNA binding activity was assessed by colorimetric assay. Data are expressed in percentage of DNA binding in control cells (Nx) and correspond to the mean value ± 1 SD from three independent experiments. (C and D) PARP-1, caspase 3 (CASP3), and p53 were detected in total cell extracts by Western blot analysis, using specific antibody. α-Tubulin and β-actin were used to assess the total amount of proteins loaded on the gel. t0 = time zero.
Etoposide induced the stabilization and the DNA binding activity of p53 already after 2 hours of incubation (Figure 1, B and D). The DNA binding activity of p53 continued to rise up to 4 hours of incubation. For longer incubation periods, the DNA binding activity remained constant. Hypoxia led to a decrease in the etoposide-induced increase in p53 abundance and in its DNA binding activity, which was more important for longer incubation periods. In addition, hypoxia by itself decreased p53 abundance and its DNA binding activity (Figure 1C). Because p53 is a proapoptotic factor, the regulation of p53 by hypoxia could be one of the causes of cell resistance to etoposide-induced apoptosis in these conditions.
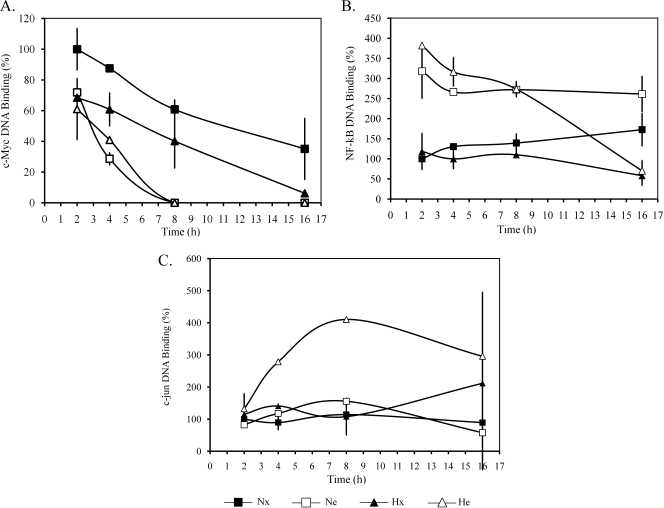
We observed that c-myc DNA binding activity decreased over time for all conditions, and this effect was amplified when cells were incubated in the presence of etoposide and/or under hypoxia (Figure 2A). The absence of serum during the incubation could possibly explain this decrease. The DNA binding activity of NF-κB (p65) was already increased by etoposide from 2 hours of incubation, regardless of the Po2 (Figure 2B). No differential effect according the Po2 was observed on the etoposide-induced DNA binding of NF-κB before the onset of the effect of hypoxia on the etoposide-induced caspase 3 activity. Therefore, we also ruled out a participation of NF-κB.
Figure 2.
Effect of hypoxia and/or etoposide on the DNA binding activity of c-myc, NF-κB, and c-jun. HepG2 cells were incubated under normoxic (Nx; 21% O2) or hypoxic (Hx; 1% O2) conditions with (Ne-He; 50 µM) or without etoposide (Nx-Hx) for increasing periods from 2 to 16 hours. The DNA binding activity of c-myc (A), NF-κB (B), or c-jun (C) was assessed by a colorimetric assay. Data are expressed in percentage of DNA binding in control cells (Nx) and correspond to the mean value ± 1 SD from three independent experiments.
Differences in c-jun DNA binding activity according to conditions were also observed. The DNA binding activity of c-jun was not modified by etoposide alone or by hypoxia alone. However, etoposide increased the DNA binding activity of c-jun under hypoxia already after 4 hours of incubation (Figure 2C). Therefore, the increase in c-jun DNA binding activity might be required for the hypoxia-induced resistance to etoposide. According to these data, we decided to investigate the involvement of p53 and c-jun in the hypoxia-induced resistance to etoposide.
Effect of p53 Silencing on the Etoposide-Induced Apoptosis
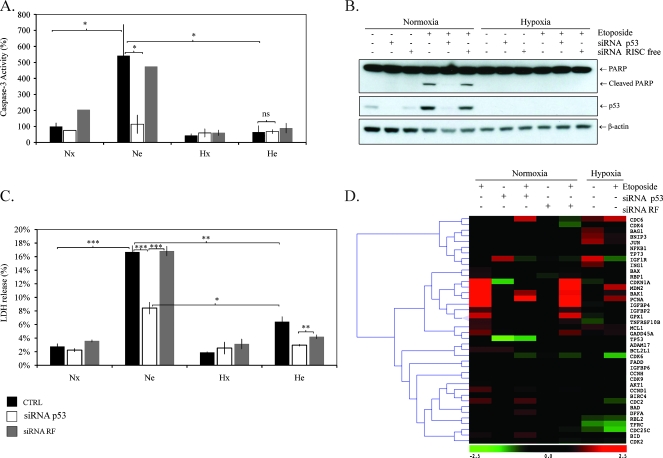
Hypoxia decreased p53 stabilization as well as p53 DNA binding activity. DNA damage-like double-strand break induced the stabilization and the activation of p53 through posttranslational modifications. Thereafter, p53 contributes to induce cell cycle arrest or apoptosis [14]. We observed that etoposide induced the stabilization and the activation of p53. Hypoxia could therefore protect HepG2 cells to etoposide-induced apoptosis by down-regulating p53. To evidence a putative role of p53 in the etoposide-induced apoptosis and in its modulation by hypoxia, we inhibited p53 expression using specific siRNA. Cells were transfected with siRNA targeting p53 or negative control siRNA during 24 hours. At the end of the transfection, transfection medium was replaced by fresh medium. Twenty-four hours after the end of the transfection, p53 mRNA and protein levels were assessed. The knock down of p53 by an siRNA smartpool at a final concentration of 50 nM reached more than 80% at both levels (Figure W1). The consequence of p53 silencing on apoptosis and cell death was then investigated by following the activity of caspase 3 and LDH release, respectively. HepG2 cells were incubated during 16 hours with or without etoposide under normoxia or under hypoxia. Caspase 3 activity was assayed using fluorogenic substrate Ac-DEVD-AFC and by following the cleavage of one of its main endogenous substrates, PARP-1 (Figure 3, A and B). Hypoxia inhibited the effect of etoposide on the activity of caspase 3 and had little effect per se in the absence of etoposide. Under normoxia, p53 siRNA inhibited the etoposide-induced caspase 3 activity as well as PARP-1 cleavage but had no effect on caspase 3 activity when cells were incubated without etoposide. Neither etoposide nor siRNA influenced caspase 3 activity under hypoxia, whereas hypoxia per se slightly decreased caspase 3 activity. Negative control siRNA had little or no effect on caspase 3 activity and PARP-1 cleavage whatever the conditions. To assay cell death, LDH release was measured after 40 hours of incubation. Etoposide strongly increased the LDH release under normoxia and, to a lesser extent, under hypoxia (Figure 3C). Under normoxia, p53 siRNA reduced by 50% the etoposide-induced LDH, whereas negative control siRNA had no effect. Hypoxia did not influence LDH release, and p53 siRNA had very limited effect on etoposide-induced LDH release under hypoxia compared with that observed under normoxia. All together, these results indicated that p53 was necessary for etoposide-induced apoptosis and cell death under normoxia. Moreover, the effect of p53 siRNA on etoposide-induced apoptosis under normoxia was very similar to the one of hypoxia.
Figure 3.
Effect of p53 silencing on etoposide-induced apoptosis. HepG2 cells were transfected with 50 nM p53 siRNA or negative control siRNA for 24 hours. Eight hours later, cells were incubated under normoxic (Nx) or hypoxic (Hx) conditions with (Ne-He) or without etoposide (Nx-Hx) for 16 hours (A, B, D) or 40 hours (C). (A) Caspase 3 activity was assayed by measuring fluorescence intensity associated to free AFC released from the cleavage of the caspase 3 substrate Ac-DEVD-AFC. Results are normalized by fluorescence intensity of control cells (Nx) and are expressed in percentages as means ± 1 SD (n = 3). (B) Total extracts were analyzed by Western blot analysis for PARP-1 and cleaved 85-kDa fragment using a specific anti-PARP-1 antibody. p53 protein level was assessed using specific anti-p53 antibody, and α-tubulin was used to assess the total amount of proteins loaded on the gel. (C) LDH release was assessed after 40 hours of incubation. Results are normalized by the LDH release of control cells and are presented in percentages as means ± 1 SD (n = 3). (D) Effect of p53 silencing on cell cycle and apoptosis-related genes expression. Biotinylated complementary DNA were generated by reverse transcription and hybridized on DualChip microarray as described in the Materials and Methods section. Data are logarithm base 2 of the mean of three ratio values calculated from three independent experiments. Data are presented on a tree obtained by applying a hierarchical clustering method (metric = Pearson correlation; method = complete linkage) to the expression data. Statistical analysis was carried out with the Student's t test. ns indicates nonsignificant; * .05 > P > .01; ** .01 > P > .001; ***P < .001.
Effect of p53 Silencing on Gene Expression
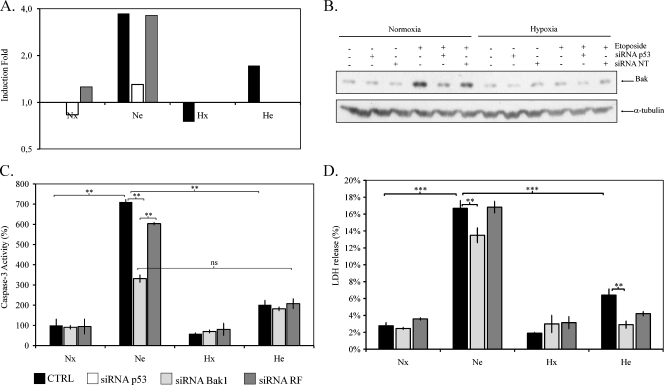
To better understand the role of p53 on the etoposide-induced apoptosis, we assessed the effect of p53 silencing on the mRNA level of genes encoding proteins involved in the regulation of apoptosis process using a low-density DNA microarray. The level of several transcripts was increased after incubation in the presence of etoposide. Etoposide-induced genes could be divided into two categories: the genes that are expressed in a p53-dependent manner and the others. Unlike AKT1, CCND1, or DFFA, the expression of some genes was dependent on the presence of p53 because they were not induced by etoposide in cells transfected with p53 siRNA. Among them were well-known p53 target genes such as CDKN1A [15], MDM2 [16], Bax [17], PCNA [18], IGFBP2 [19], GPX1 [20], TNFRSF10B [21], and GADD45A [22] (Figure 3D). Moreover, Bak1, encoding a proapoptotic protein, displayed an expression profile that was similar to the one of p53 target genes. These data suggested that Bak1 could be a p53 target gene. Conversely, IGF1R seemed to be negatively regulated by p53 because it was downregulated by etoposide and upregulated if p53 was absent because of p53 siRNA or under hypoxia. Overall, the negative control siRNA had little or no effect on the expression of genes studied regardless of the condition. Hypoxia reduced the expression of p53 target genes and Bak1 even if they were induced by etoposide. Among the etoposide-induced genes in a p53-independent manner, some of them were also downregulated by hypoxia such as CDC2, whereas others such as CCND1, DFFA, and Bid were not. Hypoxia per se affected the expression of some genes: CDC6, BAG1, Jun, and BNIP3 were upregulated, whereas RBL2 and TFRC were downregulated. The efficiency of p53 siRNA was confirmed by the sharp decline of p53 mRNA level after transfection with p53 siRNA, whereas negative control siRNA had no effect. Moreover, if etoposide induced a slight increase in p53 mRNA level, it was not affected by hypoxia. The fact that our data are in good agreement with previous studies reporting modifications in hypoxia-responsive genes (BNIP3, JUN [23]) during hypoxia as well as in p53 target genes (CDKN1A, Bax, Gadd45A, etc.) in the presence of etoposide already validates the DNA microarray used in this study. The data obtained for Bak (Figure 4A) as well as for other genes (data not shown) were confirmed by SYBR Green quantitative real-time polymerase chain reaction. To study whether the modulation of Bak mRNA was also observed at the protein level, the abundance of Bak was assessed by Western blot analysis. We observed that there was a very good correlation between mRNA and protein levels: etoposide increased Bak1 protein and mRNA levels under normoxia, whereas hypoxia and p53 siRNA inhibited the effect of etoposide (Figure 4B). In addition, hypoxia decreased Bak level. According to these results, we postulated that Bak is a protein by which p53 can induce apoptosis in response to etoposide.
Figure 4.
Effect of p53 silencing on Bak1 expression and effect of Bak1 silencing on the etoposide-induced apoptosis. HepG2 cells were transfected with 50 nM Bak1 siRNA, 50 nM p53 siRNA, or negative control siRNA for 24 hours. Eight hours later, cells were incubated under normoxic (Nx; 21% O2) or hypoxic (Hx; 1% O2) conditions with (Ne-He; 50 µM) or without etoposide (Nx-Hx) for 16 hours (A, B, C) or 40 hours (D). The effect of p53 silencing on Bak mRNA was confirmed by real-time reverse transcription-polymerase chain reaction (A), and the Bak1 protein level was assessed by Western blot using a specific antibody (B). α-Tubulin was used to assess the total amount of proteins loaded on the gel. One experiment of two independent ones is shown here (A, B). The effect of Bak1 silencing on the etoposide-induced apoptosis was assessed by measuring the caspase 3 activity (C) and the LDH release (D). (C) Caspase 3 activity was assayed by measuring fluorescence intensity associated to free AFC released from the cleavage of the caspase 3 substrate Ac-DEVD-AFC. Results are normalized by fluorescence intensity of control cells (Nx) and are expressed in percentages as means ± 1 SD (n = 3). (D) LDH release was assessed after 40 hours of incubation. Results are presented in percentages as means ± 1 SD (n = 3). Statistical analysis was carried out with the Student's t test. ns indicates nonsignificant; * .05 > P > .01; ** .01 > P > .001; ***P < .001.
Effect of Bak Silencing on the Etoposide-Induced Apoptosis
To study the involvement of Bak in the etoposide-induced apoptosis through a p53-dependant pathway under normoxia, Bak expression was silenced through RNA interference. Bak protein and mRNA levels were reduced to less than 20% when Bak siRNA was transfected (Figure W2). To evaluate caspase 3 activity, cells were incubated with or without etoposide under normoxia or hypoxia during 16 hours. Bak siRNA decreased by more than 50% the etoposide-induced caspase 3 activity under normoxia, whereas negative control siRNA had no effect (Figure 4C). Under hypoxia, the etoposide-induced caspase 3 activity was much lower than under normoxia and was not affected by Bak siRNA. The abundance of cleaved PARP-1 was investigated to confirm the results. Under normoxia, Bak siRNA reduced the etoposide-induced PARP-1 cleavage, whereas negative control siRNA had no effect (data not shown). To determine the effects of Bak siRNA on cell viability, LDH release was investigated after 40 hours of incubation. Bak siRNA decreased by 20% the etoposide-induced LDH release under normoxia, whereas negative control siRNA had no effect (Figure 4D). The etoposide-induced LDH release was strongly inhibited under hypoxia. Under hypoxia, Bak siRNA had very limited effects in comparison to negative control siRNA. On the basis of these data, we concluded that Bak may be involved in the etoposide-induced apoptosis under normoxia but did not seem to be the only one protein involved because the effect of Bak1 siRNA was only partial.
Effect of c-jun Silencing on the Etoposide-Induced Apoptosis
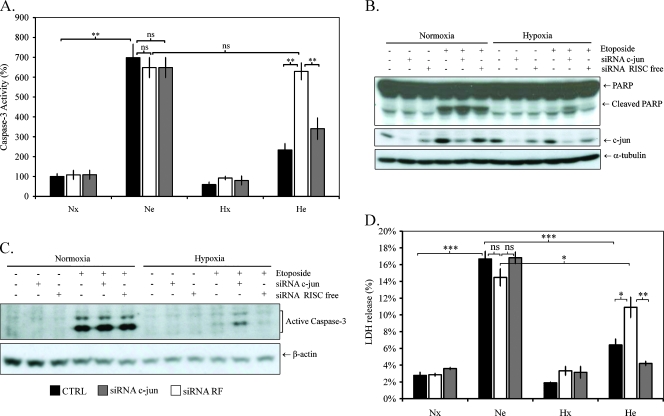
As shown in Figure 2C, incubation of cells under hypoxia in the presence of etoposide increased c-jun DNA binding activity. To determine if c-jun was implicated in the hypoxia-induced resistance to etoposide, we inhibited its expression using siRNA. We observed a lower silencing effect of c-jun siRNA than for other siRNA used (Figure W3): a decrease of 50% was observed at the mRNA level. However, this decrease was sufficient to obtain a significant decrease at the protein level. In addition, a larger decrease of the mRNA level was observed when c-jun expression was induced by hypoxia. To evaluate the effect of c-jun silencing on the etoposide-induced apoptosis and its modulation by hypoxia, cells were incubated with or without etoposide under normoxia or hypoxia during 16 hours. Under normoxia, c-jun siRNA did not influence the effect of etoposide on caspase 3 as observed by following the abundance of active caspase 3 studied by Western blot and on its activity measured by the cleavage of fluorogenic substrate Ac-DEVD-AFC and by following the cleavage of PARP-1 by Western blot (Figure 5, A–C). In addition, c-jun silencing had no consequence on etoposide-induced cell death as revealed by LDH release under normoxia when cells were incubated for 40 hours in the presence of etoposide (Figure 5D). Contrary to what was observed under normoxia, c-jun silencing partially relieved hypoxia-induced resistance to etoposide. Indeed, c-jun siRNA led to an increase in the abundance of active caspase 3. Moreover, the etoposide-induced cleavage of PARP and Ac-DEVD-AFC were higher in cells transfected with siRNA c-jun than in nontransfected cells in response to etoposide. These data were confirmed by LDH release assay after 40 hours of incubation: an increase in LDH release was observed in transfected cells (Figure 5D). Negative control siRNA had no effect neither on caspase 3 abundance and activity nor on LDH release. Together, these data indicate that c-jun was not involved in the etoposide-induced apoptosis but was required for the hypoxia-induced resistance to etoposide-induced apoptosis.
Figure 5.
Effect of c-jun silencing on hypoxia-induced resistance to etoposide-induced apoptosis. HepG2 cells were transfected with 50 nM c-jun siRNA or negative control siRNA for 24 hours. Eight hours later, cells were incubated under normoxic (Nx; 21% O2) or hypoxic (Hx; 1% O2) conditions with (Ne-He; 50 µM) or without etoposide (Nx-Hx) for 16 hours (A, B, C) or 40 hours (D). (A) Caspase 3 activity was assayed by measuring fluorescence intensity associated to free AFC released from the cleavage of the caspase 3 substrate Ac-DEVD-AFC. Results are normalized by fluorescence intensity of control cells (Nx) and are expressed in percentages as means ± 1 SD (n = 3). (B and C) Total extracts were analyzed by Western blot analysis for PARP-1 and active caspase 3 using specific anti-PARP-1 and active caspase 3 antibodies. c-jun protein level was assessed using specific anti-c-jun antibody, and α-tubulin was used to assess the total amount of proteins loaded on the gel. (D) LDH release was measured after 40 hours of incubation. Results are presented in percentages of LDH release in control cells (Nx) as means ± 1 SD (n = 3). Statistical analysis was carried out with the Student's t test. ns indicates nonsignificant; * .05 > P > .01; ** .01 > P > .001; ***P < .001.
In an attempt to understand the role of c-jun in the hypoxia-induced resistance to etoposide, we considered the involvement of several mechanisms. Hettinger et al. [24] highlighted that c-jun promoted cellular survival by negatively regulating the expression of PTEN, resulting in the concomitant activation of Akt survival pathway. To determine whether c-jun was involved in the regulation of PTEN in this model, PTEN protein level was assessed. PTEN protein level was not affected by etoposide neither under normoxia nor under hypoxia (Figure 6C). In contrast, hypoxia per se significantly reduced protein level. Neither c-jun siRNA nor negative control siRNA affected PTEN level, whatever the oxygen concentration.
Figure 6.
Effect of c-jun silencing on hypoxia-induced resistance to etoposide-induced apoptosis. HepG2 cells were transfected with 50 nM c-jun siRNA or negative control siRNA for 24 hours. Eight hours later, cells were incubated under normoxic (Nx; 21% O2) or hypoxic (Hx; 1% O2) conditions with (Ne-He; 50 µM) or without etoposide (Nx-Hx) for 16 hours. (A and B) At the end of the incubation, total RNA was extracted, submitted to reverse transcription, and then to amplification in the presence of SYBR Green and specific primers for CDKN1A and PUMA. RPL13A was used as the housekeeping gene for data normalization. One experiment of two independent ones is shown here (A, B). (C) At the same time, total extracts were prepared and analyzed by Western blot analysis for p53, PTEN, Bak1, and c-jun using specific antibodies. α-Tubulin was used to assess the total amount of proteins loaded on the gel.
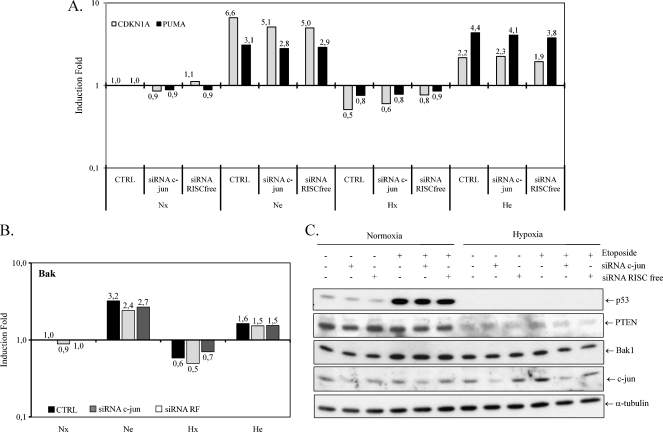
The preventive effect of c-jun over cell death by suppression of p53 has been highlighted by Eferl et al. [25]. To study the effect of c-jun on the regulation of p53, the effect of c-jun silencing on the p53 mRNA and protein levels was investigated. The etoposide-induced stabilization of p53 under normoxia and reduced abundance of p53 under hypoxia were not influenced by the inhibition of the expression of c-jun (Figure 6C). The inhibition of the expression of c-jun did not alter the mRNA level of p53 (data not shown). In addition, the p53 silencing did not influence the expression of p53 target genes, as shown for CDKN1A and PUMA (Figure 6A). We have shown previously that the expression of Bak was similar to the expression of p53 target genes. We observed that the expression of Bak was not influenced by inhibiting the expression of c-jun (Figure 6B). Therefore, the involvement of c-jun in the hypoxia-induced resistance to etoposide is independent of p53.
Discussion
Cell adaptation to hypoxia is largely mediated by gene expression regulation through the activation of HIF-1. Numerous reports showed that HIF-1 is closely linked to positive and negative regulation of the apoptotic process [26–29]. However, HIF-1-independent regulations of apoptosis by hypoxia have also been described [15,27,30,31]. Besides HIF-1, numerous transcription factors also respond to hypoxia; among them are NF-κB, p53, c-jun, c-myc, ETS-1, Egr-1, or SP-1 [3]. In this study, we investigated the involvement of four of them, namely, c-Myc, NF-κB, p53, and c-jun/AP-1, in the hypoxia-induced resistance to etoposide in HepG2 cells.
The myc protein is a transcription factor that regulates cellular processes including cell growth, proliferation, differentiation, and apoptosis. c-Myc is able to induce apoptosis by the activation of the ARF-MDM2-p53 axis [32]. In addition, c-Myc regulates the balance between proapoptotic and antiapoptotic members of Bcl-2 family by inducing the expression of Bax and the repression of Bcl-2 and Bcl-XL [33–35]. Under hypoxia, cooperation of c-myc with HIF-1 contributes to angiogenesis and to the metabolic switch [4]. However, the results of this work suggest that c-myc is not involved in the regulation of etoposide-induced apoptosis by hypoxia in HepG2 cells. If c-Myc DNA binding activity was modified by etoposide and serum deprivation, hypoxia had no effect.
Similarly, NF-κB, which favors cell survival by increasing the expression of antiapoptotic genes such as IAPs, was not a good candidate because the effect of hypoxia on NF-κB occurred much later than its inhibiting effect on caspase 3 activity. In contrast, p53 DNA binding activity and protein level were downregulated by hypoxia, and c-jun DNA binding was increased when cells were incubated in the presence of etoposide under hypoxia, both events occurring early during the incubation under hypoxia.
We observed that etoposide induced the stabilization and the DNA binding activity of p53. These data are coherent with the literature because it was widely described that p53 is rapidly stabilized by a wide array of cellular stresses such as oncogene-mediated hyperproliferation, translation perturbation, or DNA damage. In case of genotoxic damage, kinases belonging to PIKK family (ATM, ATR, DNA-PK) contribute to the stabilization and the activation of p53 [36]. p53 may induce DNA repair, cell cycle arrest, or apoptosis according to the nature of stress and the extent of the damage. The regulation of apoptosis by p53 occurs through the induction of genes encoding proapoptotic protein, such as Bax, Puma, Noxa, Fas, and FasL, and by a direct interaction with members of the Bcl-2 family. We observed that hypoxia decreased p53 abundance and p53 DNA binding activity and inhibited the effect of etoposide on its stability and its DNA binding activity. Numerous reports have evidenced positive or negative regulation of p53 by hypoxia according to the severity and the duration of hypoxia. Mild hypoxia does not influence or even decreases p53 abundance. In HepG2 cells, low abundance of p53 under mild hypoxia was explained by CK2-mediated targeting of p53 for proteasomal degradation [37]. Conversely, severe or prolonged hypoxia leads to the stabilization of p53 by an HIF-1-dependent pathway. In addition, differential effects of hypoxia on the regulation of p53 according to the cell lines have been described [8].
To test if the hypoxia-induced down-regulation of p53 contributes to the resistance to etoposide, the expression of p53 was inhibited using siRNA. We observed that p53 silencing desensitized HepG2 cells to etoposide-induced apoptosis under normoxia. Indeed, caspase 3 activity and LDH release were significantly reduced by the knock down of p53. These data indicated that p53 was required for etoposide-induced apoptosis and supported the hypothesis that postulates that hypoxia protects HepG2 cells against etoposide-induced apoptosis by down-regulating p53. To delineate the mechanisms responsible for the etoposide-induced apoptosis through p53, gene expression changes were also studied. Unsurprisingly, we observed that p53 silencing affected etoposide-induced known p53 target genes as CDKN1A, MDM2, Bax, PCNA, IGFBP2, GPX1, TNFRSF10B, or GADD45A. However, Bak1 expression profile was similar to p53 target genes because it was upregulated by etoposide and downregulated by p53 silencing and hypoxia. This result indicates that Bak1 is induced by a p53-dependent mechanism. Bak is proapoptotic protein of Bcl2 family that is permanently embedded into the outer mitochondrial membrane. In response to apoptotic stimulus, Bak is activated by conformational change, which allows its oligomerization in the outer mitochondrial membrane to induce the release of cytochrome c and other factors such as AIF and SMAC/DIABLO. Because of these data, the involvement of Bak in the etoposide-induced apoptosis was investigated. We observed that Bak silencing reduced etoposide-induced caspase 3 activity and cytotoxicity. However, the Bak silencing only had partial effect on the etoposide-induced apoptosis in contrast to what was observed with p53 siRNA. So, it seemed that Bak is involved in the etoposide-induced apoptosis but that other proteins are probably also involved.
In addition, we observed that hypoxia induced the DNA binding activity of c-jun when HepG2 cells were incubated in the presence of etoposide. As the involvement of c-jun in the positive or negative regulation of apoptosis was described, we investigated if the enhancing of c-jun DNA binding activity contributed to the hypoxia-induced resistance to etoposide. Our data indicate that c-jun was not involved in the etoposide-induced apoptosis under normoxia. However, the c-jun silencing relieved the hypoxia-induced resistance to etoposide as observed by the increase in the activity of caspase 3 and in the LDH release. These data indicate that c-jun is required for the hypoxia-induced resistance to etoposide. Hettinger et al. [24] have shown that c-jun can inhibit apoptosis through the repression of PTEN, allowing the activation of the PI3K/AKT survival pathway in human cancer cells. However, the abundance of PTEN was not affected by the silencing of c-jun whatever the conditions. It must be noted that we observed a decrease in the abundance of PTEN under hypoxia. This decrease was not affected by etoposide or by the transfection with siRNA. However, it could be envisaged that the decrease in the abundance of PTEN is favorable to the resistance of cancer cells to etoposide under hypoxia.
A negative regulation of p53 by c-jun has been highlighted by several teams. Schreiber et al. [38] have demonstrated that c-jun represses the expression of p53 by direct binding to a variant AP-1 site in the p53 promoter in 3T3 fibroblast. Conversely, an antagonizing effect of c-jun on the p53 activity has been shown on mice hepatocytes by Eferl et al. [25]. However, it was not the case in this model because the silencing of c-jun did not influence the mRNA (data not shown) and the protein levels of p53 (Figure 6C). In addition, hypoxia did not decrease the abundance of p53 mRNA unlike what is observed at the protein level (Figure 3, B and D). The inhibition of the expression of c-jun did not modify the transcriptional activity of p53 as observed on the mRNA level of CDKN1A and PUMA (Figure 6A). In the same line, the expression of Bak was not modified by the inhibition of the expression of c-jun (Figure 6B). These data indicate that the silencing of c-jun did not allow to restore, under hypoxia, the pathway by which etoposide induced apoptosis under normoxia. Therefore, the silencing of c-jun allowed the etoposide-induced apoptosis by a p53-independent pathway under hypoxia.
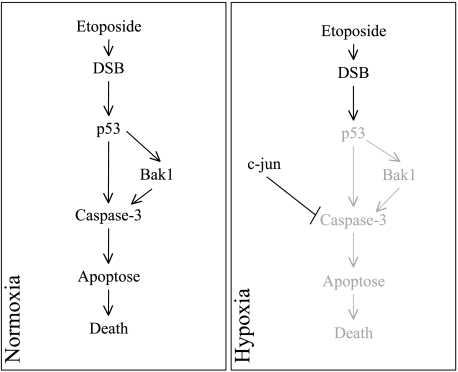
In conclusion, we showed that hypoxia protected HepG2 cells from etoposide-induced apoptosis by two independent pathways: the decrease in p53 abundance and the increase in c-jun DNA binding activity (Figure 7). The results of this study showed that hypoxia can protect cancer cells that express a functional p53 against chemotherapy by downregulating p53 at protein level. However, it seems that the regulation of p53 by hypoxia is not sufficient to induce the resistance of hypoxic tumoral cells against chemotherapy: a prosurvival pathway, for example, triggered by c-jun activity, also needs to be upregulated.
Figure 7.
Schematic representation of the hypoxia-induced mechanisms responsible for cancer cell resistance to etoposide. DSB indicates double-strand break.
Supplementary Material
Acknowledgments
C. Michiels is research director of Fonds National de la Recherche Scientifique). This article presents results of the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The responsibility is assumed by its authors.
Footnotes
This article refers to supplementary materials, which are designated by Figures W1 to W3 and are available online at www.neoplasia.com.
Competing Interests
The authors declare that they have no competing interests.
Authors' Contributions
J.P.C. and M.R. carried out all the experiments, N.N. carried the immunofluorescence studies, M.R. participated in the design of the study, and C.M. conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors have read and approved the final manuscript.
References
- 1.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 2.Cosse JP, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anticancer Agents Med Chem. 2008;8:790–797. doi: 10.2174/187152008785914798. [DOI] [PubMed] [Google Scholar]
- 3.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch. 2005;450:363–371. doi: 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- 4.Huang LE. Carrot and stick: HIF-α engages c-Myc in hypoxic adaptation. Cell Death Differ. 2008;15:672–677. doi: 10.1038/sj.cdd.4402302. [DOI] [PubMed] [Google Scholar]
- 5.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 6.Behrens A, Sibilia M, Wagner EF. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat Genet. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- 7.Potapova O, Basu S, Mercola D, Holbrook NJ. Protective role for c-Jun in the cellular response to DNA damage. J Biol Chem. 2001;276:28546–28553. doi: 10.1074/jbc.M102075200. [DOI] [PubMed] [Google Scholar]
- 8.Cosse JP, Sermeus A, Vannuvel K, Ninane N, Raes M, Michiels C. Differential effects of hypoxia on etoposide-induced apoptosis according to the cancer cell lines. Mol Cancer. 2007;6:61. doi: 10.1186/1476-4598-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piret JP, Cosse JP, Ninane N, Raes M, Michiels C. Hypoxia protects HepG2 cells against etoposide-induced apoptosis via a HIF-1-independent pathway. Exp Cell Res. 2006;312:2908–2920. doi: 10.1016/j.yexcr.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Sermeus A, Cosse JP, Crespin M, Mainfroid V, de Longueville F, Ninane N, Raes M, Remacle J, Michiels C. Hypoxia induces protection against etoposide-induced apoptosis: molecular profiling of changes in gene expression and transcription factor activity. Mol Cancer. 2008;7:27. doi: 10.1186/1476-4598-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozano J, Menendez S, Morales A, Ehleiter D, Liao WC, Wagman R, Haimovitz-Friedman A, Fuks Z, Kolesnick R. Cell autonomous apoptosis defects in acid sphingomyelinase knockout fibroblasts. J Biol Chem. 2001;276:442–448. doi: 10.1074/jbc.M006353200. [DOI] [PubMed] [Google Scholar]
- 12.Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 13.de Longueville F, Surry D, Meneses-Lorente G, Bertholet V, Talbot V, Evrard S, Chandelier N, Pike A, Worboys P, Rasson JP, et al. Gene expression profiling of drug metabolism and toxicology markers using a low-density DNA microarray. Biochem Pharmacol. 2002;64:137–149. doi: 10.1016/s0006-2952(02)01055-9. [DOI] [PubMed] [Google Scholar]
- 14.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 15.Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1α. J Exp Med. 2004;199:113–124. doi: 10.1084/jem.20030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 17.Thornborrow EC, Manfredi JJ. One mechanism for cell type-specific regulation of the bax promoter by the tumor suppressor p53 is dictated by the p53 response element. J Biol Chem. 1999;274:33747–33756. doi: 10.1074/jbc.274.47.33747. [DOI] [PubMed] [Google Scholar]
- 18.Jackson P, Ridgway P, Rayner J, Noble J, Braithwaite A. Transcriptional regulation of the PCNA promoter by p53. Biochem Biophys Res Commun. 1994;203:133–140. doi: 10.1006/bbrc.1994.2159. [DOI] [PubMed] [Google Scholar]
- 19.Grimberg A, Coleman CM, Shi Z, Burns TF, MacLachlan TK, Wang W, El-Deiry WS. Insulin-like growth factor binding protein-2 is a novel mediator of p53 inhibition of insulin-like growth factor signaling. Cancer Biol Ther. 2006;5:1408–1414. doi: 10.4161/cbt.5.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, Li S, Swaroop M, Guan K, Oberley LW, Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 21.Tomasetti M, Andera L, Alleva R, Borghi B, Neuzil J, Procopio A. α-Tocopheryl succinate induces DR4 and DR5 expression by a p53-dependent route: implication for sensitisation of resistant cancer cells to TRAIL apoptosis. FEBS Lett. 2006;580:1925–1931. doi: 10.1016/j.febslet.2006.02.054. [DOI] [PubMed] [Google Scholar]
- 22.Zhan Q, Chen IT, Antinore MJ, Fornace AJ., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol Cell Biol. 1998;18:2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster KA, Discher DJ, Bishopric NH. Induction and nuclear accumulation of fos and jun proto-oncogenes in hypoxic cardiac myocytes. J Biol Chem. 1993;268:16852–16858. [PubMed] [Google Scholar]
- 24.Hettinger K, Vikhanskaya F, Poh MK, Lee MK, de Belle I, Zhang JT, Reddy SA, Sabapathy K. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007;14:218–229. doi: 10.1038/sj.cdd.4401946. [DOI] [PubMed] [Google Scholar]
- 25.Eferl R, Ricci R, Kenner L, Zenz R, David JP, Rath M, Wagner EF. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 26.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erler JT, Cawthorne CJ, Williams KJ, Koritzinsky M, Wouters BG, Wilson C, Miller C, Demonacos C, Stratford IJ, Dive C. Hypoxia-mediated down-regulation of Bid and Bax in tumors occurs via hypoxia-inducible factor 1-dependent and -independent mechanisms and contributes to drug resistance. Mol Cell Biol. 2004;24:2875–2889. doi: 10.1128/MCB.24.7.2875-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fels DR, Koumenis C. HIF-1α and p53: the ODD couple? Trends Biochem Sci. 2005;30:426–429. doi: 10.1016/j.tibs.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Piret JP, Minet E, Cosse JP, Ninane N, Debacq C, Raes M, Michiels C. Hypoxia-inducible factor-1-dependent overexpression of myeloid cell factor-1 protects hypoxic cells against tert-butyl hydroperoxide-induced apoptosis. J Biol Chem. 2005;280:9336–9344. doi: 10.1074/jbc.M411858200. [DOI] [PubMed] [Google Scholar]
- 30.Dong Z, Wang JZ, Yu F, Venkatachalam MA. Apoptosis-resistance of hypoxic cells: multiple factors involved and a role for IAP-2. Am J Pathol. 2003;163:663–671. doi: 10.1016/S0002-9440(10)63693-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilic M, Kasperczyk H, Fulda S, Debatin KM. Role of hypoxia inducible factor-1 α in modulation of apoptosis resistance. Oncogene. 2007;26:2027–2038. doi: 10.1038/sj.onc.1210008. [DOI] [PubMed] [Google Scholar]
- 32.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eischen CM, Packham G, Nip J, Fee BE, Hiebert SW, Zambetti GP, Cleveland JL. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene. 2001;20:6983–6993. doi: 10.1038/sj.onc.1204892. [DOI] [PubMed] [Google Scholar]
- 34.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments γ irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol. 2003;23:7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell KO, Ricci MS, Miyashita T, Dicker DT, Jin Z, Reed JC, El-Deiry WS. Bax is a transcriptional target and mediator of c-myc-induced apoptosis. Cancer Res. 2000;60:6318–6325. [PubMed] [Google Scholar]
- 36.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 37.Hubert A, Paris S, Piret JP, Ninane N, Raes M, Michiels C. Casein kinase 2 inhibition decreases hypoxia-inducible factor-1 activity under hypoxia through elevated p53 protein level. J Cell Sci. 2006;119:3351–3362. doi: 10.1242/jcs.03069. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.