Abstract
Sanitization of the cellular nucleotide pools from mutagenic base analogs is necessary for the accuracy of transcription and replication of genetic material and plays a substantial role in cancer prevention. The undesirable mutagenic, recombinogenic and toxic incorporation of purine base analogs (i.e. ITP, dITP, XTP, dXTP or 6-hydroxyaminopurine (HAP) deoxynucleoside triphosphate) into nucleic acids is prevented by inosine triphosphate pyrophosphatase (ITPA). The ITPA gene is a highly conserved, moderately expressed gene. Defects in ITPA orthologs in model organisms cause severe sensitivity to HAP and chromosome fragmentation. A human polymorphic allele 94C->A encodes for the enzyme with a P32T amino acid change and leads to accumulation of non-hydrolyzed ITP. ITPase activity is not detected in erythrocytes of these patients. The P32T polymorphism has also been associated with adverse sensitivity to purine base analog drugs. We have found that the ITPA-P32T mutant is a dimer in solution, as is wild-type ITPA, and has normal ITPA activity in vitro, but the melting point of ITPA-P32T is 5 degrees C lower than that of wild-type. ITPA-P32T is also fully functional in vivo in model organisms as determined by a HAP mutagenesis assay and its complementation of a bacterial ITPA defect. The amount of ITPA protein detected by western blot is severely diminished in a human fibroblast cell line with the 94C->A change. We propose that the P32T mutation exerts its effect in certain human tissues by cumulative effects of destabilization of transcripts, protein stability and availability.
Introduction
Nucleotides of hypoxanthine and xanthine have multiple functions in all organisms. Inosine monophosphate (IMP) is synthesized in cells by enzymes of the de novo purine biosynthesis pathway, universal in all organisms and best studied in yeast and bacteria 1; 2. Hypoxanthine, xanthine, and their derivatives are also generated at different steps of the purine salvage pathway, utilizing exogenous and endogenous purines. IMP serves as a precursor for the synthesis of adenine and guanine nucleotides, required for energy metabolism and biosynthesis of DNA, RNA, cofactors, signal messengers, and hormones. The triphosphate forms of hypoxanthine (dITP) or xanthine (dXTP) are undesirable, because they could be incorporated into nucleic acids instead of canonical nucleotides and cause genetic damage 3; 4. One source of intracellular ITP is the activation of IMP by machinery used for other nucleoside monophosphates 2. Another source of hypoxanthine nucleotides in the cell is spontaneous or induced (e.g. by oxidative stress or inflammation) deamination of adenine nucleotides 5; 6; 7.
As structural analogues of dATP and dGTP, the non-canonical dITP and dXTP nucleotides are incorporated into DNA during DNA replication. Incorporation of dITP from the pools opposite template C is non-mutagenic, similar to the incorporation of dUTP 8; misincorporation of dXTP impedes the progression of replication 9. On the other hand, on the DNA level, the deamination of adenine to hypoxanthine or guanine to xanthine results in elevated mutagenesis 10; 11, because hypoxanthine pairs as G 12, and xanthine is a DNA replication blocking lesion. Irrespective of the route of appearance in DNA, both inosine and xanthine are recognized in most organisms by a specialized repair system initiated by the orthologs of endonuclease V 13 and elicit DNA repair reactions that lead to DNA fragmentation and genomic instability when the level of analogues is high 3; 14; 15.
The efficient system for the control of the concentration of hypoxanthine and xanthine from bacteria to mammalian cells is based on the action of specific triphosphatases. The prominent enzyme sanitizing the DNA precursor pool is inosine triphosphatase (ITPA), which prevents the accumulation of ITP, XTP, dITP and dXTP in the cell. ITPA catalyzes the hydrolysis of these triphosphates to corresponding nucleoside monophosphates and inorganic pyrophosphate. The genes encoding ITPase are conserved in evolution from bacteria and archaea to humans 3; 16. All investigated proteins of the ITPase family have similar biochemical properties (see 17 and references therein).
Interestingly, the inactivation of genes encoding ITPA leads to different phenotypes in different species. Phenotypic characteristics of ITPase-defective mutants were described long before they were linked to the gene encoding ITPase. In yeast S. cerevisiae, ITPase is encoded by the HAM1 gene. Mutations in the HAM1 gene were found in the screen for elevated sensitivity of yeast strains to the toxic and mutagenic effects of the base analogue 6-hydroxylaminopurine (HAP)18. The gene disruption mutation of the HAM1 gene is viable, does not lead to elevated spontaneous mutagenesis, and has no other phenotypes beside HAP-hypersensitivity 19. In E. coli, ITPase is encoded by the rdgB gene. The rdgB mutants were first identified by their lethal effect on the recA200 (Ts) background at the non-permissive temperature 20. Inactivation of the rdgB gene alone does not lead to increased sensitivity to HAP, because of an additional powerful HAP-protecting system dependent on the molybdenum cofactor in bacteria 21; 22. The effect of the rdgB inactivation on HAP sensitivity is clear in strains defective in the biosynthesis of the molybdenum cofactor 4. In these strains rdgB mutations lead to increased recombination, DNA fragmentation, and SOS induction 3; 4; 15. The link between the phenotypes in microorganisms described previously and ITPase deficiency was established when the crystal structure of the protein encoded by the gene Mj0226 from a thermostable bacterium M. jannaschii homologous to rdgB and HAM1 was determined 23. Mj0226 was found to be a dimer of identical subunits with some structural features in common with the MutT and MTH1 triphosphate pyrophosphohydrolases, suggesting that ITPA is a member of a new subclass of dNTPases. Biochemical studies confirmed that ITPase from E. coli, yeast, mouse and human hydrolyzes deoxy- and ribonucleosides of hypoxanthine, xanthine and HAP much more effectively than canonical nucleotides 24; 25; 26; 27; 28.
In humans, ITPase is encoded by the ITPA gene located on the short arm of chromosome 20 25. The ITPA gene is orthologous to microbial genes Mj0226, rdgB and HAM1. Several allelic variants of the ITPA gene have been described in different human populations: the coding sequence ITPA 94C->A causing a P32T change in the corresponding protein and changes in introns, ITPA IVS2 + 21A->C, and most recently found ITPA IVS2 + 68T->C and ITPA IVS2 + 68T->G alleles 29; 30; 31; 32; 33; 34; 35. All of these variants are associated with different ranges of decreased ITPase activity. It was shown that the concentration of ITP is dramatically increased to a readily detectable level in erythrocytes of ITPase-deficient patients. No pathological conditions have been found so far in those individuals. Several reports connect low or zero ITPase activity in red blood cells of 94C->A allele carriers and adverse drug reactions. For example, there is intolerance of therapeutic thiopurines (6-mercaptopurine and azathioprine) that are often used in treatment of inflammatory diseases, acute lymphoblastic leukemia, and for prevention of organ rejection after transplantation 36; 37; 38; 39; 40. This association was questioned 41 and prompted further population studies 42. More recently, a significantly higher probability of severe febrile neutropenia was observed in patients with a variant ITPA allele among patients whose dose of mercaptopurine had been adjusted for the thiopurine S-methyltransferase genotype 40. A new role of ITPase in regulation of levels of ATP for proper muscle contraction was proposed based on recent finding of heart failure in Itpa knockout mice 43. Because of the strong connection between ITPase deficiency and certain medical conditions it is important to understand the mechanisms of substrate specificity of ITPase and its variants in direct biochemical experiments.
One obvious question is the basis of ITPase activity loss in patients with a 94C->A allele variant of the ITPA gene. Recently, a crystal structure of human ITPA has been solved 17; 44. The P32 residue is located in the loop connecting helix α1 and β-strand 2 (Fig 1A), which is located away from the active site of the ITPase and is not conserved among species (13, 20). The change could affect the catalysis by an allosteric mechanism and disrupt the proper folding of the whole protein. Another hypothesis is that P32T substitution disrupts the proper assembly or cross-talk between the two subunits 17. This idea is consistent with the observation that homozygotes for the 94C->A mutation have no ITPAse activity in red blood cells, whereas in heterozygotes the ITPase is retained at about 22.5% of the wild-type activity 32. The result is predicted by random combinations of the two subunits of the enzyme, where only homodimers of the wild-type subunits are active. It is also possible that NTP hydrolysis in the two monomers is coupled across the dimer interface. Examination of the wild-type ITPase structure indicates that the loop with the P32T change is not part of the dimer interface (Fig. 1A), thus the mutation is unlikely to disrupt the interaction between subunits, but could exert a dominant-negative effect on the ITPase reaction. The third possible reason for the absence of ITPase activity in 94C->A patients is a defect in mRNA processing 34. None of the proposed mechanisms have been rigorously tested experimentally.
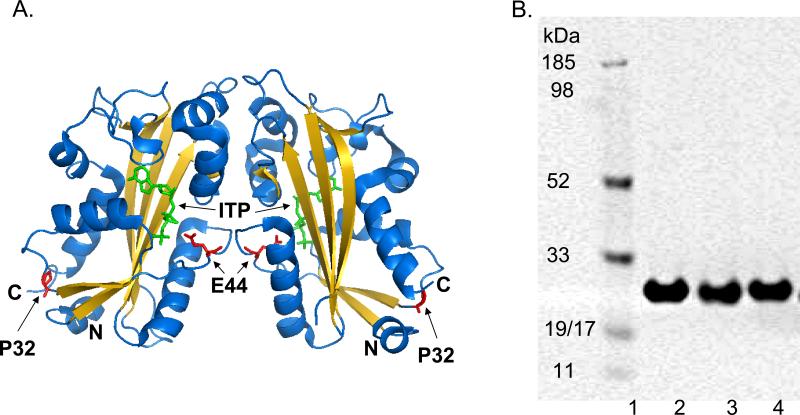
Fig. 1. Human ITPA and its variants.
A. The location of P32T and E44L mutations on the crystal structure of human ITPase. For the dimer structure, P32 and E44 residues are in red and marked with arrows, ITP is in green. The secondary structure of ITPase is shown with a ribbon diagram colored with β-strands yellow and all other elements blue. The location of N- and C-termini are labeled. The dimer interface is located in the center of the figure.
B. Purified proteins separated on SDS-PAGE gel and visualized with Coomassie stain. Lane 1 – MultiMark (Invitrogen) molecular weight marker, lane 2 – wild-type ITPA, lane 3 – ITPA-P32T, lane 4 – ITPA-E44L
We have found that purified human ITPA-P32T mutant protein is dimeric in solution and possesses pyrophosphatase activity comparable to the wild-type protein. The 94C->A allele compensates HAP-sensitivity of ham1 and rdgB mutations in S. cerevisiae and E. coli similar to the wild-type ITPA. We propose that the reason of complete ITPase deficiency in humans with a P32T change is catastrophic coincidence of several moderate changes of gene expression and protein properties.
Results
ITPase activity studies
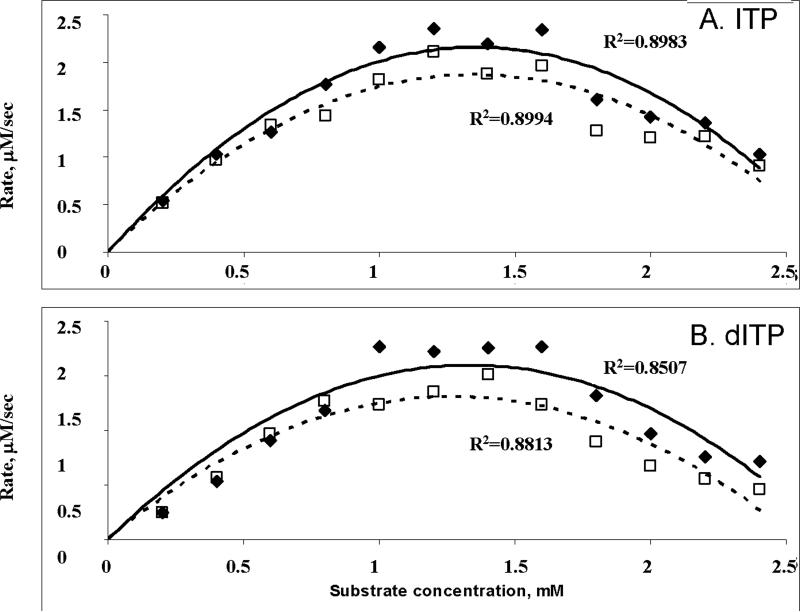
To measure ITPase activity of purified proteins (Fig. 1B) on their presumed natural substrates, dITP and ITP, we used the colorimetric assay described in the previous section. Because ITPase is known to be inhibited by high concentrations of substrates (from 0.25 to 3.5 mM in different studies) 25; 28, we first determined the concentration dependence of the ITPase reaction. Under our experimental conditions (see Materials and Methods) we observed substrate inhibition of both wild type and P32T mutant proteins at a concentration of ITP above 1 mM (Fig 2). For dITP, ITPA-P32T was more sensitive to the substrate inhibition (ANOVA, P<0.01, Fig.2B). There is no difference between mutant and wild-type enzymes for ITP (Fig. 2A). Protein with the amino acid change E44L, removing the carboxylate that from the crystal structure is expected to abolish Mg2+ binding, was inactive (data not shown). The lack of activity of ITPA-E44L, which is unable to perform a catalytic reaction, suggests that our affinity column purification protocol is specific to the His-tagged human enzyme and does not result in co-purification of bacterial ITPase. For kinetic constant measurements we used 12 concentration points. The resulting values are presented in Table 1. Surprisingly, the P32T protein had almost the same activity as normal ITPase.
Fig. 2. The dependence of ITPase activity on the concentrations of substrates, ITP and dITP.
Activity assay was performed as described in Materials and Methods. High R2 values close to 1 confirm the validity of approximation.
Filled diamonds – wild-type ITPA, open squares – ITPA-P32T
Table 1.
Kinetic parameters of human wild-type ITPA and ITPA-P32T.
| Protein | Substrate | Km (mM) | kcat (s−1) | kcat/ Km (mM−1 s−1) |
|---|---|---|---|---|
| Wild type | dITP | 0.35±0.05 | 71±10 | 200±38 |
| ITP | 0.40±0.07 | 91±23 | 228±72 | |
| P32T | dITP | 0.21±0.03* | 38±7* | 178±38 |
| ITP | 0.48±0.09 | 103±24 | 216±65 |
The values are averaged from at least three experiments with SE.
indicates the value that is statistically different from the other values in the same column as determined by Student's t-test (P<0.05).
Dimerization status of ITPA P32T in solution
We used size-exclusion chromatography – multi-angle light scattering (SEC-MALS) to examine the molecular weight of WT and P32T-ITPA in solution. Monomers of WT and P32T-ITPA have an approximate molecular weight of 21.73 kDa, thus giving an expected dimer weight of 43.46 kDa. WT and P32T-ITPA were eluted as a single peak with a molecular weight consistent with a dimer (Suppl. Fig. 1AB, upper panels). SDS-PAGE followed by silver staining confirmed that the peak contained a single species running at a molecular weight consistent with monomeric ITPA (Suppl. Fig. 1AB, lower panels). If the P32T mutation affected the ability of ITPA to dimerize it would be expected that, especially at low concentration, ITPA-P32T would exist, at least partially, as a monomer. Wild-type ITPA and ITPA-P32T were examined over a 50-fold concentration range and invariably eluted as a single species that had a molecular weight consistent with a dimer (Table 2). Our data with different concentrations of ITPA demonstrate that ITPA-P32T is efficient at dimerization.
Table 2.
Molecular weight in solution of wild-type ITPA and ITPA-P32T as determined by SEC-MALS.
| WT | P32T | ||
|---|---|---|---|
| Concentration | Molecular Weight | Concentration | Molecular Weight |
| 10.08 mg/mL | 45.92 ± 0.37 kDa | 8.74 mg/mL | 43.75 ± 0.09 kDa |
| 1.01 mg/mL | 46.47 ± 0.19 kDa | 0.87 mg/mL | 44.00 ± 0.22 kDa |
| 0.20 mg/mL | 42.33 ± 0.85 kDa | 0.17 mg/mL | 44.07 ± 0.88 kDa |
Decreased thermostability of ITPA-P32T
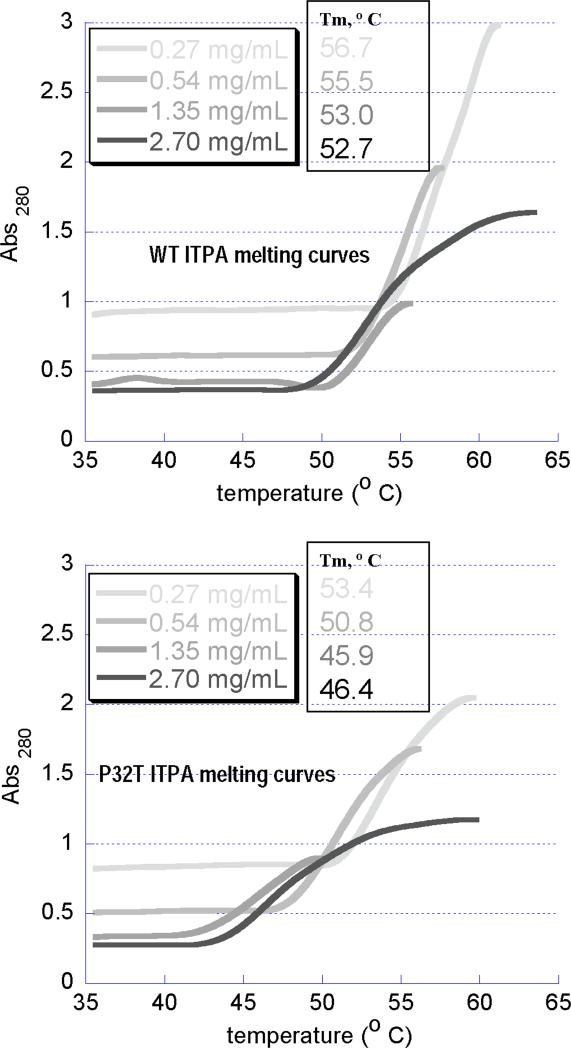
We have examined the temperature-induced denaturation profile of ITPA and its P32T variant and found that the P32T variant is less thermostable than the wild-type protein (Fig. 3). Complete denaturation of the wild-type ITPA is achieved at 53−57°C, depending on concentration (Fig 3A). P32T-ITPA consistently denatures at about 5°C lower temperatures over the concentration range tested (Fig 3B).
Fig. 3. UV thermal denaturation curves of purified wild-type ITPA and ITPA-P32T.
Solutions of wild-type ITPA (A) or ITPA-P32T (B), at the indicated concentrations were heated while monitoring the absorbance at 280 nm. Plots are truncated at the peaks, after which precipitation of the protein led to a decline in absorbance. Melting temperatures, which occur at the inflection point of the plots, were determined by identifying the temperature at the peaks in first derivative plots.
Expression of wild-type ITPA and ITPA-P32T in bacteria and yeast
In biochemical tests in vitro we have shown that the amino acid substitution P32T does not reduce ITPase activity. This result is not expected from human population studies, which show that individuals homozygous for the 94C->A mutation result in null ITPase activity in erythrocytes. To test the hypothesis that mutation leading to P32T amino acid change may reduce ITPase activity in vivo we studied the effect of heterologous expression of the ITPA gene in bacteria E. coli and yeast S. cerevisiae. From previous studies it is known that the ITPase encoded by the yeast HAM1 gene and human ITPA gene retains their enzymatic activity when expressed in E. coli 28; 45. Here we tested the ability of the wild type and mutant alleles of thehuman ITPA genes encoding for P32T and E44L amino acid changes to protect from HAP-induced killing and mutagenesis and compensate for the inactivation of microbial orthologs rdgB from E. coli and HAM1 from S. cerevisiae.
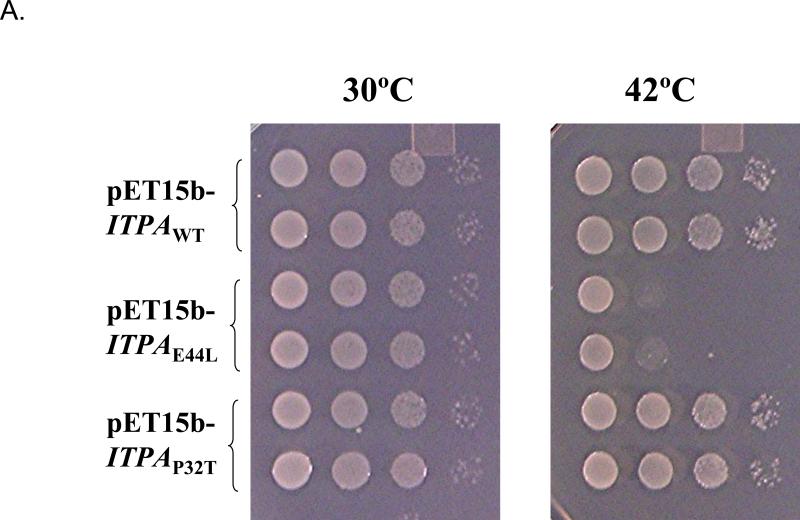
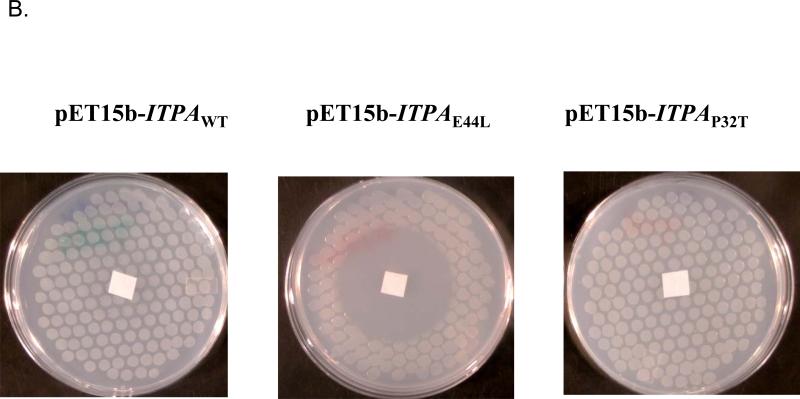
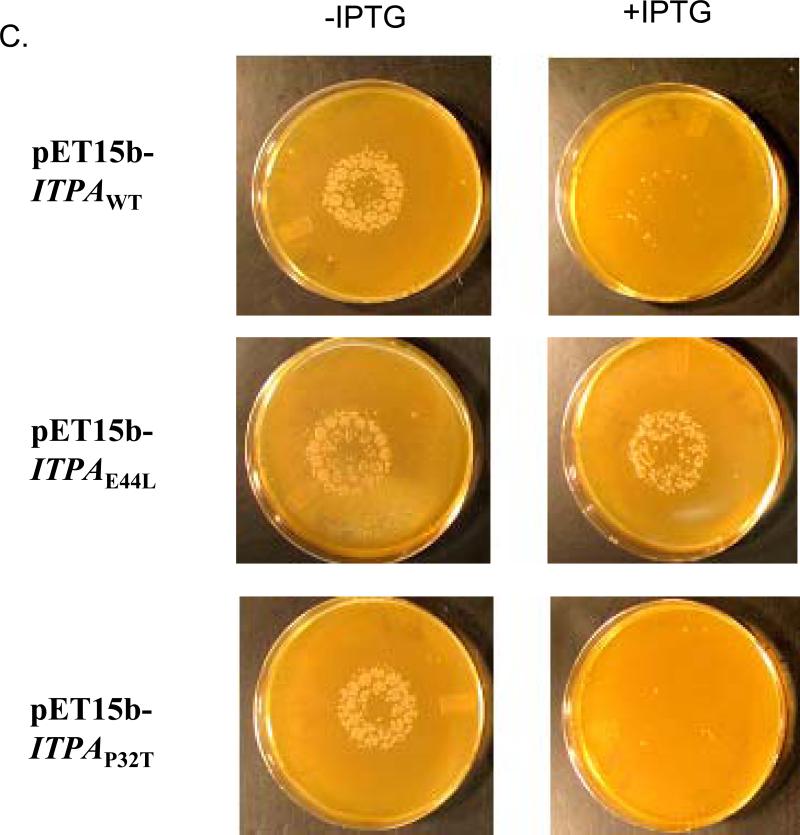
In bacteria, mutations of the rdgB gene are incompatible with the recA gene defect 20. We used the E. coli strain EK5 described before (Materials and Methods). This strain has two mutations, rdgB allele ΔyggV62 and recA200(Ts) and does not grow at the non-permissive temperature, 42 °C. We expected the expression of functional ITPase in this strain to complement the yggV and rescue the temperature sensitivity. We plated serial dilutions of EK5 transformants by pET15b vector with the wild-type ITPA gene or mutant alleles encoding for P32T and E44L changes onto minimum media with IPTG for induction of ITPA expression. Then we grew them at permissive and non-permissive temperatures (Fig. 4A). All transformants grew well at 30°C (left panel). The expression of the wild-type ITPA gene completely rescues the temperature sensitivity of the EK5 (two top clones on right panel). Mutant allele ITPA-E44L encoding for catalytically dead ITPA-E44L does not compensate for the lethal effect of a double mutation combination (two middle rows). Transformants with plasmids with the mutant allele encoding for P32T were indistinguishable from the wild type (two bottom rows). Our data show that human ITPase and its P32T variant, when produced in E. coli, are similarly functionally active in the ability to compensate the rdgB defect. The result is consistent with the data on the catalytic proficiency of the purified P32T protein. We confirmed this observation in two additional tests in bacteria with the use of HAP. In one test we have studied the ability of the human ITPA gene to suppress HAP-induced killing of E. coli Rosetta strain. The Rosetta strain is HAP hypersensitive likely due to the presence of the gal deletion encompassing some part of the mod operon, which disturbs molybdenum uptake and leads to a molybdenum cofactor deficiency 22; 46. As is seen in Fig. 4B, wild type ITPA and ITPA-P32T mutants of the ITPA gene protect the Rosetta strain from HAP. The plasmid with allele encoding for the E44L change within the active site of ITPA, which results in the abrogation of catalytic activity (the results of biochemical study of ITPA), does not affect the hypersensitivity of Rosetta to HAP. In the other test, we studied the effects of ITPA expression on HAP-induced mutagenesis in the wild-type bacterial strain EK1. In this test, HAP was spotted onto the center of the plate with a lawn of bacteria (as in Fig. 4B) and after 24 hours of incubation plates were replica-plated onto LB plates with antibiotic rifampicin. HAP is a potent inducer of forward mutations to rifampicin resistance as judged by the appearance of numerous resistant colonies surrounding the spot of application of the mutagen. The expression of wild type ITPA suppressed the mutagenic effect almost completely (Fig 4C, upper row, compare plate without and with IPTG). The expression of the ITPA-E44L did not affect HAP-induced mutagenesis at all (middle row), while the expression of the ITPA-P32T led to a similar level of protection to the expression of wild-type ITPA (bottom row). Taken together, the data presented in Fig. 4 are consistent with the full functionality of ITPA P32T, both in regard to natural substrates and in regard to HAP toxicity and mutagenesis.
Fig 4. ITPA P32T is functional in bacterium E. coli.
A. Sensitivity to high temperature of double mutant ΔrdgB recA200(Ts) strain (EK5) transformed with vector expressing the human wild-type ITPA gene and its mutant variants ITPA-P32T and ITPA-E44L . Five μl of the serial dilutions of the overnight cultures grown at 30°C were spotted on a minimal medium plates containing 50 μM IPTG. The plates were incubated overnight at 30°C or at 42°C.
B. The ability of ITPA-P32T to compensate for the HAP-sensitivity of the Rosetta strain. Cells were spotted on a minimal medium plate containing 50 μM IPTG using a multi-prong replicator device, and 50 μg of HAP was spotted onto the filter paper at the center of each plate. The plates were incubated overnight at 37°C.
C. Protection of the wild type strain EK1 from HAP-induced mutagenesis by human ITPA and its variants. Spot-tests were performed as in Fig. 4B on a minimal medium with or without ITPG. The cells were then replica-plated onto LB plates containing rifampicin and incubated overnight at 37°C.
Expression of ITPA wt and ITPA-P32T mutant alleles in yeast
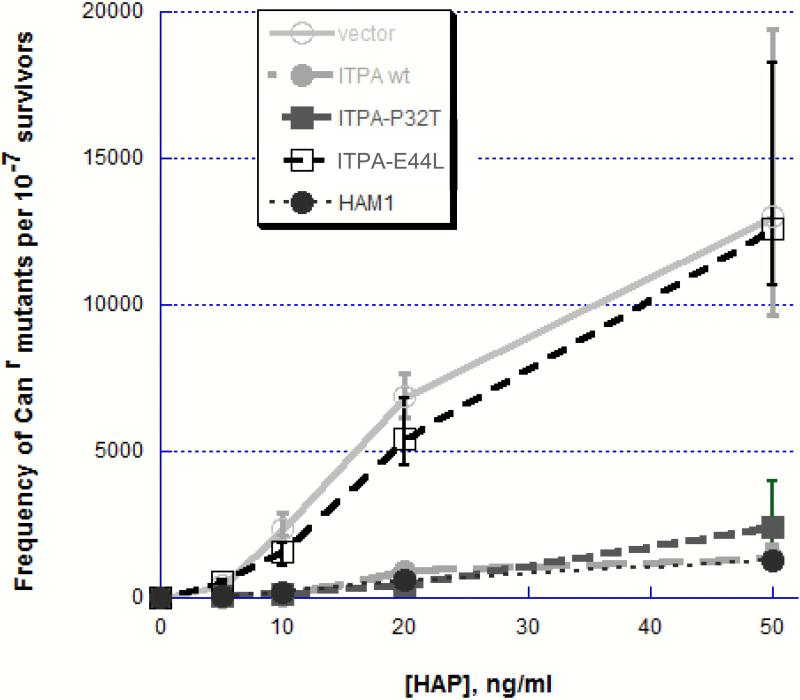
We expressed human alleles encoding for wild-type ITPA and ITPA with amino acid substitutions P32T and E44L under a galactose promoter in pESC-URA vector in yeast and have shown that the ITPA wild type and the ITPA-P32T complement HAP-induced mutability of a yeast ham1 mutant (Fig 5). A yeast strain with the ham1 mutation and with empty vector or with the vector with the ITPA-E44L allele encoding for inactive ITPase is hypersensitive to the mutagenic effect of HAP. Plasmids with the yeast HAM1 gene, human ITPA or ITPA-P32T all protected this strain from HAP-induced mutagenesis to the same level – the dose response curves are identical.
Fig. 5. ITPA-P32T is functional in yeast Saccharomyces cerevisiae.
Suppression of HAP-induced mutagenesis in ham1 mutant transformed with plasmid expressing the yeast HAM1 gene, human ITPA gene and its mutant alleles ITPA-P32T and ITPA-E44L. Transformants with empty vector pESC-URA were used as a control. For frequency determination we have used six independent cultures. The experiment was repeated three times and the data appeared to be homogenous. Medians of mutant frequencies for each data point were then found. Error bars represent 95% confidence limits.
Low levels of ITPA-P32T in human fibroblasts
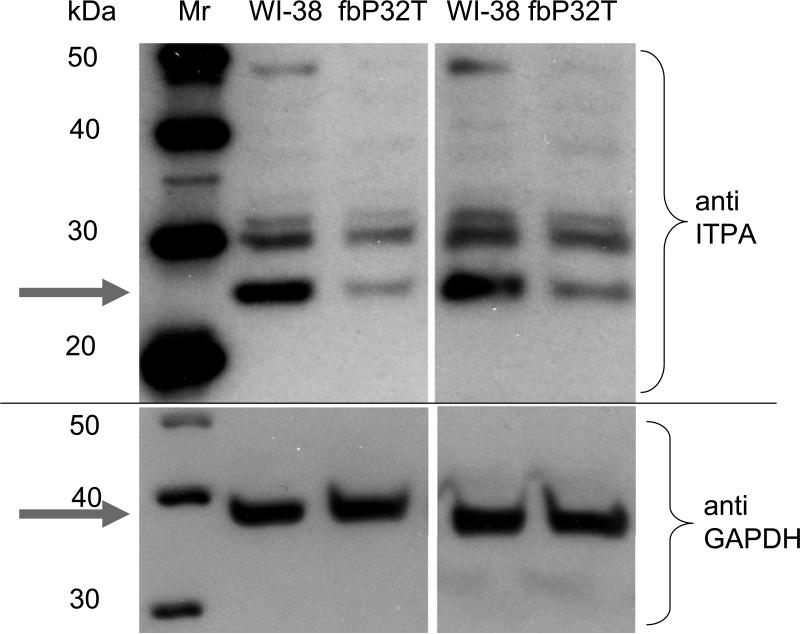
We compared the levels of soluble immunoreactive ITPase in normal WI-38 and in P32T fibroblasts (Fig. 6, two repeats are presented). The level of ITPA is almost 10-fold lower in P32T fibroblasts (gray arrow, upper part), while the levels of GAPDH were essentially the same (gray arrow, bottom part).
Fig. 6. Diminished levels of P32T ITPA in human fibroblasts.
Upper panel represents the results of two independent western blots with extracts of human fibroblasts with anti-ITPA antibodies. Bottom panel serves as a loading control and illustrates that the level of unrelated enzyme, GAPDH, is similar in the two cell lines.
Discussion
In the present study we evaluated a recently recognized system of protection from base analogs by ITPase. The ITPA gene is important to study from both medical and basic science points. Cleansing of potentially mutagenic nucleotide analogs from the precursor dNTP pool is an important prerequisite for high fidelity DNA replication 47; 48; 49. Inaccurate DNA replication and repair lead to cancer and other diseases 50; 51; 52. Environmental exposures and biochemical reactions during oxidative stress and inflammation damage natural nucleoside triphosphates 53; 54; 55; 56; 57. Such mutagenic contaminants in dNTP pools lead to elevated mutation rates and a risk of cancer and other diseases 58. ITPA is involved in the regulation of the quality of the pool of purine nucleotides 4. Disturbance of this enzyme leads to cellular accumulation of non-canonical nucleoside triphosphates such as ITP, XTP and others, which instigate elevated chromosome fragmentation and mutagenesis. ITPA has pharmacogenetic significance because of adverse drug reactions in patients with decreased ITPA activity, which is associated with a point mutation leading to a P32T amino acid change 40. Here we examined the possible mechanisms of these effects.
We have unexpectedly found that ITPA-P32T is very similar (but not identical) to wild-type IPTA. This conclusion is based on the kinetic characteristics of the protein, its melting profile, and the ability of corresponding alleles to compensate for the ITPase defects in model organisms.
There are just a few papers describing the kinetic characteristics of human ITPA. There is some variability of estimates. Km values vary ∼10-fold, for example, for dITP it is 32.5 μM in 28 and is 310 μM in 25. The Km values we observed in our study for both ITP and dITP are very close to those of Lin et al. 25. This might relate to slightly different reaction conditions such as buffer used and pH. As for the kinetic parameters of the mutant enzyme, we observed a significant difference between P32T and the wild type only for dITP (Table 1). The enzyme kinetics did not fit the Michaelis-Menten equation because of substrate inhibition. Therefore, we have used for the estimation of kinetic parameters a more adequate special model built for such cases (Materials and Methods). The use of this model allowed us to better describe the differences in substrate inhibition between wild type and mutant ITPA. It is interesting that though the kcat value for wild-type ITPA is almost as twice as high than for P32T, there is no significant difference between the corresponding kcat/Km values. This may reflect the stronger sensitivity of the mutant enzyme to the substrate inhibition (Fig. 2). Unlike the other kinetic parameters, kcat/Km connects the reaction rate with the concentration of free enzyme, thus characterizing the V-function for low concentrations of substrate. Then, the inhibition is still almost negligible. So we can conclude that ITPA-P32T in vitro can effectively catalyze the reaction of pyrophosphohydrolysis, though it is slightly (our estimate is 1.3 times) more sensitive to substrate inhibition by dITP. The ITPA P32T protein is a dimer in solution, like wild type ITPA. These observations do not support the hypothesis that the basis of ITPase deficiency in ITPA-P32T homozygous patients is in the defect of cross-talk between ITPase subunits and the consequent loss of the enzymatic activity 17. We propose a new, “synthetic” model explaining the ITPA-P32T allele phenotypes. Opposite of the idea of “yes or no” effects of ITPA-P32T, we propose that the null phenotype is a result of a catastrophic coincidence of several relatively mild (when alone) changes. The defect of mRNA splicing 34 decreases the amount of normal mRNA available to generate active protein by approximately 50%. In individuals with the mutation, a smaller amount of protein is synthesized in all tissues. Next, the P32T protein is less stable as suggested by the UV thermal denaturation studies and therefore might be a more preferable target for proteolytic degradation than the wild-type protein. The efficiency of this degradation could be tissue-specific. Indeed, we have found that the level of ITPA in fibroblasts of individual with the 94C->A allele is severely diminished. The enzyme/substrate ratio in blood of such patients is elevated and ITPase is inhibited by substrate. In addition, the P32T protein may be intrinsically more susceptible to substrate inhibition. The net result is a thermally unstable, substrate-inhibited ITPase that is present at such low levels that it is ineffective in protecting patients from purine base analogs. It is also likely that the levels of this unstable but catalytically active P32T ITPA are varying in different tissues from zero (in erythrocytes) to almost normal (presumably in heart). This will explain the discrepancy between the relatively mild effects of the ITPA-P32T mutation in humans in comparison to drastic effect on heart development in the Itpa knockout in mice 43.
Materials and Methods
E. coli strains and growth conditions
As a starting material we have used E. coli strains AB1157 (used as a wild-type control) and JB30 (ΔyggV62 recA200(Ts) srlC300::Tn10) kindly provided by Dr. A. Kuzminov 3. The yggV is another name of rdgB. We modified the strains by introducing T7 RNA polymerase gene with the Novagen λDE3 Lysogenization Kit. The kit allowed us site-specific integration of λDE3 prophage into an E. coli host chromosome. The lysogenized host could be used for controlled expression of target genes cloned in T7 expression pET vectors. The derivative of AB1157 was named EK1 and the derivative of JB30 was named EK5. We have used LB medium supplemented with the appropriate antibiotics to propagate bacteria and Vogel-Bonner based minimal medium supplemented with the appropriate nutrients 21 and 0.05 mM IPTG when T7 promoter induction was required 44.
Plasmid Constructs
For the human ITPA overexpression in E.coli and for purification of the wild-type ITPA, we have used the vector pET15b-ITPA described previously 44. This plasmid was used for engineering of nucleotide changes encoded for the P32T and E44L changes in ITPA by site-directed mutagenesis (QuikChange kit from Stratagene). To generate the mutations we have used the following primers (codon sequence encoding the change underlined):
- for ITPA-P32T :
- P32T-F 5’ GGAGATAAGTTTACCTGCACTTTGGTGGC
- P32T-R 5’GCCACCAAAGTGCAGGTAAACTTATCTCC
- for ITPA-E44L:
- E44L-F 5’CAGAAAATTGACCTGCCGTTGTACCAGGGGGAGCCG
- E44L-R 5’CGGCTCCCCCTGGTACAACGGCAGGTCAATTTTCTG
All results of site-directed mutagenesis were verified by DNA sequencing using the primers ITPA_sN 5’TCATTGGTGGGGAAGAAGATC and ITPA-sC 5’AAGCTGCCAAACTGCCAAA. To study the effects of ITPA expression in yeast we cloned the ITPA gene and its mutant alleles amplified from corresponding pET15b vectors into XhoI-HindIII sites of the yeast expression vector pESC-URA (Stratagene). This generated N-terminal fusion of the ITPA with the sequence encoding for the c-myc epitope and put the expression of the resulting gene under the GAL1 promoter. The yeast HAM1 gene has been cloned to the pESC-URA vector into BglII-SacI restriction sites, generating N-terminal fusion of the HAM1 with the sequence encoding for the FLAG epitope under the GAL10 promoter.
ITPA protein purification
The genes encoding for recombinant 6-His-tagged human wild type ITPA and its mutant forms (with substitutions P32T and E44L) were expressed in the E. coli Rosetta2(DE3) strain (Novagen) and purified by affinity chromatography as described previously 44. To check the purity of extracted proteins we loaded samples of the peak fraction from a nickel column on SDS-PAGE (Fig. 1B). Purity of all proteins was approximately 95%. The average yield of three protein variants was the same, about 5 mg of protein from 1 g of bacteria. The protein from peak fractions was dialyzed against BICINE-NaCl buffer and stored at −20°C in a similar buffer with glycerol and DTT (50 % glycerol, 20 mM BICINE pH 8.5, 5 mM DTT, 50 mM NaCl, and 1 mM EDTA). These samples were used for studies of enzymatic activity. For biophysical characterization of ITPA, the His tag was cleaved and the enzyme was further purified by ion exchange chromatography as described 44.
Protein activity assay
To measure enzymatic constants for purified proteins we determined the amount of inorganic pyrophosphate released at different concentrations of natural substrates (ITP or dITP). First, we performed the enzymatic reaction in the reaction mix (10 μL) containing 50 mM Tris-HCl, pH 9, 1 mM DTT, 10 mM MgCl2, varying concentrations of substrate (0.05 to 5 mM), and 15 nM ITPA. We added the enzyme to a pre-warmed mixture and incubated it for five more minutes at 37°C. Reactions were stopped by adding 1 μL of 10 mM EDTA. To measure the concentration of the released pyrophosphate we used 4 μL of reaction mixture and PiPer™ Pyrophosphate Assay Kit (Invitrogen). ITPA is sensitive to strong substrate inhibition. Therefore, we used special equations 59 to determine kinetic parameters:
V=Vmax*S/(Ks + S + S2/K's), where Vmax is a maximum velocity of the enzymatic reaction, S is a substrate concentration, Ks is a constant for the formation of active enzyme complex and K's is a constant of the formation of an inactive complex.
Km=Vmax/Vo, where Km is the Michaelis constant and Vo is an initial velocity of the enzymatic reaction.
First, we solved equation (1) to determine the Vmax value. To solve this equation for three unknown parameters we compiled a special program in the MatLab system (MatLab 6.5, The MathWorks Inc.). The program simultaneously uses the four groups of values with three values of substrate concentrations and reaction velocity in each group. Then we used the Vmax value to determine Km via equation (2). The Vo value was calculated as a derivative of the V-function for S=0.
Size-exclusion chromatography – Multi-angle light scattering (SEC-MALS)
Dynamic light scattering (DLS) helps the investigator understand the size distribution, stability, and aggregation state of macromolecules in solution 60. Here we used a more sophisticated procedure to examine the molecular weight of ITPA molecules, which involves size-exclusion chromatography and analysis of resulting fractions by multi-angle static light scattering 61 to determine absolute molecular weight of the proteins in solution. SEC-MALS experiments were performed by loading 100 μL of protein at the indicated concentrations onto a Superose 6 10/300 GL column (Amersham Biosciences) with a bed volume of approximately 24 mL, an exclusion limit of 4×104 kDa, and an optimal separation range of 5 to 5×103 kDa. Protein concentrations were determined by measuring the absorption at 280 nm with a NanoDrop® ND-1000 Spectrophotometer, blanked with elution buffer, and using a molar extinction coefficient of 19,900 M−1cm−1 and a molecular weight of 21.73 kDa as determined by DNASTAR software. A flow rate of 0.3 mL/min was maintained with an Agilent HPLC instrument. The column was eluted with 20 mM BICINE, pH 8.5, 20 mM NaCl, 2 mM β-ME. Downstream from the column, a UV detector (Agilent), a miniDAWN triple-angle light scattering detector (LS, Wyatt Technology), and an Optilab DSP interferometric refractometer (IR, Wyatt Technology) were connected in series. The refractometer provided a continuous index of protein concentration. A dn/dc (specific refractive index increment) value of 0.185 mL/mg was used. Bovine serum albumin was used as an isotropic scatterer for detector normalization. The intensity of light scattered by a protein is directly proportional to its weight-average molecular mass and concentration. Therefore, molecular masses (MW) were calculated from light scattering and IR concentration data using ASTRA software. Fractions (0.1 mL) were collected and analyzed by SDS-PAGE followed by silver staining.
UV denaturation curves of ITPA
The purified protein with cleaved His tag 39 was concentrated using an Amicon Ultra 5MWCO spin concentrator (Millipore) and adjusted to 2.2−2.7 mg/mL, then examined using dynamic light scattering (DynaPro MS/X, Protein Solutions, Inc.) to confirm monomodality and monodispersity before the UV denaturation experiments. Preparations showing bimodal peaks were further polished by use of a size exclusion column (Superdex 200, GE Healthcare), rechecked for monomodality, and re-concentrated as above. UV temperature-induced denaturation curves of ITPA were performed using a Perkin Elmer UV-Vis Spectrophotometer Lambda10. Refining the UV denaturation curve data required several experiments involving a range of protein concentrations. To achieve this without exceeding the detection limits of the spectrophotometer we employed varied light pathlength cuvettes (1, 2, 5, and 10 mm) which allowed up to a 10-fold concentration change while maintaining the same A280 (≈0.4). The samples were loaded into these cuvettes and all were assayed at 35−65°C with a heating rate of 0.2°C/minute. Response was measured and recorded every 0.2 second at 280 nm absorbance. The data were smoothed using a Savitzky-Golay filter, implemented in the freely available MacroBundle for Microsoft Excel (http://www.bowdoin.edu/~rdelevie/excellaneous/) and plotted as A280 vs. temperature. Plots of the first derivative with respect to temperature were generated with the same software. Melting temperatures, TMS, were determined by taking the temperature at the maximum of the first derivative plots.
Antibodies against ITPA
We have sent 4 mg of pure ITPA with His tag cleaved to Rockland Biosciences. Two rabbits were immunized twice at their facility, blood drawn and serum sent back to us. The serum was run on Protein G resin at the UNMC monoclonal Antibody facility to yield pure polyclonal antibody against ITPA.
Temperature-sensitivity test in E. coli
The cultures of transformants of the temperature sensitive strain EK5 (see above) by pET15b vector with wild-type or mutated ITPA were grown in LB in a permissive temperature (30°C). Serial dilutions were spotted on IPTG-containing plates and plates were incubated at 30°C or 42°C (non-permissive) overnight.
HAP mutagenesis test in bacteria
We performed a spot test by replica-plating of VB plates with growing cells and spotted HAP onto LB plates with rifampicin as described 22.
HAP sensitivity assay in yeast
We have used yeast strain yo699 62, which was modified as follows. First, it was made Ade+ by transformation with a PCR fragment of the ADE2 gene. Second, we introduced the disruption of the HAM1 gene by one-step disruption with the DNA fragment released from HLAM plasmid 19. The strain was transformed by the pESC-URA vector with human ITPA and its alleles to Ura+. Individual, independent transformants were grown in synthetic medium without uracil. Forward Canr mutant frequencies in the presence of HAP were determined as described previously 63.
Human cell culture
The normal diploid human lung fibroblast cell line, WI-38, (ATCC CCL-75) was kindly provided by Dr. Vera Gorbunova (University of Rochester, NY). The human fibroblast cell line, abbreviated as fbP32T, which is homozygous for a C->A transversion at nucleotide 94 (94C->A) in exon 2 of the ITPA gene resulting in a proline to threonine substitution at codon 32 (P32T) was obtained from the Coriell Institute Biorepository (GMO1617). According to the description provided by the vendor, the patient who donated the cell line, is a 29 year-old Caucasian female homozygous for the 94 C->A mutation resulting in no detectable ITPA activity in red blood cells and 20% of normal ITPase activity in lymphoblasts. We confirmed the presence of this DNA sequence change by sequencing of appropriate PCR product and have also detected the accumulation of low level of 10 μmol/L of ITP in the cell line extract, which was not detectable in the control fibroblasts.
These untransformed cell lines were cultivated as monolayers in Minimal Eagle's Medium with non-essential amino acids (Invitrogen), containing 10% fetal bovine serum (GIBCO) and the antibiotics penicillin, streptomycin and gentamycin (Invitrogen) to a total final concentration of 1%. The growth medium was further supplemented with 1mM sodium pyruvate (Invitrogen). The cells were incubated in a humidified incubator at 37°C and 5% CO2. Cells at early passages (<25 passages) were used in all experiments to avoid complications with senescence because WI-38 cells have a mean lifespan of approximately 45 to 60 population doublings.
Western Blot Analysis
WI-38 and P32T fibroblasts were cultivated as monloayers in 100-mm dishes (5×105 cells per plate) until subconfluence. The cells were harvested by washing thrice with phosphate buffered saline (PBS) and the pellet was collected by centrifugation. The pellets were resuspended in 100 L of lysis buffer (PBS, pH 7.4, containing protease inhibitor cocktail, Roche Biochemicals, IN) and mechanically sheared using a pestle. The lysate was cleared by centrifugation and protein content determined by the Bradford method. Lysate equivalent to 100 g of protein was boiled in Laemmli's buffer (Invitrogen) containing β-mercaptoethanol (Sigma-Aldrich). The protein samples were resolved on a 10−20% Tris-Glycine gel (Invitrogen) by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked overnight in commercially available blocking buffer. Membranes were then incubated in 1:1500 dilutions of primary antibody against ITPA (described above) and GAPDH (Cell Signalling, #2118) for one hour at room temperature. The membranes were then washed with commercially available washing buffer (Thermo Scientific) five times for 10 mins each. This was followed by incubation with secondary antibody (1:1500 dilution, Cell Signaling, #7074) for 40 mins at room temperature. This was followed by washing (5 times for 10 mins each) and detection by an ECL system (Thermo Scientific) according to the manufacturer's instructions. Images on film were scanned and the resulting TIFF files were analyzed using Image Quant TL software.
Supplementary Material
Supplemental Fig. 1. SEC-MALS analysis suggests that wild-type ITPA (A) and ITPA-P32T (B) are dimers in solution.
Upper panels - Elution profile
Bottom panels - SDS-PAGE followed by silver staining confirmed that the peak contained a single species
Acknowledgements
We are grateful to Dr. P. Shcherbakova and to Dr. S. Kozmin for critical reading of the manuscript. We thank Dr. A. Kuzminov for bacterial strains. The work was supported by NE DHHS LB506 grant to YP and UNMC Eppley Cancer Center 010107 Pilot Grant to GB and YP and National Cancer Institute Eppley Cancer Center Support Grant P30CA036727. The work of ES was supported by Post-Doctoral Fellowship provided by CRDF and Ministry of Education of Russian Federation. MM was supported by UNMC graduate student fellowship.
Abbreviations
- ITPase
generic name of enzymes hydrolyzing ITP
- ITPA
human ITP pyrophosphatase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones EW, Fink G. Regulation of amino acid and nucleotide biosynthesis in yeast. In: N. SJ, W. JE, R BJ, editors. Molecular biology of the yeast Saccharomyces. Metabolism and gene expression. Cold Spring Harbor Press; Cold Spring Harbor: 1982. pp. 181–299. [Google Scholar]
- 2.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt FC, editor. Escherichia coli and Salmonella, cellular and molecular biology. ASM press; Washington DC: 1996. pp. 561–579. [Google Scholar]
- 3.Bradshaw JS, Kuzminov A. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol Microbiol. 2003;48:1711–25. doi: 10.1046/j.1365-2958.2003.03540.x. [DOI] [PubMed] [Google Scholar]
- 4.Burgis NE, Brucker JJ, Cunningham RP. Repair system for noncanonical purines in Escherichia coli. J Bacteriol. 2003;185:3101–10. doi: 10.1128/JB.185.10.3101-3110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerit I, Filipe P, Meunier P, Auclair C, Freitas J, Deroussent A, Gouyette A, Fernandes A. Clastogenic activity in the plasma of scleroderma patients: a biomarker of oxidative stress. Dermatology. 1997;194:140–6. doi: 10.1159/000246083. [DOI] [PubMed] [Google Scholar]
- 6.Delaney CA, Eizirik DL. Intracellular targets for nitric oxide toxicity to pancreatic beta-cells. Braz J Med Biol Res. 1996;29:569–79. [PubMed] [Google Scholar]
- 7.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Second Edition edit ASM Press; Washington, DC: 2006. [Google Scholar]
- 8.Budke B, Kuzminov A. Hypoxanthine incorporation is nonmutagenic in Escherichia coli. J Bacteriol. 2006;188:6553–60. doi: 10.1128/JB.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T, Yoshida M, Yamada M, Ide H, Kobayashi M, Kanaori K, Tajima K, Makino K. Misincorporation of 2'-deoxyoxanosine 5'-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry. 1998;37:11592–8. doi: 10.1021/bi980971f. [DOI] [PubMed] [Google Scholar]
- 10.Orgel LE. The chemical basis of mutation. Adv Enzymol Relat Areas Mol Biol. 1965;27:289–346. doi: 10.1002/9780470122723.ch6. [DOI] [PubMed] [Google Scholar]
- 11.Hill-Perkins M, Jones MD, Karran P. Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat Res. 1986;162:153–63. doi: 10.1016/0027-5107(86)90081-3. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi A, Kitaoka M, Hayashi K. Analyses of PCR products using DNA templates containing a consecutive deoxyinosine sequence. Nucleic Acids Symp Ser (Oxf) 2004:225–6. doi: 10.1093/nass/48.1.225. [DOI] [PubMed] [Google Scholar]
- 13.Moe A, Ringvoll J, Nordstrand LM, Eide L, Bjoras M, Seeberg E, Rognes T, Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–31. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukas L, Kuzminov A. Chromosomal fragmentation is the major consequence of the rdgB defect in Escherichia coli. Genetics. 2006;172:1359–62. doi: 10.1534/genetics.105.051144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozmin SG, Schaaper RM, Shcherbakova PV, Kulikov VN, Noskov VN, Guetsova ML, Alenin VV, Rogozin IB, Makarova KS, Pavlov YI. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat Res. 1998;402:41–50. doi: 10.1016/s0027-5107(97)00280-7. [DOI] [PubMed] [Google Scholar]
- 17.Stenmark P, Kursula P, Flodin S, Graslund S, Landry R, Nordlund P, Schuler H. Crystal structure of human inosine triphosphatase. Substrate binding and implication of the inosine triphosphatase deficiency mutation P32T. J Biol Chem. 2007;282:3182–7. doi: 10.1074/jbc.M609838200. [DOI] [PubMed] [Google Scholar]
- 18.Pavlov Iu I. [Mutants of Saccharomyces cerevisiae supersensitive to the mutagenic effect of 6-N-hydroxylaminopurine]. Genetika. 1986;22:2235–43. [PubMed] [Google Scholar]
- 19.Noskov VN, Staak K, Shcherbakova PV, Kozmin SG, Negishi K, Ono BC, Hayatsu H, Pavlov YI. HAM1, the gene controlling 6-N-hydroxylaminopurine sensitivity and mutagenesis in the yeast Saccharomyces cerevisiae. Yeast. 1996;12:17–29. doi: 10.1002/(SICI)1097-0061(199601)12:1%3C17::AID-YEA875%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 20.Clyman J, Cunningham RP. Escherichia coli K-12 mutants in which viability is dependent on recA function. J Bacteriol. 1987;169:4203–10. doi: 10.1128/jb.169.9.4203-4210.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozmin SG, Pavlov YI, Dunn RL, Schaaper RM. Hypersensitivity of Escherichia coli Δ(uvrB-bio) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J Bacteriol. 2000;182:3361–7. doi: 10.1128/jb.182.12.3361-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozmin SG, Leroy P, Pavlov YI, Schaaper RM. YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol Microbiol. 2008;68:51–65. doi: 10.1111/j.1365-2958.2008.06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang KY, Chung JH, Kim SH, Han YS, Cho Y. Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nat Struct Biol. 1999;6:691–6. doi: 10.1038/10745. [DOI] [PubMed] [Google Scholar]
- 24.Chung JH, Back JH, Park YI, Han YS. Biochemical characterization of a novel hypoxanthine/xanthine dNTP pyrophosphatase from Methanococcus jannaschii. Nucleic Acids Res. 2001;29:3099–107. doi: 10.1093/nar/29.14.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin S, McLennan AG, Ying K, Wang Z, Gu S, Jin H, Wu C, Liu W, Yuan Y, Tang R, Xie Y, Mao Y. Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene. J Biol Chem. 2001;276:18695–701. doi: 10.1074/jbc.M011084200. [DOI] [PubMed] [Google Scholar]
- 26.Behmanesh M, Sakumi K, Tsuchimoto D, Torisu K, Ohnishi-Honda Y, Rancourt DE, Nakabeppu Y. Characterization of the structure and expression of mouse Itpa gene and its related sequences in the mouse genome. DNA Res. 2005;12:39–51. doi: 10.1093/dnares/12.1.39. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J, Singh VK, Jia Z. Identification of an ITPase/XTPase in Escherichia coli by structural and biochemical analysis. Structure. 2005;13:1511–20. doi: 10.1016/j.str.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Burgis NE, Cunningham RP. Substrate specificity of RdgB protein, a deoxyribonucleoside triphosphate pyrophosphohydrolase. J Biol Chem. 2007;282:3531–8. doi: 10.1074/jbc.M608708200. [DOI] [PubMed] [Google Scholar]
- 29.Marsh S, King CR, Ahluwalia R, McLeod HL. Distribution of ITPA P32T alleles in multiple world populations. J Hum Genet. 2004;49:579–81. doi: 10.1007/s10038-004-0183-y. [DOI] [PubMed] [Google Scholar]
- 30.Sumi S, Marinaki AM, Arenas M, Fairbanks L, Shobowale-Bakre M, Rees DC, Thein SL, Ansari A, Sanderson J, De Abreu RA, Simmonds HA, Duley JA. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360–7. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 31.Cao H, Hegele RA. DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency. J Hum Genet. 2002;47:620–2. doi: 10.1007/s100380200095. [DOI] [PubMed] [Google Scholar]
- 32.Marinaki AM, Sumi S, Arenas M, Fairbanks L, Harihara S, Shimizu K, Ueta A, Duley JA. Allele frequency of inosine triphosphate pyrophosphatase gene polymorphisms in a Japanese population. Nucleosides Nucleotides Nucleic Acids. 2004;23:1399–401. doi: 10.1081/NCN-200027641. [DOI] [PubMed] [Google Scholar]
- 33.Shipkova M, Lorenz K, Oellerich M, Wieland E, von Ahsen N. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clin Chem. 2006;52:240–7. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 34.Arenas M, Duley J, Sumi S, Sanderson J, Marinaki A. The ITPA c.94C>A and g.IVS2+21A>C sequence variants contribute to missplicing of the ITPA gene. Biochim Biophys Acta. 2007;1772:96–102. doi: 10.1016/j.bbadis.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 35.von Ahsen N, Oellerich M, Armstrong VW. Characterization of the inosine triphosphatase (ITPA) gene: haplotype structure, haplotype-phenotype correlation and promoter function. Ther Drug Monit. 2008;30:16–22. doi: 10.1097/FTD.0b013e318161a21a. [DOI] [PubMed] [Google Scholar]
- 36.Marinaki AM, Ansari A, Duley JA, Arenas M, Sumi S, Lewis CM, Shobowale-Bakre el M, Escuredo E, Fairbanks LD, Sanderson JD. Adverse drug reactions to azathioprine therapy are associated with polymorphism in the gene encoding inosine triphosphate pyrophosphatase (ITPase). Pharmacogenetics. 2004;14:181–7. doi: 10.1097/00008571-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 37.Marinaki AM, Duley JA, Arenas M, Ansari A, Sumi S, Lewis CM, Shobowale-Bakre M, Fairbanks LD, Sanderson J. Mutation in the ITPA gene predicts intolerance to azathioprine. Nucleosides Nucleotides Nucleic Acids. 2004;23:1393–7. doi: 10.1081/NCN-200027639. [DOI] [PubMed] [Google Scholar]
- 38.von Ahsen N, Armstrong VW, Behrens C, von Tirpitz C, Stallmach A, Herfarth H, Stein J, Bias P, Adler G, Shipkova M, Oellerich M, Kruis W, Reinshagen M, Schutz E. Association of inosine triphosphatase 94C>A and thiopurine S-methyltransferase deficiency with adverse events and study dropouts under azathioprine therapy in a prospective Crohn disease study. Clin Chem. 2005;51:2282–8. doi: 10.1373/clinchem.2005.057158. [DOI] [PubMed] [Google Scholar]
- 39.Zelinkova Z, Derijks LJ, Stokkers PC, Vogels EW, van Kampen AH, Curvers WL, Cohn D, van Deventer SJ, Hommes DW. Inosine triphosphate pyrophosphatase and thiopurine s-methyltransferase genotypes relationship to azathioprine-induced myelosuppression. Clin Gastroenterol Hepatol. 2006;4:44–9. doi: 10.1016/j.cgh.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Stocco G, Cheok MH, Crews KR, Dervieux T, French D, Pei D, Yang W, Cheng C, Pui CH, Relling MV, Evans WE. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;85:164–72. doi: 10.1038/clpt.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dieren JM, van Vuuren AJ, Kusters JG, Nieuwenhuis EE, Kuipers EJ, van der Woude CJ. ITPA genotyping is not predictive for the development of side effects in AZA treated inflammatory bowel disease patients. Gut. 2005;54:1664. [PMC free article] [PubMed] [Google Scholar]
- 42.Duley JA, Marinaki AM, Arenas M, Florin TH. Do ITPA and TPMT genotypes predict the development of side effects to AZA? Gut. 2006;55:1048–9. [PMC free article] [PubMed] [Google Scholar]
- 43.Behmanesh M, Sakumi K, Abolhassani N, Toyokuni S, Oka S, Ohnishi YN, Tsuchimoto D, Nakabeppu Y. ITPase-deficient mice show growth retardation and die before weaning. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.53. in press, e-publ. June 2009. [DOI] [PubMed] [Google Scholar]
- 44.Porta J, Kolar C, Kozmin SG, Pavlov YI, Borgstahl GE. Structure of the orthorhombic form of human inosine triphosphate pyrophosphatase. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62:1076–81. doi: 10.1107/S1744309106041790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozmin SG, Leroy P, Pavlov YI. Overexpression of the yeast HAM1 gene prevents 6-N-hydroxylaminopurine mutagenesis in Escherichia coli. Acta Biochim Pol. 1998;45:645–52. [PubMed] [Google Scholar]
- 46.Kozmin SG, Schaaper RM. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in Escherichia coli is independent of MobA function. Mutat Res. 2007;619:9–15. doi: 10.1016/j.mrfmmm.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutat Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 48.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–9. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 49.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol. 1992;174:6321–5. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–21. [PubMed] [Google Scholar]
- 51.Jackson AL, Loeb LA. The mutation rate and cancer. Genetics. 1998;148:1483–90. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat. Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 53.Colussi C, Parlanti E, Degan P, Aquilina G, Barnes D, Macpherson P, Karran P, Crescenzi M, Dogliotti E, Bignami M. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr Biol. 2002;12:912–8. doi: 10.1016/s0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- 54.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 55.Hakim IA, Harris RB, Brown S, Chow HH, Wiseman S, Agarwal S, Talbot W. Effect of increased tea consumption on oxidative DNA damage among smokers: a randomized controlled study. J Nutr. 2003;133:3303S–3309S. doi: 10.1093/jn/133.10.3303S. [DOI] [PubMed] [Google Scholar]
- 56.Polidori MC, Mecocci P, Stahl W, Sies H. Cigarette smoking cessation increases plasma levels of several antioxidant micronutrients and improves resistance towards oxidative challenge. Br J Nutr. 2003;90:147–50. doi: 10.1079/bjn2003890. [DOI] [PubMed] [Google Scholar]
- 57.Simandan T, Sun J, Dix TA. Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochem J. 1998;335:233–40. doi: 10.1042/bj3350233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekiguchi M, Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21:8895–904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- 59.Fersht A. Enzyme structure and mechanism. 2nd edit W.H. Freeman; New York: 1985. [Google Scholar]
- 60.Borgstahl GE. How to use dynamic light scattering to improve the likelihood of growing macromolecular crystals. Methods Mol Biol. 2007;363:109–29. doi: 10.1007/978-1-59745-209-0_6. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Weiss W. F. t., Roberts CJ. Characterization of high-molecular-weight nonnative aggregates and aggregation kinetics by size exclusion chromatography with inline multi-angle laser light scattering. J Pharm Sci. 2009;12:12. doi: 10.1002/jps.21726. [DOI] [PubMed] [Google Scholar]
- 62.Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle. 2008;7:3026–36. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stepchenkova EI, Kozmin SG, Alenin VV, Pavlov YI. Genome-wide screening for genes whose deletions confer sensitivity to mutagenic purine base analogs in yeast. BMC Genet. 2005;6:31. doi: 10.1186/1471-2156-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. SEC-MALS analysis suggests that wild-type ITPA (A) and ITPA-P32T (B) are dimers in solution.
Upper panels - Elution profile
Bottom panels - SDS-PAGE followed by silver staining confirmed that the peak contained a single species