Abstract
The evolution of genome size as well as structure and organization of genomes belongs among the key questions of genome biology. Here we show, based on a comparative analysis of 30 genomes, that there is generally a tight correlation between the number of genes per chromosome and the length of the respective chromosome in eukaryotic genomes. The surprising exceptions to this pattern are placental mammalian genomes. We identify the number and, more importantly, the uneven distribution of gene deserts among chromosomes, i.e., long (>500 kb) stretches of DNA that do not encode for genes, as the main contributing factor for the observed anomaly of eutherian genomes. Gene-rich placental mammalian chromosomes have smaller proportions of gene deserts and vice versa. We show that the uneven distribution of gene deserts is a derived character state of eutherians. The functional and evolutionary significance of this particular feature of eutherian genomes remains to be explained.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-009-9251-4) contains supplementary material, which is available to authorized users.
Keywords: Genome evolution, Gene deserts, Genome size, Mammalia
Introduction
One of the major challenges of genome biology is the understanding of how genomes are organized and how genes are distributed in genomes of different sizes. This question was brought to the fore and attracted interest after it was discovered that genomes of vastly different sizes often contain relatively similar numbers of genes (Bork and Copley 2001; Gregory 2005; Lynch 2007). Recent comparative genomic studies have uncovered some general trends in the evolution of genome size. For example, it appears that larger genomes contain proportionally fewer genes compared with smaller ones (see, e.g., Gregory 2005); however, they are characterized by higher numbers of transposable elements (TEs; Kidwell 2002; Lynch and Conery 2003). It has further been shown that the total number of genes in a genome is strongly correlated with the length of the protein-coding sequence in both prokaryotic and eukaryotic genomes, suggesting that gene length is highly conserved within each of these two anciently diverged lineages (Xu et al. 2006, which includes most of the taxa reported herein). However, whereas gene number and genome size are correlated in prokaryotes (Gregory and DeSalle 2005), such a trend does not exist in eukaryotic genomes, in which only a small fraction of the nuclear DNA is protein-coding. Finally, the whole genome sequencing of mammals during the last years (see, e.g., Lander et al. 2001; Lindblad-Toh et al. 2005; Venter et al. 2001) showed that genes are not evenly distributed in a given genome and that substantial fractions of mammalian genomes— ≤25% in the case of Homo sapiens—are made up of so-called gene deserts, i.e., long regions >500 kb in length that are devoid of any genes (Venter et al. 2001). Here we show that in all eukaryotic genomes analyzed, except in placental mammalian ones, the number of genes per chromosome is strongly correlated with chromosome length, irrespective of genome size, chromosome number, and taxonomy. We then tested whether particular genomic features, such as repetitive elements or gene deserts, may account for this difference. We found that the distinctiveness of placental mammal genomes can be best explained by the uneven distribution of gene deserts.
Materials and Methods
Data Mining
We obtained chromosome length data from the University of California Santa Cruz (UCSC) genome browser (Karolchik et al. 2003) and the European Molecular Biology Laboratory (EMBL) European Bioinformatics Institute (EBI) Web site (http://www.ebi.ac.uk). Coding genes and their number by chromosome were obtained from the EBI website for the following species chosen to be representative for the major lineage of organismal diversity (in alphabetical order): Arabidopsis thaliana (thale cress), Aspergillus fumigatus, Bos taurus (domestic cow), Caenorhabditis briggsae (roundworm), Candida glabrata (candida yeast), H. sapiens (human), Leishmania major, Mus musculus (house mouse), Oryza sativa (domestic rice), Ostreococcus lucimarinus, Plasmodium falciparum, Saccharomyces cerevisiae (baker’s yeast), and Vitis vinifera (common grape wine). We also used the UCSC genome browser for data from Anopheles gambiae (mosquito), C. elegans (roundworm), Canis lupus familiaris (dog), Ciona intestinalis (vase tunicate), Danio rerio (zebrafish), Drosophila melanogaster (fruit fly), Equus caballus (horse), Gallus gallus (chicken), Gasterosteus aculeatus (three-spine stickleback), Macaca mulatta (rhesus monkey), Monodelphis domestica (gray short-tailed opossum), Ornithorhynchus anatinus (platypus), Oryzias latipes (Japanese killifish), Pan troglodytes (chimpanzee), Rattus norvegicus (brown rat) and Tetraodon nigroviridis (green spotted puffer fish). In addition we used National Center for Biotechnology Information GenBank and the Maize Genetics and Genomics Database for estimations for Zea mays (maize) (http://www.maizegdb.org). All data were downloaded in March 2008.
Data for the number of TEs, long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and simple repeats were obtained from the UCSC genome browser (Karolchik et al. 2003) for A. gambiae, B. taurus, C. briggsae, C. elegans, C. lupus familiaris, C. intestinalis, D. rerio, D. melanogaster, E. caballus, G. gallus, G. aculeatus, H. sapiens, M. mulatta, M. domestica, M. musculus, O. anatinus, O. latipes, P. troglodytes, R. norvegicus, and T. nigroviridis, and from the Arabidopsis information resource (i.e., TAIR) for A. thaliana. All data were downloaded in March 2008.
We used the information provided by the UCSC genome browser to identify intergenic regions in A. gambiae, B. taurus, C. briggsae, C. elegans, C. lupus familiaris, C. intestinalis, D. rerio, D. melanogaster, E. caballus, G. gallus, G. aculeatus, H. sapiens, M. mulatta, M. domestica, M. musculus, O. anatinus, O. latipes, P. troglodytes, R. norvegicus, S. cerevisiae, T. nigroviridis, and V. vinifera. Data for P. falciparum were obtained from the Plasmodium genome database (Kissinger et al. 2002). No information on the size of intergenic regions was available for A. thaliana, A. fumigatus, L. major, O. sativa, and Z. mays. However, it has already been shown that the larger genome of some plant species (e.g., Z. mays) is due to the expansion in number of transposable elements (Kidwell 2002; Messing et al. 2004). We follow the definition provided by Venter et al. (2001), who classified all intergenic regions >500 kb as gene deserts (see Table 1 for a list of taxa, their genome sizes, and the number and percentage fraction of gene deserts). We note that other investigators have applied a modified classification in mammals, defining only the 3% longest intergenic intervals as gene deserts (Ovcharenko et al. 2005). However, this strategy was not suitable for our approach comparing a variety of organisms because it would have led to the classification of very short intergenic regions as gene deserts in nonvertebrate genomes (e.g., 12 to 70 kb in C. elegans or 2.5 to 80 kb in S. cerevisiae). In addition, this approach would require an a priori acceptance of the existence of gene deserts in any given genome.
Table 1.
Organisms used in this study and information about their genomesa
| Taxon | Assembly size (Mb) | Deserts (n) | Deserts (%) | 18S GenBank accession number |
|---|---|---|---|---|
| P. troglodytes | 3175.6 | 949 | 36.80 | AC183378 |
| H. sapiens | 3047.0 | 915 | 38.33 | NR_003286 |
| M. mulatta | 2863.7 | 810 | 30.44 | CN805008 |
| M. musculus | 2654.9 | 895 | 34.26 | NR_003278 |
| R. norvegicus | 2718.9 | 743 | 23.19 | X01117 |
| C. lupus familaris | 2445.1 | 552 | 20.14 | DQ287955 |
| B. taurus | 2422.9 | 149 | 4.26 | DQ222453 |
| E. caballus | 2367.1 | 573 | 23.13 | AJ311673 |
| M. domestica | 3431.4 | 1098 | 27.77 | AJ311676 |
| O. anatinus | 1843.0 | 161 | 8.37 | AJ311679 |
| G. gallus | 1031.9 | 200 | 16.13 | AF173612 |
| D. rerio | 1277.1 | 129 | 7.10 | XR_045186 |
| O. latipes | 724.2 | 29 | 6.06 | AB105163 |
| G. aculeatus | 400.9 | 0 | 0.00 | DW607648 |
| T. nigroviridis | 217.4 | 5 | 1.17 | AJ270032 |
| C. intestinalis | 938.1 | 0 | 0.00 | AB013017 |
| A. gambiae | 228.2 | 0 | 0.00 | AM157179 |
| D. melanogaster | 120.4 | 0 | 0.00 | EU188739 |
| C. elegans | 100.3 | 0 | 0.00 | AY284652 |
| C. briggsae | 91.2 | 0 | 0.00 | U13929 |
| A. fumigatus | 29.4 | 0 | 0.00 | NT_166520 |
| C. glabrata | 12.3 | 0 | 0.00 | AF114470 |
| S. cervisiae | 12.1 | 0 | 0.00 | J01353 |
| Z. mays | 1979.4 | 0 | 0.00 | NC_008332 |
| A. thaliana | 119.2 | 0 | 0.00 | X16077 |
| O. sativa | 370.8 | 0 | 0.00 | AY120865 |
| V. vinifera | 303.1 | 0 | 0.00 | AF321270 |
| O. lucimarinus | 13.2 | 0 | 0.00 | DQ007077 |
| L. major | 32.8 | 0 | 0.00 | NC_007268 |
| P. falciparum | 22.9 | 0 | 0.00 | NC_004325 |
Deserts (n)—number of gene deserts (>500 kB) in a given genome; deserts (%)—size fraction of gene deserts (>500 kB) in a given genome; 18S acc. no
aGenBank accession numbers of 18S sequences used for regression equation mapping are also given. Data were obtained from GenBank. Note that no information on the size of intergenic regions was available for A. thaliana, A. fumigatus, L. ajor, O. sativa, and Z. mays. Also note that assembly size does not necessarily equal actual genome size
Analysis of Genome Data
To test the hypothesis that gene number and chromosome length are correlated, we first plotted the number of genes per chromosome (NG) versus chromosome length (LC) for the genomes mentioned previously. Some limitations exist in the inference of genome and chromosome sizes based only on sequence data (Gregory 2005), which is in part caused by the fact that genome sequences are rarely complete. However, these slight differences in completeness among the sequenced genomes should not affect our analyses. We used only genomes for which a continuous genome assembly, in the form of individual chromosomes, was available; therefore, a satisfactory coverage of these genomes can be assured. In addition, no systematic a priori bias in genome completeness can be assumed. We used the square of the correlation coefficient (R2) to describe the goodness-of-fit of the data to the hypothesized correlation between NG and LC. In addition, we performed pairwise comparison using Tukey-Kramer method for unplanned comparisons among a set of regression coefficients to identify those pairs of genomes that show significantly different correlations of NG/LC.
We then plotted the total number of TEs, LINEs, SINEs, simple repeats, and gene deserts against genome size for those organisms for which these data were available. The same procedure was followed with the numbers of TEs, LINEs, SINEs, simple repeats, and gene deserts per chromosome. To further evaluate the contribution of gene deserts to the distribution of genes on chromosomes in mammalian genomes, we plotted the relative proportion of genes per chromosome (NG/TG) plus the relative proportion of gene deserts (ND/TD) versus chromosome length (LC), where NG is the number of genes per chromosome; TG is the total number of genes in a genome; ND is the number of gene deserts per chromosome; and TD is the total number of gene deserts in a genome. For mammals, we also plotted the length-corrected sum of the relative proportion of genes per chromosome plus the relative proportion of gene deserts ((NG/TG + ND/TD)/LC) for each chromosome. In addition, to test whether the number of LINEs, SINEs, LTRs, DNA transposons, simple repeats, or gene deserts compensates best for varying gene densities in mammalian chromosomes, we performed a partial regression analysis based on NG and LC.
Regression Equation Mapping
We mapped the number of gene deserts as well as their relative proportion in the respective genome onto a phylogeny based on 18S rRNA sequences (see Table 1 for GenBank accession numbers) that was in concordance with recent phylogenomic studies (see, e.g., Delsuc et al. 2006; Dunn et al. 2008; Lartillot et al. 2007). We performed a maximum likelihood analysis and 100 maximum likelihood bootstrap replicates with PHyML (Guindon and Gascuel 2003) using the TrN + Γ model of sequence evolution according to ModelGenerator (Keane et al. 2006). To determine whether there is a correlation between phylogenetic position and number and relative proportion of gene deserts, we used an independent contrast analysis (Garland and Ives 2000) as implemented in the phenotypic diversity analysis program in the Mesquite package (Maddison and Maddison 2004).
Results
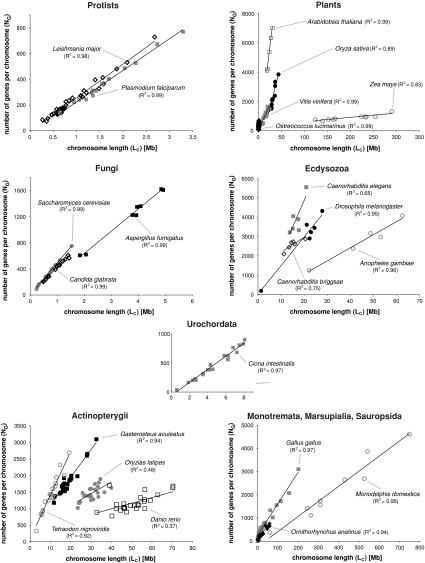
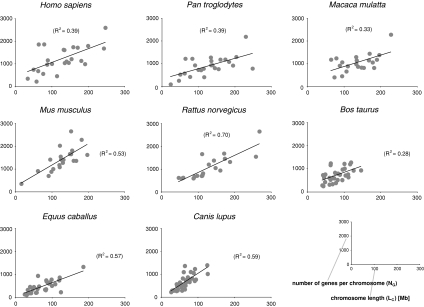
The plot of the number of genes per chromosome against chromosome length showed very strong correlations for nonmammalian genomes as well as for platypus (monotremes) and opossum (marsupials) (Fig. 1), whereas placental mammalian (eutherian) genomes deviate from this eukaryote-wide trend (Fig. 2). R2 values for the plot of NG/LC typically are approximately ≥0.9 in noneutherian genomes, whereas they are approximately ≤0.6 in mammals. There are only few exceptions to this pattern: The only two nonmammalian species with R2 < 0.6 are medaka (O. latipes; R = 0.48) and zebrafish (D. rerio; R = 0.37). These relatively low R2 values appear to be due to the relative equal size of chromosomes in O. latipes and by two outlier chromosomes in D. rerio (if those two outlier chromosomes are removed in D. rerio, R2 is <0.76). The two nematodes (C. briggsae and C. elegans) also have relatively small R2 values, which might be due to their small number of chromosomes and the small range of sizes of such. The only placental taxon showing R2 > 0.6 is the rat (R. norvegicus; R2 = 0.70).
Fig. 1.
The relationship between the number of genes per chromosome (NG) over chromosome length (LC) shows a strong correlation in nonmammals, irrespective of genome size, chromosome number and taxonomy. In noneutherian genomes, the slope of the trend-line can be interpreted as measurement for genome-compactness. The small R2-value in zebrafish (Danio rerio; 0.37) can be explained by two outlier chromosomes, that of medaka (Oryzias latipes; 0.48) by the relatively equal size of its chromosomes and variance in the number of genes. The somewhat smaller R2-value observed in Caenorhabditis elegans (R2 = 0.65) is due to the relatively even length of its chromosomes (Nelson et al. 2004), which makes it difficult to test for a linear relationship between NG and LC
Fig. 2.
The correlation between NG and LC is weak in genomes of placental mammals. In general, larger chromosomes also tend to have more genes in mammals; however, many chromosomes significantly deviate from a constant NG/LC ratio, rendering the genome-wide trend much weaker in placental mammalian genomes than in all other genomes. The highest R2 value in a mammal was found for rat (R2 = 0.70) whose genome contains the smallest relative fraction of gene deserts
The trend of linear correlation of NG/LC in nonplacental mammals but not in placental mammals was further supported by the pairwise comparison of regression coefficients by means of Tukey-Kramer method for unplanned comparisons. This test showed significant differences in most comparisons between eutherian and noneutherian genomes. The exceptions concerned all pairwise comparisons with both Caenorhabditis species and R. norvegicus as well as some comparisons involving D. rerio and O. latipes. None of the pairwise comparisons between two noneutherian (with the exception of D. rerio and O. latipes as explained previously) or two eutherian genomes showed significant differences in their regression coefficients, thus pointing to a deviation from a linear relation between NG and LC only in eutherian genomes.
Repetitive elements could be one factor to explain the nonlinearity of NG/LC in mammals. However, we found that the number of TEs, LINEs, and SINEs in a genome is correlated with genome size but not with the number of genes in a particular genome. The R2 values for the plots of TEs, LINEs, SINEs, and long terminal repeats (LTRs) against genome size were 0.93, 0.84, 0.87, and 0.82, respectively. When plotted against the number of genes, the R2 values were all <0.04. The numbers of TEs, LINEs, and SINEs per chromosome also correlate with chromosome length. For example, regarding the human genome—the most complete and best-assembled genome of all—the corresponding R2 values were 0.95 for TEs, 0.96 for LINEs, and 0.80 for SINEs (Supplementary Fig. 1). In addition, the number of DNA transposons (R2 = 0.94), LTRs (R2 = 0.94), and simple repeats (R2 = 0.97) per chromosome was correlated with chromosome size. Thus, because each class of repetitive elements correlates with chromosome length, none of these genomic features seems to account for the uneven gene density on eutherian chromosomes.
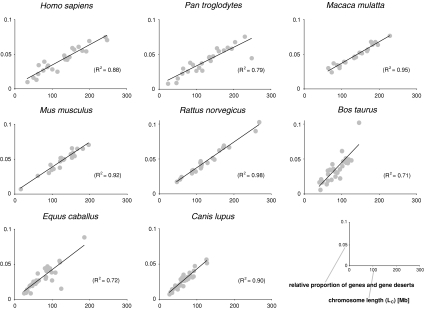
The situation appears different when gene deserts (intergenic regions >500 kb in size) are considered. That gene deserts counterbalance the number of genes on eutherian chromosomes is best illustrated by plotting the sum of the proportion of genes per chromosome plus the proportion of gene deserts per chromosome against chromosome size ((NG/TG + ND/TD)/LC) (Fig. 3). This factoring of (ND/TD) into the equation (NG/TG/LC) leads to strong correlations in placental genomes with R2 values between 0.71 (B. taurus) and 0.98 (R. norvegicus). That gene deserts counterbalance gene densities is further supported by the observation that the length-corrected sum of the relative proportion of genes per chromosome plus the relative proportion of gene deserts ((NG/TG + ND/TD)/LC) appears relatively constant for each chromosome, except for the Y chromosome (data not shown). Most importantly, the partial regression analysis showed that in the genomes of placental mammals, gene deserts, together with SINEs, have the highest partial regression coefficients with highly significant p values (<0.01) (Table 2). This suggests that of all the genomic features analyzed, gene deserts are the ones that best account for and contribute to the large variation in gene densities among mammalian chromosomes. In some genomes, LINEs and simple repeats also showed p values <0.01.
Fig. 3.
Gene deserts counterbalance the number of genes on mammalian chromosomes. The sum of the proportion of genes per chromosome plus the proportion of gene deserts per chromosome is plotted against chromosome length
Table 2.
Results from the partial regression analysisa
| Feature | H. sapiens | P. troglodytes | M. mulatta | M. musculus | R. norvegicus | C. familiaris | B. taurus | E. caballus |
|---|---|---|---|---|---|---|---|---|
| LINEs | 0.330 | 0.260 | 0.156 | 0.081 | 0.149 | −0.441* | 0.537* | 0.132 |
| SINEs | 0.618* | 0.901* | 0.322 | 0.743* | 0.816* | 0.832* | 0.666* | 0.3562* |
| LTRs | −0.0218 | 0.034 | 0.001 | 0.452 | 0.121 | −0.095 | −0.620 | −0.2172 |
| DNA transp. | −0.233 | −0.303 | 0.001 | 0.132 | 0.164 | 0.140 | 0.395 | 0.305 |
| Simple repeats | −0.132 | −0.156 | 0.127 | −0.374 | 0.041 | −0.339* | −0.542* | 0.186 |
| Gene deserts | 0.533* | 0.883* | 0.532* | 0.353 | 0.603* | 0.837* | 0.941* | 0.513* |
aThe partial regression coefficients for the respective contribution to NG/LC is given for LINEs, SINEs, LTRs, DNA transposons, simple repeats, and gene deserts. The highest coefficient for each genome is shown in bold, and significant values (p > 0.01) are marked with an asterisk
The mapping of the number of gene deserts as well as their relative proportion in the respective genome onto the phylogeny showed a substantial expansion of the number of gene deserts in the lineage leading to the placental mammals. Independent contrast analysis showed that the observed trend is statistically significant (p > 0.01).
Discussion
We first tested—in 30 plant, fungal, and animal genomes for which this information was available—whether or not a linear correlation exists between NG and LC on which they are located. When plotting the number of genes per chromosome versus chromosome length, we observed a strong linear correlation in eukaryotic genomes, with the notable exception of placental mammals (Fig. 1). R2 values were typically >0.9, suggesting relatively constant ratios of gene number per chromosome length for all chromosomes in a given genome. However, the genomes of placental mammals did not show constant chromosomal gene densities (Fig. 2), and R2 values were much lower and only ranged from 0.28 to 0.70.
The difference in NG/LC between nonplacental and placental genomes is further substantiated by a pairwise comparison of regression coefficients by means of Tukey-Kramer method for unplanned comparisons, which showed significant differences in most pairwise comparisons between mammalian and nonmammalian genomes. The exceptions concerned all pairwise comparisons with both Caenorhabditis species and R. norvegicus as well as some comparisons involving D. rerio and O. latipes. This could be explained by the similar size of the chromosomes in O. latipes as well as in both Caenorhabditis species (which also seems to be responsible for the comparably low R2 values in the plot of NG/LC of 0.65 and 0.75); the relatively small and evenly distributed gene deserts in R. norvegicus (leading to the highest R2 value of 0.70 among mammals); and two outlier chromosomes in D. rerio. Importantly, none of the pairwise comparisons between two nonplacental or placental genomes showed significant differences in their regression coefficients, which strongly points to a deviation from a linear relation between NG and LC only in eutherian genomes.
We hypothesized that this characteristic in the genomes of placental mammals might be due to particular genomic features of this group. We therefore examined whether the distribution of repeats, TEs, subclasses thereof (LINEs, SINEs), or gene deserts might be responsible for the unexpected variation in gene densities on different mammalian chromosomes. To this end, we plotted the number of TEs, LINEs, SINEs, simple repeats, and gene deserts against genome size and chromosome length. We found that although the number of TEs, LINEs, and SINEs is strongly correlated with genome size itself (as already shown by, e.g., Kidwell 2002; Lynch and Conery 2003) and also with chromosome length, none of these classes of repeat elements contributes particularly strongly to the observed pattern (Supplementary Fig. 1). Instead, it appears that the uneven distribution of gene deserts on mammalian chromosomes accounts for the deviations from otherwise constant ratios of NG/LC.
The plot of the sum of the relative proportion of genes per chromosome plus the relative proportion of gene deserts per chromosome versus chromosome length showed a strong correlation in placental mammals, with R2 values ranging from 0.71 (cow) to 0.98 (rat) (Fig. 3). This suggests that the distribution of genes and gene deserts counterbalance one another and predicts that, in eutherians, chromosomes with fewer genes have proportionally more gene deserts and vice versa. Indeed, such an observation has already been reported from the human genome, in which the proportion of gene deserts in the gene-rich chromosomes 17, 19, and 22 is less than half compared with the gene-poor chromosomes 4, 13, and 18 (Venter et al. 2001). In addition, partial regression analysis showed that the number of gene deserts, together with SINEs, contribute most to the lack of fit between the number of genes and the length of the respective chromosomes in placental mammals (Table 2). Note that in some genomes, LINEs and simple repeats also showed p values <0.01. This is not surprising, given that SINEs, LINEs, and simple repeats are not completely independent from gene deserts, which have been shown to be enriched with such repetitive elements (Ovcharenko et al. 2005). However, because these repetitive elements are in general evenly distributed across genomes (see Supplementary Fig. 1 for human), they are unlikely to account for the uneven distribution of gene numbers on eutherian chromosomes.
A randomization test for phylogenetic signal showed that both the number of gene deserts and their relative proportion in a genome are significantly associated with the organisms’ phylogenetic position (p < 0.01). This suggests that the genomic organization of placental mammals, characterized by a much higher number and an uneven distribution of gene deserts, is a derived state among the studied genomes. The number of gene deserts in a genome is most likely dependent on genome size, and it will be interesting to see how many gene deserts can be identified in very large genomes, such as lungfish or salamander, and whether per-chromosome gene densities also vary in these genomes. As we show here, the distribution of gene deserts does not appear to be dependent on genome size. This is best illustrated by the genome sizes of platypus (O. anatinus; 3.0 Gb) and gray short-tailed opossum (M. domestica; 3.4 Gb), which show strong correlations between NG/LC (R2 = 0.94 and R2 = 0.95, respectively) and which lie within the range of the sizes of the placental mammalian genomes analyzed here ranging from 3.1 Gb (horse and dog) to 3.6 Gb (chimpanzee). Hence, the uneven distribution of gene deserts is the most likely explanation for the deviation from constant chromosome gene density in placental mammalian genomes. We are aware that, thus far, only a limited number of genomes are available for such kinds of analyses. It remains to be elucidated whether or not the uneven distribution of genes and gene deserts is also found in other large noneutherian genomes, such as salamanders of lungfishes. The inclusion of larger genomes from other lineages seems crucial to test our hypothesis.
The uneven distribution of gene deserts is not the only peculiarity of eutherian genomes. The eutherian karyotype seems to be extensively rearranged compared with the ancestral vertebrate karyotype (Ferguson-Smith and Trifonov 2007). This genomic rearrangement occurred after the divergence from marsupials because the genome of the opossum is more syntenic to the chicken genome than it is to the human one (Ferguson-Smith and Trifonov 2007; Mikkelsen et al. 2007). Our results, which demonstrate the similarity of overall genome organization found in chicken, platypus, and opossum (again to the exclusion of eutherian mammals) are in agreement with this previous finding. Although such chromosome rearrangements might explain the overall similarity in the organization of placental mammalian genomes, they cannot explain the origin of the uneven distribution of gene deserts therein. Fusion or fission of ancestral chromosomes with an even distribution of gene deserts along these chromosomes would necessarily lead to an even distribution of gene deserts in the newly rearranged chromosomes. Within-genome differences in chromosome length do not seem to contribute to the distribution of gene deserts, either: Micro-chromosomes (as observed in chicken or platypus) or giant chromosomes (such as chromosome 1 in the gray short-tailed opossum) show strong correlations in chromosome gene densities.
More than 20 years ago, Ohno (1985) postulated the desertification of the euchromatic region of the higher vertebrates’ genomes owing to continuous gene duplication events followed by degeneration of newly emerged gene copies in their evolutionary history. Only with the first release of the complete sequence of the human genome in 2001 was Ohno’s prediction of the existence of such deserts confirmed (Lander et al. 2001; Venter et al. 2001). Another mechanism to generate imbalance of gene deserts between chromosomes might be large intra-chromosomal duplications, from which some mammalian-specific gene deserts appear to have evolved (Itoh et al. 2005). However, the functional or evolutionary significance of gene deserts is not yet fully understood. At first glance, gene deserts seem to be devoid of biologic functions because of their lack of protein-coding DNA. Despite this, some gene deserts have been shown to contain important regulatory and sometimes ultraconserved regions for neighboring genes that function over large distances (Bejerano et al. 2004; Nobrega et al. 2003; Sandelin et al. 2004). In addition, the observation of stable gene deserts with homologous flanking genes (often transcription factors), which are maintained for long evolutionary durations (Ovcharenko et al. 2005), as well as the existence of numerous conserved nongenic sequences in mammalian genomes (Dermitzakis et al. 2005; Siepel et al. 2005) suggest that gene deserts are not just genomic junkyards but instead might be of functional significance. However, some gene deserts can be deleted without noticeable phenotypic effects (Nobrega et al. 2004). Thus, it remains unclear whether the uneven accumulation of gene deserts in eutherian chromosomes, which appears to be the strongest causal agent for the observed relative lack of a NG/LC relation in the genomes of placental mammals, is simply a byproduct of their genome evolution and possibly the long-term decrease in population-size (Lynch and Conery 2003), as would be suggested by the enrichment of gene deserts along the evolutionary lineage leading to mammals. Upcoming detailed reconstructions of ancestral vertebrate genomes will help clarify these points.
Alternatively, this particular architectural feature of eutherian genomes might be linked to some of their morphologic, physiologic, neurologic, and cognitive evolutionary innovations, possibly by regulating genes with essential functions in development (e.g., see de la Calle-Mustienes et al. 2005; Taylor 2005). It has recently been hypothesized that regulatory elements in gene deserts function in the regulation of core vertebrate genes (Bejerano et al. 2004; de la Calle-Mustienes et al. 2005; Lindblad-Toh et al. 2005; Taylor 2005). Furthermore, approximately 20% of conserved noncoding elements are eutherian-specific (Mikkelsen et al. 2007). Thus, the uneven distribution of gene deserts could itself be caused by an underlying pattern of uneven distribution of some core genes in placental genomes. This hypothesis should be tested in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported by grants from the Landesstiftung Baden-Württemberg GmbH (W. S.), the Center for Junior Research Fellows of the University of Konstanz (W. S.), the National Sciences and Engineering Research Council of Canada (D. S.), and the German Science Foundation (A. M.). We also thank two anonymous reviewers and I. Nanda for valuable comments.
Abbreviations
- kb
Kilobase
- LINE
Long interspersed nuclear element
- SINE
Short interspersed nuclear element
- TE
Transposable element
Footnotes
W. Salzburger, D. Steinke and I. Braasch contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-009-9251-4) contains supplementary material, which is available to authorized users.
Contributor Information
Walter Salzburger, Email: walter.salzburger@unibas.ch.
Dirk Steinke, Email: dsteinke@uoguelph.ca.
Ingo Braasch, Email: ingo.braasch@biozentrum.uni-wuerzburg.de.
Axel Meyer, Email: axel.meyer@uni-konstanz.de.
References
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (2004) Ultraconserved elements in the human genome. Science 304:1321–1325 [DOI] [PubMed]
- Bork P, Copley R (2001) The draft sequences. Filling in the gaps. Nature 409:818–820 [DOI] [PubMed]
- de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A, Allende ML, Gomez-Skarmeta JL (2005) A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res 15:1061–1072 [DOI] [PMC free article] [PubMed]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H (2006) Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439:965–968 [DOI] [PubMed]
- Dermitzakis ET, Reymond A, Antonarakis SE (2005) Conserved non-genic sequences—an unexpected feature of mammalian genomes. Nat Rev Genet 6:151–157 [DOI] [PubMed]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD et al (2008) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749 [DOI] [PubMed]
- Ferguson-Smith MA, Trifonov V (2007) Mammalian karyotype evolution. Nat Rev Genet 8:950–962 [DOI] [PubMed]
- Garland TJ, Ives AR (2000) Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am Nat 155:346–364 [DOI] [PubMed]
- Gregory RT (2005) Synergy between sequence and size in the study of genomes. Nat Rev Genet 6:699–708 [DOI] [PubMed]
- Gregory RT, DeSalle R (2005) Comparative genomics in prokaryotes. In: Gregory RT (ed) The evolution of the genome. Elsevier, San Diego, CA, pp 585–675
- Guindon S, Gascuel O (2003) PhyML – A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704 [DOI] [PubMed]
- Itoh T, Toyoda A, Taylor TD, Sakaki Y, Hattori M (2005) Identification of large ancient duplications associated with human gene deserts. Nat Genet 37:1041–1043 [DOI] [PubMed]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ et al (2003) The UCSC Genome Browser Database. Nucleic Acids Res 31:51–54 [DOI] [PMC free article] [PubMed]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29 [DOI] [PMC free article] [PubMed]
- Kidwell MG (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63 [DOI] [PubMed]
- Kissinger JC, Brunk BP, Crabtree J, Fraunholz MJ, Gajria B, Milgram AJ, Pearson DS, Schug J, Bahl A, Diskin SJ et al (2002) The Plasmodium genome database. Nature 419:490–492 [DOI] [PubMed]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed]
- Lartillot N, Brinkmann H, Philippe H (2007) Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7(Suppl. 1):S4 [DOI] [PMC free article] [PubMed]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ 3rd, Zody MC et al (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819 [DOI] [PubMed]
- Lynch M (2007) The origins of genome architecture. Sinauer, Sunderland, MA
- Lynch M, Conery JS (2003) The origins of genome complexity. Science 302:1401–1404 [DOI] [PubMed]
- Maddison WP, Maddison DR (2004) Mesquite: a modular system for evolutionary analysis. www.mesquiteproject.org
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KF et al (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci U S A 101:14349–14354 [DOI] [PMC free article] [PubMed]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A et al (2007) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447:167–177 [DOI] [PubMed]
- Nelson CE, Hersh BM, Carroll SB (2004) The regulatory content of intergenic DNA shapes genome architecture. Genome Biol 5:R25 [DOI] [PMC free article] [PubMed]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413 [DOI] [PubMed]
- Nobrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM (2004) Megabase deletions of gene deserts result in viable mice. Nature 431:988–993 [DOI] [PubMed]
- Ohno S (1985) Dispensable genes. Trends Genet 1:160–164 [DOI]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L (2005) Evolution and functional classification of vertebrate gene deserts. Genome Res 15:137–145 [DOI] [PMC free article] [PubMed]
- Sandelin A, Bailey P, Bruce S, Engstrom PG, Klos JM, Wasserman WW, Ericson J, Lenhard B (2004) Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5:99 [DOI] [PMC free article] [PubMed]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S et al (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15:1034–1050 [DOI] [PMC free article] [PubMed]
- Taylor J (2005) Clues to function in gene deserts. Trends Biotechnol 23:269–271 [DOI] [PubMed]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed]
- Xu L, Chen H, Hu X, Zhang R, Zhang Z, Luo ZW (2006) Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol Biol Evol 23:1107–1108 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.