Summary
Kinetochores of mitotic chromosomes are coupled to spindle microtubules in ways that allow the energy from tubulin dynamics to drive chromosome motion. Most kinetochore-associated microtubule ends display curving “protofilaments,” strands of tubulin dimers that bend away from the microtubule axis. Both a kinetochore “plate” and an encircling, ring-shaped protein complex have been proposed to link protofilament bending to poleward chromosome motion. Here we show by electron tomography that slender fibrils connect curved protofilaments directly to the inner kinetochore. Fibril-protofilament associations correlate with a local straightening of the flared protofilaments. Theoretical analysis reveals that protofilament-fibril connections would be efficient couplers for chromosome motion, and experimental work on two very different kinetochore components suggests that filamentous proteins can couple shortening microtubules to cargo movements. These analyses define a ring-independent mechanism for harnessing microtubule dynamics directly to chromosome movement.
Introduction
Mitotic spindle microtubules (MTs) and their associated proteins (MAPs) interact with kinetochores to ensure the accurate segregation of eukaryotic chromosomes. The interface between MTs and a kinetochore provides a firm attachment that can generate force for chromosome motion, either toward or away from the associated spindle pole. This interface is also a site where MTs can either add or lose tubulin subunits as chromosomes move, so it is significant for normal mitosis (Rieder and Salmon, 1998). Identification of the proteins that provide these functions is now well advanced in budding yeast (Westermann et al., 2007) and higher eukaryotes (Cheeseman and Desai, 2008; Liu et al., 2006). Several motor enzymes contribute to chromosome segregation, but MT depolymerization in vitro, even in the absence of soluble nucleotides, can mimic chromosome-to-pole motion in vivo (Coue et al., 1991; Koshland et al., 1988). Moreover, minus-end directed motors are dispensable for poleward chromosome motion in yeasts (Grishchuk and McIntosh, 2006; Tanaka et al., 2007), suggesting that the root of chromosome movement lies in MT dynamics, not motor activity.
MT shortening can generate force because tubulin dynamics are associated with GTP hydrolysis. Tubulin-bound GTP is hydrolyzed shortly after polymerization, so most of the MT wall is GDP-tubulin. Surprisingly, GDP-tubulin will not polymerize, probably because its shape does not fit the MT lattice (Wang and Nogales, 2005). Assembled GDP-tubulin is therefore strained by interactions with its neighbors in the MT wall. This strain is relieved during depolymerization when strands of tubulin dimers, called protofilaments (PFs), become flared at the MT end (Mandelkow et al., 1991); presumably this PF shape reveals the minimum energy configuration of GDP tubulin, whereas GTP tubulin PFs are comparatively straight (Chretien et al., 1995; Muller-Reichert et al., 1998). Thus, the morphology of a MT end in vitro reflects its polymerization state.
PF bending during MT shortening has been proposed to do mechanical work (Koshland et al., 1988). Indeed, microbeads coupled to MTs by static links, e.g. a MAP or a biotin-streptavidin bond, experience a brief tug during PF bending (Grishchuk et al., 2005). Cargo that is bound to MTs by either motor enzymes (Lombillo et al., 1995b) or an encircling protein complex (Westermann et al., 2006) will move processively during MT shortening in vitro, even without ATP. Thus, if kinetochores were harnessed to MTs with the right “couplers”, the energy liberated by tubulin depolymerization could drive chromosome-to-pole motion (Efremov et al., 2007; Hill, 1985; Molodtsov et al., 2005). This raises the question of how kinetochores take advantage of the depolymerization machinery to facilitate chromosome segregation.
Kinetochore structure has been studied for years, but most kinetochores are so small that informative early work has been done either by immuno light microscopy or electron microscopy. Recent light microscopy has localized tagged kinetochore components along the spindle axis with almost nanometer precision (Joglekar et al., 2008; Schittenhelm et al., 2007), but electron microscopy has defined the image of kinetochores that most scientists consider. With this method, vertebrate chromosomes show kinetochore-associated MTs (KMTs) penetrating a darkly staining “outer plate” (Rieder and Salmon, 1998), which has commonly been interpreted as the principal MT-chromosome connection. Electron tomography of well-preserved kinetochores in PtK1 cells has refined this description as a meshwork of fibers that connect adjacent KMTs to one another and to nearby chromatin (Dong et al., 2007). PFs at the ends of many KMTs flare as they penetrate this plate and approach the chromatin (VandenBeldt et al., 2006). The fraction of KMTs with flared ends appears greater during anaphase than metaphase, perhaps a reflection of increased MT depolymerization in anaphase. However, ~70% of metaphase KMTs have flared ends, which poses an enigma: why do so many KMTs appear to be depolymerizing when their length is, on average, constant? Is KMT PF flare a simple reflection of dynamic state, as in vitro, or do other factors affect PF curvature in vivo?
To address these questions and to develop a more complete picture of the kinetochore-MT interface, we have used electron tomography to study the 3D structure of individual PFs at the plus ends of MTs from mitotic PtK1 cells, obtaining quantitative information about the curvature of PFs from several kinds of MTs. Approximately half of the PFs at KMT plus ends curve differently from those on either polymerizing or depolymerizing MTs in vitro. Our data suggest that these shapes do not result simply from MT dynamics but from associations between PFs and kinetochore-associated fibrils. These fibrils appear to impede PF bending, suggesting a mechanism for converting the energy of MT depolymerization into chromosome movement. This hypothesis is supported by a theoretical analysis of the effects of force on PF curvature and by observations that implicate two fibrillar, kinetochore-associated proteins as couplers between cargo and MT depolymerization in vitro.
Results
Electron tomography provides sufficient resolution to characterize PF shape at MT ends
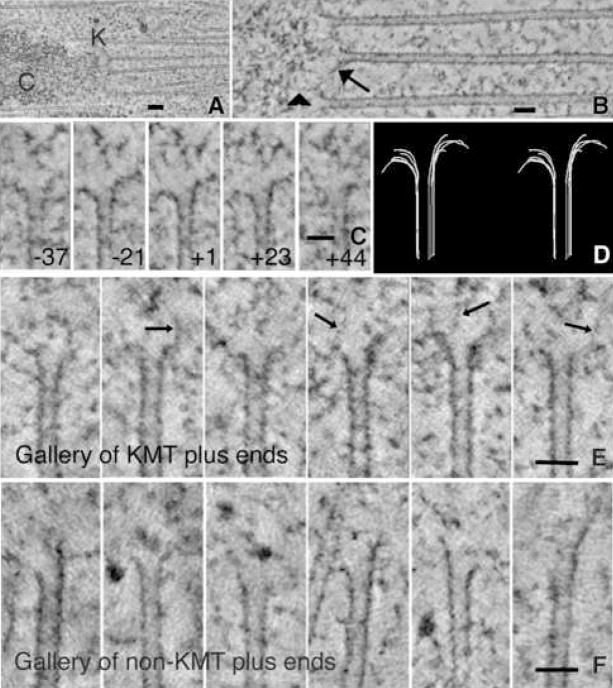
The structure of a kinetochore and its relationship to spindle MTs can be visualized in electron tomograms (Fig 1A), which permit clear distinctions between KMTs and other MTs that do not associate with kinetochores (nonKMTs). Most PFs at the plus ends of KMTs show “flare”, i.e., they bend away from the MT axis (O’Toole et al., 1999; 2003; VandenBeldt et al., 2006; Fig 1B and Video 1). We have examined 4–8 PFs at the end of each MT within the ~1 μm3 of cellular volume reconstructed in each of 23 tomograms (2,325 spindle PFs in total). Figs 1C, D portray a single prometaphase KMT plus end in tomographic slices cut at different orientations through its axis; the flares of its PFs are evident. Additional examples of KMTs from cells in prometaphase (Fig 1E and Video 2), metaphase, and anaphase (Fig S1, Videos 3,4) demonstrate that PF flare is extensive throughout mitosis.
Figure 1. Microtubules from mitotic PtK1 cells.
A. Average of 15 consecutive tomographic slices, total thickness ~30 nm, analogous to a conventional thin section. Chromosome (C) and kinetochore (K) show characteristic staining. MTs were identified as KMTs when they ended in a cluster near a chromosome. Bar = 0.1μm.
B. KMTs from the same cell but seen in a ~2 nm slice cut parallel to their axes with the “Slicer” feature of IMOD. Arrow identifies a bending PF; arrowhead indicates a fibril that runs from a PF to the chromatin. Bar = 0.05μm.
C. Multiple image planes that contain the MT axis but are oriented at the angles stated relative to the plane of section; PFs in each view differ in length and extent of flare. Bar = 0.05μm.
D. Stereo pair of a 3D model of all the PFs traced on the MT shown in Fig 1C. Use wall-eyed viewing.
E. Gallery of different KMT ends from the same cell. PF flare is variable; fibrillar material is associated with some PFs (arrows). Bar = 0.05μm.
F. Gallery of non-KMT ends from the polar region of a metaphase spindle; no chromosomes are near. Some MT ends are flared, others not; fibrils are not seen. Bar = 0.05μm.
The plus ends of KMTs are initially like those of non-KMTs, but during mitosis their PFs become longer and less curved
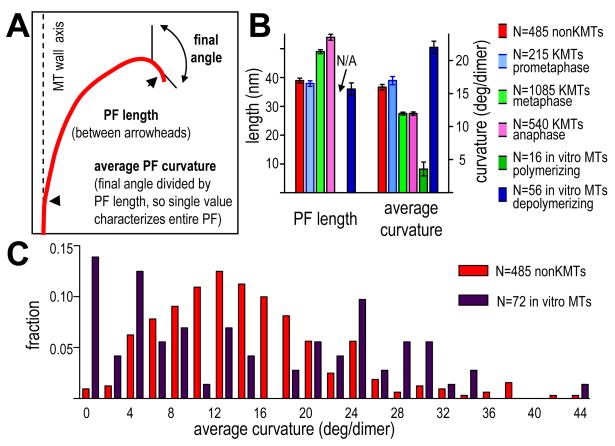
To ask whether MT ends showed systematic variations in shape we have compared tomograms of different places in spindles and times in mitosis. The plus ends of non-KMTs displayed a mixture of flared and straight PFs (Fig 1F). To compare these MTs with KMTs, we determined the length of the flare on each PF, the orientation of its final segment, and its average curvature (Fig 2A). With any of these measures, PF flare on a single MT was variegated (Table 1): all KMTs contained some PFs that were widely flared, and most included others that were rather straight. The variability among the PFs from all the MTs of a single kinetochore was similar to that of PFs from a single MT (Table 1). Variation in PF flare is not likely to be an artifact of our sample preparation techniques (Sup. Text), but it was sufficiently great that large number of PFs were required to identify statistically significant differences between MTs from different biological contexts (Sup. Text and Tables S1-3,S7). Thus, we have compared PFs by using the mean values for all the PFs in one category. With this approach, mean PF structure from KMT plus ends in early mitosis was similar to that of non-KMTs, but as mitosis proceeded, KMT PFs became longer, and their average curvatures decreased (Fig 2B).
Figure 2. Quantitative analysis of average PF shape.
A. Diagram of a traced PF and the ways the howflared program identified its length, final angle, and average curvature.
B. Average properties of PFs from non-KMTs, KMTs at several mitotic stages, and MTs in vitro. N=number of PFs for each MT class. N/A=not applicable.
C. Histogram of average PF curvature for non-KMTs (red) and pooled samples of P- and D-MTs in vitro (dark red). N=number of PFs in each class.
Table 1.
Typical average PF structures
| Eight PFs from one prometaphase KMT numbers are parameters for each PF | ||||
|---|---|---|---|---|
| MT# | PF# | Length (nm) | Final Ang (deg) | Avg. Curv. (deg/dimer) |
| 1 | 1 | 48 | 114 | 20 |
| 2 | 34 | 125 | 29 | |
| 3 | 35 | 64 | 15 | |
| 4 | 61 | 159 | 21 | |
| 5 | 13 | 35 | 21 | |
| 6 | 21 | 38 | 14 | |
| 7 | 97 | 178 | 15 | |
| 8 | 23 | 76 | 26 | |
| Mean | 41 | 98 | 20 | |
| SD | 27 | 53 | 5.5 | |
| PFs from prometa. KMTs (63 PFs from 9 MTs) numbers are means for all PFs from each MT | ||||
| 1 | 1–8 | 41 | 98 | 20 |
| 2 | 1–6 | 36 | 77 | 35 |
| 3 | 1–8 | 35 | 83 | 21 |
| 4 | 1–8 | 35 | 82 | 19 |
| 5 | 1–6 | 44 | 104 | 19 |
| 6 | 1–7 | 36 | 100 | 22 |
| 7 | 1–8 | 33 | 84 | 21 |
| 8 | 1–6 | 33 | 75 | 20 |
| 9 | 1–6 | 33 | 64 | 17 |
| Means for all PFs | 36 | 85 | 21.2 | |
| SDs for all PFs | 16 | 36 | 14.5 | |
Many PFs on spindle MTs are different from those on MTs formed in vitro
Changes in mean flare during mitotic progression suggested that PFs might play a direct role in chromosome segregation, so we compared PF flares in vivo with those of MTs with known dynamic state, using images of MTs that had been grown from pure tubulin in vitro and described in the literature. Most PFs on polymerizing MTs (P-MTs) do not project beyond their neighbors and do not flare, so these MT ends are commonly blunt (Mandelkow et al., 1991). A few growing PFs are longer than their neighbors, extending by highly variable amounts that depend on conditions (Chretien et al., 1995; Muller-Reichert et al., 1998). The mean (±SD) of their average curvatures were, however, reasonably consistent: 3.6±4.2°/dimer. PFs on “D-MTs”, i.e., those depolymerized by either subunit dilution or a drop in temperature (Chretien et al., 1995; Mandelkow et al., 1991) showed a mean curvature of 22 ± 7°/dimer and a mean length of 36 ± 16 nm (Fig 2B).
Previous studies of KMT ends in vivo have described PFs as similar to those in vitro (VandenBeldt et al., 2006), but our quantitative methods have found differences. Whereas PF lengths in prometaphase and metaphase were similar to those of D-MTs, their mean average curvatures fell between those of P- and D-MTs in vitro (Fig 2B). This might, of course, be because in vivo populations are mixtures of growing and shrinking MTs. However, a histogram of average PF curvatures for pooled P- and D-MTs in vitro showed two peaks, while comparable data from non-KMTs in vivo showed only one broad distribution (Fig 2C). Thus, the structure of MT ends in mitotic cells differs from that seen in vitro.
PF fine structure as an indicator of MT dynamics
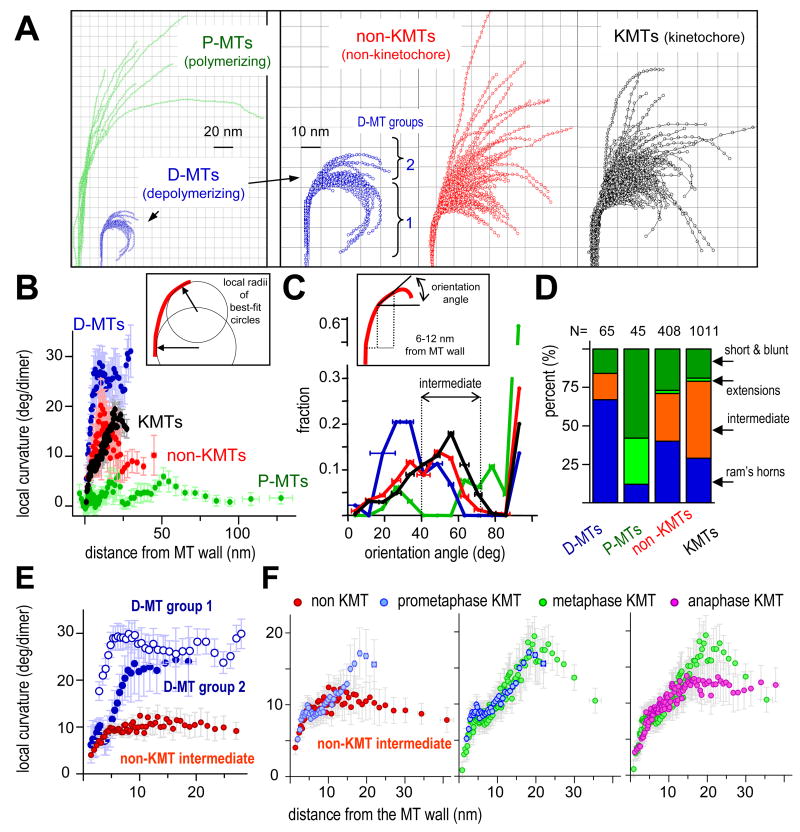
Since the unique features of mitotic MT end structure might reflect function in chromosome segregation, we worked to understand the details of PF flare, not just its average characteristics. We developed procedures and software to visualize and analyze the local shapes of individual PFs (Fig 3 and Sup. Mat. Part 3). Coordinates along each PF were extracted as X,Y values in the plane of the PF, and multiple PFs were displayed in a single frame of reference. Fig 3A compares PFs from MTs in vitro (Mandelkow et al., 1991) with non-KMTs and KMTs from PtK1 cells in mitosis. Most PFs in vivo have shapes that are intermediate between the PFs of P- and D-MTs. To quantify local PF curvature, we fit a circle to each region along the PF and took the reciprocal of its radius (Fig 3B, box). The distributions of mean local PF curvature for MTs in vivo also lie between the comparable values of P- and D-MTs, confirming the differences between these populations (Fig 3B, graph).
Figure 3. Quantitative analysis of local PF shape.
A. PFs from different MT ends: Polymerizing (green) and depolymerizing (blue) MTs formed from pure tubulin in vitro (Mandelkow et al., 1991). Non-KMT PFs (red) are from plus MT ends near the pole of a metaphase PtK1. KMT PFs (black) represented by ~25% of all metaphase and anaphase KMTs, selected at random. Scale for P-MTs is ½ that for other MT ends. PFs for D-MTs are shown at both scales for comparison.
B. Determination and distributions of local curvatures. Box diagrams use of a running average of 10 consecutive points along a PF to determine best-fit circles. Graph displays local curvatures as functions of distance from the MT wall; colors as in A. Error bars here and on all graphs are SEMs.
C. Assessing PF orientation. Box diagrams a strategy for measuring the orientation of PFs in any interval of distances from the MT wall. Graph shows normalized distributions of average PF orientations between 6 and 12 nm from the MT wall for four dataset, colors as in A. Error bars are SEMs of all PFs in that bin. Most P-PFs are nearly straight, so their greatest fraction is at about 90°, represented with a broken scale.
D. PFs were sorted into four groups based on their orientation angles (see text). N is the number of PFs in each dataset.
E. Mean local curvature as a function of distance from MT wall. PFs from D-MTs fall into two groups (3A). Different local curvature developments as a function of distance from MT wall are consistent with the model that some adjacent PFs adhere close to the wall (group 2).
F. Mean local curvatures as a function of distance from MT wall for “intermediate” PFs from metaphase non-KMTs and KMTs from prometaphase, metaphase, and anaphase. The same data for non-KMT PFs are plotted on E and F, for easier comparison
Given the diversity in local PF shape, we sought new measures that would allow statistically meaningful comparisons between PFs from different MT groups. The angle between a line perpendicular to the MT axis and the PF segment 6–12 nm from the MT wall was chosen as a good descriptor (Fig 3C, box) because this was the place nearest the MT wall that showed clear differences between P- and D-MTs. With this metric the distribution of PF orientations on D-MTs showed a peak at relatively small angles (26°–34°), as expected for highly curved PFs (Fig 3C, graph). The PFs of P-MTs were predominately straight, so an angle of ~90° was common, but there were also two small peaks: one at ~72°, which corresponded to the gently curving PFs, and one at ~26°, which resembled D-MTs. The latter may represent PFs that were depolymerizing at the instant of freezing.
We used these orientations to sort PFs objectively into four distinct groups (Fig 3D). Two groups (“Short & blunt” PFs with orientations ~90° and “extensions”, 72° – 90°) comprise ~90% of the PFs on P-MTs. It is therefore plausible that PFs with this shape in vivo (21% of the PFs on KMTs and 29% on non-KMTs) were polymerizing at the time of freezing. A third group, called “ram’s horns” (orientation < 40°), is likely to reflect depolymerization, because ~70% of the PFs on D-MTs in vitro had this orientation. Only 29% of the PFs on KMTs and 40% of those on non-KMTs had these angles. The remaining “intermediate” group (orientations from 40° to 72°) included 17% of PFs from D-MTs and none from P-MTs, but it contained 30% of the PFs from non-KMTs and ~50% from KMTs (Figs 3C,D). These representations demonstrate that the intracellular environment modifies the structure of many MTs ends so their PFs differ from those in vitro. Moreover, there are measurable differences between PFs on KMTs and non-KMTs.
Treating KMT and non-KMT PFs as classes provided sufficient numbers to permit statistically meaningful distinctions, but it obviated consideration of MTs as individuals. It would have been useful to characterize each MT as polymerizing, depolymerizing, or in some other state, but when we used the objectively sorted PFs to try to classify individual MTs, the results only confirmed the variability of PF shape (Sup. Text).
Differences between PF shapes in vivo and in vitro are not due to variation in PF adhesion
The results from looking at PFs by MT class motivated a quest for factors that might make PFs in living cells different from those in vitro. The distribution of D-MT PFs displayed a shoulder containing ~17% of this class (Fig 3C graph); these PFs comprised a group whose local curvatures grew more slowly than the rest (Group 2 on Figs 3A and E), resulting in “intermediate” orientations. These MTs were assembled from pure tubulin, so this fraction could not have arisen from MAPs or local environment. After considering several possible explanations (Sup. Text), we concluded that they probably resulted from a combination of the way the data were collected and a tendency for PFs in vitro to adhere as they begin to flare. It followed that greater PF adhesion in vivo might account for the larger number of intermediate PFs on spindle MTs, but direct examination of this hypothesis has rejected it (Sup. Text); PF adhesion cannot explain the differences between PF shape in vivo and in vitro.
MT-associated proteins could account for the shapes of non-KMT PFs, but the factors that shape KMT PFs are more complex
Most spindle MTs bind MAPs, including “tip-interacting proteins” (Morrison, 2007). There are currently only limited data with which to assess the effects of these proteins on PF curvature (Arnal et al., 2004), but it is reasonable to assume that such binding would change local PF curvature consistently throughout its flare. Indeed, the local curvature of non-KMT PFs with “intermediate” orientation was markedly constant as a function of distance from the MT wall (Fig 3E), consistent with the notion that MAPs bind flaring PFs on these MTs and modify their minimum energy shape.
The local curvature of intermediate PFs from KMTs, however, varied with distance from the MT wall, particularly before anaphase (Fig 3F). Proximal to the MT wall, these PFs were slightly less curved than those from non-KMTs; farther out, their local curvatures increased, although not to equal that of a D-MT (Fig 3E,F). These variations in local curvature suggested that kinetochores provide a special environment that modifies PF structure in complex ways.
PF shapes change little from metaphase to anaphase
Unlike previous reports, our analysis of average PF curvature demonstrated changes during the establishment of proper KMT attachment (prometaphase vs. metaphase) but not between metaphase and anaphase (Fig 2B). This was surprising, given that metaphase KMTs are, on average, growing at the kinetochore, while anaphase MTs are shortening. We therefore reexamined the issue, looking at local PF structure. The fractions of KMT PFs that resembled P- and D-MTs were very similar in metaphase and anaphase, so we compared the shapes of the intermediate PFs from these stages (Fig 3F). Far from the MT wall, the curvature of anaphase intermediate KMT PFs was both smaller and more constant than that seen in metaphase. Close to the MT wall, however, local PF shapes were very similar. The orientation angles of KMT PFs in metaphase and early anaphase were almost superimposable, but as anaphase proceeded, more PFs with orientations like D-MTs appeared (Fig S2B). Since more MTs are shortening during anaphase than metaphase, the shape of KMT PFs cannot be a simple reflection of dynamic state; other factors must contribute.
The curvature of KMT PFs is low and constant near the MT wall, but it becomes greater and more variable where PFs encounter kinetochore-associated fibrils
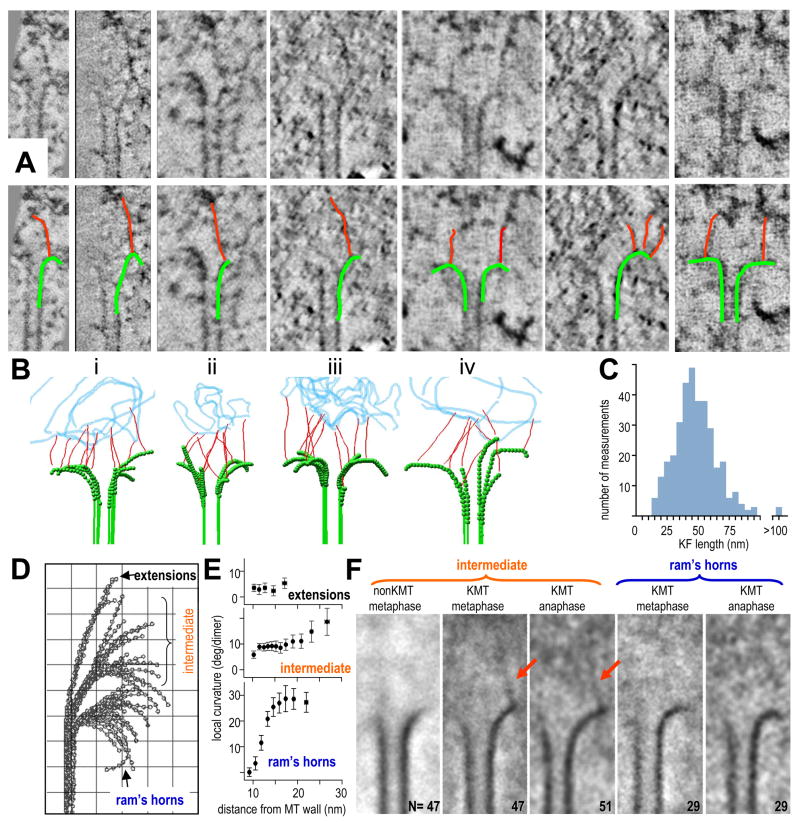
We next examined the vicinity of KMT PFs, seeking structures that might account for their unusual curvatures. A mat of fibers, like that seen by Dong et al., is visible near the tips of KMTs (Fig S1), but its clarity depends on the thickness of the slices viewed (Fig S1B,B′). Our study of PFs was carried out on 2 – 4 nm slices in which this fibrous mat is subtle (Video 1), and much of it lies farther from the chromatin than the flared MT end, so it seemed unlikely to account for intermediate PF curvatures. We also sought, but did not find, MT-encircling rings. Flared PFs did, however, frequently associated with fibrils 2–4 nm in diameter; these ran from the PFs to places deeper in the kinetochore (Fig 4A). In our tomographic slices, the material above and below the fibrils was removed from view, so they appeared with good contrast, but their sinuous trajectories made it necessary to step though multiple slices to follow them (Fig 4B). The fibrils were neither robust nor periodic, and their distribution of lengths was broad (Fig 4C). Only in favorable cases was much fibril visible in a single view. Some fibrils linked with straight MT walls, but most connected to flaring PFs. We counted the number of fibrils associated with each PF from 10 KMTs in each of two metaphase and two anaphase cells, a total of 317 PFs: the average fibril/PF ratio was 1.35 ± 0.17; values for metaphase and anaphase were not detectably different. Similar structures were seen on KMTs at all stages of mitosis (e.g., Figs 1B,E, S1), but they were absent from non-KMTs (Fig 1F). We therefore refer to them as kinetochore fibrils (KFs).
Figure 4. Fibrils associate with PFs at places that correlate with changes in local PF curvature.
A. Tomographic slices of KMT ends. The same gallery is also shown with PFs and their associated KFs indicated by graphic overlays.
B. Models of metaphase (i,ii) and anaphase (iii,iv) KMT ends; PFs traced in green, KFs in red; a representation of chromatin is in blue.
C. Histogram of KF lengths based on data from 40 KMTs chosen at random from 4 cells.
D,E. PFs from all the KMTs reconstructed by electron tomography from one metaphase kinetochore. PFs were grouped by their orientation angles. Mean local curvatures of each group are plotted in (E). The intermediate group of PFs is conspicuous for its consistent local curvature near the MT wall and its curvature variation farther from the wall. Grid=10nm.
F. Averages of multiple tomographic slices containing PFs from different groups (numbers as indicated). Alignments maximized overlap for the PFs curving right. Intermediate group PFs from metaphase non-KMTs average well, but no KF is evident. Intermediate group KMT PFs from one metaphase and one anaphase cell show 45 – 54 nm KFs, attached to the averaged PF and extending toward the chromatin. Averages of the ram’s-horn groups from metaphase and anaphase cells showed no additional electron density at PF tips.
To study the relationship between KFs and PF shape, we sorted PFs by their orientation angles, using KMTs and non-KMTs from of our two best-preserved metaphase and anaphase cells. 52 of the 98 KMT PFs from a metaphase cell fell in the intermediate class (Fig 4D). Their initial segments were rather straight and made modest angles with the MT axis, but farther from the MT wall the variations in PF curvature grew large (Fig 4E). To ask if these PFs were associated with any specific structures we aligned their images to achieve maximum overlap in their straight and consistently curving parts, then averaged them to reveal any additional consistent features (Fig 4F). The straight and consistently curving parts of the PFs generated good contrast, but the more distal PF parts looked diffuse, as expected. Average images of the intermediate PFs from both metaphase and anaphase KMTs showed a KF extending from near the tip of the well-aligned part of the PF toward the chromatin. No clear KFs were seen in the vicinity of non-KMTs or KMTs from the “ram’s horn” groups, suggesting that KFs with well-defined positions and orientations are specific to KMT PFs with intermediate shape. These results led us to hypothesize that the shapes of intermediate PFs result from interplay between the tendency of GDP-tubulin to curve and forces exerted by KFs that opposed this bending. Such interactions would straighten the PF segments near the MT wall but leave the more distal PF parts in diverse configurations. The reaction to this force would be a poleward pull on the inner kinetochore, potentially accounting for the ability of MT depolymerization to move chromosomes.
Bending PFs can act through KFs to generate processive and efficient chromosome movement
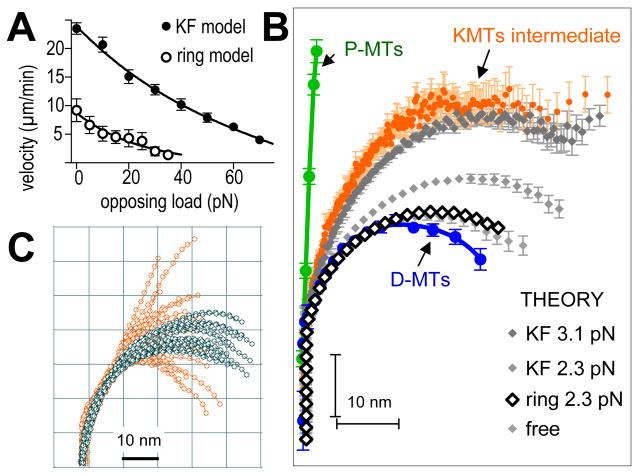
To ask whether a KF could in principle couple MT depolymerization to chromosome movement, we used our molecular-mechanical model for depolymerizing MTs (Molodtsov 2005), modified to include elastic elements (KFs) that connected the tubulin in flaring PFs to “cargo” (e.g., a chromatid) (Sup. Mat. Part 2). Briefly, each PF is modeled as a linear array of tubulins that interact with defined energy relationships. KFs are assumed to bind at random places along the PF but not to soluble tubulin. As PFs tend toward their minimum energy conformation (~23° bend/dimer), a bound KF opposes this tendency. When tubulin depolymerizes from the tip of a bending PF, any associated KF is released and joins the pool of fibrils that interact randomly with KF binding sites. We examined the role that such fibrils might play in chromosome motion by varying the KF-tubulin association constant (parameter k+ in Sup. Mat. Part 2). If binding was weak (k+ < 25 sec−1), only a few of the MT’s 13 PFs bound KFs at any time. As the PFs curled, they pulled on the bound KFs; PFs that failed to form lasting KF connections peeled away from their load, so it stalled (Video 5, Fig S3A). When PF-KF binding was stronger (k+ > 50 sec−1), “cargo” moved steadily, driven by PF power strokes (Video 6). Thus, fibrous connections to bending PFs can theoretically couple MT depolymerization to processive cargo motion, even against a significant load. In fact, fibrillar coupling can move a larger load than can be moved by rings formed by Dam1 complexes, which are thought to perform this function in yeasts (Fig 5A). A ring that binds MTs strongly enough to provide processive coupling impedes MT shortening and reduces the work that MT depolymerization can accomplish. In contrast, KFs undergo cycles of detachment/reattachment, so the efficiency of the depolymerization motor is not limited (Fig 5A and Sup. Mat. Part 2).
Figure 5. PFs as mechanical elements that can do work as they bend.
A. Force velocity curves for motions of various loads, due to shortening MTs acting through two types of couplers. Data for ring coupler is from (Efremov et al., 2007).
B. Average PF shapes from P-MTs (N=45) or D-MTs (N=65) compared with the average shape of intermediate KMT PFs (N=505). Curves in different grays show theoretical PFs under various tensions applied by randomly attaching KFs. Bars are SEMs. PFs interacting with a Dam1-like ring (blue) are experiencing 2.3 pN/PF. Larger forces stall motion and induce ring detachment (A) (Efremov et al., 2007).
C. Comparison of intermediate PFs from one metaphase kinetochore (orange, same set as Fig 4B) with a family of theoretical PFs (grey) under an average tension of 3.1 pN/PF. The model describes PF straightening close to the MT wall quite well, but the spread in the data farther from the wall indicates the presence of unidentified factors that modify these PF parts.
The shapes of “intermediate” KMT PFs could result from their interaction with fibrils but not with MT-encircling structures
To assess the ability of forces acting through either fibrils or rings to account for structures of KMT PFs, we compared observed PF morphologies with those calculated for PFs under various tensions applied either by randomly attaching KFs or by a MT-encircling ring. Increased tension from KFs straightened the PFs (Fig 5B); the average shape of model PFs experiencing 3–4 pN per PF matched well the average shape of intermediate KMT PFs. Processive movement under this load would result from 1.35±0.15 KFs per PF, a ratio almost identical to that seen in tomograms (above). The theoretical and experimental PFs were also similar in both their gentle and consistent curvature near the MT wall and their more variable behavior in distal regions (Fig 5C).
When the same MT model was used to predict PF shape as tension was applied via a MT-encircling ring, like the Dam1 complex described theoretically in Efremov et al. (2007), the fit to observed PF shape was poor (Fig 5B, Sup. Mat. Part 2). Such rings associate with the MT wall in front of the PF flare, so applied tension has little effect on PF shape, even when it is the maximum that the ring can withstand. This suggested that rings or other MT-encircling structures are not likely to explain KMT PF shape and may therefore not be involved in coupling PF-generated forces to kinetochores in PtK1 cells.
Fibrillar, kinetochore-localized proteins can couple MT depolymerization to movement of beads in vitro
Some evidence from the literature supports the idea that filamentous proteins might enable processive cargo motion at the ends of depolymerizing MTs. Chromosomes will move with MT depolymerization in vitro when soluble nucleotide is <1 nM (Coue et al., 1992). Domain-specific antibodies to the neck of CENP-E (a kinetochore-associated kinesin) block this motion (Lombillo et al., 1995a). Recent work on the structure of CENP-E has shown that a highly flexible, ~200 nm coiled coil separates the MT-binding and kinetochore-binding domains of this protein (Kim et al., 2008), reminiscent of the longest KFs seen here. This fibrous, processive, plus end directed motor may therefore couple kinetochores to MTs in ways that contribute to minus end-directed motion with the ends of shortening MTs.
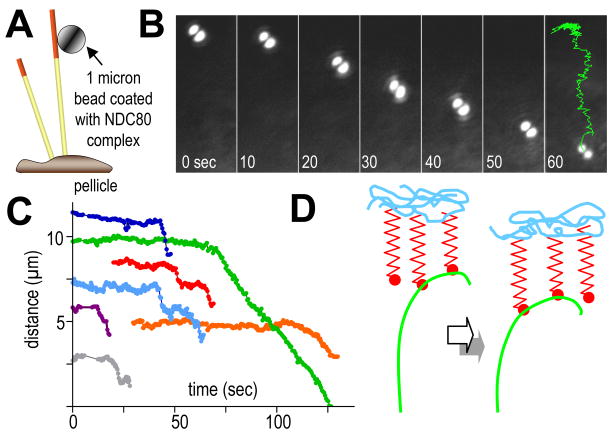
Another candidate for fibrous coupling is the ~57 nm long, 4-subunit Ndc80 complex (Hec1), which is both highly conserved and essential for kinetochore-MT interactions in many cell types (Davis and Wordeman, 2007). Recent work has reconstituted the Ndc80 complex from C. elegans in vitro (Cheeseman et al., 2006), so we asked whether it could support the motion of microbeads with depolymerizing MTs. Beads coated with Ndc80 bound readily to MTs (Fig 6A). When MT depolymerization was induced, most beads detached, but ~10% moved towards the minus MT ends (N=211). The mean distance traveled was 2.1 μm, so these motions were not very processive, although a few beads moved as much as 9 μm (Figs 6B,C, Video 7). The probability that these motions were random is < 0.05 (Fig. S4). Their average speed was 18±3 μm/min, about the same as the rate of MT shortening in our system. Together with the CENP-E data cited above, the behavior of this fibrous protein complex shows that cargos can be coupled to MT depolymerization by a ring-independent mechanism, although it remains to be seen whether their movement is like that predicted in Video 6. Thus, the structure of the kinetochore-MT interface, a rigorous theory of depolymerization-dependent motion, and experiments with appropriately shaped proteins all support the idea that fibrous coupling may be important in chromosome motion.
Figure 6. Beads coated with Ndc80 complex moving with depolymerizing MTs in vitro.
A. Experimental design (not to scale). Bovine brain MTs were capped with GMPCPP-assembled, rhodamine-labeled tubulin, so they would break up upon illumination with green light, permitting MT depolymerization.
B. Bead position at times shown (sec); last time shows trajectory of bead center for the entire motion.
C. Distances from bead to pellicle as a function of time for 7 separate experiments. Movement began shortly after the MT cap was removed.
D. Two consecutive times during the life of a PF from a depolymerizing MT that is stably attached to a “load”. The propagation of depolymerization, reflected in a progression of PF position, allows recycling KFs to transmit force to the load without a noticeable change in PF curvature where the KFs attach.
Discussion
Our new methods for visualizing and analyzing PFs have demonstrated structural differences between the ends of MTs in vivo and in vitro, and they have drawn particular attention to the complexity of the PFs on KMTs.
Our data extend published evidence on the structure of chromosome-MT coupling
Previous studies of kinetochores from vertebrate cells have emphasized an outer plate that surrounds KMTs and appears attached to chromatin (reviewed in Rieder and Salmon, 1998 and exemplified by Dong et al., 2007). Our tomographic slices of rapidly frozen, freeze-substitution fixed mitotic PtK1 cells confirm the existence of a fibrillar mat at the outer kinetochore (Figs 1, S1 and Video 1), but its clarity depends on mitotic stage and the thickness of the slice being viewed. Our images instead have emphasized slender fibrils (KFs) that connect MT ends with a deeper region of the kinetochore. KFs have not previously been reported, perhaps because of differences between our methods for specimen and image preparation and those used in published work (Sup. Text). More likely to be significant, however, was our interest in links between KMTs and chromosomes, rather than an overall description of the kinetochore.
Kinetochore composition identifies many candidates for chromosome-MT coupling
Several kinetochore-MT attachment factors have been identified; these protein complexes come in at least two shapes: rings and fibers (Davis and Wordeman, 2007). The Dam1/DASH complex, which oligomerizes to form rings around MTs in vitro (Miranda et al., 2005; Westermann et al., 2005), is required for proper chromosome segregation in budding yeast (Cheeseman et al., 2002); it also works as a processive coupler in vitro (Asbury et al., 2006; Grishchuk et al., 2008a). In fission yeast, however, an homologous complex is not essential for normal spindle function (Gachet et al., 2008; Sanchez-Perez et al., 2005), and close Dam1 homologs have not yet been found in other organisms. These results point to other MT-associated, kinetochore-localized proteins, like EB1, XMAP215, Clip 170, CLASP, INCENP, CENP-E, CENP-F, KNL1, and Ndc80/Hec1, all of which have been implicated in MT-kinetochore attachment and/or the regulation of KMT dynamics (reviewed in Morrison, 2007). Of these, CENP-E, XMAP215, and the Ndc80 complex are filamentous, CENP-F and KNL1 are likely to be, and other proteins may associate to make fibers. We hope that our evidence for fibrous couplers will draw attention to this shape and encourage structural work on all these candidate couplers.
The Ndc80 complex is a particularly interesting member of this group because it is both ubiquitous and essential for kinetochore-MT interaction (DeLuca et al., 2006; McAinsh et al., 2006). It is, however, too short to account for all the KFs seen, and its demonstrated binding is to the MT wall, not the polymer’s end. Our structural evidence suggests that KFs may bind PFs on a portion of the tubulin dimer that normally faces the MT lumen, a region not commonly explored in MT binding studies. The resolution of our images is not sufficient to say exactly where KFs and PFs meet, so this question deserves further study.
KMT end structure emphasizes the importance of fibrils, but rings too may be important
Our analyses of ring-shaped and fibrillar couplings (Fig. 5) suggest that fibrils account better than rings for PF structure at the kinetochore-MT interface. Although rings have not yet been seen on KMTs in vivo, they may still be present and important for some aspect of mitosis. They may contribute to chromosome-MT coupling in some organisms and they may regulate MT dynamics (Grishchuk et al., 2008b), a critical issue when there is only one MT per kinetochore, as in budding yeast. The MT-encircling fibrous mat in some kinetochores may also work as a polymerization regulator, keeping the dynamics of multiple KMTs in harmony so they can all function effectively for kinetochore motion. As shown above and in Sup. Mat., however, fibrils offer advantages of efficiency when multiple MTs are available to effect reliable chromosome segregation (Fig 5A).
PF-KF binding explains aspects of spindle physiology
It has been proposed that inhibiting PF bending slows MT shortening (Franck et al., 2007; Grishchuk et al., 2005; McIntosh et al., 2002). If PFs are prevented from bending by KF tension, for example when a KF is attached to a metaphase kinetochore whose sister is being pulled toward the opposite pole, then the rate at which KMTs lose tubulin will be reduced. This explains both the intermediate curvature of metaphase KMT PFs and the greater stability of KMTs relative to non-KMTs. It can also rationalize aspects of KMTs “flux”, i.e., their slow, poleward movement while kinetochores are on average at rest (Ganem and Compton, 2006). Flux requires that KMTs add subunits at their plus ends in spite of PF flare. We propose that KMT plus end structure reflects a steady state in which KFs inhibit PFs from attaining the curvature at which depolymerization becomes rapid. The shortening of such MTs is continuously being rescued, so polymerization can continue, even though most PFs are flared.
Some details of PF shape change during mitosis are also rationalized by the idea that KF tension modifies PF curvature. Both the length and average curvature of PFs change as proper chromosome attachments are formed during prometaphase; this may reflect increased tension at sister kinetochores as chromosome attachments mature. However, average PF structure changes little with anaphase onset. Metaphase chromatids are obviously under tension, but many early anaphase chromosomes may likewise experience drag from their sisters as chromatid arms disengage. Kinetochores at both stages may therefore be pulling a considerable load. By mid-anaphase, however, the force necessary for chromosome movement should be reduced to viscous drag and action against any “governor” that regulates chromosome speed. This may explain why PF orientation is different in early and mid-anaphase and why PF curvature in anaphase is relatively constant as a function of distance from the MT wall (Fig 3B).
A PF with constant curvature can exert a processive force, thanks to the ability of KFs to detach as tubulin depolymerizes and recycle to PF regions nearer the MT wall (Fig 6D). Our results do suggest, however, that the tension developed by PF bending is rather high, and it is not yet clear why this force does not cause chromosome to move faster. Perhaps, as suggested above, other kinetochore complexes, like Dam1 rings or the outer kinetochore mat, are governors that regulate the rate at which PFs can bend as chromosomes move. Then tension would not be rate limiting unless load became large, e.g. from a persistent connection between sisters.
Experimental Procedures
Cell Culture
PtK1 cells were grown by standard methods and plated on either chips of plastic or PET track-etched membranes (Morphew and McIntosh, 2003). When ~80% confluent, cells were cryo-immobilized in an HPM-010 high-pressure freezer (Bal-Tec AG, Liechtenstein) then fixed by freeze-substitution at −90°C in acetone containing glutaraldehyde and tannic acid, then OsO4 and uranyl acetate; embedding was in Epon-Araldite. The reliability of these methods was tested by comparison with alternative methods (Sup. Mat., Part 1, Tables S3 vs. S4, S5 vs. S6).
Microtomy, Microscopy, and 3D Reconstruction
Samples were serially sectioned on an UltraCut microtome (Leica Microsystems, Vienna). Sections were picked up on “slot” grids with a 1×2 mm hole covered by Formvar, then post-stained in 2% uranyl acetate and lead citrate for 15 and 3 min, respectively. Cellular regions were selected and imaged with either a JEM-1000 (Japanese Electron Optical Laboratories, Tokyo, JP) or an F-30 (FEI Co., Einhoven, NL), using Gatan CCD cameras to record serial tilts from ±60° or 70° in increments of 1° – 1.5°. Each section was imaged in 2 tilt series around orthogonal axes, then assembled into a single reconstruction (Mastronarde, 1997).
Acquiring images of MT ends
Spindle MTs were traced, and ends that lay within the reconstructed volume were studied further. Special purpose programs (mtrotlong and rotallends from IMOD (Kremer et al., 1996) were used to find and excise rectangular solids, ~140×120×300 nm, that included the plus end of each MT and the region around it. The Slicer feature of IMOD was then used to examine this volume by making its axis of rotation parallel to the excised MT, adjusting the thickness of the displayed slice to 4nm, and then tipping the orientation of the slice to angles as high as ±60°. We searched a range of tilts to find an orientation that displayed one or more clearly curling PFs. The two MT walls and any flaring PFs were then traced by hand, a process that involved some judgment. For example, any place along a possible PF that showed a sharp change in curvature (e.g., 90° in a few pixels) or an obvious change in thickness or stain density was interpreted as the association of that PF with some other object, not a continuation of the PF.
After two PF traces on one MT end had been stored, the orientation of the sampling plane was changed by 28°±6° to find additional PFs on the same MT (360°/13=27.7; ±6° allows for variation, given the distortions of plastic sections). Most MT ends yielded 6 satisfactory PF tracings.
Quantitative Structural Characterization of Microtubule Ends
The average structure of each PF trace was assessed by the howflared program in IMOD. This returns a table of numbers that describe the average path of the PF from the point it diverged from the MT wall to its end. The average curvature of each PF was expressed as the average angle between adjacent 8 nm segments, i.e., degrees/dimer. To examine variation along a PF, we used the 2-D coordinates of each PF, mapped to a single frame of reference that allowed both sorting by shape and quantification of local curvature as a function of distance from the MT wall, using the “Curvature calculation program” (Figs 4, S5 and Sup. Mat. Part 3). This software was also used to compute PF orientation angles, and to calculate the mean local curvatures of each PF group.
To create average images of the PFs and their surroundings, we used Metamorph 6.4 (Universal Imaging) to align all images of a given PF class so there was maximal overlap in the straight and consistently curving parts of the PFs. The aligned images were averaged then contrasted using PhotoShop.
Mathematical model of fibrillar coupler is described in Sup. Mat., Part 2.
In vitro assay of MT-depolymerization dependent motility
Bead motility was examined as described in (Grishchuk et al., 2008a). Briefly, MTs were grown from coverslip-attached Tetrahymena pellicles, using purified bovine brain tubulin. MT ends were stabilized with rhodamine-labeled tubulin, assembled with GMPCPP, so depolymerization could be induced by photo-dissolution of the caps. C. elegans Ndc80 complex was purified as described (Cheeseman et al., 2006). Polystyrene 1μm beads coated with streptavidin (Bangs Laboratories) were incubated with biotinylated pentaHIS-antibodies (Qiagen), followed by the Ndc80 complex, in which the Spc24 subunit was tagged with 6HIS. Successful conjugation was confirmed with anti-Ndc80 antibodies. The conjugation density was ~ 5×104 Ndc80 complexes per bead, enough for up to 40 fibrils to contact one MT end. Images were acquired and analyzed using Metamorph.
Supplementary Material
Acknowledgments
We are grateful to I. Spiridonov and other members of McIntosh and Ataullakhanov labs for help and advice, and to A. Vorobjev for support. This work was supported in part by grants from the NIH (GM033787 and RR000592 to JRM, who has been a Research Professor of the American Cancer Society), from the NIH and the Human Frontiers Science Program to AD, CGP2006B#2863, from the CRDF to FIA and JRM, and from MCB RAS, Russia.
Abbreviations
- MT
Microtubule
- MAP
MT-associated protein
- KMT
kinetochore-associated MT
- P-MT
polymerizing MT
- D-MT
depolymerizing MT
- PF
protofilament
- KF
kinetochore fibril
- Sup. Mat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnal I, Heichette C, Diamantopoulos GS, Chretien D. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr Biol. 2004;14:2086–2095. doi: 10.1016/j.cub.2004.11.055. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. JCell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TN, Wordeman L. Rings, bracelets, sleeves, and chevrons: new structures of kinetochore proteins. Trends Cell Biol. 2007;17:377–382. doi: 10.1016/j.tcb.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci U S A. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, Gonen T, Davis TN, Asbury CL. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y, Reyes C, Courtheoux T, Goldstone S, Gay G, Serrurier C, Tournier S. Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring dam1 and klp2. Mol Biol Cell. 2008;19:1646–1662. doi: 10.1091/mbc.E07-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. Functional roles of poleward microtubule flux during mitosis. Cell Cycle. 2006;5:481–485. doi: 10.4161/cc.5.5.2519. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. Embo J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci U S A. 2008a;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, Gudimchuk N, Westermann S, Drubin D, Barnes G, McIntosh JR, Ataullakhanov FI. The Dam1 ring is an efficient and processive transducer of depolymerization energy because its binding to microtubules is tight. Proc Natl Acad Sci U S A. 2008b;105 in press. [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. ProcNatlAcadSciUSA. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008 doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995a;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995b;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo- electron microscopy study. JCell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. JStructBiol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. Embo J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, West RR. Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Molodtsov MI, Grishchuk EL, Efremov AK, McIntosh JR, Ataullakhanov FI. Force production by depolymerizing microtubules: a theoretical study. Proc Natl Acad Sci U S A. 2005;102:4353–4358. doi: 10.1073/pnas.0501142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphew MK, McIntosh JR. The use of filter membranes for high-pressure freezing of cell monolayers. J Microsc. 2003;212:21–25. doi: 10.1046/j.1365-2818.2003.01231.x. [DOI] [PubMed] [Google Scholar]
- Morrison EE. Action and interactions at microtubule ends. Cell Mol Life Sci. 2007;64:307–317. doi: 10.1007/s00018-007-6360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Reichert T, Chretien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha, beta)methylenediphosphonate. Proc Natl Acad Sci U S A. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, McDonald KL, Mantler J, McIntosh JR, Hyman AA, Muller-Reichert T. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J Cell Biol. 2003;163:451–456. doi: 10.1083/jcb.200304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen B. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Current Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann SA, Avila-Sakar Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17(2):277–90. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.