Abstract
Diversity in neuronal signaling is a product not only of differential gene expression, but also of alternative splicing. However, although recognized, the precise contribution of alternative splicing in ion channel transcripts to channel kinetics remains poorly understood. Invertebrates, with their smaller genomes, offer attractive models to examine the contribution of splicing to neuronal function. In this study we report the sequencing and biophysical characterization of alternative splice variants of the sole voltage-gated Na+ gene (DmNav, paralytic), in late-stage embryos of Drosophila melanogaster. We identify 27 unique splice variants, based on the presence of 15 alternative exons. Heterologous expression, in Xenopus oocytes, shows that alternative exons j, e, and f primarily influence activation kinetics: when present, exon f confers a hyperpolarizing shift in half-activation voltage (V1/2), whereas j and e result in a depolarizing shift. The presence of exon h is sufficient to produce a depolarizing shift in the V1/2 of steady-state inactivation. The magnitude of the persistent Na+ current, but not the fast-inactivating current, in both oocytes and Drosophila motoneurons in vivo is directly influenced by the presence of either one of a pair of mutually exclusive, membrane-spanning exons, termed k and L. Transcripts containing k have significantly smaller persistent currents compared with those containing L. Finally, we show that transcripts lacking all cytoplasmic alternatively spliced exons still produce functional channels, indicating that splicing may influence channel kinetics not only through change to protein structure, but also by allowing differential modification (i.e., phosphorylation, binding of cofactors, etc.). Our results provide a functional basis for understanding how alternative splicing of a voltage-gated Na+ channel results in diversity in neuronal signaling.
INTRODUCTION
The emergence of appropriate behavior is a consequence of the correct development and maturation of neuronal circuits that, together, constitute the nervous system. The individual neurons that comprise these circuits must establish appropriate synaptic connectivity and acquire characteristic electrical properties, including the expression of a correct mix and number of voltage-gated ion channels. Although significant progress has been made toward our understanding of axon guidance and synaptogenesis (Brose and Tessier-Lavigne 2000; Charron and Tessier-Lavigne 2007; Goodman and Shatz 1993; Schnorrer and Dickson 2004), the same cannot be said of mechanisms that shape electrical development (Marder and Prinz 2002; Siegel et al. 1994; Spitzer 2006; Spitzer et al. 2002). This is unfortunate because aberrations of these mechanisms likely contribute to clinically significant neurological disorders, including epilepsy and schizophrenia (Levitt et al. 1997; Romano and Harvey 1996; Stanwood et al. 2001).
Neuronal diversity results, at least in part, from differential regulation of gene expression in individual neurons. However, diversity of mature electrical properties goes beyond genes because additional mechanisms such as alternative splicing, RNA editing, and protein modification (i.e., phosphorylation) also make significant contributions. The isolation of variant transcripts encoding vertebrate voltage-gated Na+ (VgNa+) channel genes provided initial evidence for alternative splicing in this important class of channels (Auld et al. 1988; Belcher et al. 1995; Gustafson et al. 1993; Lu and Brown 1998; Plummer et al. 1998). VgNa+ channels are responsible for the rising phase of the action potential and, as such, are key regulators of neuronal excitability (Catterall 2000). The Na+ channel consists of a pore-forming α-subunit and one or two accessory β-subunits. At least ten different mammalian α-subunit genes have been identified and partially characterized (Nav1–10; Goldin 2001). By contrast, insects contain only one confirmed Na+ channel gene, termed paralytic (para, DmNav) (Feng et al. 1995; Mee et al. 2004; Miyazaki et al. 1996). The lack of redundancy, coupled with a structure that is highly homologous to its mammalian counterparts (Loughney et al. 1989), means that DmNav is an advantageous model to fully explore the contribution of splicing of Na+ channels to neuronal signaling.
The cloning of multiple variant transcripts by Loughney et al. (1989) was among the first evidence for the existence of alternative splicing of the para gene. Subsequent studies in Drosophila, Musca, and cockroach have identified 11 alternatively spliced exons in this gene (Davies 2007; Lee et al. 2002; O'Donnell Olson et al. 2008; Tan et al. 2002). Importantly, many of these alternative exons are also spliced in mammalian VgNa+ channels (Diss et al. 2001; Oh and Waxman 1998; Plummer et al. 1997). That these spliced exons are conserved across such evolutionarily diverged species is strongly indicative of physiological importance.
In this study we have isolated and sequenced 50 open reading frames (ORFs) of DmNav from late-stage Drosophila embryos and have identified a number of more common splice variants. Heterologous expression in Xenopus oocytes shows that the presence of specific spliced exons does not affect the amplitude of the fast-inactivating (transient) current, but are critical for both voltage-dependent activation and voltage-dependent inactivation. Moreover, we identify a pair of mutually exclusive exons (k and L) that significantly alter the magnitude of the slowly inactivating (persistent) current exhibited by the expressed channel. When L is present the persistent current is large (∼8% of the transient current), but this falls to about 2% when k is substituted. We verify this conclusion, in vivo, by comparing both the transient and persistent Na+ currents present in identified motoneurons in wild-type and in pasilla (ps) mutant embryos. Previous work (Park et al. 2004) reported that the expression of exon k is increased in Drosophila S2 cells following targeted knockdown (RNAi) of ps. We show here that exon k is also up-regulated, at the expense of exon L, in whole CNS in ps mutant embryos and, moreover, that the persistent Na+ current in motoneurons is reduced as a consequence. Exons k and L form part of the voltage sensor in homology domain III of the channel and our results are indicative that this region contributes to the ubiquitous, yet poorly understood, persistent Na+ current. Combined, our results show that splicing of a single VgNa+ channel is likely sufficient to generate considerable diversity in neuronal signaling.
METHODS
Cloning of full-length embryonic DmNav cDNA
Total RNA was extracted from whole late-stage 17 Canton-S embryos as described in Muraro et al. (2008). cDNA synthesis was carried out in a total volume of 20 μl (RevertAid First-Strand cDNA Synthesis kit; Fermentas, York, UK). A primer specific to part of the 3′ untranslated region (UTR) of DmNav (5′-AAT ACT CGC GTG CAT CTT GG-3′) (0.2 μg) was mixed with RNA (final concentration, 250 ng/μl) and made ≤11 μl with RNase-free water. The mix was incubated at 65°C for 5 min to denature RNA, followed by incubation on ice for 2 min. Reaction buffer (4 μl; in mM: 250 Tris-HCl, 250 KCl, 20 MgCl2, 50 DTT), 2 μl of 10 mM dNTPs, and 3 μl of RNase-free water were added and the mix was incubated at 37°C for 5 min. Following addition of 1 μl of RevertAid M-MuLV (monkey murine leukemia virus) reverse transcriptase (Fermentas), the reaction was first incubated at 42°C for 60 min followed by 65°C for 15 min. To amplify the DmNav ORF, polymerase chain reaction (PCR) primers were designed to anneal to flanking sequences in the 5′ and 3′ UTRs (forward primer: 5′-ATC GTT GGC CGC ATA GAC AAT GAC AG-3′; reverse primer: 5′-AAT ACT CGC GTG CAT CTT GGA GGG-3′). The PCR mixture consisted of 1 μl of cDNA, 2 μl Elongase Enzyme Mix (Invitrogen, Paisley, UK), dNTPs at a final concentration of 0.2 mM each, 7.6 pmol of each primer, and 1× PCR buffer with a final Mg2+ concentration of 1.3 mM. Cycling conditions were as follows: initial denaturation at 94°C for 30 s; 40 cycles of 94°C for 30 s, 63.5°C for 30 s, and 68° for 7 min; a final extension step at 68°C for 10 min. The PCR product was excised from an agarose gel (0.7%) following electrophoresis and purified using the QIAquick Gel Extraction Kit (Qiagen, Crawley, UK). The PCR product was polyadenylated and ligated into pGEM-T Easy (Promega, Southampton, UK) using the manufacturer's protocol. The ligation mix was used to transform MAX Efficiency DH5α Competent Cells (Invitrogen) according to the manufacturer's protocol, but incubating transformants at 30 instead of 37°C. DNA was prepared using the QIAprep Spin Miniprep Kit (Qiagen), and the DmNav ORF was sequenced using Big Dye Terminator Version 3.1 Chemistry and the primers described in Table 1, and run on an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems, Warrington, UK). Sequences were aligned and analyzed using Clone Manager 7 (Scientific and Educational Software, Cary, NC). Exon composition was identified by comparing the cDNA sequences to the genomic sequence (Accession number: FBgn0260993).
TABLE 1.
Primers used to sequence the complete DmNav ORF
| Sequence (5′ to 3′) | Exon |
|---|---|
| CTCACTATAGGGCGAATTGG | T7 promoter |
| TACGTCGTGTGGCCATTTAC | 6 |
| TCAAGAAGTTCCCGCTGGAC | 9 |
| AGAAGGCCGAAGAAGAAGAG | 10 |
| TACCCGGTAGCGATCGTAAG | 13 |
| TATCGAGCCCGTCCAGACAC | 15 |
| CATTATCGTGGCCCTATCGC | 18 |
| TCTTGGCCACCGTTGTCATC | 20 |
| ACGACACTGCCAGCATTAAC | 23 |
| CGTGGTGTTGGCTCGATTTC | 24 |
| CACCTTCAAAGGCTGGATAC | 27 |
| ACGCGGTCCTAGACTATCTC | 29 |
| TCCGGGCAATTGTGGTTCAG | 31a |
| GTTAACGGTACTGCAGAAGG | 31b |
Construction of exon-specific splice variants for heterologous expression
Although our PCR-based approach isolated full ORFs, it was clear from sequencing that each full-length clone contained about 10 PCR-induced errors. Without a full understanding of a structure–function relationship for DmNav, we felt it unwise to use these clones for expression studies because of the unknown effect these mutations might have. Therefore to ensure expression constructs were free of point mutations, we used a single, previously published and characterized clone, DmNav10/pGH19 (para13-5) (Warmke et al. 1997) as a backbone. Using restriction enzyme mapping of DmNav10, we divided the para ORF into two major fragments, termed S and T. Fragment S contains exons j, i, a, and b; fragment T contains exons c/d, e, f, h, and k/L (see Supplemental Fig. S1).1 Fragment S was subcloned from DmNav10 into pBluescript II KS (Stratagene, La Jolla, CA). By replacing the Kpn I/Bgl II or Bgl II/Bst XI fragments, we made S2, S3, and S4, which contain different combinations of exons j, i, a, and b. T1 is DmNav10 lacking the Sac I/Sac I fragment. By changing the Sac I/Stu I, Stu I/Apa I, or Apa I/Bam HI fragments, we constructed T2 to T10. Finally, we subcloned the Sac I fragment of S into T/pGH19 to make full-length ORFs. By using different combinations of intermediate S and T, we made error-free DmNav isoforms. All clones were checked by sequencing prior to expression analysis. Escherichia coli cultures were grown in a low-salt Luria–Bertani broth (0.5% NaCl, 1.0% tryptone, and 0.5% yeast extract) at 30°C.
Semiquantitative analysis of k and L exon-containing DmNav transcripts in late-stage Drosophila embryos
RNA was extracted from 20 Canton-S (wild-type [WT]) and pasilla (ps2) embryos using the RNeasy Micro kit (Qiagen) with final elution in 12 μl RNase-free water. Because DmNav expression is relatively low, RNA (10 μl) was reverse transcribed using a RevertAid First-Strand cDNA Synthesis kit (Fermentas) with a combination of a reverse DmNav gene-specific primer (exon 27, 5′-GTG TGA AAA AGG ATC CAA ATA TGA-3′) and random hexamer primers (Fermentas) in a total volume of 20 μl. cDNA (1 μl) was amplified in a 20-μl reaction mixture containing 20 pmol of gene-specific primer pairs, 200 μM each of dNTPs, 16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2, and 2.5 U of BIOTAQ DNA polymerase (Bioline, London, UK). Reactions were run for 25 cycles of amplification (10 s at 94°C, 20 s at 66.5°C, and 30 s at 72°C), followed by a 10-min extension at 72°C. The PCR products were resolved by 2% TAE-agarose gel electrophoresis. Primer sequences were: k exon-specific forward primer, 5′-AAT TAA TTT GGC CGC GGT CTG-3′ and L exon-specific forward primer, 5′-GCT TAT CAA CTT CGT TGC TTC ACTTGT-3′ and reverse primer, 5′-ATT GCG ATT TGG TAT GAT CTC GTG-3′. Gel images were quantified using the UN-SCAN-IT gel Version 5.1 software (Silk Scientific, Orem, UT).
Expression of DmNav sodium channels in Xenopus oocytes
Xenopus laevis (large adults) ovaries were dissociated by incubating in Ca2+-free ND96 saline (in mM: 96 NaCl, 2.0 KCl, 5.0 HEPES, 1.0 MgCl2, 2.5 Na-pyruvate, and 0.5 Theophylline, adjusted to pH 7.5 with NaOH) containing 1 mg/ml collagenase (Sigma-Aldrich, Poole, UK) for 45 min at room temperature. Following a wash in ND96 saline (as before, but with the addition of 1.8 mM CaCl2) oocytes were incubated at 20°C for ≥1 h. Oocytes at stage V or stage VI were used for defolliculation and then coinjected with 10 ng each of capped DmNavX and tipE cRNA, which was in vitro synthesized with T7 polymerase (mMESSAGE mMACHINE kit, Ambion, Austin, TX). Injected oocytes were incubated in ND96 at 20°C for 3 days.
Electrophysiology and analysis of expressed channels in oocytes
Sodium currents were recorded from oocytes bathed in ND96 saline at room temperature (22–24°C) by two-electrode voltage clamp using an Axoclamp 2B amplifier (Molecular Devices, Sunnyvale, CA). Thin-walled borosilicate glass electrodes (GC150TF-10, Harvard Apparatus, Kent, UK) were pulled to resistance of about 0.5 to 1.0 MΩ when filled with 3 M KCl. Voltage protocols were controlled and data recorded using pClamp (v8, Molecular Devices). Data were filtered at 10 kHz and sampled at 20 kHz. Linear leak currents were subtracted using a P/N protocol within pClamp. No currents were observed in water-injected control oocytes and all currents observed were abolished following application of tetrodotoxin (TTX, 1 μM) (data not shown).
The voltage dependence of activation of the transient Na+ current was calculated by dividing individual currents elicited at each test voltage (−80 to +5 mV in 5-mV increments) by the peak current to produce I/Imax. This value was then plotted (Origin 7.5, MicroCal, Northampton, MA) against test voltage and half-activation voltage (V1/2) calculated from that plot. We adopted this approach, rather than calculating conductance values (G), because we were not able to directly measure reversal potentials (Erev).
The voltage dependence of channel inactivation was determined by applying 100-ms inactivating prepulses (−80 to +5 mV in 5-mV increments) followed by a test potential of −10 mV (50 ms). Current amplitudes at the test potential were normalized to the maximum peak current obtained. V1/2 of inactivation was derived from plots of I/Imax against test voltage. To calculate the persistent to transient current ratio, the voltage steps eliciting the largest peak transient current and the largest persistent current (recorded at 100 ms after onset of the voltage step) were used. These were not necessarily from the same test potential. All values are shown as means ± SE. Significance was determined using a nonpaired Student's t-test. Results were deemed significant at either *P < 0.05 or **P < 0.01.
Electrophysiology in embryos
Late-stage 17 (19–21 h after egg laying at 25°C) embryos were dissected and central neurons were accessed for electrophysiology as described by Baines and Bate (1998). Embryos were first dechorionated using 50% bleach for 2 min; the vitelline membrane was then manually removed using sharp tungsten wires. The embryo was visualized using a water-immersion lens (total magnification, ×600) combined with Normarski optics (BX51W1 microscope; Olympus Optical, Tokyo). Na+ currents were recorded as described in Muraro et al. (2008). Pasilla2 mutant embryos (Seshaiah et al. 2001) were identified by lack of a GFP-containing balancer chromosome (TM3::KrGFP): ps2 mutants are embryonic lethal but develop to late-stage 17.
RESULTS
Alternative splicing of full-length cDNA clones
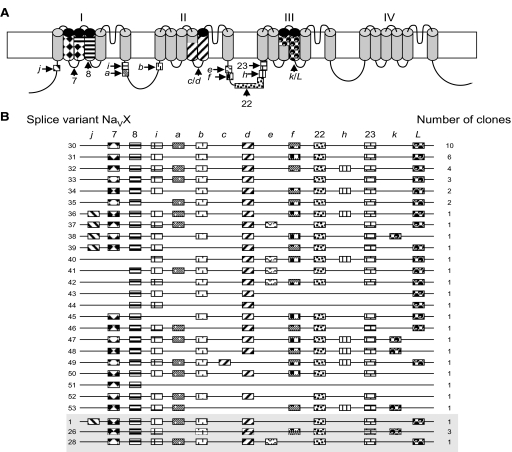
To determine the most common splice isoforms of DmNav transcripts present in late-stage 17 embryos (the final stage prior to hatching), RT-PCR was used to isolate and clone 50 complete ORFs. Comparison of exon composition of these clones revealed 27 unique splice variants. This degree of variability is remarkably similar to the 29 splice types (identified from 64 clones) recently sequenced from adult heads (O'Donnell Olson et al. 2008). To avoid confusion and maintain consistency, we have adopted the nomenclature used by O'Donnell Olson et al. (2008) and have numbered our novel splice variants DmNav30–53. Surprisingly, only three splice variants (termed DmNav1, 26, and 28) are present in both developmental stages. Of these, only DmNav26 is present in more than one copy in embryo, whereas DmNav1, which is present at high copy number in adult (13/64 clones), is present as only a single clone in embryo.
Exon usage within embryonic splice variants and their frequencies is presented in Figs. 1 and 2. A schematic showing the full intron/exon structure of DmNav is shown in Supplemental Fig. S2. Of the spliced exons, two pairs (c/d and k/L) are mutually exclusive. Both pairs are membrane spanning, contributing to IIS4–5 and IIIS3–4, respectively. The remaining 11 spliced exons (j, 7, 8, i, a, b, e, f, 22, h, 23) are independent and cytoplasmic. Seven splice variants (DmNav26, 30–35) are present more than once, indicative of increased occurrence within the CNS. The remaining 20 variants (DmNav1, 28, 36–53) are each represented by a single clone. However, most of these single clones differ only at two loci: exons a and h. The absence of exons previously reported as being constitutively spliced from certain isoforms (e.g., 7, 8, 22, and 23) suggests novel putative sites of alternative splicing. Thus exon 7 is predicted to encode parts of IS2–3 and is homologous to the alternatively spliced exon designated K in cockroach BgNav (Song et al. 2004). This exon was absent from five clones. One isoform lacks exon 8, which is predicted to encode parts of IS3–4. Two isoforms lack both exons 22 and 23, which together encode part of the II–III linker, and the start of IIIS1. These two exons have both been previously shown to use multiple 5′ splice acceptor sites, resulting in the inclusion or exclusion of two forms of regions f and h (Thackeray and Ganetzky 1994, 1995). This is the first reported instance of the downstream regions being excluded as well. Finally, three clones lack both of the mutually exclusive exons k and L that encode IIIS3–4.
FIG. 1.
Characterization of 27 open reading frames (ORFs) of DmNav isolated from late-stage 17 embryos. A: schematic of the predicted 4-homology domain (I, II, III, and IV) structure of DmNav protein with approximate locations of spliced exons shown. Exons j, 7, 8, i, a, b, e, f, 22, h, 23 are optional, whereas exons c/d and k/L are mutually exclusive. B: exon usage of the 27 individual clones isolated. The variant number (DmNavX) and frequency of clones are indicated. The clones in the shaded box (i.e., 1, 26, and 28) are also found in adult heads (O'Donnell Olson et al. 2008).
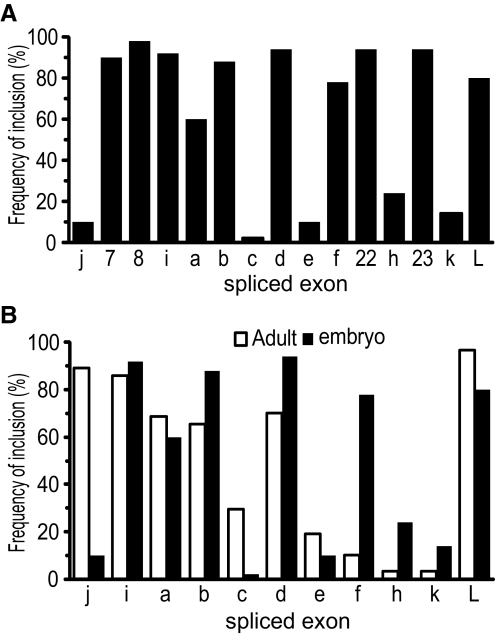
FIG. 2.
Frequency of alternative exon usage. A: analysis of exon usage across all 27 DmNav ORFs shows that exons 7, 8, i, b, d, f, 22, 23, and L are significantly more common (present in >75% of clones) than j, c, e, h, and k. Exon a exhibits a more variable frequency of inclusion, being present in 60% of clones. B: comparison of exon frequency in embryo and adult CNS for those exons that have been analyzed at both developmental stages. The data for adults are taken from O'Donnell Olson et al. (2008).
Exons i, b, d, f, and L are present in >75% of clones, whereas exons j, c, e, h, and k are uncommon in isoforms of this early developmental stage, being present in <25% clones (Fig. 2A). Exon a exhibits a more variable frequency of inclusion, being present in 60% of clones. Our statistical analysis (not shown) suggests that inclusion of any specific alternatively spliced exon is independent of either the presence or the absence of other spliced exons. Thus we can conclude that the embryonic CNS contains a wide diversity of splice forms of DmNav and that these forms are not present, for the most part, in the adult (O'Donnell Olson et al. 2008). This difference is largely due to the absence of exon j and inclusion of exon f in embryo and vice versa in adult (Fig. 2B). The physiological significance of this is unclear, although it is indicative that the signaling requirements of embryonic neurons differ from those in the adult.
Electrophysiological properties of common splice variants in oocytes
The Xenopus oocyte has been extensively used for characterization of ion channel properties, including VgNa+ channels. We have thus used this heterologous expression system to characterize the properties of selected DmNav splice variants. We chose not to use our original PCR-derived clones (used to determine exon composition) because sequencing showed them to contain on average 10 point mutations per ORF. We believe that these mutations are random PCR-induced errors and are not consistent with RNA editing. To remove these point mutations, we constructed identical clones from the same DmNav cDNA backbone (para13-5; see methods) by standard cloning. All recombinant clones were sequenced and verified not to contain any point mutations prior to heterologous expression. Thus the recombinant clones represent identical copies of naturally occurring splice variants in which PCR-induced errors are corrected. Expression of all recombinant variants tested resulted in robust Na+ currents that were abolished in the presence of TTX and control injections of water resulted in no currents (see methods; data not shown). By contrast, expression of some of the original PCR-derived clones (which contain point mutations induced by PCR) failed to express in oocytes, presumably due to amino acid substitutions (data not shown, but see O'Donnell Olson et al. 2008). For the analysis described in the following text, all splice variants that are found in either embryonic (Fig. 1) or adult (O'Donnell Olson et al. 2008) CNS are denoted by a number (i.e., DmNav1–53). To verify our conclusions made from analysis of such naturally occurring variants, we generated artificial splice variants that differed by the inclusion of one additional specific spliced exon. Where used, these are denoted by both a number (relating to the naturally occurring variant) and a letter (denoting which additional exon is present).
To characterize the channel properties of the most common embryonic splice variants, we coexpressed each recombinant isoform, together with the presumed auxiliary subunit TipE (Derst et al. 2006) and recorded VgNa+ current. Analysis of peak transient current revealed a range of amplitudes (∼1.3 to 6.3 μA) that were seemingly independent of exon usage, but instead probably resulting from slight differences in expression efficiency. Thus the values shown in Table 2 do not segregate according to exon composition. This is important because it suggests that the differences in channel properties that we do observe (see following text) are not due to differences in efficiency of expression.
TABLE 2.
Averaged peak transient current for clones expressed
| Variant |
Exon |
Average Transient Current, nA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| j | i | a | b | c/d | e | f | h | k/L | ||
| DmNav29 | i | b | d | e | L | 1,283 ± 97 | ||||
| DmNav10f | j | i | b | d | e | f | L | 1,643 ± 267 | ||
| DmNav26 | i | b | d | f | k | 1,723 ± 309 | ||||
| DmNav1 | j | i | a | b | d | L | 1,726 ± 431 | |||
| DmNav49 | i | a | b | c | f | h | L | 1,738 ± 97 | ||
| DmNav10h | j | i | b | d | e | h | L | 1,845 ± 217 | ||
| DmNav3 | j | i | b | d | L | 1,882 ± 285 | ||||
| DmNav33 | i | a | b | d | L | 2,095 ± 242 | ||||
| DmNav30 | i | a | b | d | f | L | 2,181 ± 277 | |||
| DmNav10 | j | i | b | d | e | L | 2,562 ± 366 | |||
| DmNav15 | j | i | b | c | e | L | 2,638 ± 358 | |||
| DmNav34 | i | b | d | f | h | L | 2,942 ± 413 | |||
| DmNav10k | j | i | b | d | e | k | 3,091 ± 362 | |||
| minimal | d | L | 3,145 ± 382 | |||||||
| DmNav31 | i | b | d | f | L | 3,472 ± 509 | ||||
| DmNav49k | i | a | b | c | f | h | k | 3,529 ± 307 | ||
| DmNav32 | i | a | b | d | f | h | L | 6,293 ± 1,045 | ||
A minimum of eight oocytes were used to calculate the average and SE of peak transient current for each clone expressed. DmNav variants denoted by numbers (i.e., 29) are naturally occurring splice variants expressed in either adult (DmNav1–29) or embryonic (DmNav30–53) CNS. DmNav variants denoted by numbers and a letter (i.e., 10f) are artificial splice variants that have not been isolated from CNS of either developmental stage. DmNavminimal is an artificial splice variant lacking exons j, i, a, b, e, f, and h.
Voltage-dependent activation
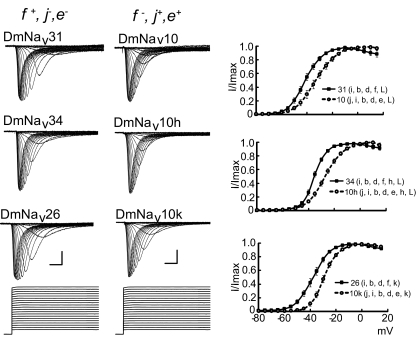
In contrast to peak transient current, sorting of voltages observed for V1/2 activation reveals two distinct clusters (Table 3). The first cluster, which shows hyperpolarized V1/2 values (including the naturally occurring variants DmNav26, 30, 31, 32, 34, and 49), are common in that they all include exon f. The second cluster, which exhibits more depolarized V1/2 values (including the naturally occurring variants DmNav10 and 15), mostly contain exons j and e. The presence of exons j and e, however, is seemingly predominant to the additional presence of f. Thus the V1/2 activation of DmNav10f (an artificial clone not found in either adult or embryo CNS) containing exons j, e, and f is more similar to variants containing just j and e compared with those that contain just f (see Table 3). To independently verify the contribution of these exons to voltage-dependent activation, we used cloning to construct pairs of identical clones that consistently differ only in that one of the pair contains exons j and e, whereas the other contains exon f (Fig. 3). Three such different comparisons (DmNav10 and 31, 10h and 34, 10k and 26) clearly show that the clones containing exon f have a statistically significant hyperpolarized V1/2 activation compared with that of its exon j and e–containing counterpart. Although it is difficult to transpose these findings to functional neurons, our data are indicative that neurons expressing splice variants containing exon f should be more easily activated (i.e., fire action potentials at more negative membrane potentials) than those that express j and e.
TABLE 3.
Activation kinetics of DmNav variants
| Variant |
Exon |
50% Activation, mV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| j | i | a | b | c/d | e | f | h | k/L | ||
| minimal | d | L | −40.7 ± 1.1 | |||||||
| DmNav49 | i | a | b | c | f | h | L | −43.9 ± 1.6 | ||
| DmNav31 | i | b | d | f | L | −42.4 ± 1.2 | ||||
| DmNav26 | i | b | d | f | k | −38.3 ± 1.4 | ||||
| DmNav32 | i | a | b | d | f | h | L | −37.8 ± 1.5 | ||
| DmNav49k | i | a | b | c | f | h | k | −36.3 ± 1.9 | ||
| DmNav34 | i | b | d | f | h | L | −35.5 ± 0.7 | |||
| DmNav30 | i | a | b | d | f | L | −34.9 ± 2.1 | |||
| DmNav15 | j | i | b | c | e | L | −34.8 ± 2.3 | |||
| DmNav10 | j | i | b | d | e | L | −32.0 ± 2.0 | |||
| DmNav3 | j | i | b | d | L | −30.7 ± 1.5 | ||||
| DmNav33 | i | a | b | d | L | −30.4 ± 1.1 | ||||
| DmNav10k | j | i | b | d | e | k | −29.4 ± 1.1 | |||
| DmNav10f | j | i | b | d | e | f | L | −29.2 ± 1.4 | ||
| DmNav1 | j | i | a | b | d | L | −28.1 ± 1.8 | |||
| DmNav29 | i | b | d | e | L | −27.8 ± 1.3 | ||||
| DmNav10h | j | i | b | d | e | h | L | −27.3 ± 1.2 | ||
A minimum of eight oocytes were used to calculate the average and SE of V1/2 activation for each clone expressed. DmNav variants denoted by numbers (i.e., 49) are naturally occurring splice variants expressed in either adult (DmNav1–29) or embryonic (DmNav30–53) CNS. DmNav variants denoted by numbers and a letter (i.e., 49k) are artificial splice variants that have not been isolated from CNS of either developmental stage. DmNavminimal is an artificial splice variant lacking exons j, i, a, b, e, f, and h.
FIG. 3.
Voltage dependence of activation is affected by inclusion of exons f, j, and e. Activation curves (means ± SE for n ≥ 8) are shown for 3 paired comparisons (DmNav31 vs. 10, 34 vs. 10h, and 26 vs. 10k) in which identical clones differ by either containing f+, j−, e−, or f−, j+, e+. Inclusion of exon f and loss of exons j and e result in a hyperpolarizing shift in half-activation voltage (V1/2). See Table 3 for actual V1/2 values. All paired comparisons are significantly different at P ≤ 0.01. Example traces are shown on the left. Scale bars are 5 ms and 0.5 μA, respectively. Voltage dependence of activation was determined by 100-ms depolarizing potentials from −80 to +5 mV (in 5-mV steps) applied from a holding prepulse potential of −90 mV.
Voltage-dependent inactivation
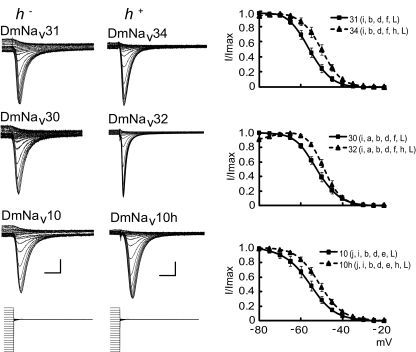
Sorting splice variants by V1/2 inactivation voltage reveals a small cluster that all contain exon h (Table 4). Inclusion of this exon results in splice variants (e.g., DmNav32 and 34) that inactivate at more depolarized membrane potentials compared with variants lacking h. Comparing identical clone pairs that consistently differ only by the inclusion of exon h confirms that inclusion of h results in a depolarized shift in V1/2 inactivation voltages (Fig. 4). Thus comparison of the naturally occurring variants DmNav31 and 34, 30 and 32 show that the latter of each pair, containing exon h, has a more depolarized inactivation voltage. A third comparison between the naturally occurring DmNav10 and its artificial variant 10h, further validates that inclusion of exon h is sufficient to shift inactivation to more depolarized values.
TABLE 4.
Inactivation kinetics of DmNav variants
| Variant |
Exon |
50% Inactivation, mV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| j | i | a | b | c/d | e | f | h | k/L | ||
| DmNav26 | i | b | d | f | k | −57.4 ± 1.7 | ||||
| DmNav31 | i | b | d | f | L | −55.7 ± 1.3 | ||||
| DmNav10 | j | i | b | d | e | L | −54.9 ± 1.5 | |||
| DmNav3 | j | i | b | d | L | −54.5 ± 1.3 | ||||
| DmNav49 | i | a | b | c | f | h | L | −53.8 ± 1.3 | ||
| DmNav15 | j | i | b | c | e | L | −53.6 ± 2.1 | |||
| DmNav1 | j | i | a | b | d | L | −52.8 ± 0.9 | |||
| DmNav10k | j | i | b | d | e | k | −52.7 ± 0.5 | |||
| DmNav10f | j | i | b | d | e | f | L | −52.4 ± 0.8 | ||
| DmNav30 | i | a | b | d | f | L | −52.1 ± 1.3 | |||
| DmNav10h | j | i | b | d | e | h | L | −51.1 ± 1.2 | ||
| DmNav49k | i | a | b | c | f | h | k | −50.8 ± 2.0 | ||
| DmNav34 | i | b | d | f | h | L | −50.6 ± 1.4 | |||
| DmNav33 | i | a | b | d | L | −49.9 ± 0.7 | ||||
| minimal | d | L | −49.1 ± 1.3 | |||||||
| DmNav32 | i | a | b | d | f | h | L | −48.6 ± 1.0 | ||
| DmNav29 | i | b | d | e | L | −48.1 ± 1.3 | ||||
A minimum of eight oocytes were used to calculate the average and SE of V1/2 steady-state inactivation for each clone expressed. DmNav variants denoted by numbers (i.e., 26) are naturally occurring splice variants expressed in either adult (DmNav1–29) or embryonic (DmNav30–53) CNS. DmNav variants denoted by numbers and a letter (i.e., 10k) are artificial splice variants that have not been isolated from CNS of either developmental stage. DmNavminimal is an artificial splice variant lacking exons j, i, a, b, e, f, and h.
FIG. 4.
Voltage dependence of steady-state inactivation is influenced by inclusion of exon h. Inactivation curves (means ± SE for n ≥ 8) are shown for 3 paired comparisons (DmNav31 vs. 34, 30 vs. 32, and 10 vs. 10h), in which identical clones differ by either containing h+ or h−. Inclusion of exon h results in a depolarizing shift in V1/2 of inactivation. See Table 4 for actual V1/2 values. All paired comparisons are significantly different at P ≤ 0.05. Example traces are shown on the left. Scale bars are 5 ms and 0.5 μA, respectively. Voltage dependence of steady-state inactivation was determined by 100-ms depolarizing prepotentials from −80 to +5 mV (in 5-mVsteps) followed by a test potential of −10 mV for 50 ms.
A similar comparison of pairs also indicates that exon j influences inactivation. Thus comparison of DmNav1 with 33 and DmNav10 with 29 shows that inclusion of j (present in DmNav1 and 10) shifts V1/2 inactivation voltages more negative (−52.8 vs. −49.9 mV and −54.9 vs. −48.1 mV, respectively; see Table 4). In conclusion, it might be expected that neurons expressing h-containing channel variants will inactivate slower and would thus be predicted to fire action potentials for longer than neurons that primarily contain channels lacking this exon. Inclusion of exon j might, by contrast, be predicted to generate a quicker termination of excitation. In this regard it is interesting to note that DmNav10h (which contains both exon j and exon h) has a V1/2 inactivation voltage that is intermediate between clones containing either exon j or exon h, but not both (Table 4).
Persistent Na+ current
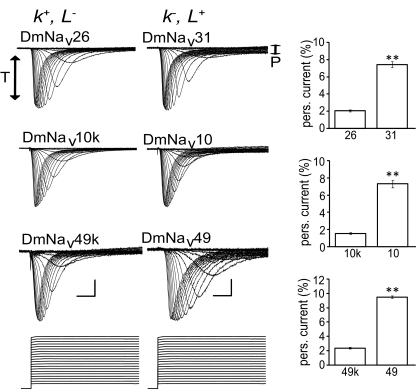
In addition to the well-characterized rapidly inactivating (transient) voltage-gated Na+ current, the existence of a slowly inactivating (persistent) current component has been widely observed. The persistent fraction of Na+ current is activated in the subthreshold voltage range and is believed to contribute to plateau generation, pacemaker activity, and increased firing frequencies (Li Y and Bennett 2003; Li Y et al. 2004; Nikitin et al. 2006; Tazerart et al. 2008). Significantly, this current component is also a principal target for antiepileptic drugs (Kuo et al. 1997; Lampl et al. 1998; Spadoni et al. 2002). Despite the importance to neuronal signaling, the origin of the persistent current remains controversial (Huguenard 2002; Liu et al. 2004; see discussion). To determine whether the persistent Na+current is also affected by exon usage in DmNav, we measured the ratio of persistent to transient current obtained of each splice variant expressed (Table 5). This analysis clearly shows that splice variants containing membrane-spanning exon k have a significantly reduced persistent current compared with variants containing the L form of this exon (i.e., compare the naturally occurring variants 26 and 31 in Table 5). To verify the contribution of exons k and L to persistent current, we again constructed identical pairs of splice variants that consistently differed by inclusion of either exon (Fig. 5). By comparing these clones (DmNav26 and 31, 10k and 10, 49k and 49, the former containing k and the latter L), it is clear that inclusion of k, at the expense of L, is sufficient to significantly reduce the percentage of persistent current compared with the transient component (Fig. 5).
TABLE 5.
Persistent to transient current of DmNav variants
| Variant |
Exon |
P/T Current, % | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| j | i | a | b | c/d | e | f | h | k/L | ||
| DmNav10k | j | i | b | d | e | k | 1.5 ± 0.1 | |||
| DmNav26 | i | b | d | f | k | 2.0 ± 0.1 | ||||
| DmNav49k | i | a | b | c | f | h | k | 2.4 ± 0.1 | ||
| DmNav33 | i | a | b | d | L | 4.1 ± 0.2 | ||||
| DmNav10h | j | i | b | d | e | h | L | 4.7 ± 0.3 | ||
| DmNav3 | j | i | b | d | L | 4.9 ± 0.4 | ||||
| DmNav32 | i | a | b | d | f | h | L | 5.0 ± 0.4 | ||
| DmNav10f | j | i | b | d | e | f | L | 5.4 ± 0.3 | ||
| DmNav15 | j | i | b | c | e | L | 5.5 ± 0.5 | |||
| DmNav34 | i | b | d | f | h | L | 5.5 ± 0.3 | |||
| DmNav29 | i | b | d | e | L | 6.2 ± 0.3 | ||||
| DmNav1 | j | i | a | b | d | L | 6.5 ± 0.4 | |||
| DmNav10 | j | i | b | d | e | L | 7.3 ± 0.4 | |||
| DmNav30 | i | a | b | d | f | L | 7.3 ± 0.2 | |||
| DmNav31 | i | b | d | f | L | 7.4 ± 0.3 | ||||
| minimal | d | L | 8.1 ± 0.3 | |||||||
| DmNav49 | i | a | b | c | f | h | L | 9.5 ± 0.2 | ||
A minimum of eight oocytes were used to calculate the average and SE of percentage of persistent current for each clone expressed. DmNav variants denoted by numbers (i.e., 26) are naturally occurring splice variants expressed in either adult (DmNav1–29) or embryonic (DmNav30–53) CNS. DmNav variants denoted by numbers and a letter (i.e., 10k) are artificial splice variants that have not been isolated from CNS of either developmental stage. DmNavminimal is an artificial splice variant lacking exons j, i, a, b, e, f, and h.
FIG. 5.
Amount of persistent current is influenced by exons k and L. Measurement of the percentage of persistent (P) current relative to transient current (T) in 3 paired comparisons (DmNav 26 vs. 31, 10k vs. 10, and 49k vs. 49), in which identical clones differ by either containing k+, L− or k−, L+. Inclusion of exon L results in a marked increase in persistent relative to transient current. Actual values are listed in Table 5. Example traces are shown on the left and averaged values (%) of persistent current (means ± SE for n ≥ 8) on the right (P < 0.01 for all comparisons). For ease of analysis, all example traces have been scaled to have the same amplitude of transient current. Scale bars are 5 ms and 0.5 μA, respectively. Voltage commands were the same as those for activation voltage determination (see Fig. 3). Persistent current was measured at the end of a depolarizing command voltage. The largest persistent current was normalized by division with the largest transient current obtained (i.e., not necessarily from the same voltage step).
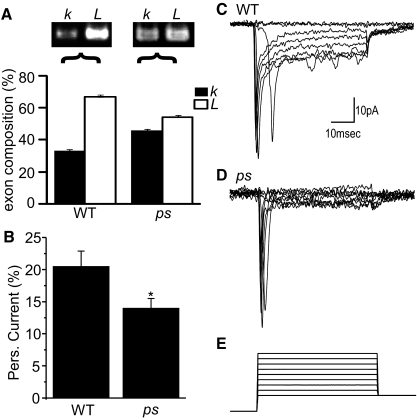
A screen for proteins sufficient to alter alternative splicing in Drosophila S2 cells identified Pasilla (Ps) to be sufficient to alter the k/L ratio in expressed DmNav (Park et al. 2004). In untreated cells roughly 95% of transcripts contain L (termed exon N in the original study); <0.5% of transcripts contained exon k (termed exon O in the study), with the remainder lacking both k and L. Significantly, RNAi knockdown of ps resulted in a greatly increased expression of k at the expense of L. However, whether Ps has the same activity in CNS has not been reported. Thus we examined the expression of exons k and L in the CNS of both WT (Canton-S) and ps mutants (ps2). Our analysis of DmNav, isolated from whole CNS, predictably shows that exon L predominates (67%) over k (33%) in WT (Fig. 6A). However, in the absence of ps (i.e., a similar, but likely stronger, reduction compared with RNAi knockdown), expression of exon k (46%) is increased relative to that of L (54%) (Fig. 6A). Thus as in S2 cells, the removal of ps is sufficient to increase exon k–containing DmNav transcripts at the expense of those containing L. We also see an increase in DmNav transcripts lacking both k and L in ps mutant embryos (data not shown).
FIG. 6.
The presence of exon k reduces persistent Na+ current in Drosophila motoneurons. A: comparison of expression of DmNav transcripts containing either exon k or L in both wild-type (WT, Canton-S) and pasilla (ps) mutant CNS in late-stage 17 embryos. Semiquantitative polymerase chain reaction (PCR) shows that, in WT, expression of L-containing DmNav predominates (66.8 ± 0.9%) relative to inclusion of exon k (33.2 vs. 0.9%). By contrast, in the absence of ps, exon frequencies are more similar (54.0 ± 0.8 vs. 46.0 ± 0.8%, L vs. k, respectively). The differences in exon composition between WT and ps are significant at P < 0.01. Example images of PCR products are shown for both WT and ps (images have been manipulated using Photoshop) and the average of 5 independent PCR experiments are shown (means ± SE). B: whole cell patch recordings from identified motoneurons (aCC and RP2) show that the percentage persistent Na+ current, relative to the transient component, is significantly reduced in the absence of ps (20.5 ± 2.4 vs. 14.0 ± 1.5%, WT vs. ps, respectively; P ≤ 0.05, n ≥ 10, mean ± SE). Persistent current was measured at the end of a depolarizing command voltage. The largest persistent current was normalized by division with the largest transient current obtained (i.e., not necessarily from the same voltage step). C and D: example whole cell traces of Na+ currents obtained from aCC/RP2 motoneurons in both WT (C) and ps (D) late-stage 17 embryos. The 2 traces were chosen because the peak transient amplitude was the same in both recordings (i.e., the 2 traces are not scaled). E: voltage steps (50 ms) from −60 to +20 mV (in 10-mV steps) were applied from a holding prepulse potential of −90 mV.
To determine whether the persistent Na+ current is also affected in ps mutants we used whole cell patch recordings from two identified motoneurons (termed aCC and RP2; see Muraro et al. 2008). Our results, which fully agree with our predictions based on heterologous expression in oocytes (see earlier text), show that the percentage of persistent Na+ current is significantly reduced in the absence of ps (i.e., increased expression of exon k; Fig. 6, B–E). By contrast, the VgNa+ transient current is not affected by loss of ps (107 ± 5.5 vs. 120 ± 11 pA, WT and ps, respectively, P > 0.05). The peak persistent Na+ current in WT aCC and RP2 (which is the same in both neurons; Muraro et al. 2008) is about 20% of the transient current component. In the absence of ps, this drops to 14% (Fig. 6B). These data fully validate our analysis of DmNav transcripts in oocytes and, moreover, indicate that the choice of whether exon L or k is present is, at least partially, dependent on the presence of the Ps protein in Drosophila CNS.
Cytoplasmic alternate exons are not required for channel function
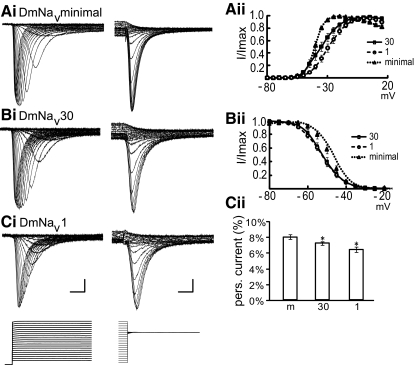
A key question we sought to ask was whether the presence of cytoplasmic alternate exons (j, i, a, b, e, f, and h) are absolutely required for channel function or whether their presence is required to facilitate only a change in channel kinetics. To answer this we constructed a DmNav clone that lacked all cytoplasmic spliced exons (termed DmNavminimal). Heterologous expression clearly shows that DmNavminimal is capable of producing functional Na+ channels (Fig. 7A). This outcome is highly indicative that cytoplasmic alternate exons evolved to facilitate diversity in channel properties.
FIG. 7.
Cytoplasmic spliced exons are not required for channel function. Removal of all cytoplasmic spliced exons (j, i, a, b, e, f, and h) does not abolish channel function, indicating that these exons are required to modify only gating properties. The minimal clone expressed contains only spliced exons d and L. Ai and Aii: comparison of DmNaVminimal with that of the most common embryonic (DmNaV30) and adult (DmNaV1) splice variant shows removal of all cytoplasmic spliced exons results in a significant hyperpolarization of V1/2 activation (see Table 3 for precise values). Bi and Bii: DmNaVminimal also exhibits a significantly depolarized V1/2 inactivation compared with either DmNaV1 or 30. Ci and Cii: DmNaVminimal exhibits the greatest degree of persistent current compared with either DmNaV1 or 30. Example traces for activation and inactivation are shown on the left and averaged values (means ± SE for n ≥ 8) on the right (DmNaVminimal is different from either DmNaV1 or 30 at P < 0.05 for all comparisons).
To begin to address how inclusion of cytoplasmic spliced exons alter channel properties, we compared the channel kinetics of DmNavminimal to that of both the predominant embryonic (DmNav30) and adult (DmNav1) DmNav splice variants. Figure 7 shows that DmNavminimal has a significantly hyperpolarized V1/2 activation voltage (−40.7 mV) compared with that of either DmNav30 (−34.9 mV) or DmNav1 (−28.1 mV). Thus it might be predicted that if neurons were to express DmNavminimal then they would exhibit increased membrane excitability (i.e., fire action potentials more easily). Interestingly, DmNavminimal also has a significantly depolarized V1/2 inactivation voltage (−49.1 mV) compared with that of either DmNav30 (−52.1 mV) or DmNav1 (−52.8 mV). This alteration would, again, be predicted to further increase membrane excitability (i.e., allowing action potential firing to continue for longer). Analysis of the persistent current component shows that this current, the presence of which increases membrane excitability, is also largest in DmNavminimal compared with that in DmNav30 or DmNav1 (8.1, 7.3, and 6.5%, respectively, Fig. 7). Taken together, the kinetics suggests that cells expressing DmNavminimal would likely be highly excitable and that the inclusion of cytoplasmic spliced exons has evolved primarily to reduce excitability to suit the requirements of neural signaling.
DISCUSSION
This study describes the cloning and characterization of complete splice isoforms of DmNav present in Drosophila embryos. Critically, it is during this stage that neurons first form the circuits that subsequently underpin larval behavior. The frequencies we observe for alternatively spliced exons a, b, c/d, e, and f are similar to those previously reported by Thackeray and Ganetzky (1994), indicating that our results are based on a sufficiently large number of clones to accurately represent the proportions of different transcripts in vivo. By analyzing the complete ORF, it has become apparent that exons j, h, and k are rarely used in the embryo, whereas exons i, f, and L are frequently included. Comparing our data to the frequency of spliced exons in DmNav ORFs recently isolated from adult CNS by O'Donnell Olson et al. (2008) reveals some striking differences. Although the number of splice isoforms isolated is remarkably similar (27 vs. 29), only 3 variants were isolated from both stages. The main source of disparity appears to be the utilization of exons j (which increases from 10% inclusion in the embryo to 89% in the adult) and f (which decreases from 78% inclusion in the embryo to 10% in the adult). Indeed, if these two loci are overlooked, then the most common splice isoform (i, a, b, d, and L) is the same in the embryo and the adult. Other differences between adult and embryo are a decrease in the inclusion of exon h (3 vs. 24%) and an increase in the usage of exon L (97 vs. 80%) in adult compared with embryo.
We also observe alternative splicing of several exons that were previously believed to be constitutive: exons 7, 8, 22, and 23 were found to be absent from the ORF of one or more clones, suggesting that their exclusion may be regulated splicing events, rather than isolated splicing catastrophes. Exon 7 is absent in 10% of clones, all of which have otherwise unique combinations of exons. Its exclusion is not predicted to disrupt the ORF and, consequently, it would be expected that these splice isoforms would encode an almost complete protein. However, these proteins would be missing regions of IS2–3 and, as such, it seems unlikely that they would be able to form functional channels. Indeed, para clones lacking the homologous sequence (exon K) in cockroach (BgNav) fail to form functional channels when expressed in Xenopus oocytes (Song et al. 2004). Exclusion of exons 8, 22, and 23 would result in a frame shift of the coding region and subsequent premature termination of translation. It is conceivable that the resulting truncated proteins serve functional roles, perhaps in modulating the activity of the complete protein. However, because the exclusion of these exons was found to be infrequent and in the case of exons 22 and 23, not previously reported, it seems probable that these isoforms most likely result from errors of the splicing process. Testing of these possibilities will require expression and analysis of these splice variants, both in isolation and in combination with other clones known to produce functional channels.
Three splice isoforms lacking both membrane-spanning exons k and L were also isolated. The function of the encoded protein is unclear because the channel would be missing IIIS3–4, a region critical for coupling depolarization across the membrane to activation and inactivation of the channel (Elinder et al. 2007). Isoforms of DmNav without this region have also been reported by Park et al. (2004). A splice isoform (termed Δ18) of the vertebrate voltage-gated sodium channel Nav1.6 lacking the homologous region has also been identified (see following text; Oh and Waxman 1998; Plummer et al. 1997). In the latter case, it has been speculated that any resulting channels may act as a “fail-safe” mechanism to prevent the expression of functional channels in nonneuronal cells (Plummer et al. 1997). We also identified one splice isoform lacking exon 8 (DmNav40), which encodes the equivalent voltage-sensing region in domain IS3–4 (see Fig. 1). Again, splice variants encoding vertebrate VgNa+ channels have been identified without the homologous region (exon 5) to DmNav exon 8 (Auld et al. 1988; Belcher et al. 1995; Gustafson et al. 1993; Lu and Brown 1998; Plummer et al. 1998; Sarao et al. 1991), suggesting that removal of these regions is evolutionarily conserved and, as such, may have functional implications. Moreover, splicing has also been reported in this region of mammalian VgNa+ channels with exon 5A being more common in adult brain and exon 5N in neonates (Gustafson et al. 1993).
Heterologous expression of DmNav clones clearly shows that splicing is sufficient to alter gating properties. Examination of multiple clones strongly suggests that spliced exons impart specific attributes to channel kinetics. In particular, the presence of exon f results in a hyperpolarizing shift in the V1/2 of activation indicative of increased excitability for those neurons that express f-containing variants. By the same analysis, the inclusion of exons j and e results in depolarized V1/2 activation voltages and possibly reduced excitability. Neurons expressing h-containing variants are predicted to be more excitable, given that this exon, when present, shifts V1/2 inactivation to more depolarized voltages. It is important to reiterate here that our splice variants were checked to ensure that they did not contain any PCR-induced errors that, if present, have been shown to impart diversity between identical clones equal to, or greater than, that between different splice variants (O'Donnell Olson et al. 2008).
DmNav clones expressed in oocytes and VgNa+ currents recorded in Drosophila neurons (Mee et al. 2004) exhibit a persistent (noninactivating) Na+ current. The identity of this current component in mammalian neurons remains unclear, with possibilities including an as yet unidentified channel subtype or a noninactivating mode of the transient (fast-inactivating) current component (Liu et al. 2004). By contrast, it has been known for many years that the insect Na+ current, now known to be encoded by a single gene, exhibits both transient and persistent components, thereby favoring the second hypothesis: a noninactivating mode of the transient current component (Mee et al. 2004; Vais et al. 2000). Additional support for the persistent current being a mode change in gating of the conventional transient current has recently been reported in cockroach, in which a U-to-C RNA edit (causing an F-to-S transition at AA 1919) greatly increases the persistent current component (Liu et al. 2004). Our data are also consistent with, and indeed indicative of, a switch in gating modes rather than a requirement for an unknown additional subtype. This is because we clearly show that splicing is sufficient to influence just the persistent-current component (without affecting the transient current). Splice variants containing membrane-spanning exon k have a significantly reduced persistent current compared with variants containing exon L. Most human Na+ channels show just over 50% identity to exons k and L, including a region of 11 residues (RTLRALRPLRA), that is conserved in all insect and vertebrate sodium channels (Fig. 8). These regions, termed exons 18A and 18N, are alternatively spliced in Nav1.1 and 1.6 (Diss et al. 2001; Oh and Waxman 1998; Plummer et al. 1997). Exon 18A predominates in adult brain and 18N in the embryo and in nonneuronal tissues. Significantly, 18N includes an in-frame stop codon and, as such, any resulting proteins would be truncated from this region onward. A third isoform, lacking both 18A and 18N, was also identified (Δ18) that is also present early in development and in nonneuronal tissues.
FIG. 8.
DmNav exons k and L are conserved in human voltage-gated Na+ channels. Mutually exclusive DmNav spliced exons k and L differ by 16/41 residues (shown in gray boxes in the exon k sequence). Analysis of exon 18 of human Nav1.1–1.9 shows that it is more similar to DmNav exon L (identical residues shown in black) than to exon k.
A screen of RNA-binding proteins identified Pasilla (Ps) to be sufficient to regulate splicing of mutually exclusive exons k and L (Park et al. 2004). We now confirm and extend this study by showing that Ps is sufficient to alter splicing of these exons in vivo. We show that in the absence of this RNA-binding protein, the inclusion of k in DmNav transcripts is increased at the expense of L. As a consequence, the persistent Na+ current present in two identified Drosophila motoneurons is reduced, validating our conclusions concerning the roles of these two mutually exclusive exons drawn from heterologous expression in oocytes. The function of Ps is thus likely to be involved in the spliceosome that regulates splicing at this region of DmNav. Ps is highly similar to the human Nova-1 and Nova-2 proteins, which are known neuron-specific alternative splicing factors (Ule et al. 2006). A binding motif (YCAY clusters) has been identified as being required for Nova binding in premessenger RNA and, significantly, such motifs are also present in the intron downstream of exon L (Park et al. 2004). Because the persistent current is an important target for many antiepileptic drugs, further analysis of the contribution of IIIS3–4, exons k and L in particular, and the regulation of splicing in this region are likely to be of both fundamental and clinical benefit.
It remains to be determined how inclusion of specific spliced exons changes the gating properties of DmNav. With the exception of exons c/d and k/L, all spliced exons are cytoplasmic. This fact alone is indicative that inclusion of spliced exons likely influences channel gating by facilitating posttranslational modifications and/or binding of cofactors. It is already known that phosphorylation can alter Na+ channel conductivity in both mammalian (Smith and Goldin 1997) and Drosophila neurons (Baines 2003). We used the prediction server NetPhos (http://www.cbs.dtu.dk/services/NetPhos) to predict 13 potential sites of phosphorylation within DmNav (Davies et al. 2007 predicts 11 such sites using DISPHOS 1.3). As noted, exons i and a are targets for phosphorylation (Loughney et al. 1989; O'Dowd et al. 1995). Other alternatively spliced regions predicted to contain targets of phosphorylation are j, e, k, and L. Modulation by phosphorylation at these regions may be responsible for the decrease in sodium current density observed on the elevation of protein kinase A (PKA) activity in Drosophila motoneurons (Baines 2003). It is equally conceivable that differential inclusion of exon sequences encoding phosphorylation sites may generate proteins with varying sensitivity to modulation by PKA, an area open to future investigation.
Phosphorylation of vertebrate and insect Na+ channels by protein kinase C (PKC) also leads, in most cases, to a reduction of current (Numann et al. 1991; West et al. 1991; Wicher 2001). Furthermore, a requirement for phosphorylation by PKC prior to phosphorylation by PKA has been identified (Li M et al. 1993). Unfortunately, no PKC-specific phosphorylation sites have yet been described in DmNav (but see Thimmapaya et al. 2005). West et al. (1991) showed that the intracellular loop between domains III and IV, of rat brain Na+ channels, likely contains a PKC phosphorylation site. However, this region lacks spliced exons in Drosophila embryos and adults (O'Donnell Olson et al. 2008). Additional posttranslational modifications include glycosylation of extracellular loops, specifically between S5 and S6 in homology domains I, II, and III (Davies et al. 2007). Loughney et al. (1989) identified a potential N-linked glycosylation site within a 5.5-kb portion of the para ORF by searching for the consensus recognition sequence N–X–(S/T) (X can be any amino acid apart from proline) (Hubbard and Ivatt 1981). This revealed that exon d contains one potential glycosylation site, whereas its equivalent, exon c, contained two such sites. How these modifications influence channel function remains unknown.
In summary, heterologous expression indicates that the inclusion of specific exons imparts characteristic gating properties to individual splice variants of DmNav channels. These differences likely contribute to, and may even explain, the diversity in action potential firing between different neurons observed in the Drosophila CNS. Verification of this will require the identification of exon usage in single, identified neurons that are accessible for electrophysiological recordings.
GRANTS
This work was supported by the Biotechnology and Biological Sciences Research Council (United Kingdom).
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey Warmke (Merck Research Laboratories, Rahway, NJ) for the para13-5 construct, Dr. Martin Williamson (Rothamsted Research, Harpenden, UK) for the tipE clone, and Dr. Ke Dong (Michigan State University, East Lansing, MI) for the pGH19 vector, Dr. Deborah Andrew (The Johns Hopkins University) for the gift of pasilla mutant flies, and the members of the Baines group and Dr. Kevin Moffat (University of Warwick) for help and advice during the course of this work.
Footnotes
The online version of this article contains supplemental data.
REFERENCES
- Auld 1988.Auld VJ, Goldin AL, Krafte DS, Marshall J, Dunn JM, Catterall WA, Lester HA, Davidson N, Dunn RJ. A rat brain Na+ channel alpha subunit with novel gating properties. Neuron 1: 449–461, 1988. [DOI] [PubMed] [Google Scholar]
- Baines 2003.Baines RA. Postsynaptic protein kinase A reduces neuronal excitability in response to increased synaptic excitation in the Drosophila CNS. J Neurosci 23: 8664–8672, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines 1998.Baines RA, Bate M. Electrophysiological development of central neurons in the Drosophila embryo. J Neurosci 18: 4673–4683, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher 1995.Belcher SM, Zerillo CA, Levenson R, Ritchie JM, Howe JR. Cloning of a sodium channel alpha subunit from rabbit Schwann cells. Proc Natl Acad Sci USA 92: 11034–11038, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose 2000.Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr Opin Neurobiol 10: 95–102, 2000. [DOI] [PubMed] [Google Scholar]
- Catterall 2000.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26: 13–25, 2000. [DOI] [PubMed] [Google Scholar]
- Charron 2007.Charron F, Tessier-Lavigne M. The Hedgehog, TGF-beta/BMP and Wnt families of morphogens in axon guidance. Adv Exp Med Biol 621: 116–133, 2007. [DOI] [PubMed] [Google Scholar]
- Davies 2007.Davies TG, Field LM, Usherwood PN, Williamson MS. A comparative study of voltage-gated sodium channels in the Insecta: implications for pyrethroid resistance in Anopheline and other Neopteran species. Insect Mol Biol 16: 361–375, 2007. [DOI] [PubMed] [Google Scholar]
- Derst 2006.Derst C, Walther C, Veh RW, Wicher D, Heinemann SH. Four novel sequences in Drosophila melanogaster homologous to the auxiliary Para sodium channel subunit TipE. Biochem Biophys Res Commun 339: 939–948, 2006. [DOI] [PubMed] [Google Scholar]
- Diss 2001.Diss JK, Archer SN, Hirano J, Fraser SP, Djamgoz MB. Expression profiles of voltage-gated Na(+) channel alpha-subunit genes in rat and human prostate cancer cell lines. Prostate 48: 165–178, 2001. [DOI] [PubMed] [Google Scholar]
- Elinder 2007.Elinder F, Nilsson J, Arhem P. On the opening of voltage-gated ion channels. Physiol Behav 92: 1–7, 2007. [DOI] [PubMed] [Google Scholar]
- Feng 1995.Feng G, Deak P, Chopra M, Hall LM. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell 82: 1001–1011, 1995. [DOI] [PubMed] [Google Scholar]
- Goldin 2001.Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol 63: 871–894, 2001. [DOI] [PubMed] [Google Scholar]
- Goodman 1993.Goodman CS, Shatz CJ. Developmental mechanisms that generate precise patterns of neuronal connectivity. Cell Suppl 72: 77–98, 1993. [DOI] [PubMed] [Google Scholar]
- Gustafson 1993.Gustafson TA, Clevinger EC, O'Neill TJ, Yarowsky PJ, Krueger BK. Mutually exclusive exon splicing of type III brain sodium channel alpha subunit RNA generates developmentally regulated isoforms in rat brain. J Biol Chem 268: 18648–18653, 1993. [PubMed] [Google Scholar]
- Hubbard 1981.Hubbard SC, Ivatt RJ. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem 50: 555–583, 1981. [DOI] [PubMed] [Google Scholar]
- Huguenard 2002.Huguenard JR. Sodium channels: grit, determination, and persistence. Neuron 33: 492–494, 2002. [DOI] [PubMed] [Google Scholar]
- Kuo 1997.Kuo CC, Chen RS, Lu L, Chen RC. Carbamazepine inhibition of neuronal Na+ currents: quantitative distinction from phenytoin and possible therapeutic implications. Mol Pharmacol 51: 1077–1083, 1997. [DOI] [PubMed] [Google Scholar]
- Lampl 1998.Lampl I, Schwindt P, Crill W. Reduction of cortical pyramidal neuron excitability by the action of phenytoin on persistent Na+ current. J Pharmacol Exp Ther 284: 228–237, 1998. [PubMed] [Google Scholar]
- Lee 2002.Lee SH, Ingles PJ, Knipple DC, Soderlund DM. Developmental regulation of alternative exon usage in the house fly Vssc1 sodium channel gene. Invert Neurosci 4: 125–133, 2002. [DOI] [PubMed] [Google Scholar]
- Levitt 1997.Levitt P, Eagleson KL, Chan AV, Ferri RT, Lillien L. Signaling pathways that regulate specification of neurons in developing cerebral cortex. Dev Neurosci 19: 6–8, 1997. [DOI] [PubMed] [Google Scholar]
- Li 1993.Li M, West JW, Numann R, Murphy BJ, Scheuer T, Catterall WA. Convergent regulation of sodium channels by protein kinase C and cAMP-dependent protein kinase. Science 261: 1439–1442, 1993. [DOI] [PubMed] [Google Scholar]
- Li 2003.Li Y, Bennett DJ. Persistent sodium and calcium currents cause plateau potentials in motoneurons of chronic spinal rats. J Neurophysiol 90: 857–869, 2003. [DOI] [PubMed] [Google Scholar]
- Li 2004.Li Y, Gorassini MA, Bennett DJ. Role of persistent sodium and calcium currents in motoneuron firing and spasticity in chronic spinal rats. J Neurophysiol 91: 767–783, 2004. [DOI] [PubMed] [Google Scholar]
- Liu 2004.Liu Z, Song W, Dong K. Persistent tetrodotoxin-sensitive sodium current resulting from U-to-C RNA editing of an insect sodium channel. Proc Natl Acad Sci USA 101: 11862–11867, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney 1989.Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 58: 1143–1154, 1989. [DOI] [PubMed] [Google Scholar]
- Lu 1998.Lu CM, Brown GB. Isolation of a human-brain sodium-channel gene encoding two isoforms of the subtype III alpha-subunit. J Mol Neurosci 10: 67–70, 1998. [DOI] [PubMed] [Google Scholar]
- Marder 2002.Marder E, Prinz AA. Modeling stability in neuron and network function: the role of activity in homeostasis. Bioessays 24: 1145–1154, 2002. [DOI] [PubMed] [Google Scholar]
- Mee 2004.Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci 24: 8695–8703, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki 1996.Miyazaki M, Ohyama K, Dunlap DY, Matsumura F. Cloning and sequencing of the para-type sodium channel gene from susceptible and kdr-resistant German cockroaches (Blattella germanica) and house fly (Musca domestica). Mol Gen Genet 252: 61–68, 1996. [PubMed] [Google Scholar]
- Muraro 2008.Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci 28: 2099–2109, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin 2006.Nikitin ES, Kiss T, Staras K, O'Shea M, Benjamin PR, Kemenes G. Persistent sodium current is a target for cAMP-induced neuronal plasticity in a state-setting modulatory interneuron. J Neurophysiol 95: 453–463, 2006. [DOI] [PubMed] [Google Scholar]
- Numann 1991.Numann R, Catterall WA, Scheuer T. Functional modulation of brain sodium channels by protein kinase C phosphorylation. Science 254: 115–118, 1991. [DOI] [PubMed] [Google Scholar]
- O'Donnell Olson 2008.O'Donnell Olson R, Liu Z, Nomura Y, Song W, Dong K. Molecular and functional characterization of voltage-gated sodium channel variants from Drosophila melanogaster. Insect Biochem Mol Biol 38: 604–610, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dowd 1995.O'Dowd DK, Gee JR, Smith MA. Sodium current density correlates with expression of specific alternatively spliced sodium channel mRNAs in single neurons. J Neurosci 15: 4005–4012, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh 1998.Oh Y, Waxman SG. Novel splice variants of the voltage-sensitive sodium channel alpha subunit. Neuroreport 9: 1267–1272, 1998. [DOI] [PubMed] [Google Scholar]
- Park 2004.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA 101: 15974–15979, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer 1998.Plummer NW, Galt J, Jones JM, Burgess DL, Sprunger LK, Kohrman DC, Meisler MH. Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics 54: 287–296, 1998. [DOI] [PubMed] [Google Scholar]
- Plummer 1997.Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J Biol Chem 272: 24008–24015, 1997. [DOI] [PubMed] [Google Scholar]
- Romano 1996.Romano AG, Harvey JA. Prenatal exposure to cocaine disrupts discrimination learning in adult rabbits. Pharmacol Biochem Behav 53: 617–621, 1996. [DOI] [PubMed] [Google Scholar]
- Sarao 1991.Sarao R, Gupta SK, Auld VJ, Dunn RJ. Developmentally regulated alternative RNA splicing of rat brain sodium channel mRNAs. Nucleic Acids Res 19: 5673–5679, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer 2004.Schnorrer F, Dickson BJ. Axon guidance: morphogens show the way. Curr Biol 14: R19–R21, 2004. [DOI] [PubMed] [Google Scholar]
- Seshaiah 2001.Seshaiah P, Miller B, Myat MM, Andrew DJ. pasilla, the Drosophila homologue of the human Nova-1 and Nova-2 proteins, is required for normal secretion in the salivary gland. Dev Biol 239: 309–322, 2001. [DOI] [PubMed] [Google Scholar]
- Siegel 1994.Siegel M, Marder E, Abbott LF. Activity-dependent current distributions in model neurons. Proc Natl Acad Sci USA 91: 11308–11312, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith 1997.Smith RD, Goldin AL. Phosphorylation at a single site in the rat brain sodium channel is necessary and sufficient for current reduction by protein kinase A. J Neurosci 17: 6086–6093, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song 2004.Song W, Liu Z, Tan J, Nomura Y, Dong K. RNA editing generates tissue-specific sodium channels with distinct gating properties. J Biol Chem 279: 32554–32561, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadoni 2002.Spadoni F, Hainsworth AH, Mercuri NB, Caputi L, Martella G, Lavaroni F, Bernardi G, Stefani A. Lamotrigine derivatives and riluzole inhibit INa, P in cortical neurons. Neuroreport 13: 1167–1170, 2002. [DOI] [PubMed] [Google Scholar]
- Spitzer 2006.Spitzer NC. Electrical activity in early neuronal development. Nature 444: 707–712, 2006. [DOI] [PubMed] [Google Scholar]
- Spitzer 2002.Spitzer NC, Kingston PA, Manning TJ, Conklin MW. Outside and in: development of neuronal excitability. Curr Opin Neurobiol 12: 315–323, 2002. [DOI] [PubMed] [Google Scholar]
- Stanwood 2001.Stanwood GD, Washington RA, Shumsky JS, Levitt P. Prenatal cocaine exposure produces consistent developmental alterations in dopamine-rich regions of the cerebral cortex. Neuroscience 106: 5–14, 2001. [DOI] [PubMed] [Google Scholar]
- Tan 2002.Tan J, Liu Z, Nomura Y, Goldin AL, Dong K. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J Neurosci 22: 5300–5309, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazerart 2008.Tazerart S, Vinay L, Brocard F. The persistent sodium current generates pacemaker activities in the central pattern generator for locomotion and regulates the locomotor rhythm. J Neurosci 28: 8577–8589, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray 1994.Thackeray JR, Ganetzky B. Developmentally regulated alternative splicing generates a complex array of Drosophila para sodium channel isoforms. J Neurosci 14: 2569–2578, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray 1995.Thackeray JR, Ganetzky B. Conserved alternative splicing patterns and splicing signals in the Drosophila sodium channel gene para. Genetics 141: 203–214, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmapaya 2005.Thimmapaya R, Neelands T, Niforatos W, Davis-Taber RA, Choi W, Putman CB, Kroeger PE, Packer J, Gopalakrishnan M, Faltynek CR, Surowy CS, Scott VE. Distribution and functional characterization of human Nav1.3 splice variants. Eur J Neurosci 22: 1–9, 2005. [DOI] [PubMed] [Google Scholar]
- Ule 2006.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586, 2006. [DOI] [PubMed] [Google Scholar]
- Vais 2000.Vais H, Williamson MS, Goodson SJ, Devonshire AL, Warmke JW, Usherwood PN, Cohen CJ. Activation of Drosophila sodium channels promotes modification by deltamethrin. Reductions in affinity caused by knock-down resistance mutations. J Gen Physiol 115: 305–318, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke 1997.Warmke JW, Reenan RA, Wang P, Qian S, Arena JP, Wang J, Wunderler D, Liu K, Kaczorowski GJ, Van der Ploeg LH, Ganetzky B, Cohen CJ. Functional expression of Drosophila para sodium channels. Modulation by the membrane protein TipE and toxin pharmacology. J Gen Physiol 110: 119–133, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West 1991.West JW, Numann R, Murphy BJ, Scheuer T, Catterall WA. A phosphorylation site in the Na+ channel required for modulation by protein kinase C. Science 254: 866–868, 1991. [DOI] [PubMed] [Google Scholar]
- Wicher 2001.Wicher D. Peptidergic modulation of an insect Na(+) current: role of protein kinase A and protein kinase C. J Neurophysiol 85: 374–383, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.