Abstract
Protein Kinase C alpha (PKCα) has been implicated in cancer but the mechanism is largely unknown. Here we show that PKCα promotes head and neck squamous cell carcinoma (SCCHN) by a feed forward network leading to cell cycle deregulation. PKCα inhibitors decrease proliferation in SCCHN cell lines and xenografted tumors. PKCα inhibition or depletion in tumor cells decreases DNA synthesis by suppressing ERK phosphorylation and cyclin E synthesis. Additionally, PKCα down-regulates miR-15a, a microRNA that directly inhibits protein synthesis of cyclin E as well as other cell cycle regulators. Furthermore, both PKCα and cyclin E protein expression are increased in primary tumors, and PKCα inversely correlates with miR15a expression in primary tumors. Finally, PKCα is associated with poor prognosis in SCCHN. These results identify PKCα as a key regulator of HNSCC tumor cell growth by a mechanism involving activation of MAP kinase, an initiator of the cell cycle, and suppression of miR-15a, an inhibitor of DNA synthesis. Although the specific components may be different, this type of feed forward loop network, consisting of a stimulus that activates a positive signal and removes a negative brake, is likely to be a general one that enables induction of DNA synthesis by a variety of growth or oncogenic stimuli.
INTRODUCTION
The protein kinase C (PKC) serine/threonine kinases [classical (α, β, and γ), novel (δ, ε, η and Θ) and atypical (ζ and Ι/λ)] are critical mediators of a multitude of cell signaling events (1). Despite their integral participation in cellular physiology, relatively little is known regarding their expression and function in malignant disease such as squamous cell carcinoma of head and neck (SCCHN).
PKCα and PKCζ are among the isoforms expressed in normal adult human keratinocytes and SCCHN (2). We have previously demonstrated that PKCζ activation is required in epidermal growth factor (EGF)-stimulated MAP kinase signaling and proliferation in SCCHN cell lines, and its expression increases with tumor progression in SCCHN tissues (2). PKCα is implicated in the progression of other epithelial-derived tumors including breast and lung (3), and is associated with poor prognosis in hepatocellular carcinoma (4). Alterations in keratinocyte differentiation markers caused by oncogenic Ras and characteristic of late stage papilloma development are also mediated by (5). Although late stage keratinocyte differentiation markers are induced by PKCα. and oncogenic Ras, the cells do not undergo the normal apoptotic processes characteristic of envelope cornification (5). These data suggest that PKCα in the context of an active oncogene does not effectively promote terminal keratinocyte differentiation. Consistent with these results, a broad spectrum PKC inhibitor that is most potent against classic and novel isoforms, chelerythrine chloride, reduced tumor growth in a xenograft model of SCCHN (6). These studies suggest that PKCα plays a key role in the growth of SCCHN.
The cell cycle is a tightly controlled process that involves transient expression of specific cyclins in association with cyclin dependent kinases (cdks), leading to transcription factor activation (7). The G1-S phase transition requires initial expression of cyclin D complexed with cdk 4/6 followed by induction of cyclin E complexed with cdk 2; each cyclin-activated kinase phosphorylates Rb, releasing E2F1-3 transcription factors that promote DNA synthesis.
MicroRNAs, 21-23 nucleotide RNAs that regulate the stability or translational efficiency of target mRNAs, have been implicated in diverse cellular processes relevant to cancer including cell cycle (8, 9). Specific miRs, including miR-17-5p and miR-20a, regulate E2F family members (10-14). The miR-16-1 family, including miR-15-a, affects cell cycle progression (11).
In the present study, we demonstrate that PKCα controls DNA synthesis via a feed forward network involving miR-15a regulation of cyclin E. Tissue array analysis of PKCα and cyclin E protein expression revealed a progressive increase from normal oral mucosa and dysplasia to SCCHN. Finally, in a cohort of patients with SCCHN, higher PKCα gene expression was significantly associated with lower miR-15a levels and adverse outcome, highlighting the relevance of this signaling cascade.
EXPERIMENTAL PROCEDURES
Immunohistochemistry
Immunohistochemistry was performed as previously described and scored on a 0- 3+ scale (2). Specific antibodies employed were PKCα rabbit primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), phosphoPKCα rabbit primary antibody (catalog #9375, Cell Signaling, Beverly, MA), and PKCβ mouse primary antibody (catalog #P2584, Sigma, Saint Louis, MO).
Growth of Human Tumor Xenografts
SQ20B tumor cells were injected into the right hind limbs of female athymic nude mice as described (6).
DNA microarray analyses
Biotin-labeled RNA was hybridized onto Affymetrix Human Genome U133 Plus 2.0 GeneChip. Raw data containing approximately 55,000 Affymetrix probe sets (representing 31,000 genes) were normalized and statistical significance determined using Significance Analyses of Microarrays (SAM) (false discovery rate < 10%) as previously described (15, 16).
RNA Isolation and Real-time PCR from Human Tissue
All tissue samples, collected under a Vanderbilt University Institutional Review Board-approved protocol, were macrodissected to obtain at least 70% tumor cells (17). The RNA was purified using PureLink Total RNA Purification System and miRNA Isolation Kit (Invitrogen, Carlsbad, CA). miR-15a quantitative RT-PCR used TaqMan miRNA assays (ABI, Foster City, CA) normalized to U6 CT values.
Additional procedures are described in Supplementary Material.
RESULTS
Inhibition of PKCα reduces DNA synthesis and cell growth in multiple SCCHN cell lines in vitro and tumor growth in vivo
Since PKCα is expressed in all SCCHN cell lines examined (2), we determined whether PKCα is required for DNA synthesis. Gö6976 is an inhibitor of the classic PKC isoforms (α, β, γ) with a reported IC50 of 100 nM (18); since SCCHN cell lines do not express the β and γ PKC isoforms (2), this inhibitor primarily targets PKCα in these cells. Serum-starved SCC61, CAL27, HN31, and SQ20B cells were pretreated with 100 nM Gö6976 for 24 hours prior to serum-induced BrdU incorporation into DNA. Dose response analysis showed 50-100 nM Gö6976 inhibited SQ20B DNA synthesis, and 100 nM Gö6976 suppressed DNA synthesis in all cell lines (Fig. 1A). Time course analysis revealed complete inhibition of SQ20B DNA synthesis by 9-12 hours of Gö6976 treatment. FACS analysis of SQ20B cells confirmed that Gö6976-treated cells arrested in S phase (Supplemental Fig. 1A). To confirm that PKCα is required for DNA synthesis, two cell lines (SCC61 and SQ20B) were transfected with either control or PKCα siRNA prior to serum-stimulated BrdU incorporation (Fig. 1B). Protein immunoblotting confirmed siRNA specificity for PKCα (Supplemental Fig. 1B). These results indicate that PKCα is required for DNA synthesis in SCCHN tumor cells.
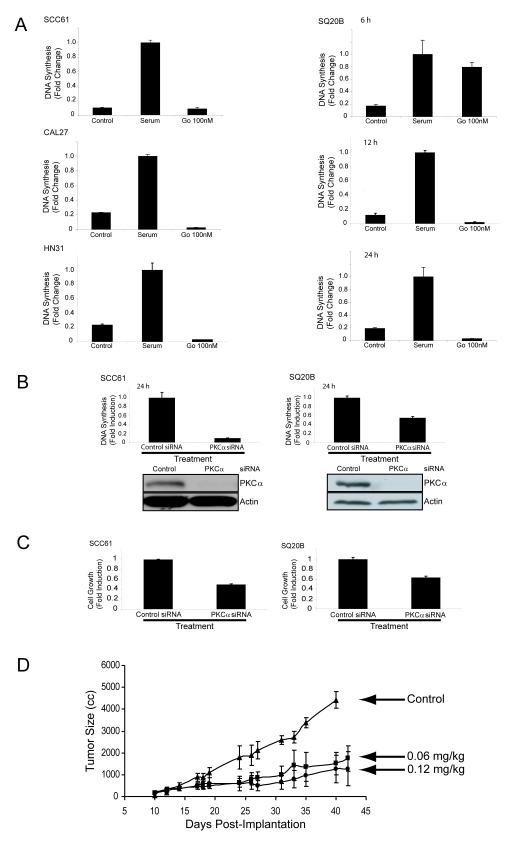
Figure 1. PKC alpha inhibition reduces DNA synthesis, cell viability, and tumor growth in vivo.
A) SCC61, SQ20B, CAL27, and HN31 cell lines were incubated in serum-free media (control), serum alone (serum), or serum with 100 nM of Gö6976 (Gö 100nM) for up to 24 hours and DNA synthesis was quantitiated for BrdU incorporation as described in Experimental Procedures. B) SCC61 (left) and SQ20B (right) cells were transfected with control or PKC alpha siRNA. DNA synthesis was quantitiated for BrdU incorporation as described in Experimental Procedures. All BrdU incorporation results are the mean +/- S.E. of three independent experiments. Immunoblotting for PKCα and actin was performed in conjunction with BrdU incorporation after control and PKCα siRNA transfection as described in Experimental Procedures. C) SCC61 (left) and SQ20B (right) cells were transfected with control or PKCα siRNA and cell survival was quantitated by MTT assay as described in Experimental Procedures. All MTT results are the mean +/- S.E. of three independent experiments. D) SQ20B tumor xenografts were established in female athymic nude mice. After reaching an average size of 150 mm3 the mice (n = 5) were treated with 0.1% DMSO in PBS (control) or Gö6976 (n = 10 per group) at 0.06 mg/kg/week or 0.12 mg/kg/week by intravenous tail vein injection. Values represent mean tumor size +/- S.E.
To assess whether inhibition of decreases cell growthin SCC61 and SQ20B cells transfected with control or PKCα siRNA, we used the MTT assay that measures changes in metabolism. The inhibition was generally less robust than for DNA synthesis, possibly due to differences in kinetics or assays, but in both cell lines PKCα depletion reduced cell growth (Fig. 1C). Pretreatment with Gö6976 reduced proliferating SCC61, CAL27, HN31, and SQ20B cells in a dose-dependent manner with almost complete inhibition at 100 nM (Supplemental Fig. 1C). These results demonstrate that PKCα is a critical mediator of SCCHN cell proliferation.
To determine whether PKCα inhibitors affect SCCHN tumor growth in vivo, hind limb SQ20B tumor xenografts were established in female athymic nude mice. Administration of Gö6976 dramatically retarded growth of the SQ20B xenografts (Fig. 1D). Although we cannot rule out effects on other cells or drug targets within the microenvironment, these results are consistent with the inhibition of SCCHN tumor cell growth in culture by PKCα.
PKCα Inhibition Suppresses Genes and Proteins that Regulate the Cell Cycle
Previous studies have shown that PKCα as well as other PKC isoforms such as PKCζ stimulate the MAPK signaling cascade via Raf activation (2, 19, 20), and MAPK activation is required for DNA synthesis in SQ20B cells (2). Consistent with this observation, cell stimulation with serum or a PKC inducer, PDBu (phorbol 12,1h3-dibutylate), activates ERK, and PKCα siRNA decreases ERK activation (Fig. 2A). PKCα siRNA also reduces baseline ERK activation in PDBu and serum-treated SQ20B cells. Thus, under these conditions, PKCα is both necessary and sufficient for MAPK signaling.
Figure 2. PKC alpha inhibition induces changes in MAPK activity and regulates cell cycle genes.
A) Decreased ERK activity in SQ20B cells with PKCα inhibition. SQ20B cells were transfected with control (siControl) or PKCα (siPKCalpha) siRNA. After 48 hours in serum-free media, cells were incubated with increasing concentrations of PDBu (left panels) or serum (right panels) for 30 minutes. Whole cell lysates were probed for phosphorylated and total ERK, and blots were quantified as described in Experimental Procedures. Graphs represent quantification of the ratio of phospho-ERK to total ERK protein. Representative experiments are shown for at least two independent experiments. B) Gene array heat map of cell cycle genes in the SQ20B cell line with decreased expression at 24 hours upon exposure to 100nM Gö6976 (left panel). Right panel shows RT-PCR expression of selected cell cycle genes at 0 (black bars), 12 (gray bars), and 24 (white bars) hours shown as fold change normalized to untreated control. C) Protein immunoblotting and immunoprecipitation of selected cell cycle genes upon exposure to 100nM Gö6976 at 0, 12, and 24 hours. SQ20B cells were serum-starved for 48 hours, followed by treatment in serum with 100 nM Gö6976 for 0, 12, and 24 hrs. Total protein was extracted from cells and immunoblotted for cyclin E, Cdk2, E2F2, cyclin A, MCM6 and γ or α-tubulin as described in Experimental Procedures (left panel). Cell lysates were immunoprecipitated with an anti-Cdk2 antibody and the immunoprecipitates were examined by immunoblotting to detect cdk2-complexed cyclin E (cdk2 IP/cyclin E IB, right panel). Total protein lysates were also probed for cyclin E and cdk2 antibodies. Representative data from three separate experiments are shown. D) E2F 1-3 protein levels in SQ20B cells. SQ20B cells were serum-starved for 48 hours, followed by treatment in serum with 100 nM Gö6976 for 0, 12, and 24 hrs. Total protein lysates were probed with the respective antibodies. Lysate from untreated U2OS cells was used as a positive control for E2F1.
Although MAPK activation initiates tumor cell proliferation, additional mechanisms sustain the growth response. To identify potential effectors of PKCα that regulate DNA synthesis in SCCHN cells, we inhibited PKCα and analyzed changes in gene expression. Serum-starved SQ20B cells were treated with 100 nM Gö6976 for 0, 12, or 24 hours, and gene expression changes determined by DNA microarrays. To detect differential expression of low abundance regulatory genes, data were queried using 171 probes related to cell cycle regulation (21). Fifty-five probes (39 unique genes) were differentially expressed upon comparison of the 0 versus 24 hour data sets (Supplemental Fig. 2). Eight of the 39 genes had decreased expression between 12-24 hours following Gö6976treatment (Fig. 2B, left panel).
Inhibition of four genes (cyclin E1, E2F2, MCM6 and PCNA) was confirmed by quantitative RT-PCR (Fig. 2B, right panel). Among E2F2-regulated genes are cyclins E1 and E2, E2F2 itself, PCNA (22, 23) and the MCM proteins (24). Cyclins E1 and E2 interact with cdk2 to form a serine/threonine kinase holoenzyme complex that phosphorylates the Rb protein, relieving repression of E2F2-mediated transcription (7). MCMs, minichromosome maintenance proteins, are DNA helicases required for replication (24), and PCNA, a DNA polymerase cofactor, recruits key factors to the replication fork (25).
Analysis of cyclin E and E2F2 protein levels by immunoblotting similarly revealed a decrease in response to the PKCα inhibitor. Cell treatment with 100 nM Gö6976 for 0, 12, and 24 hours significantly reduced cyclin E and E2F2 protein levels (Fig. 2C). No loss of cyclin E prior to 10 hours treatment, consistent with the DNA synthesis results, was observed (data not shown). In contrast, no change in MCM6, cdk2, or cyclin A protein levels was detected. The cell cycle transition to S phase requires phosphorylation by cyclin E complexed with cdk 2. Immunoprecipitation of cdk2 revealed a dramatic decrease in cyclin E associated with cdk2 following Gö6976 treatment, indicative of cdk2 kinase inactivation (Fig. 2C). Although PCNA was similarly regulated, expression levels for the other E2F family members (1 and 3) were not significantly altered by PKCα inhibition (Fig. 2D). These results suggest that lowered cyclin E and possibly E2F2 protein expression caused by PKCα inhibition accounts for the decrease in DNA synthesis.
We then tested whether cyclin E or E2F2 are required for DNA synthesis in SCCHN cells. SQ20B cells were depleted of human cyclin E by transfection with siRNA (Fig. 3A). Analysis of BrdU incorporation into DNA indicates that cyclin E expression is required for DNA synthesis. Conversely, expression of cyclin E was sufficient to significantly rescue BrdU incorporation into DNA following 100 nM Gö6976 treatment (Fig. 3B). E2F2 was also required for DNA synthesis as shown by siRNA depletion (Fig. 3A). However, in contrast to Gö6976 treatment, depletion did not significantly decrease E2F2 protein levels. These differences between drug and siRNA effects could reflect differences in kinetics or mechanism of PKCα inactivation, or alternatively, a role for other PKC isoforms or enzymes in mediating the drug’s effects on E2F2. Furthermore, E2F2 overexpression only induced a limited recovery of DNA synthesis in Gö6976 treated cells (Fig. 3B) consistent with the presence of other E2Fs in the cell. Finally, these results confirm that siRNA depletion of PKCα leads not only to inhibition of DNA synthesis but also to loss of cyclin E expression (Fig. 3A). Thus, induces cyclin E expression that is necessary for SCCHN cell cycle progression; furthermore, cyclin E is sufficient to overcome the cell cycle block generated by loss of PKCα activity.
Figure 3. Cyclin E mediates PKCα induction of DNA synthesis in SQ20B cells.
A) Effect of siRNA on DNA synthesis. SQ20B cells were transfected with control, PKCα, cyclin E, or E2F2 siRNA and protein expression was determined as described in Experimental Procedures (left panel). SQ20B cells were incubated in serum-free media (starved), serum alone (serum), serum with control siRNA, serum with 100nM Gö6976 (Gö6976), or serum with respective siRNAs for 24 hours (right panel). DNA synthesis was quantitiated for BrdU incorporation as described in Experimental Procedures and expressed as fold change compared with serum alone. B) Rescue of DNA synthesis with cyclin E and E2F2 expression in Gö6976 treated cells. SQ20B cells were mock transfected or transfected with cyclin E and E2F2 expression vectors and protein expression was determined as described in Experimental Procedures (left panel). SQ20B cells were incubated in serum-free media (starved), serum alone (serum), serum with Gö6976 (Gö6976), or serum with 100nM Gö6976 and the respective transfection vector(s) for 24 hours. DNA synthesis was quantitiated for BrdU incorporation as described in Experimental Procedures and expressed as fold change compared with serum alone (right panel). All BrdU incorporation results are the mean +/- S.E. of three independent experiments. C) Change in rate of cyclin E protein synthesis. SQ20B cells were either untreated or pretreated with Gö6976 (100 nM) for 12h and then exposed to MG132 (30 μM) for the times indicated. Graph represents quantification of cyclin E levels from the protein immunoblot. Cyclin E values were normalized to α-tubulin levels. Results are representative of three independent experiments.
PKCα regulation of cyclin E protein levels results from changes in protein synthesis or degradation. To test the former possibility, SQ20B cells were left untreated or pretreated with the PKCα inhibitor Gö6976 for 12 hours followed by addition of MG132, a proteosome inhibitor, to prevent proteolysis. Although the cyclin E level eventually plateaus due to resumed degradation, inhibition of PKCα significantly decreased the initial rate of cyclin E protein synthesis relative to that in untreated tumor cells (Fig. 3C). To test whether the proteosome is a target of PKCα, SQ20B cells were pretreated with Gö6976 and then exposed to cyclohexamide to block protein synthesis. However, no consistent increase in the rate of cyclin E degradation was observed (data not shown). These results indicate that inhibition of PKCα suppresses cyclin E protein synthesis.
PKCα Regulates Cyclin E Synthesis through miR-15a
Our previous results established that PKCα increased DNA synthesis via enhancement of cyclin E protein levels. Both gene expression arrays and qRT-PCR analyses showed decreases in mRNA levels between 12-24 hours of treatment by Gö6976 (Fig. 2B). However, time course experiments reveal that inhibition of DNA synthesis was maximal at 12 hours (Fig. 1A). In addition, our results confirm that PKCα inhibition caused maximal suppression of cyclin E protein expression by 12 hours, prior to subsequent changes in transcription (Fig. 2B, C). Thus, PKCα regulation of transcription rates cannot account for the kinetics of cyclin E protein enhancement.
A recently described mechanism for regulating protein synthesis involves inhibition of mRNA translation by miRs (8). To investigate whether cyclin E protein loss in response to PKCα inhibition results from induction of miRs, we searched the PicTar database (http://pictar.bio.nyu.edu/) and found that miR-15a could theoretically bind to two regions within the 3′ untranslated region (3′-UTR) of the cyclin E mRNA (Fig. 4B). Quantitative RT-PCR confirmed that PKCα inhibition by Gö6976 treatment or PKCα depletion by siRNA significantly increased miR-15a expression (Fig. 4A). The kinetics of inhibition are different since drug inhibition of kinase activity occurs more rapidly than elimination of PKCα expression. Similarly, a different siRNA that had slower kinetics for PKCα depletion required a consistent shift in the kinetics to induce miR-15a expression by 2-fold (Supplemental Fig. 3).
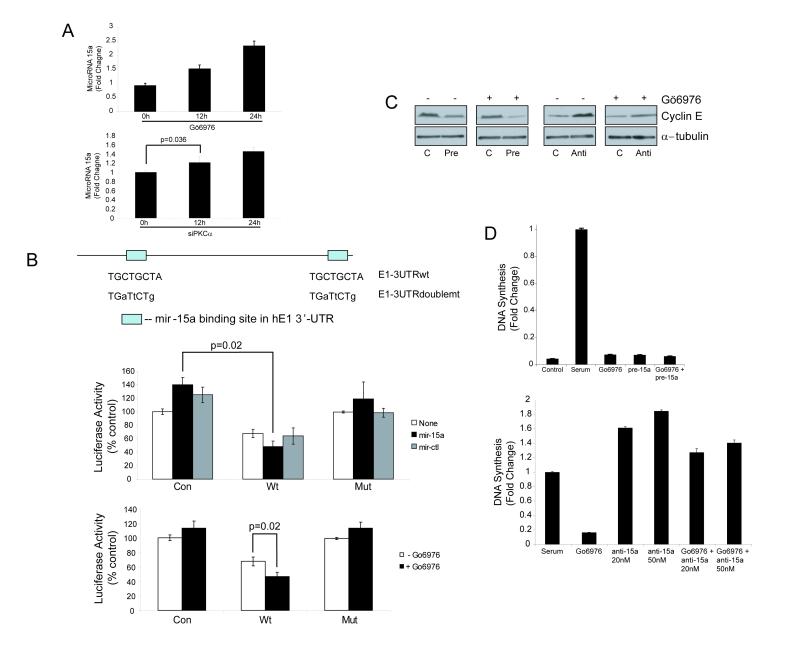
Figure 4. PKC alpha regulates cyclin E expression and DNA synthesis through miR-15a.
A) Detection of mature miR-15a expression in SQ20B cells by quantitative real-time PCR. SQ20B cells were untreated or treated with Gö6976 (100nM, top panel) or PKCα siRNA (bottom panel) for 12 and 24 hours. After miRNA extraction, qRT-PCR was performed in triplicate as described in Experimental Procedures. Bars represent relative miRNA levels +/- S.E normalized to no treatment. B) miR-15 and PKCα regulate protein synthesis via miR-15 binding sites in the 3′UTR of cyclin E. SQ20B cells were transfected with a luciferase reporter expressed in conjunction with control vector (Con), wild type (Wt) 3′ UTR of cyclin E or doubly mutated (Mut) 3′ UTR of cyclin E. The reporter-expressing cells were either transfected with pre-miR-15a or control vector (middle panel), or pretreated with Gö6976 (100 nM) for 24 hrs (lower panel). The upper panel shows the sites of miR-15a binding in the 3′UTR of cyclin E and the mutations that were introduced. P values were determined using 2-tailed Student’s T-test. C) Effect of pre-miR-15a and anti-miR-15a on cyclin E levels in the absence or presence of Gö6976. SQ20B cells were transfected with 20 nM of pre-miR-15a (Pre), 50 nM of anti-miR-15a (anti), or control precursor (C) and treated (+) or untreated (-) with Gö6976 (100 nM) for 24 hrs. Cyclin E level was determined by protein immunoblotting. Each blot of paired lanes represents an independent experiment and was not performed simultaneously with the others. D) Effect of miR-15a and anti-miR-15a on DNA synthesis. Top panel: SQ20B cells were transfected with either 20 nM of control precursor or pre-miR-15a RNA by electroporation. Cells were grown in serum-free medium (control) or serum containing medium (all others) and exposed to Gö6976 (100 nM), pre-miR-15a miRNA (pre15a), or both. BrdU incorporation was quantified and expressed as fold change normalized to serum. Bottom panel: SQ20B cells were transfected with 20 nM or 50 nM of anti-miR-15a or Gö6976 (100 nM) and grown in serum containing medium. BrdU incorporation assays were performed and levels were expressed as fold change normalized to serum. All BrdU incorporation results are the mean +/- S.E. of three independent experiments.
We therefore determined whether miR-15-a directly inhibits cyclin E translation. The 3′-UTR region of the human cyclin E1 gene containing intact (Wt) or doubly mutated miR-15-a binding sites (Mut) was cloned into Rellina luciferase reporter vectors. SQ20B cells were transfected with control (Con) or cyclin E 3′-UTR-containing luciferase vectors and co-transfected with either control or miR-15-a precursor microRNA (Con or pre-miR-15a). Treatment with pre-miR-15a reduced cyclin E1-dependent luciferase activity in cells transfected with the Wt cyclin E 3′UTR (Fig. 4B). By contrast, no reduction was observed with the Mut cyclin E 3′ UTR, confirming that miR-15a directly inhibits cyclin E1 translation via binding to its 3′ UTR at the predicted sites. Moreover, treatment with Gö6976 further reduced luciferase activity in cells transfected with the Wt but not the Mut cyclin E 3′ UTR reporters (Fig. 4B), confirming that regulation of cyclin E1 by its 3′ UTR is both and miR-15a-dependent.
These 3′ UTR results suggest that miR-15a inhibits cyclin E protein expression. Transfection with pre-miR-15a confirmed that miR-15a reduces cyclin E protein levels in SQ20B cells; this decrease occurred both in cells that were untreated or pretreated with the PKCα inhibitor Gö6976 (Fig. 4C). Furthermore, expression of an inhibitor of miR-15-a, anti-miR-15a, increases cyclin E protein expression and partially rescues the effects of Gö6976 (Fig. 4C).
Similar results were obtained for regulation of DNA synthesis by miR-15-a. Transfection of pre- miR-15a into SQ20B cells inhibits DNA synthesis to a similar extent as Gö6976 (Fig. 4D). Conversely, anti-miR-15a enhances DNA synthesis and antagonizes the anti-proliferative effect of Gö6976 (Fig. 4D). Consistent with a miRNA-dependent mechanism, washout experiments demonstrated that the effects of Gö6976 treatment on cyclin E protein levels are reversible (data not shown). These results demonstrate that miR-15a is suppressed by PKCα and inhibits cyclin E expression and DNA synthesis.
Enhanced PKCα and cyclin E expression in malignant oral epithelium
If PKCα is a critical mediator of tumor cell growth, we should observe increased expression in SCCHN tumors. Furthermore, if PKCα upregulates cyclin E expression, then a corresponding increase in cyclin E protein expression would be observed in SCCHN. We examined PKCα and cyclin E expression in normal human oral mucosa, dysplastic oral mucosa, and head and neck tumor biopsies by immunohistochemistry. Phosphorylation of the PKCα at the turn motif site maintains catalytic competence and is required for kinase activity (26, 27); therefore, to assess expression of enzymatically competent PKCα, samples were also immunostained with anti-phospho antibody directed against Thr638. Analysis of the relative distribution of staining intensity reveals that both PKCα and phospho-PKCα expression increased progressively from normal to dyplastic to malignant tissue (Fig. 5A, B p<0.0001 by Cuzick’s trend test). Both normal and dysplastic tissue show predominantly nuclear staining for both anti-PKCα and anti-phospho-PKCα. In contrast, analysis of the malignant tissue reveals prominent cytoplasmic staining that is not present in the neighboring stroma. These results indicate that PKCα expression correlates with progression to SCCHN.
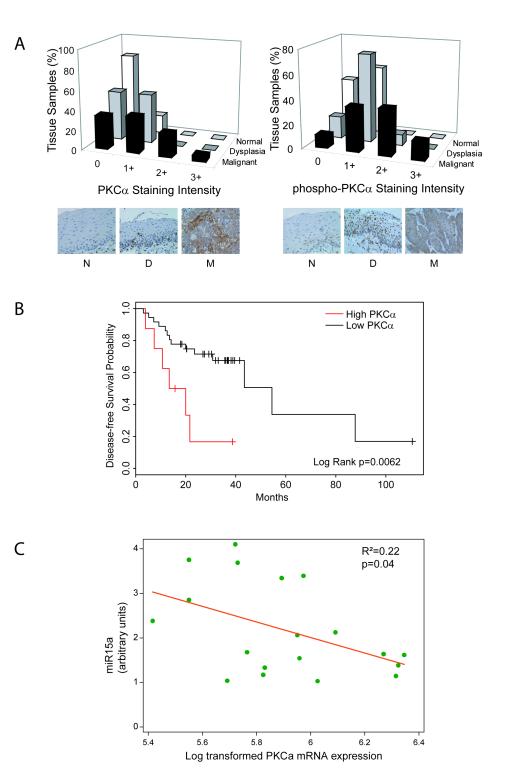
Figure 5. Tissue expression of total and phosphorylated PKC alpha and association of PKC alpha gene expression with miR-15a expression and tumor recurrence.
A) Left panel: Proportion of normal, dysplastic, or malignant specimens classified by staining intensity with anti-PKCα antibody. Representative sections of normal (N), dysplastic (D) and malignant (M) tissue sections stained with anti-PKCα antibody. Right panel: Proportion of normal, dysplastic, or malignant specimens classified by staining intensity with anti-phospho-PKCα antibody. Representative sections of normal (N), dysplastic (D), and malignant (M) tissue sections stained with anti-phospho-PKCα antibody. Staining intensity is expressed on a 4-point scale from 0 to 3+ (see Experimental Procedures). B) Kaplan-Meier curve of disease-free survival probability as a function of high (red) versus low (black) PKCα gene expression by Affymetrix array. Statistical significance was determined by the log rank test. C) PKCα transcript and miR-15a levels inversely correlate in individual SCCHN tumor samples (R2=0.22). mRNA and miRNA were isolated from 19 SCCHN tumor samples and analyzed for expression of PKCα transcripts and miR-15a by RT-PCR as described in Experimental Procedures. The results were plotted and analyzed by linear regression analysis for an inverse correlation. The p value reveals statistical significance.
The phospho-PKCα antibody also detects phosphorylated PKCβII at a complementary site (Thr641). Therefore, we assessed expression of PKCβII in the same tissue microarray and observed no staining in normal and dysplastic tissue. The SCCHN samples expressed low levels of PKCβII (17% were 1+, 3% were 2+, 0 were 3+, data not shown) suggesting that expression detected using the phospho-antibody stems mostly from PKCα. Moreover, samples staining for PKCα were highly correlated with the phospho-antibody (Spearman’s Rho correlation = 0.48, probability that PKCα and p-PKCα are independent < 0.00001).
Cyclin E staining displayed a similar pattern as PKCα. No expression in normal and dysplastic tissue was detected in tissue microarrays stained with anti-cyclin E antibody whereas 18% of SCCHN samples stained positively (10% were 1+, 6% were 2+, 2% were 3+; p<0.0001, Supplemental Fig. 4A). The relatively low staining intensity in all tissues likely reflects the transient expression of cyclin E in proliferating cells, the heterogeneity in aggressiveness of the malignant tissues examined, and the limited sensitivity of the antibody employed. Nevertheless, the increased expression of PKCα and cyclin E in SCCHN tumor tissue mirrors the PKCα induction of these proteins in the radioresistant, highly proliferative SCCHN cell line SQ20B.
To assess whether PKCα expression had prognostic significance, disease-free survival (DFS) analysis was performed in a previously described cohort of 44 patients treated for SCCHN using expression measurements for a PKCα probe on Affymetrix microarray (15). Median follow-up time was 27 months (range: 3-111 months) and events were recorded as either disease recurrence or death. Since PKCα expression is measured as a continuous variable, we examined the data using a Cox proportional hazards model using PKCα expression as a predictor and DFS as outcome measure. In this model, lower PKCα expression was a significant predictor of longer DFS (p=0.027). For visualization of the data, patient samples were divided based on PKCα expression (threshold = 0.15) and Kaplan-Meier curves were plotted using DFS as the outcome measure (Fig. 5B). Log rank analysis reveals that high PKCα expression was associated with a significantly higher probability of disease recurrence or death (p=0.0062). In addition, overall survival was also significantly shortened in the high PKCα expression group (p=0.0028 by log rank test or p=0.017 by Cox proportional hazards model, data not shown). This evidence further substantiates the regulation seen in vitro and suggests that PKCα mediates SCCHN tumor progression.
Since PKCα negatively regulates mir-15a in vitro we examined expression of miR-15a in 29 different SCCHN primary patient tumor samples. Ten of the 29 samples were selected based on availability of tissue for miRNA isolation and relative PKCα gene expression (Supplemental Fig. 4B) from a previously published data set (15). An inverse relationship was noted between miR-15-a expression and PKCα transcript levels in tumor samples (p=0.09, Supplemental Fig. 4C). To determine whether this relationship exists within individual SCCHN tumors, an additional 19 samples were analyzed for both PKCα and miR-15-a expression by qRT-PCR (Fig. 5C). The results confirm that PKCα and miR-15-a expression are inversely related (slope -1.75, p=0.04). This evidence further substantiates the regulation seen in vitro and suggests that PKCα mediates its oncogenic effects, at least in part, through miR-15a. In addition, since PKCα is prognostic for progression in primary SCCHN tumors, these results suggest that miR-15a expression should correlate with disease-free survival.
DISCUSSION
These results implicate PKCα as a key mediator of SCCHN proliferation through activation of MAPK and negative regulation of miR-15-a, an inhibitor of cyclin E expression, leading to increased synthesis of cell cycle proteins and enhanced DNA synthesis. Our findings fit a coherent type 4 feed-forward loop (FFL) network (28) (Fig. 6). The FFL is initiated by PKCα with one arm comprising a series of activating reactions and the second arm consisting in part of two successive inhibitory reactions that together promote cyclin E expression and DNA synthesis. In the present case, PKCα activates a “driver”, MAPK, as well as releasing a “brake”, miR-15a (Fig. 6B). The key characteristics of FFLs are a delay in the activation of the system upon stimulation as well as a rapid shut-off upon loss of the stimulus. Both of these features represent key elements that ensure the integrity of DNA synthesis by enabling initiation only when two input conditions are satisfied, and causing rapid cessation when either input is lost. However, in cancer cells, the stimulus is constitutively activated so that there is no initial delay in the activation of the cell cycle. It is likely that this type of coherent type 4 FFL network, consisting of a stimulus that activates a positive signal and removes a negative brake, are general characteristics of cell cycle progression in both normal and tumor cells.
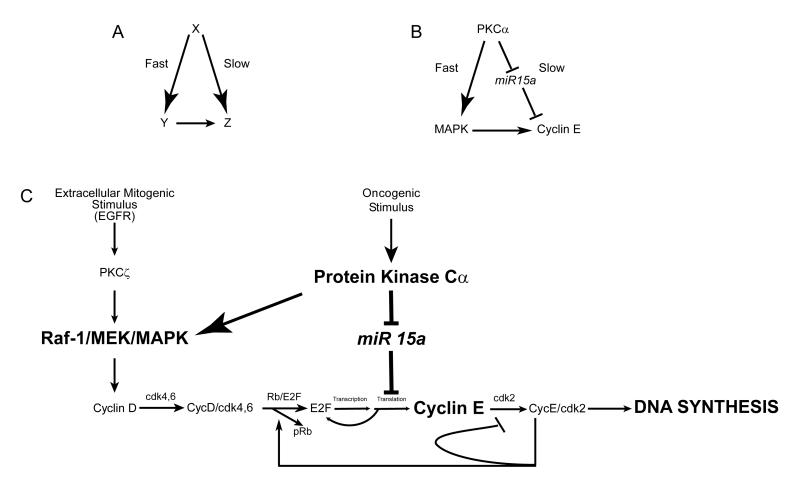
Figure 6.
Schema illustrating a feed-forward loop initiated by PKCα that regulates DNA synthesis. A) Illustration of a typical feed-forward loop with X as the initiation signal that stimulates Y activation rapidly and Z activation slowly. Y also stimulates Z resulting in maximal activation of the network. B) The feed-forward loop involves PKCα that activates MAPK as well as cyclin E translation via inhibition of mir-15a. Note that the “slow” arm of the loop involves negative regulation of a suppressor resulting in forward flow. C) Specific components of the feed-forward loop. Normal G1-S phase transition in response to mitogenic extracellular stimuli such as EGF induces activation of the Raf/MEK/MAPK cascade. We have previously shown that PKCζ mediates EGF activation of MAPK (2). Here we show that PKCα can also activate MAPK. This signaling cascade leads to expression of cyclin D complexed with the cyclin-dependent kinases cdk 4/6 followed by induction of cyclin E complexed with cdk 2; each cyclin-activated kinase in turn phosphorylates Rb. In concert with this, PKCα activity induced by oncogenic stimuli deregulates cyclin E expression by suppressing miR-15a, an inhibitor of cyclin E. The net effect of PKCα activation is to trigger a positive feedback loop resulting in upregulated cyclin E expression and DNA synthesis.
Since both PKCα and cyclin E have been implicated in diverse cancers, these findings could have broader relevance. The mechanism described for upregulating cyclin E involves inhibition of miR-15-a by activated PKCα. miR-15-a and miR-16-1 are located in the 13q14 locus, deleted in the majority of chronic lymphocytic leukemias, consistent with tumor suppressor function (29). Further evidence demonstrated that miR-15a negatively regulates Bcl-2 and thus induces apoptosis in a leukemic cell line (30). In pituitary adenomas, suppression of miR-15a and miR16-1 has been associated with tumor growth (31). However, in endocrine pancreatic tumors the same miRs appear to be overexpressed underscoring the likelihood that their role is tissue specific (32). A functional screen of the miR16 family that includes miR-15a in HCT116, HeLa, and TOV21G cell lines demonstrated that these miRs negatively regulate cell cycle progression (11). The miR16 family in these cells functioned primarily in feedback regulation, and its inhibition of DNA synthesis was mediated by its cumulative effect on multiple cell cycle targets. By contrast, we demonstrate here that, in SCCHN, miR-15a regulation of cyclin E expression is sufficient to account for the negative regulation of DNA synthesis by PKCα. Our results suggest that miR-15a is a positive prognostic marker based on its relationship with PKCα expression and may function as a tumor suppressor that negatively regulates cell proliferation.
It is noteworthy that inhibition of PKCα with siRNA did not mimic all the effects of Gö6976 in vitro suggesting that other PKCs, such as PKCε, could also play a role in proliferation. Thus, the in vivo effects observed in response to Gö6976 could result in part from inhibition of other PKC isoforms in tumor cells, mouse stromal tissue or immune cells. PKCβ has been implicated as a mediator of angiogenesis through inhibition of GSK3β (33) but a recent study using Gö6976 and other PKC inhibitors concluded that PKCα rather than PKCβ promotes angiogenesis (34). PKCs also regulate immune cell function (35-37); thus, the contribution of specific isoforms with respect to tumor microenvironment and immune system needs to be further elucidated.
PKC has been shown to regulate cyclins or E2Fs in other cell types but the outcomes can differ depending upon the specific system. For example, overexpression or TPA activation of PKCα in late G1 phase was shown to inhibit rather than activate E2F activity and DNA synthesis in rat 3Y1 fibroblasts (38). Conversely, constitutively activated PKCα enhances both cyclins D and E promoter activity in NIH3T3 cells (39). In human keratinocytes, activation of PKCα has been implicated in cell cycle arrest or terminal differentiation (40-42) while in some cancers PKCα expression is decreased or possesses tumor suppressor function (3). This is not necessarily contradictory to our findings since similar mechanisms may be utilized in other tissues but other pathways may also be present that counteract the effect of PKCα or function as feedback regulators. In SQ20B cells, cyclin E gene expression and proliferation was significantly altered by PKCα. It is also important to note that the cell lines we characterized were derived from rapidly proliferating, radiation-resistant, highly EGFR expressing tumors, and the signaling cascade elucidated may not be characteristic of less aggressive SCCHN cancers.
Previous work from our laboratory showed that EGF-stimulated PKCζ regulates SCCHN DNA synthesis by contributing to Raf-1/MAPK activation (2). Numerous studies have shown that other PKC isoforms such as PKCα also stimulate Raf-1 activation. Here we demonstrate a complementary mechanism for the oncogenic effects of PKCα, namely, stimulation of MAPK together with inhibition of miR-15a, leading to upregulation of cyclin E synthesis. However, unlike PKCζ PKCα is activated independently of the EGF receptor (data not shown). The mechanism leading to constitutive activation and overexpression in tumors remains to be determined.
Our results highlighted cyclin E as the key target of PKCα regulation. Cyclin E was able to substantially rescue the inhibition of DNA synthesis upon PKCα kinase inactivation in our cells. In addition to its role in complex with cdks, cyclin E was recently found to be required for loading the MCM replicative helicase onto replicative origins (replicative licensing) and for transformation by Ras in a manner that is independent of cdks (43). Thus, cyclin E performs dual functions that are both cdk-dependent and cdk-independent, and loss of cyclin E should rapidly prevent DNA synthesis.
Cyclin E overexpression or deregulation has been associated with a number of highly aggressive tumors with poor prognosis but the mechanisms that have been described differ from the one elucidated here (reviewed in (44)). In laryngeal squamous cell carcinomas (LSCC), cyclin E overexpression alone was a prognostic marker for early stage LSCC, and tumors with a combination of high cyclin E and PCNA expression yielded the poorest prognoses (45, 46). Cyclin E overexpression in tumors has been attributed to a number of mechanisms including loss or mutation of ubiquitin ligases, decreasing the rate of degradation, cyclin E gene amplification, and mutation of oncogenes upstream of the cyclin D-Rb-E2F pathway that normally regulate cell cycle progression (44). Our results contribute an additional mechanism for cyclin E overexpression involving regulation of synthesis by PKCα via mir-15a.
This study is the first demonstration that PKCα is a mediator of SCCHN proliferation and a marker of progression and prognosis. Our results indicate that PKCα is a primary driver of the cancer phenotype that promotes SCCHN development and thus represents a novel therapeutic target. These results carry important clinical implications by providing a rationale for the advancement of PKC inhibitors that are currently being developed or investigated through clinical trials. Taken together, this evidence suggests that PKCα inhibition will yield efficacy in a variety of cancers.
Supplementary Material
Acknowledgments
The authors thank Philippe Cluzel and Uri Alon for stimulating discussions, and Xinmin Li, Michael Kubal, Carolyn Pierce, Jing Liu, John Mote and Shawn Levy for technical assistance.
Financial Support: This project was funded by NIH (RO1-CA109278-01 (MRR), P50 DE11921-0551 (EEWC), DE12322 (MWL), DE00470 (MWL) and RO1-DE017982-01 (CHC)); American Society of Clinical Oncology (EEWC); Francis L. Lederer Foundation (EEWC); Damon-Runyon Cancer Research Foundation (CI-28-05 (CHC)); and Cornelius Crane Trust for Eczema Research (MRR).
REFERENCES
- 1.Dempsey EC, Newton AC, Mochly-Rosen D, et al. Protein kinase C isozymes and the regulation of diverse cell responses. Am J Physiol Lung Cell Mol Physiol. 2000;279:L429–38. doi: 10.1152/ajplung.2000.279.3.L429. [DOI] [PubMed] [Google Scholar]
- 2.Cohen EE, Lingen MW, Zhu B, et al. Protein kinase C zeta mediates epidermal growth factor-induced growth of head and neck tumor cells by regulating mitogen-activated protein kinase. Cancer Res. 2006;66:6296–303. doi: 10.1158/0008-5472.CAN-05-3139. [DOI] [PubMed] [Google Scholar]
- 3.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7:281–94. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 4.Wu TT, Hsieh YH, Wu CC, Hsieh YS, Huang CY, Liu JY. Overexpression of protein kinase C alpha mRNA in human hepatocellular carcinoma: a potential marker of disease prognosis. Clin Chim Acta. 2007;382:54–8. doi: 10.1016/j.cca.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Dlugosz AA, Cheng C, Williams EK, Dharia AG, Denning MF, Yuspa SH. Alterations in murine keratinocyte differentiation induced by activated rasHa genes are mediated by protein kinase C-alpha. Cancer Res. 1994;54:6413–20. [PubMed] [Google Scholar]
- 6.Chmura SJ, Dolan ME, Cha A, Mauceri HJ, Kufe DW, Weichselbaum RR. In vitro and in vivo activity of protein kinase C inhibitor chelerythrine chloride induces tumor cell toxicity and growth delay in vivo. Clin Cancer Res. 2000;6:737–42. [PubMed] [Google Scholar]
- 7.Sclafani RA, Holzen TM. Cell Cycle Regulation of DNA Replication. Annu Rev Genet. 2007 doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–60. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 10.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linsley PS, Schelter J, Burchard J, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 13.Sylvestre Y, De Guire V, Querido E, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 14.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 15.Slebos RJ, Yi Y, Ely K, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–9. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 16.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung CH, Parker JS, Ely K, et al. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-kappaB signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res. 2006;66:8210–8. doi: 10.1158/0008-5472.CAN-06-1213. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro BA, Ray S, Jung E, Allred WT, Bollag WB. Putative conventional protein kinase C inhibitor Godecke 6976 [12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)py rrolo(3,4-c)-carbazole] stimulates transglutaminase activity in primary mouse epidermal keratinocytes. J Pharmacol Exp Ther. 2002;302:352–8. doi: 10.1124/jpet.302.1.352. [DOI] [PubMed] [Google Scholar]
- 19.Cai H, Smola U, Wixler V, et al. Role of diacylglycerol-regulated protein kinase C isotypes in growth factor activation of the Raf-1 protein kinase. Mol Cell Biol. 1997;17:732–41. doi: 10.1128/mcb.17.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berra E, Diaz-Meco MT, Lozano J, et al. Evidence for a role of MEK and MAPK during signal transduction by protein kinase C zeta. EMBO J. 1995;14:6157–63. doi: 10.1002/j.1460-2075.1995.tb00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YY, Wang L, Lu CD. An E2F site in the 5′-promoter region contributes to serum-dependent up-regulation of the human proliferating cell nuclear antigen gene. FEBS Lett. 2003;544:112–8. doi: 10.1016/s0014-5793(03)00485-x. [DOI] [PubMed] [Google Scholar]
- 23.Noya F, Chien WM, Wu X, et al. The promoter of the human proliferating cell nuclear antigen gene is not sufficient for cell cycle-dependent regulation in organotypic cultures of keratinocytes. J Biol Chem. 2002;277:17271–80. doi: 10.1074/jbc.M112441200. [DOI] [PubMed] [Google Scholar]
- 24.Ohtani K, Iwanaga R, Nakamura M, et al. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene. 1999;18:2299–309. doi: 10.1038/sj.onc.1202544. [DOI] [PubMed] [Google Scholar]
- 25.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Paramio P, Cabrerizo Y, Bornancin F, Parker PJ. The broad specificity of dominant inhibitory protein kinase C mutants infers a common step in phosphorylation. Biochem J. 1998;333:631–6. doi: 10.1042/bj3330631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirai T, Chida K. Protein kinase Czeta (PKCzeta): activation mechanisms and cellular functions. J Biochem (Tokyo) 2003;133:1–7. doi: 10.1093/jb/mvg017. [DOI] [PubMed] [Google Scholar]
- 28.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 29.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005;204:280–5. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- 32.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 33.Goekjian PG, Jirousek MR. Protein kinase C inhibitors as novel anticancer drugs. Expert Opin Investig Drugs. 2001;10:2117–40. doi: 10.1517/13543784.10.12.2117. [DOI] [PubMed] [Google Scholar]
- 34.Bokhari SM, Zhou L, Karasek MA, Paturi SG, Chaudhuri V. Regulation of skin microvasculature angiogenesis, cell migration, and permeability by a specific inhibitor of PKCalpha. J Invest Dermatol. 2006;126:460–7. doi: 10.1038/sj.jid.5700071. [DOI] [PubMed] [Google Scholar]
- 35.Cataisson C, Pearson AJ, Torgerson S, Nedospasov SA, Yuspa SH. Protein kinase C alpha-mediated chemotaxis of neutrophils requires NF-kappa B activity but is independent of TNF alpha signaling in mouse skin in vivo. J Immunol. 2005;174:1686–92. doi: 10.4049/jimmunol.174.3.1686. [DOI] [PubMed] [Google Scholar]
- 36.Suh KS, Tatunchak TT, Crutchley JM, Edwards LE, Marin KG, Yuspa SH. Genomic structure and promoter analysis of PKC-delta. Genomics. 2003;82:57–67. doi: 10.1016/s0888-7543(03)00072-7. [DOI] [PubMed] [Google Scholar]
- 37.Sun Z, Arendt CW, Ellmeier W, et al. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–7. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 38.Nakaigawa N, Hirai S, Mizuno K, Shuin T, Hosaka M, Ohno S. Differential effects of overexpression of PKC alpha and PKC delta/epsilon on cellular E2F activity in late G1 phase. Biochem Biophys Res Commun. 1996;222:95–100. doi: 10.1006/bbrc.1996.0703. [DOI] [PubMed] [Google Scholar]
- 39.Soh JW, Weinstein IB. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J Biol Chem. 2003;278:34709–16. doi: 10.1074/jbc.M302016200. [DOI] [PubMed] [Google Scholar]
- 40.Lee YS, Dlugosz AA, McKay R, Dean NM, Yuspa SH. Definition by specific antisense oligonucleotides of a role for protein kinase C alpha in expression of differentiation markers in normal and neoplastic mouse epidermal keratinocytes. Mol Carcinog. 1997;18:44–53. [PubMed] [Google Scholar]
- 41.Tibudan SS, Wang Y, Denning MF. Activation of protein kinase C triggers irreversible cell cycle withdrawal in human keratinocytes. J Invest Dermatol. 2002;119:1282–9. doi: 10.1046/j.1523-1747.2002.19625.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang LC, Ng DC, Bikle DD. Role of protein kinase C alpha in calcium induced keratinocyte differentiation: defective regulation in squamous cell carcinoma. Journal of cellular physiology. 2003;195:249–59. doi: 10.1002/jcp.10248. [DOI] [PubMed] [Google Scholar]
- 43.Geng Y, Lee YM, Welcker M, et al. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–39. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 44.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 45.Anton RC, Coffey DM, Gondo MM, Stephenson MA, Brown RW, Cagle PT. The expression of cyclins D1 and E in predicting short-term survival in squamous cell carcinoma of the lung. Mod Pathol. 2000;13:1167–72. doi: 10.1038/modpathol.3880215. [DOI] [PubMed] [Google Scholar]
- 46.Dong Y, Sui L, Tai Y, Sugimoto K, Hirao T, Tokuda M. Prognostic significance of cyclin E overexpression in laryngeal squamous cell carcinomas. Clin Cancer Res. 2000;6:4253–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.