Abstract
Crystallization has proven to be the most significant bottleneck to high-throughput protein structure determination using diffraction methods. We have used the large-scale, systematically generated experimental results of the Northeast Structural Genomics Consortium to characterize the biophysical properties that control protein crystallization. Datamining of crystallization results combined with explicit folding studies lead to the conclusion that crystallization propensity is controlled primarily by the prevalence of well-ordered surface epitopes capable of mediating interprotein interactions and is not strongly influenced by overall thermodynamic stability. These analyses identify specific sequence features correlating with crystallization propensity that can be used to estimate the crystallization probability of a given construct. Analyses of entire predicted proteomes demonstrate substantial differences in the bulk amino acid sequence properties of human versus eubacterial proteins that reflect likely differences in their biophysical properties including crystallization propensity. Finally, our thermodynamic measurements enable critical evaluation of previous claims regarding correlations between protein stability and bulk sequence properties, which generally are not supported by our dataset.
Keywords: protein crystallization, protein thermodynamics, crystallization mechanism, surface entropy, datamining, structural genomics
The ability to determine the atomic structures of macromolecules represents a great achievement in molecular biology because of the unparalleled value of this information in understanding the fundamental chemistry of life1–5. While nuclear magnetic resonance represents an invaluable source of structural information, especially for small proteins, most macromolecular structures are determined using x-ray crystallography. Capitalizing on the recent proliferation of genomic sequence data, “structural genomics” consortia have been organized worldwide to develop methods and infrastructure for high-throughput protein structure determination. These groups have contributed to improvements in expression and structure determination methods6, and the four largest U.S. consortia accounted for 45% of all novel structures deposited in the Protein Data Bank (PDB) in 20077. While these efforts contribute to the impressive progress of the structural biology community in characterizing the full repertoire of protein structures, the rate of growth in sequence information nonetheless far out-paces that of structural information. Given the ongoing acceleration of whole-genome sequencing, the gap between the two will continue to expand without a breakthrough in macromolecular structure determination methods.
The systematic efforts of structural genomics projects show that crystallization is the major bottleneck to protein structure determination using diffraction methods. Analyses of results scored by visual inspection of crystallization reactions have shown that only ~35% of purified proteins form objects resembling crystals7. However, a substantially smaller proportion form crystals of sufficient quality for structure determination. Among structural genomics consortia, ~12% of protein preparations yield crystal structures, with exact frequency varying based on protein source and the quality control imposed during sample production7. However, even limiting consideration to biochemically well-behaved proteins, the vast majority of constructs do not yield crystals of sufficient quality for structure determination. This unfortunate fact about naturally evolved proteins poses a severe technical obstacle in myriad projects. Drug-discovery efforts are particularly handicapped by a lack of structural data because it prevents use of structure-based ligand optimization methods8, 9. The lack of understanding and control of the protein crystallization process represents a fundamental problem impacting many areas of biomedical research.
To address this problem, we investigated the physical properties that control protein crystallization. Mechanistic hypotheses were evaluated using materials and results from the Northeast Structural Genomics Consortium (NESG). The availability of hundreds of biochemically well-behaved proteins produced and evaluated using consistent methodology should reduce the influence of sporadic factors affecting crystallization outcome and allow sensitive detection of causally related properties. Experimental results and sequence parameters were correlated with whether a protein yielded a crystal of sufficient quality for determination of its atomic structure (i.e., a PDB deposition), rather than the more diffuse criterion of forming an object visually identified as a crystal.
To form a high quality crystal, a protein must be immobilized in a lattice in a consistent conformation with limited dynamic motion. Thus, thermodynamic stability could play an important role in determining crystallization behavior. We evaluated this possibility using large-scale experimental studies of protein-folding equilibria. Protein surface properties could also play a determining role in controlling crystallization behavior because formation of tight, geometrically precise inter-molecular contacts is required for lattice stability. While this premise seems conceptually obvious10, 11, limited information was available on the specific features involved and whether primary sequence analysis could detect them.
Previous research suggests that conformationally dynamic amino acid sidechains inhibit crystallization based on the entropic cost of immobilizing them in stable interprotein contacts. The “surface entropy reduction” method replaces high sidechain entropy, surface-exposed lysine, glutamate, and glutamine residues with lower entropy residues, especially alanine12–17. Such substitutions have promoted improved crystallization of some proteins, although a significant fraction of these mutant proteins have solubility problems that can hinder crystallization screening14, 17. Several papers have also reported significant correlations between bulk sequence properties and protein crystallization propensity18–22. These studies have demonstrated that low average hydrophobicity18, 21, high isoelectric point18, 21, and long stretches of backbone disorder18, 19 reduce crystallization probability. However, these studies have not addressed the mechanistic origin of observed correlations. Other than the protein engineering results described above, few available data relate protein surface properties to crystallization propensity.
The large-scale experimental and datamining studies presented in this paper provide new insight into the physical properties controlling protein crystallization. We present evidence that the prevalence of low entropy, well-ordered surface features is the principal mechanistic determinant of protein crystallization behavior, and our analyses suggest approaches to rational mutational re-engineering of proteins to improve crystallization propensity. Our data also permit evaluation of relationships between various biophysical and sequence properties of proteins, allowing critical evaluation of trends previously proposed but not rigorously tested.
Results
Does protein stability influence crystallization?
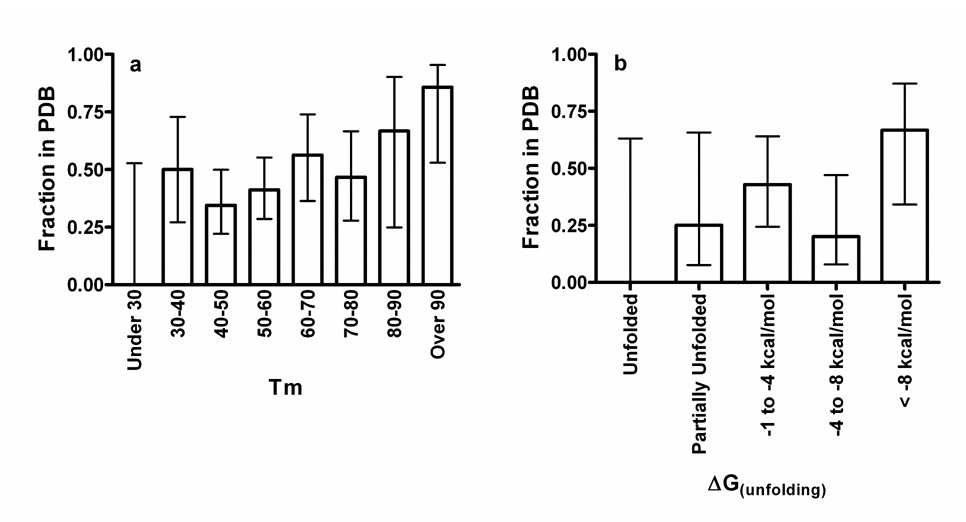
We used several biophysical techniques to evaluate the relationship between protein stability and crystallization outcome. Thermal denaturation experiments were carried out on 117 monodisperse proteins that had gone through the NESG crystallization pipeline. In these experiments, melting temperature (Tm) was determined via the fluorescence of the dye SYPRO Orange, which is enhanced upon partitioning into the hydrophobic regions of denatured proteins23 (Fig. 1a). Chemical denaturation of 36 proteins by guanidinium hydrochloride was monitored using circular dichroism spectroscopy (Fig. 1b). Finally, 121 proteins were subjected to calibrated limited proteolysis using trypsin and proteinase K (Supplementary Fig. 1). Results from these three assays are consistent (Supplementary Fig. 2 & Supplementary Table 1) and show no significant relationship between overall stability and successful crystal structure determination if unfolded and hyperstable proteins are omitted (Fig. 1 & Supplementary Fig. 1). Thermal denaturation data (Fig. 1a) show a statistically significant correlation if all proteins are included in the analysis (p=0.008), but the significance is lost if proteins with Tm’s under 30°C or over 90°C are excluded (p=0.4). Therefore, partially or fully unfolded proteins may yield crystal structures less frequently, while hyperstable proteins probably yield crystal structures somewhat more frequently. However, overall thermodynamic stability is not a major determinant of crystallization propensity and may not have any influence across the broad range of stabilities typical of folded mesophilic proteins (See Supplementary Notes for further discussion).
Figure 1. Protein stability does not strongly influence success in crystal structure solution.
Denaturation experiments were performed on biochemically well-behaved NESG proteins drawn from the set of 679 used in the datamining studies reported below (as described in detail in the Supplementary Methods). The graphs show the fraction of proteins for which crystal structures were successfully determined and deposited in PDB in each stability bin. The error bars represent 95% confidence limits calculated from counting statistics using the numbers in each bin. (a) Crystallization success vs. thermal denaturation midpoint temperature (Tm) determined via fluorescence enhancement of the hydrophobic reporter dye SYPRO Orange23. Logistic regression analyses of the probability of the observed relationships occurring at random indicate P = 0.0076 for all proteins (N = 117), P = 0.19 for those with Tm ≤ 90°C (N = 110), P = 0.020 for those with Tm ≥ 30°C (N = 114), and P = 0.40 for those with Tm ‘s between 30°C and 90°C (N = 107). (b) Crystallization success vs. ΔG of unfolding from guanidinium hydrochloride denaturation experiments monitored by circular dichrosism spectroscopy (P = 0.2, N=36 for all proteins).
A detailed discussion of the large-scale proteolysis results is presented in the Supplementary Notes. Most saliently, the size of the dominant protected fragment in proteolysis studies, likely a measure of the total content of disordered loops, significantly positively correlates with crystallization success (Supplementary Fig. 1). Our large-scale experimental analyses also allow evaluation of potential correlations between thermodynamic and sequence properties of biochemically well-behaved proteins (see Supplementary Notes; Supplementary Fig. 2; Supplementary Fig. 3; Supplementary Table 1). Most notably, our data on primarily bacterial proteins do not support Uversky’s conclusion24 that specific combinations of hydrophobicity and net charge reliably identify natively unfolded proteins. Finally, our data show insignificant correlation between fluorescence enhancement of the hydrophobic reporter dye bis-ANS and either the folding state or crystallization propensity of a set of NESG proteins (see Supplementary Notes; Supplementary Fig. 4).
Influence of other biophysical properties
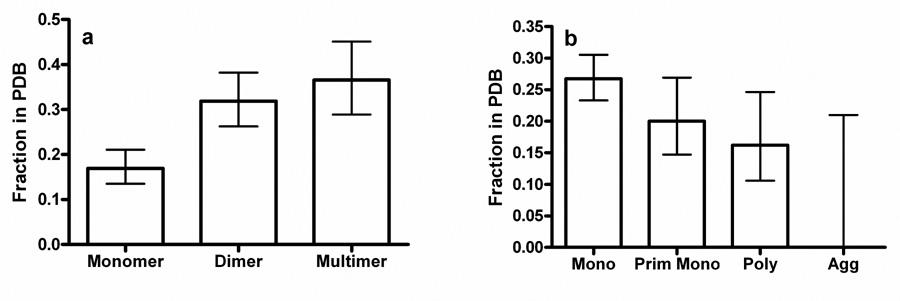
The hydrodynamic properties of all NESG proteins are characterized using analytical gel filtration chromatography monitored by static light scattering and refractive index detectors. These data, acquired from a flash-frozen aliquot of the crystallization stock, provide a rigorous description of the distribution of oligomeric species in each sample. Theoretical considerations suggest that protein oligomers should crystallize more readily than monomers because a single high quality packing epitope on the surface of one protomer can make repeated contacts, which is especially beneficial in a lattice sharing the symmetry of the oligomer25. This inference has been supported by a study where disulfide-crosslinking was used to produce non-native dimers, which crystallized more avidly than the corresponding monomer26. Statistical analysis of NESG large-scale crystallization results proves this inference, showing that monomers yield solvable crystals at a significantly lower rate than dimers or larger oligomers (Fig. 2a).
Figure 2. Hydrodynamic properties strongly influence success in crystal structure solution.
The graphs show the fraction of proteins for which crystal structures were successfully determined and deposited in PDB, with the error bars representing 95% confidence limits calculated from counting statistics using the numbers in each bin. (a) Crystallization success vs. oligomeric state (P = 0.0005 for monomers compared to dimers, P = 0.0002 for monomers compared to larger multimers, and P = 0.000007 for monomers compared to all oligomers). (b) Crystallization success vs. aggregation status (P = 0.01 for monodisperse compared to all other classes and P = 0.03 for at least predominantly monodisperse compared to polydisperse and aggregated).
The hydrodynamic homogeneity of the stock significantly correlates with successful crystal structure determination (Fig. 2b). Monodisperse proteins (≥90% in a single hydrodynamic species) more frequently yield crystal structures than predominantly monodisperse or polydisperse proteins (70–90% or <70%, respectively). The few aggregated proteins that entered the NESG pipeline during the period analyzed here failed to yield structures (Fig. 2b). Preliminary analysis of results from a substantially larger dataset containing over 100 aggregated proteins shows an equivalent trend (data not shown). Therefore, while formation of specific, homogeneous oligomers promotes successful crystallization (Fig. 2a), heterogeneous self-association inhibits successful crystallization (Fig. 2b).
Finally, the number of unvalidated crystallization hits observed in a 1536-well robotic high-throughput microbatch screen27 correlates strongly with crystal structure solution (Supplementary Fig. 5). We hypothesize that proteins crystallizing more promiscuously possess more potential interprotein interaction sites on their surfaces, or more strongly interacting sites, and that at least one of these properties is a significant determinant of the ability to form a high quality crystal lattice under some condition. The fact that structures were determined for only ~6% of proteins failing to give a hit in initial high-throughput screening, in spite of performing ≥500 additional vapor diffusion reactions, demonstrates that crystallization propensity is an intrinsic protein property even though it can be influenced by solution contents (see Supplementary Notes).
Datamining historical crystallization results
We examined the relationship between bulk sequence properties of NESG proteins and success in depositing their crystal structures into the PDB. The dataset comprised 679 strongly expressed and well-behaved proteins, predominantly but not entirely from bacterial organisms, of which 157 of yielded crystal structures. These proteins were taken through the entire NESG pipeline6 including quality-control assays and crystallization screening. Proteins with predicted transmembrane α-helices or >20% low complexity sequence were excluded from the pipeline. The dataset included just one construct for each protein target and excluded proteins identified by static light-scattering as aggregated in their crystallization stocks. Samples yielding crystals of insufficient quality for structure determination were considered failures even if diffraction was observed, as was true for 39 of the 679. Analyses presented in Supplementary Figure 20 retrospectively justify this strategy by showing that some key sequence features in these 39 proteins are more similar to proteins not yielding diffracting crystals than proteins yielding crystal structures. Target selection and classification are described in Supplementary Methods.
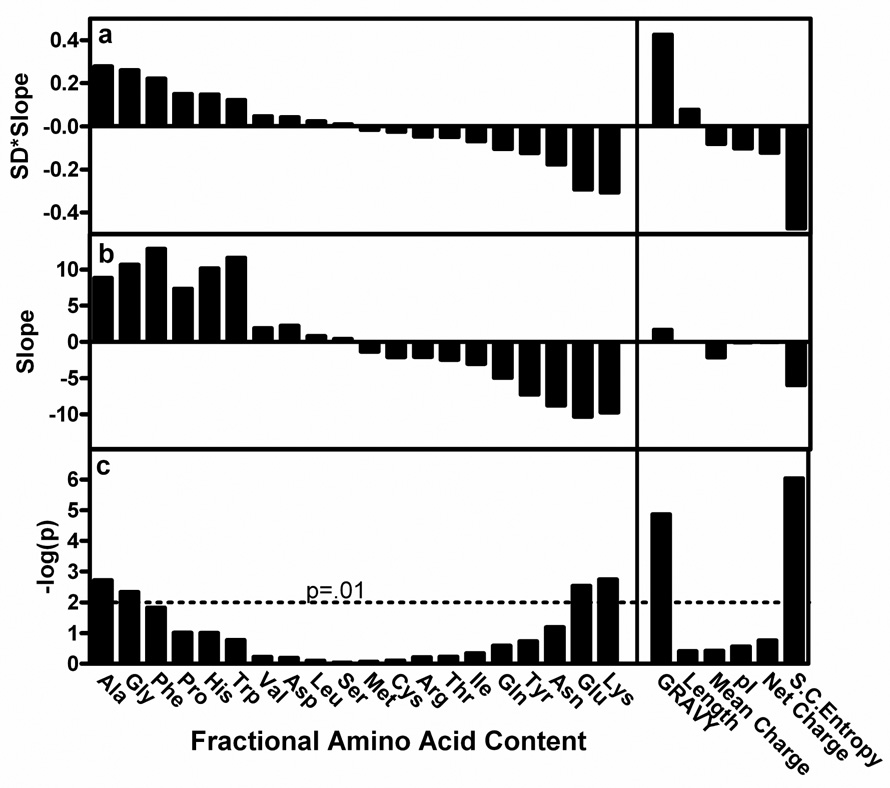
Sequence properties that were analyzed include the frequency of each amino acid, mean hydrophobicity (GRAVY28 – GRand AVerage of hydropathY), mean sidechain entropy33 (<SCE>), total and net electrostatic charge, isoelectric point (pI), the fraction of residues predicted disordered by DISOPRED229, and chain length. Logistic regressions, explained briefly in the Supplementary Notes, were performed to evaluate the dependence between the continuous variable representing the bulk sequence parameter and the binary outcome of the crystallization effort, i.e., success or failure in depositing the construct’s crystal structure into the PDB. The p-value, regression slope, and predictive value of each variable are presented in Fig. 3.
Figure 3. Correlations between sequence characteristics and success in crystal structure solution.
Logistic regressions based on success in crystal structure determination (i.e., PDB deposition) were performed on a dataset comprising 679 proteins from the NESG protein expression and crystallization pipeline. Variables evaluated included the fractional content of each amino acid, mean residue hydrophobicity (GRAVY28 – GRand AVerage of hydropathY), chain length, mean charge (fraction arg+lys+asp+glu), pI, mean net charge, and mean sidechain entropy (<SCE>). (a) Predictive value of each parameter, which is defined as the product of its logistic regression slope and the standard deviation of its distribution in the dataset. (b) Logistic regression slope. (c) Negative log of logistic regression p-value.
The frequencies of five amino acids significantly correlate with successful crystal structure determination, as do several more complex sequence metrics. Ala, gly, and phe frequency positively correlate with successful structure determination, while glu and lys frequency negatively correlate (Fig. 4 and Supplementary Fig. 8). GRAVY positively correlates, while <SCE> and fraction of predicted disordered residues negatively correlate (Fig. 4), as do fractional positive (arg + lys) or negative (asp + glu) charge (Supplementary Fig. 11; Supplementary Table 3). Fractional values (i.e., normalized to chain length) are uniformly more predictive than total values for significantly correlated parameters (Supplementary Fig. 7). While fractional positive or negative charge considered independently both significantly oppose successful crystallization, neither net charge (positive minus negative residue counts) nor its absolute value are predictive, nor are any electrostatic variables without length normalization (Supplementary Table 3; Supplementary Fig. 11). Note that the current analysis has limited sensitivity in detecting the influence of rare or weakly predictive amino acids, due to the size of our dataset (see Supplementary Notes).
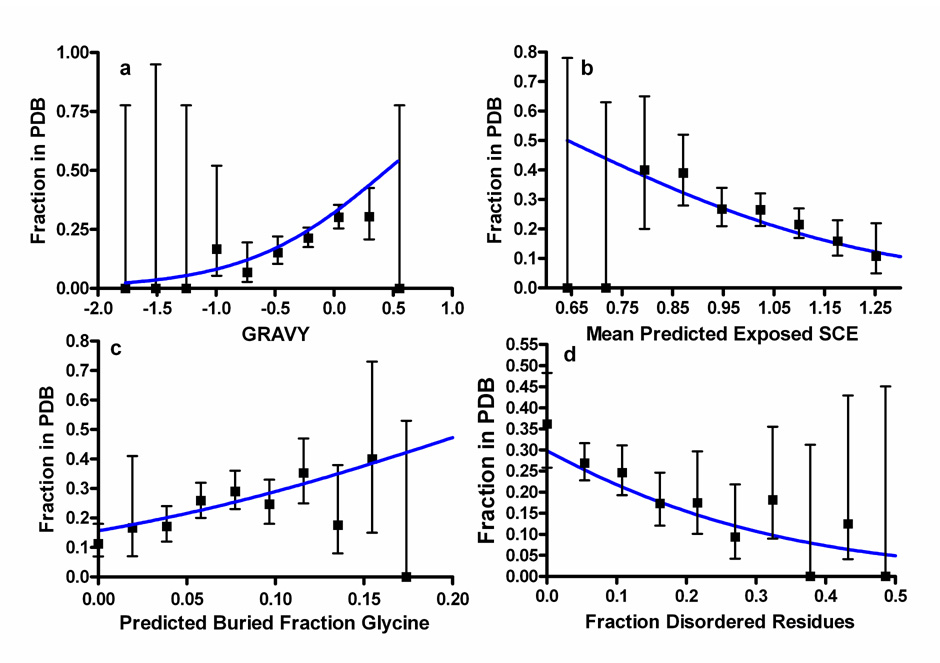
Figure 4. Four major predictors of success in crystal structure solution.
Graphs show the fraction of 679 NESG pipeline proteins for which crystal structures were successfully determined and deposited in PDB. Error bars represent 95% confidence limits calculated from counting statistics using the numbers in each bin, while gray lines show the functional forms of the optimized logistic regression equations. (a) GRAVY28 or mean residue hydrophobicity, a positive predictor (P = 0.0000135). (b) <SCE>33 of PHD/PROF31 predicted exposed residues, a negative predictor (P = 0.0001). (c) Fractional gly content of PHD/PROF31 predicted buried residues, a positive predictor (P = 0.0005). (d) Fraction of residues predicted to be disordered by DISOPRED229, a negative predictor (P = 0.0003).
Protein pI (Supplementary Fig. 9) and chain length (Supplementary Fig. 10) both show bimodal effects, with the rate of success initially increasing and later decreasing with increasing parameter values. The bimodal dependence prevents assessment of statistical significance using logistic regression, and the 95% confidence limits of all parameter bins substantially overlap in our dataset. However, similar dependencies have been reported in independent datasets18–22 and can be observed in analyses of whole genome distributions (see below), suggesting that these parameters influence crystallization outcome but not strongly enough to be significant in our dataset.
Predicted backbone disorder inhibits crystallization
We further analyzed the effect of backbone disorder in opposing successful crystallization (Fig. 4) by performing multiple logistical regression on the fraction of predicted disordered residues29 located in continuous segments at either the N- or C-terminus of the protein or at internal positions (summed together). The regression slopes in Table 1B demonstrate that predicted disorder has the same size effect opposing crystallization irrespective of location within the protein chain.
Table 1.
Single and multiple logistic regression results1.
| Regression Variable | Slope | SD*Slope | P value | |
|---|---|---|---|---|
| GRAVY | 1.68 | 0.43 | 0.0000135 | |
| <SCE> | −5.99 | −0.47 | 0.000000915 | |
| Gly | 10.685 | 0.26 | 0.0046 | |
| Pb <SCE> | −1.15 | −0.072 | 0.431 | |
| A | Pe <SCE> | −3.24 | −0.33 | 0.0001 |
| Pb GRAVY | −0.413 | −0.25 | 0.0085 | |
| Pe GRAVY | 0.744 | 0.26 | 0.0044 | |
| Pb Gly | 9.16 | 0.32 | 0.0005 | |
| Pe Gly | 3.03 | 0.089 | 0.08 | |
| DISOPRED2 | −4.21 | −0.46 | 0.0003 | |
| N/C/Internal DISOPRED2 | 0.0009 | |||
| B | N-terminal DISOPRED2 | −4.37 | −0.18 | 0.15 |
| C-terminal DISOPRED2 | −4.14 | −0.21 | 0.093 | |
| Internal DISOPRED2 | −4.19 | −0.31 | 0.013 | |
| <SCE> + GRAVY | 0.00000181 | |||
| C | <SCE> | −4.52 | −0.36 | 0.0066 |
| GRAVY | 0.73 | 0.19 | 0.16 | |
| Pb/e SCE + Amino Acids | .000085 | |||
| Pb <SCE> | 2.68 | 0.17 | 0.20 | |
| Pe <SCE> | −3.66 | −0.40 | 0.038 | |
| Pb Ala | 3.68 | 0.17 | .015 | |
| Pe Ala | 2.89 | 0.11 | .036 | |
| Pb Glu | 3.86 | 0.037 | 0.72 | |
| D | Pe Glu | −.0186 | −0.00080 | 0.99 |
| Pb Gly | 12.1 | 0.42 | 0.00020 | |
| Pe Gly | −1.51 | −0.045 | 0.71 | |
| Pb Lys | −14.7 | −0.10 | 0.38 | |
| Pe Lys | 2.0 | 0.096 | 0.50 | |
| Pb Phe | 3.71 | 0.17 | 0.10 | |
| Pe Phe | 19.5 | 0.22 | 0.02 | |
Logistic regressions evaluating success in depositing a crystal structure in the PDB as a function of various bulk sequence parameters were performed on a set of 679 biochemically well-behaved NESG proteins (157 of which yielded structures). For multiple logistic regressions, parameters for the individual component variables are shown in plain type while the overall results are shown in bold type. All p-values below 0.05 are shown in bold type. The “predictive value” is the product of the regression slope for each variable times the standard deviation of its distribution in the dataset. GRAVY represents the GRand AVerage of hydropathY28, <SCE> mean Monte Carlo sidechain entropy33, and DISOPRED229 the fraction of predicted backbone disorder predicted by this program (at a false positive rate of 5%). Pb and Pe refer to the fraction of residues predicted to be buried or surface-exposed, respectively, by the PHD/PROF algorithm as implemented by the PredictProtein server31. N-terminal and C-terminal refer to continuous stretches of amino acids at the ends of the protein, while internal refers to all internal positions combined together.
Surface entropy effects dominate other effects
GRAVY and <SCE>, which both strongly influence crystallization outcome (Fig. 3–Fig. 4; Table 1A), are anti-correlated (Supplementary Table 3). Analyzing them simultaneously in a double logistic regression demonstrates that their influence is redundant: GRAVY shows weak and insignificant additional correlation with crystallization outcome when considered simultaneously with <SCE> (Table 1C). The statistical dominance of <SCE> over GRAVY indicates that it is more strongly correlated and therefore likely to be the mechanistically significant parameter. This evidence supports Derewenda and Vekilov’s hypothesis that the thermodynamic cost of immobilizing high entropy sidechains tends to inhibit their participation in crystal-packing contacts15, 16, 30. As their prevalence on the protein surface increases and fewer well-ordered sites are available for packing, it increases the probability that thermodynamically unfavorable immobilization of high SCE residues will be needed to form a stable contact15, 16. The comparatively weak anti-correlation of some high SCE residues (e.g., arg in Fig. 3) may be attributable to an energetically favorable interaction tendency of the sidechain’s functional group partially offsetting the entropic cost of immobilizing it. (See Supplementary Notes)
This mechanistic hypothesis implies that residues influencing crystallization should be localized on the protein surface. Therefore, we segregated the amino acids in each protein sequence by their predicted location (buried vs. surface-exposed) according to PHD/PROF31 (Table 1A). This analysis strongly supports the hypothesis, showing that successful crystallization correlates with <SCE> in predicted exposed residues (<SCE>pe) but not predicted buried residues. In contrast, the correlation with GRAVY is inconsistent in direction and substantially weaker when residues are segregated by predicted location. The direction of the GRAVY correlation in exposed residues (Table 1A) is consistent with it being a surrogate for <SCE> because higher hydrophobicity correlates with lower SCE (Supplementary Table 4). Furthermore, when sets of proteins are assembled with equivalent <SCE> distributions but systematic differences in GRAVY, they show no significant differences in crystal structure determination rates (Supplementary Fig. 12). These results all reinforce the conclusion that increasing exposed sidechain entropy, rather than decreasing hydrophobicity, impedes successful crystal structure determination by inhibiting formation of stable packing contacts.
Like GRAVY, other sequence properties observed to have a significant influence on crystallization outcome strongly correlate with sidechain entropy. Ala and gly have the lowest sidechain entropies and their frequencies correlate most strongly with successful crystallization, while lys and glu have among the highest sidechain entropies and their frequencies anti-correlate most strongly. Multiple logistic regression analyses suggest that all fractional charge effects (Supplementary Table 3) and all single amino acid effects are redundant with <SCE>pe except for the frequencies of predicted buried gly and exposed phe, which remain significant when considered simultaneously with <SCE>pe (Table 1D). Furthermore, in sets of proteins assembled to have equivalent <SCE>pe distributions but systematic differences in individual amino acid frequencies, higher fractional content of gly, ala, or phe significantly increases crystal structure determination rate while higher fractional content of lys, glu, or charged residues does not significantly alter outcome (Supplementary Fig. 12). These results suggest that effects of the latter parameters represent proxies for the mechanistically dominant effect of <SCE>, while gly, ala, and phe may have mechanistically independent positive effects. These residues may preferentially mediate crystal-packing contacts. The significant effect of fractional ala content in the analysis of sets of proteins with equivalent <SCE>pe distributions (Supplementary Fig. 12) but not in multiple logistic regression (Table 1D) is likely attributable to the fact that its influence on crystallization rate does not match the logistic functional form as well as those of gly or phe (Supplementary Fig. 8).
The “buried” glycine effect
While the glycines that promote crystallization are predicted by PHD/PROF to be buried, the same program also predicts them to be preferentially localized in loops 6–15 residues in length. The Supplementary Notes present detailed analyses of this ostensibly inconsistent categorization. In brief, manual inspection of crystal structures suggests that the predicted “buried” loop category is dominated by well-ordered gly residues partially exposed on the surface of the protein, which may be favorable sites to form crystal-packing contacts.
PXS: probability of crystal structure determination
Having identified four non-redundant sequence features showing statistically significant correlation with crystallization success, we combined these into a single predictive metric (Supplementary Fig. 14):
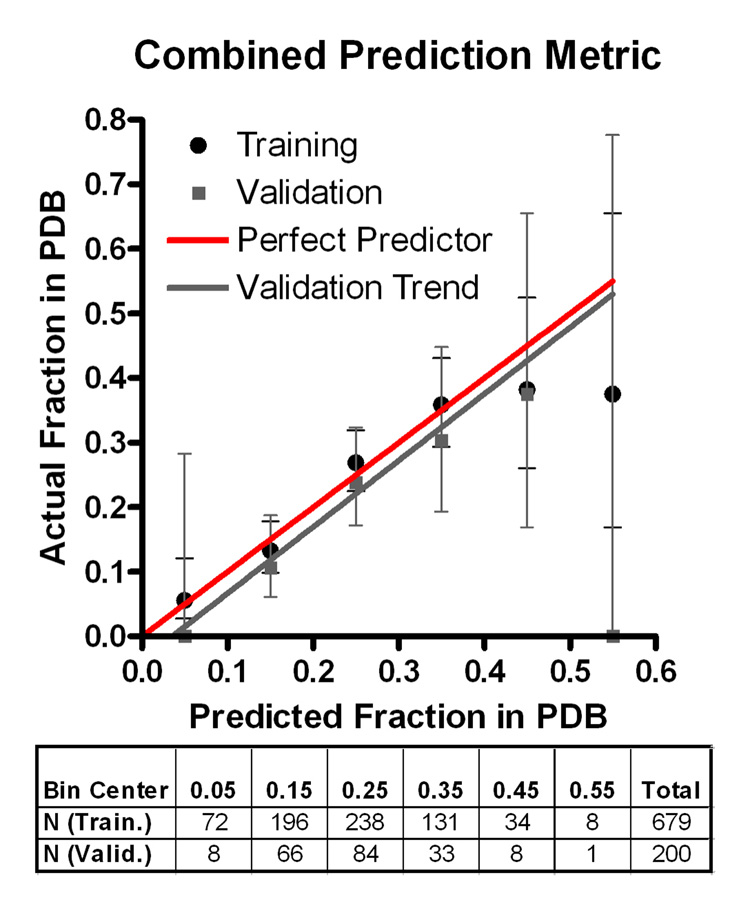
PXS represent the probability of solving a crystal (xtal) structure, Diso the fraction of residues predicted to be disordered by DISOPRED2, <SCE>pe the mean sidechain entropy of predicted exposed residues, Gpb the fraction of predicted buried gly, and F the fraction of total phe (used because this parameter predicts more strongly than exposed phe alone). This metric provides an accurate description of the behavior of the training dataset up to bins with ~35% success in depositing a crystal structure (P = 5.3×10−9) (Fig. 5). More importantly, it provides a similarly accurate description of a validation dataset comprising 200 proteins that passed through the NESG pipeline after metric development (P = 0.0014). A webserver performing this calculation is available at http://www.nesg.org/PXS/.
Figure 5. Performance of the PXS metric predicting probability of successful crystal structure determination.
Box and whisker plot shows fraction of NESG pipeline proteins in PDB with 95% confidence limits calculated from counting statistics using the numbers in each bin, binned as a function of PXS value. Black represents the 679 proteins used to develop/train the metric, while gray represents 200 proteins produced at later dates that were used for validation. The solid lines represent the functional form of the logistic regression equation describing PXS as applied to the sequences in training (dotted black) or validation (solid gray) sets. The table shows the number of proteins falling into each bin on the graph for the training and validation sets.
Genome-wide analyses of crystallization propensity
In addition to possible bias from inconsistent methods and effort being applied to different proteins, genome-wide crystallization results are systematically influenced by protein expression and solubility characteristics, factors intentionally excluded from the analyses reported above so as to isolate parameters influencing crystallization. As described in the Supplementary Notes, to begin characterizing the interplay of potentially conflicting factors influencing the successive steps required to go from gene to protein structure, and to explore the generality of PXS, we analyzed proteome-wide distributions of its value and the underlying sequence parameters. In brief, PXS is significantly predictive of the crystal structures obtained from the human and E. coli proteomes, as are all individual sequence parameters predictive in the NESG dataset, except for <SCE>pe (Supplementary Fig. 16, Supplementary Fig. 17). Sequence parameter distributions differ dramatically between E. coli and human proteins, which have on average more backbone disorder, lower GRAVY, and lower <SCE> (Supplementary Fig. 16). Human but not E. coli proteins have a very high prevalence of low SCE residues, especially gly and pro, in disordered sequences (Supplementary Fig. 18). The Supplementary Notes also present a metric developed to predict the conglated probability of expressing and determining the crystal structure of a human protein (PC-XS-Hs – Supplementary Fig. 19).
Discussion
Statistical analysis of our large-scale protein crystallization results demonstrates that the mean entropy of exposed sidechains and predicted backbone disorder both anti-correlate strongly and significantly with successful structure determination. Combining these results with the observation that stability is not a significant determinant of success leads to the conclusion that the dominant factor determining protein crystallization outcome is the prevalence of well-ordered surface epitopes capable of mediating stereochemically specific interprotein packing interactions. Beyond providing rigorous confirmation of longstanding suspicions12–16, our results provide a quantitative metric to assess crystallization propensity (PXS) and suggests possibilities for engineering protein sequences to improve outcome. While previously reported sequence correlations with crystallization propensity18–22 appear to be surrogates for surface entropy, the frequencies of gly, ala, and phe have statistically significant independent effects improving success. We hypothesize that these residues are particularly effective in mediating crystal-packing interactions, presumably via amide backbone interactions for gly and ala and hydrophobic interactions for phe. Our experimental studies demonstrate that heterogeneous self-association in a protein stock solution significantly reduces crystallization probability. Thus, successful crystallization requires minimal self-interaction in dilute aqueous buffers but strong self-interaction under the low water-activity conditions used to form a crystal, which is a non-physiological protein aggregate, albeit one with consistent intermolecular contacts and spatial organization. These requirements fundamentally tend to conflict. The charged residues lys and glu promote solubility but impede crystallization (Fig. 3), while low solubility and crystallization are both driven by low-affinity, non-physiological intermolecular interactions. Optimal crystallization eptitopes should mediate strong stereospecific interactions under low water-activity conditions without promoting promiscuous surface interaction in dilute aqueous buffers. Well-ordered, surface-exposed gly’s may be particularly efficacious in this regard compared to residues with stronger hydrophobic character, which tend to promote non-specific interactions. The Supplementary Notes present a more detailed discussion of related issues.
Materials and Methods
Protein crystallization
Protein expression, purification, and analysis methods are described in the SI. Initial high-throughput crystallization screening was conducted using the 1536 well microbatch robotic screen at the Hauptmann-Woodward Institute27. Proteins failing to yield rapidly progressing crystal leads were subjected to vapor diffusion screening, typically 250–300 conditions (Crystal Screens I & II, PEG-Ion, and Index screens from Hampton Research or equivalent screens from Qiagen) at both 4° C and 20° C, which was conducted in the presence of substrate or product compounds if commercially available. Crystal optimization, diffraction data collection at cryogenic temperatures, structure solution using single or multiple-wavelength anomalous diffraction techniques, and refinement were conducted using standard methods.
Datamining methods
Datamining analyses were conducted on native sequences with tags removed. Hydrodynamic data from the SPINE were manually verified. The frequency of each amino acid and the compound sequence metrics of charge, pI, GRAVY, SCE, length, and DISOPRED29 were individually evaluated for correlation with the binary outcome of success or failure in depositing a crystal structure of the target protein into the PDB. Charge parameters were calculated as signed or unsigned sums of the frequencies of appropriate combinations of arginine, lysine, glutamate, and aspartate residues. Isoelectric point was calculated using the EMBOSS algorithm32 at ExPASy33. GRAVY was calculated using the Kyte-Doolittle hydropathy parameters28. The Creamer scale33 was used for the SCE values of the individual amino acids34. DISOPRED scores were calculated using a locally installed copy of the DISOPRED229 program with a 5% false positive rate. Calculations of predicted burial/exposure and secondary structure were performed with the PHD/PROF algorithms from the PredictProtein server31, 35. Mean exposed SCE was calculated as the mean for all residues predicted to be exposed, while all calculations based on secondary structure class used total chain length as the denominator. In data graphs, the observed frequencies of successful PDB deposition in equally spaced parameter bins on the abscissa are plotted at the bin center, except for the terminal bin of unbounded variables which is plotted at the average parameter value in the bin.
Statistical analyses
Logistic regressions were performed in STATA (Statacorp, College Station, TX) with significance determined from Z-scores for individual variables and chi-squared distributions for models. The significance of oligomeric state, aggregation state, and the dividing line in the charge/hydrophobicity chart (Supplementary Fig. 4) were determined by evaluating contingency tables with a 2-tailed Fisher’s exact test. Counting-statistics-based 95% confidence intervals were calculated using Bayesian maximum likelihood estimates of the binomial distribution.
Supplementary Material
Acknowledgments
This work was supported by Protein Structure Initiative grants from the NIH to the Northeast Structural Genomics Consortium and the Center for High Throughput Structural Biology. The full staffs of these consortia contributed to the experimental data analyzed in this paper. WNP was supported in part by an NIH training grant to the Department of Biological Sciences at Columbia, while SKH was supported in part by an NSF grant to JFH. The authors thank Wayne Hendrickson and Liang Tong for support and advice and John Schwanoff and the New York Structural Biology Center for maintenance of the X4 beamlines at Brookhaven National Laboratory.
Literature References
- 1.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 3.Deisenhofer J, Michel H. The Photosynthetic Reaction Center from the Purple Bacterium Rhodopseudomonas viridis. Science. 1989;245:1463–1473. doi: 10.1126/science.245.4925.1463. [DOI] [PubMed] [Google Scholar]
- 4.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 5.Kendrew JC. Structure and function in myoglobin and other proteins. Fed Proc. 1959;18:740–751. [PubMed] [Google Scholar]
- 6.Acton TB, et al. Robotic cloning and Protein Production Platform of the Northeast Structural Genomics Consortium. Methods in enzymology. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Oughtred R, Berman HM, Westbrook J. Target DB: a target registration database for structural genomics projects. Bioinformatics. 2004;20:2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- 8.Blundell TL, Jhoti H, Abell C. High-throughput crystallography for lead discovery in drug design. Nat Rev Drug Discov. 2002;1:45–54. doi: 10.1038/nrd706. [DOI] [PubMed] [Google Scholar]
- 9.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta S, Iyer GH, Bryant SH, Lawrence CE, Bell JA. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 11.Janin J, Rodier F. Protein-protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- 12.Cooper DR, et al. Protein crystallization by surface entropy reduction: optimization of the SER strategy. Acta Crystallogr D Biol Crystallogr. 2007;63:636–645. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- 13.Derewenda ZS. The use of recombinant methods and molecular engineering in protein crystallization. Methods. 2004;34:354–363. doi: 10.1016/j.ymeth.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Derewenda ZS, Vekilov PG. Entropy and surface engineering in protein crystallization. Acta Crystallogr D Biol Crystallogr. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- 16.Longenecker KL, Garrard SM, Sheffield PJ, Derewenda ZS. Protein crystallization by rational mutagenesis of surface residues: Lys to Ala mutations promote crystallization of RhoGDI. Acta Crystallogr D Biol Crystallogr. 2001;57:679–688. doi: 10.1107/s0907444901003122. [DOI] [PubMed] [Google Scholar]
- 17.Mateja A, et al. The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution. Acta Crystallogr D Biol Crystallogr. 2002;58:1983–1991. doi: 10.1107/s090744490201394x. [DOI] [PubMed] [Google Scholar]
- 18.Canaves JM, Page R, Wilson IA, Stevens RC. Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: maximum clustering strategy for structural genomics. Journal of molecular biology. 2004;344:977–991. doi: 10.1016/j.jmb.2004.09.076. [DOI] [PubMed] [Google Scholar]
- 19.Oldfield CJ, Ulrich EL, Cheng Y, Dunker AK, Markley JL. Addressing the intrinsic disorder bottleneck in structural proteomics. Proteins. 2005;59:444–453. doi: 10.1002/prot.20446. [DOI] [PubMed] [Google Scholar]
- 20.Goh CS, et al. Mining the structural genomics pipeline: identification of protein properties that affect high-throughput experimental analysis. Journal of molecular biology. 2004;336:115–130. doi: 10.1016/j.jmb.2003.11.053. [DOI] [PubMed] [Google Scholar]
- 21.Overton IM, Barton GJ. A normalised scale for structural genomics target ranking: the OB-Score. FEBS Lett. 2006;580:4005–4009. doi: 10.1016/j.febslet.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Slabinski L, et al. The challenge of protein structure determination--lessons from structural genomics. Protein Sci. 2007;16:2472–2482. doi: 10.1110/ps.073037907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 24.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wukovitz SW, Yeates TO. Why protein crystals favour some space-groups over others. Nature structural biology. 1995;2:1062–1067. doi: 10.1038/nsb1295-1062. [DOI] [PubMed] [Google Scholar]
- 26.Banatao DR, et al. An approach to crystallizing proteins by synthetic symmetrization. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16230–16235. doi: 10.1073/pnas.0607674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cumbaa CA, et al. Automatic classification of sub-microlitre protein-crystallization trials in 1536-well plates. Acta crystallographica. 2003;59:1619–1627. doi: 10.1107/s0907444903015130. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of molecular biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 30.Vekilov PG. Solvent entropy effects in the formation of protein solid phases. Methods in enzymology. 2003;368:84–105. doi: 10.1016/S0076-6879(03)68006-7. [DOI] [PubMed] [Google Scholar]
- 31.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 33.Appel RD, Bairoch A, Hochstrasser DF. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends in biochemical sciences. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 34.Creamer TP. Side-chain conformational entropy in protein unfolded states. Proteins. 2000;40:443–450. doi: 10.1002/1097-0134(20000815)40:3<443::aid-prot100>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 35.Rost B. In: The Proteomics Protocols Handbook. Walker JE, editor. Totowa: Humana Press; 2005. pp. 875–901. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.