Summary
Ribonucleases H are enzymes that cleave the RNA of RNA/DNA hybrids that form during replication and repair and which could lead to DNA instability if they were not processed. There are two main types of RNases H, and at least one of them is present in most organisms. Eukaryotic RNases H are larger and more complex than their prokaryotic counterparts. Eukaryotic RNase H1 has acquired a Hybrid Binding Domain that confers processivity and affinity for the substrate, while eukaryotic RNase H2 is composed of three different proteins: the catalytic subunit (2A), similar to the monomeric prokaryotic RNase HII, and two other subunits (2B and 2C) that have no prokaryotic counterparts and as yet unknown functions, but that are necessary for catalysis. In this review, we discuss some of the most recent findings on eukaryotic RNases H1 and H2, focusing on the structural data on complexes between human RNase H1 and RNA/DNA hybrids that had provided great detail of how the Hybrid Binding- and RNase H-domains recognize and cleave the RNA strand of the hybrid substrates. We also describe the progress made in understanding the in vivo function of eukaryotic RNases H. While prokayotes and some single-cell eukaryotes do not require RNases H for viability, in higher eukaryotes RNases H are essential. Rnaseh1 null mice arrest development around E8.5 because RNase H1 is necessary during embryogenesis for mitochondrial DNA replication. Mutations in any of the three subunits of human RNase H2 cause Aicardi-Goutiéres Syndrome (AGS), a human neurological disorder with devastating consequences.
Introduction
RNases H, enzymes that hydrolyze RNA of RNA/DNA hybrids, were first described in Peter Hausen's laboratory in 1969 (1), but for many years the composition of the two RNases H they described from calf thymus remained controversial (2). Meanwhile Escherichia coli was thought to have only one RNase H enzyme, until in the late 80's a less robust and less abundant RNase H was found and called RNase HII (3). Sequences of the two E. coli proteins showed very limited similarity although it has been subsequently shown that structurally they share a similar fold and have a similar mechanism of hydrolysis. The chances that RNase HI and RNase HII described by Hausen's group corresponded in nomenclature to that of E. coli was 50:50 but, alas, they turned out to be different. To make it easier to compare the RNases H from various organisms, the names of the eukaryotic enzymes were changed to correspond to the nomenclature for E. coli RNases H but using Arabic numerals instead of Roman numeral (4;5). Thus, type 1 enzymes are designated RNase HI in prokaryotes and RNase H1 in eukaryotes, while type 2 proteins are called RNase HII and RNase H2. Human RNase H1 has 286 amino acids (MW ∼32,200) or 260 amino acids if the MTS (see below) is excluded (MW ∼ 29,400) compared with E. coli RNase HI of 155 amino acids (∼MW 17,600). Human RNase H2 has three subunits (RNase H2A − 299 amino acids ∼MW 33,400; RNase H2B −308 amino acids ∼MW 34,800; RNase H2C − 164 amino acids ∼MW 17,800). E. coli RNase HII is active is a monomeric protein of 198 amino acids (∼MW 21,500).
RNase H1 has been implicated in mitochondrial DNA replication during mouse development. Because of its essential role and its similarities in structure and enzymatic mechanism to the RNase H of HIV reverse transcriptase (see review by Champoux and Schultz in this volume), any therapeutic drugs targeted to inhibition of retroviral replication should not interfere with the endogenous enzyme. Mutations in human RNase H2 can result in Aicardi-Goutiéres Syndrome (AGS), a severe neurological disorder (6) with similar symptoms to in utero viral infections, including high levels of interferon α present in the cerebral spinal fluid. It has been proposed that AGS is an autoimmune-like disorder that results from mishandling nucleic acids (6).
RNases H1
Composition
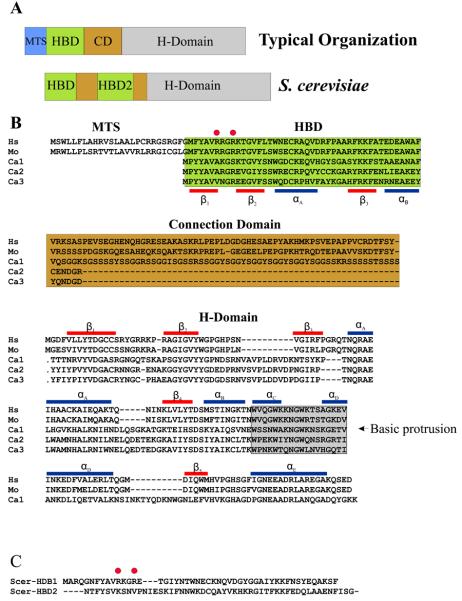
The organization of type 1 RNases H from several representative organisms is shown in Figure 1A along with the organization of Saccharomyces cerevisiae RNase H1. All eukaryotic RNases H1 have highly conserved regions at their N-and C-termini separated by a variable sequence, as exemplified by the human and mouse proteins in Figure 1B. The N-terminal region was first described in S. cerevisiae RNase H1 as two related motifs that have the ability to bind duplex RNAs (Figure 1A and C) (7). Subsequently, we have observed, as shown in Figure 1, that a single copy of the N-terminal conserved sequence is most commonly found on eukaryotic RNases H1 (8). Early studies on the N-terminal domain indicated binding to RNA/RNA and RNA/DNA duplexes, and the domain was called double-stranded RNA binding domain (dsRBD) (9) or double- stranded RNA/hybrid binding domain (dsRHbd) (10). The N-terminal domain has recently been renamed Hybrid Binding Domain as a result of our studies on the binding of this region to various nucleic acids. The HBD showed a 25 fold preference for RNA/DNA compared with the same sequence of dsRNA (11).
Figure 1.
RNase H1 organization in eukaryotes. (A) Two types of RNases H1 in eukaryotes are shown. The top “typical organization” is found in most RNases H1 from higher eukaryotes and consists of an MTS (Mitochondrial Targeting Sequence), HBD (Hybrid Binding Domain), CD (Connection Domain) and H-Domain (RNase H Domain). Not all “typical” RNases H1 have an MTS. Some, mainly fungal, RNases H1 have two HBDs as shown for S. cerevisiae. (B) Alignments of human (Hs), mouse (Mo), and three RNase H1 type proteins encoded in C. albicans (Ca1-3). The red bars represent the β-strands of human RNase H1, and the blue bars are the α-helices. The basic protrusion is shaded in grey as noted in the H-Domain alignment. (C) Alignment of the HBDs of S. cerevisiae RNase H1. Red circles in C and B are the partially conserved HBD amino acids discussed in the text.
The C-terminal RNase H domain has a remarkable similarity in structure to RNase HI of E. coli but, as discussed later, has certain distinct different enzymatic characteristics. The region connecting the N- and C-termini remains the most enigmatic. In human and mouse, the connecting region (named Connection Domain) is about 64 amino acids in length (Figure 1B) and in Drosophila melanogaster is around 150 amino acids. Only a few bacterial RNases HI have HBDs (12), and in every case the bacterial HBD is separated from the RNase H domain by a short Connection Domain, such as that found in Bacillus halodurans RNase HI (13). The presence of the HBD in these prokaryotic enzymes seems to be associated with a lack of the basic protrusion in the RNase H domain (see below). Candida albicans is unusual in that it has three genes that encode type 1 RNases H, two of which appear to be related by a duplication event. The third protein is more typical of eukaryotic RNases H1 with an approximately 60 amino acid Connection Domain that, like other fungal RNases H1, is rich in serine residues. The smaller proteins have very short Connection Domains (Figure 1B) and have a basic protrusion.
The HBD
Sequence conservation was the first indicator of which amino acids were important in the HBD, and, as expected, mutations of a few highly conserved residues on the first copy of the HBD of S. cerevisiae RNase H1 yield proteins defective in nucleic acid binding, although without discerning how the different amino acids contributed to activity (7;14). When the NMR structure of the first HBD of S. cerevisiae RNase H1 was described (15), it was clear that some of these conserved amino acids form the hydrophobic core of the domain, while others residues could be involved in substrate interactions. The recent co-crystal of the human HBD with a 12 bp hybrids (Figure 2A) revealed that in fact very few residues are in contact with the substrate, and that most of the highly conserved amino acids are either structurally important or contributed to non-specific attraction of the duplex RNA (11). From the structures of S. cerevisiae and human RNases H1 HBD, it is apparent that several of the conserved hydrophobic amino acids form the core of the domain and define its overall dimensions (11;15). The RNA/DNA hybrid in the complex exists as pure A form as it is found in dsRNA, suggesting that any changes in structure that occur upon complex formation would be in the HBD. Indeed, Nowotny et al. suggest that the differences between the structures of the HBD of S. cerevisiae and human RNases H1 could be due in part to the presence (in the human) or absence (in the yeast) of an interacting RNA/DNA hybrid (11). The loop between αA and β3 makes contacts with two consecutive 2′-OH groups of the RNA strand. Amino acids of this loop are among the least conserved in the HBD, which is reflected in the interactions occurring mainly through the carbonyl backbone, not the side chains. A shallow positively charged groove containing the highly conserved FKKF motif interacts with the DNA strand. W43 and F58, two of the invariant residues of the HBD contact two deoxyribose rings of the DNA strand and if instead of DNA, RNA were present the 2′-OH groups of the riboses would clash with the two aromatic amino acids, probably accounting for the strong preference for RNA/DNA over RNA/RNA duplexes. These contacts between the HBD and RNA/DNA are completely consistent with the non-sequence specific nature of the binding.
Figure 2.
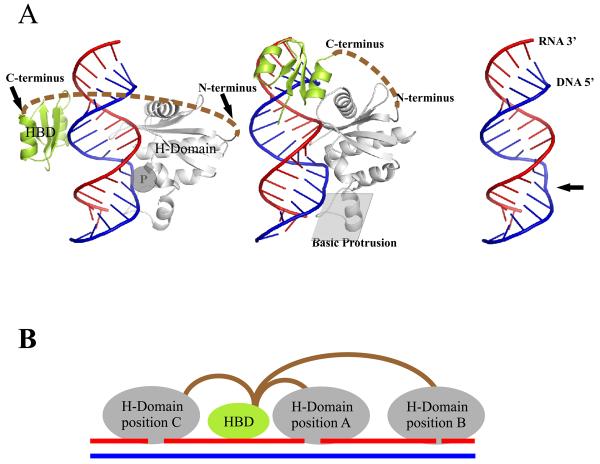
Structural model. Interplay of HBD and H-Domain is orchestrated by the connection domain. (A) Co-crystal structure of human RNase H1 HBD and RNase H domain with RNA/DNA. Left and middle panels show HBD modeled either on the opposite side of RNase H domain (left) or immediately upstream (middle). The C-terminus of the HBD is connected to the N-terminus of the RNase H domain as indicated by dashed brown lines. The phosphate binding pocket is indicated as a “P” inside a grey circle. The middle panel also highlights the position of the basic protrusion. The right panel shows the RNA/DNA with the protein removed revealing the bend in the DNA marked by an arrow. (B) Processivity of RNase H1. In the model of processivity of RNase H1, the HBD anchors the enzyme to a hybrid permitting the RNase H domain to engage with several sites on a single RNA/DNA leading to multiple cleavages from a single binding event. In position A of the H-Domain, hydrolysis will occur near the HBD while in position B cleavage will be more distal. To achieve binding and cleavage at position C, the connection domain needs to be flexible to change the relative orientation of the HBD and H-Domains although the orientation with respect to the hybrid must remain the same.
The most positively charged surface of the HBD is not directly involved in substrate binding, but rather acts as a first point of contact directing the hybrid to the formation of specific, stable interactions with the HBD that in turn will facilitate catalysis by the RNase H domain. When properly engaged, the HBD enhances the RNase H activity of the protein (16). Decreasing specific interactions either by mutagenesis or by lowering the salt concentration greatly reduce the catalytic activity of the enzyme (11;16). At low salt concentrations the non-specific charge-charge interaction between HBD and nucleic acid interfere with the RNase H activity of the protein, while at physiological salt conditions or high MgCl2 concentrations the non-specific interactions are overcome to achieve optimal RNase H activity.
Deletion of the HBD in human and mouse RNase H1 yield proteins very defective in RNase H activity, which cleave long RNA/DNA substrates distributively, similar to E. coli RNase HI (11;16). In contrast, full-length eukaryotic RNases H1 with intact HBDs process hybrids processively, likely due to the increased substrate affinity conferred by the HBD. According to this model, the HBD may first approach the RNA/DNA substrate facilitating interactions and hydrolysis by the RNase H domain, thus the increased RNase H activity when the two domains are connected. After catalysis and dissociation of the RNase H domain, the HBD may remain bound to the substrate promoting new interactions of the RNase H region at nearby sites of the hybrid and resulting in several rounds of hydrolysis per each HBD binding event, therefore behaving processively [Fig. 2B and (11)]. Proteins containing mutations of HBD amino acids involved in interacting with the DNA strand of RNA/DNA (W43, K59 and K60 in human RNase H1) have decreased binding and consequently low RNase H activity and impaired processivity, demonstrating that binding through the HBD and catalysis by the RNase H domain are related processes most likely coordinated by the sequence in between the two regions, the Connection Domain, as described below.
Interestingly, the second copy of the HBD of S. cerevisiae does not form stable complexes with duplex RNAs when alone, but it does enhance the overall binding of the protein when in combination with the first HBD (7;14). The major differences between the two copies of the HBDs of fungal RNases H1 are at their amino termini including an insertion of three amino acids in the loop between β1 and β2 (Figure 1B and C). This loop has a different position in the crystal of the HBD complex with RNA/DNA versus the structure of the HBD alone, suggesting that it may change conformation when interacting with its substrate. Insertion of the three amino acids PNI sequence from the second S. cerevisiae HBD into the corresponding loop region of the first copy (Figure 1C) made the protein incapable of binding to duplex RNAs, another indication that the length and the sequence of the loop is important (14). The HBD from human and the first HBD of S. cerevisiae have two highly conserved arginine residues (Figure 1 – red circles) important for stabilizing the helices and RNA-binding loop between αA and β3 (11). None of the “defective” HBDs from fungi closely related to S. cerevisiae has arginine residues in these two positions, and they are not conserved in any of the C. albicans enzymes. Still, it seems possible that when the “active” HBD interacts with the hybrid, the second HBD may be able to contribute to binding by assuming the correct structure when in close proximity of the RNA/DNA (i.e., an induced fit).
The HBD is also present in protein VI of cauliflower mosaic virus (CaMV) (14), a dsDNA virus that undergoes replication via an RNA/DNA hybrid intermediate. Protein VI is a multifunctional protein required for replication and translation (17). The role of the HBD may be in RNA/DNA recognition during reverse transcription to produce the DNA of the virus, and not in the translation of the CaMV polycistronic mRNA (11).
RNase H Domain
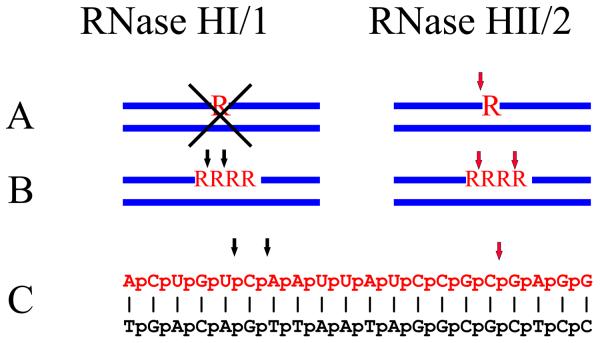
Type 1 RNases H require a substrate with at least four ribonucleotides in order for cleavage to occur (Fig. 3). Various modifications that decrease the flexibility of the DNA strand of RNA/DNA duplexes have been described to make the hybrids resistant to cleavage by RNase H1 (18;19). Thus, the enzyme must be recognizing both the RNA and DNA strands. The co-crystal structure of the C-terminal RNase H domain of B. halodurans RNase H1 in complex with RNA/DNA (13) showed that the hybrid fits in two shallow grooves in the enzyme with the 2′-OH of four consecutive ribose moieties interacting with one of the grooves. The DNA sits in the second groove, and one of the phosphates fits into a pocket requiring a distortion of the DNA backbone (Fig. 2A right: noted by an arrow), a conformational change that RNA can not accomplish. These findings confirmed that both strands contribute to the specificity of type 1 RNases H.
Figure 3.
RNase HI/1 and RNase HII/2 have distinct cleavage patterns on three different substrates. Three types of substrates are shown. (A) A single ribonucleotide in a duplex DNA that is cleaved by RNases HII/2 but not RNases HI/1 as noted by X. (B) Four consecutive ribonucleotides residues and (C) an RNA/DNA hybrid that is cleaved differently by the two classes of enzymes. Black arrows denote sites of cleavage by RNase HI/1 and red arrows represent hydrolysis by RNase HII/2.
From numerous studies on E. coli RNase HI, it is clear that four highly conserved carboxylic acid residues comprise the catalytic core and are necessary for hydrolysis (see Tadokoro and Kanaya). Structural analysis of a co-crystal between the C-terminal domain of human RNase H1 and an RNA/DNA hybrid highlighted several important features of catalysis revealing the role of each of the carboxylic catalytic residues. The two metal ion mechanism is described in the review by Tadokoro and Kanaya in this issue. The papers by Yang and Nowotny (20;21) also describe in some detail the possible movement of the metal ions after cleavage of the RNA to ensure the release of the enzyme from the hydrolyzed substrate. In the structure of human RNase H1:RNA/DNA, the protein interacts with 11 bp of the hybrid (Figure 2A) (22). There are two regions of interactions, the catalytic site and the basic protrusion. Despite the fact that the region is replete with basic amino acids, none of them directly interacts with the DNA in the co-crystal. It seems likely that the basic character of this region plays an important role by attracting the substrate to the enzyme allowing binding in the proper manner. Attraction of the hybrid by basic residues is important for engaging the substrates in both the HBD and RNase H domain. Because of the absence of the basic protrusion in the B. halodurans RNase HI, there are only 6 bp of the hybrid contacting the protein.
Crystal structures of the apo E. coli RNase HI (23) (24;25)and human RNase H1 H-domain in complex with RNA/DNA exhibit extraordinary similarity with only 34% amino acid identity. Structural differences are mainly in the basic protrusion and may reflect a movement of this region when binding to RNA/DNA (22). In eukaryotic RNases H1 a methionine often is present just 10 amino acids prior to the catalytic aspartic residue equivalent to D10 of E. coli RNase HI (Figure 1B). A protein starting at this methionine has RNase H activity that is compromised in several ways including specific activity and processivity, as described above, behaving similarly to E. coli RNase HI.
Connection domain
Because the connection between the N-terminal HBD and the C-terminal RNase H domain is so variable in length and amino acid sequence, it has been thought that it is important mainly for providing flexibility allowing the N- and C-terminal regions to move rather freely in and around the substrates. For human RNase H1, the Connection Domain is about 64 amino acids and if fully extended and completely flexible, it would permit the HBD and RNase H domain to be separated by more than 200 Å. Two configurations of interactions are shown in Figure 2A. In the left model, the C-terminus of the HBD is about 60 Å from the N-terminus of the RNase H domain. The two domains are separated by only 25 Å in the middle model. If the Connection Domain were stretched to its maximum the HBD and RNase H domain could be separated on a hybrid by about 60 nucleotides. Figure 2B depicts the two domains separated in various ways; the cartoon shows the HBD at a constant position and the RNase H domain at three different sites on the hybrid (11). With a truly flexible Connection Domain, the relative positions of the HBD and RNase H domain can be reversed as indicated in the position C of the H-Domain (Figure 2B). Dramatic differences in length and composition of the connection domain suggest that this region may also be to interact with additional proteins or complexes in higher organisms.
MTS
There are two in-frame methionine codons at the 5′-end of the RNase H1 mRNA from higher eukaryotes. Translation from the first methionine codon produces a protein with a mitochondrial targeting sequence (MTS-Fig. 1A and B) that localizes to mitochondria where it is essential during development for amplification of mitochondrial DNA (26). In general, proteins targeted to mitochondria lose the MTS upon uptake. Many, but not all, eukaryotic RNases H1 have an MTS upstream of the HBD. Two highly studied model organisms, S. cerevisiae and C. elegans, have no MTS associated with type 1 RNases H bearing the HBD. Deletion of the RNH1 gene in S. cerevisiae does not lead to loss of mitochondrial DNA (27), most likely due to mtDNA replication occurring by a mechanism that is radically different than that found in many other organisms. S. cerevisiae mtDNA is significantly larger than mammalian mtDNA and is linear instead of circular as is mammalian mtDNA (28). C. elegans mtDNA is similar to mammalian mtDNA in being circular and about 16 kbp in size. C. elegans has a second type 1 RNase H (RNase H1.1) in addition to the one with an HBD-connection domain-RNase H composition (29). RNase H1.1 has two potential start codons, the first codon will initiate a protein with a MTS. Interestingly, the abundance of the mRNA encoding RNase H1.1 increases dramatically at the same stage of development when the amount of mtDNA increases five fold over the level present initially in the embryo (30). Perhaps RNase H1.1 plays a role in C. elegans mtDNA replication.
In vivo phenotypes
In S. cerevisiae, RNH1 can be deleted showing only modest increases in sensitivity to DNA damaging agents (27). In mouse, Rnaseh1 null embryos arrest development due to failure to amplify mtDNA. RNase H1 has been suggested to be involved in generating and/or removing RNA primers for mtDNA replication but at the moment the details of its role are under investigation. The role of the nuclear isoform of RNase H1 remains unknown, and so far no human disease or disorder has been identified as related to RNase H1. However, the HIV-AIDS virus uses the RNase H domain of its reverse transcriptase for copying the viral RNA into DNA (see review by Champoux). Inhibiting the RNase H of HIV-AIDS RT could be useful for controlling replication of the virus, but because the cellular and retroviral RNases H share similar structures and mechanism of action, drugs would need to be specific for the viral enzyme.
RNase H substrates in vivo could arise from a number of different processes, and be very distinct in nature. These hybrids could originate from RNA primers of lagging strand DNA synthesis, from formation of R-loops during transcription, from some sort of RNA strand invasion of duplex DNA, from reverse transcription, from misincorporation of ribonucleotides by DNA polymerases, or in some other manner. In addition to the complexity of these putative hybrids substrates of RNase H1, the cleavage process could result in different outcomes. One in vivo role of RNase H1 might be to eliminate hybrids created accidentally during replication, thereby preventing genome instability associated with R-loops formation. Nascent RNA extruded from RNA polymerase during synthesis almost immediately interacts with proteins, the type of which depends on the particular organism (31). InE. coli, ribosomes engage the newly formed mRNA (32). In eukaryotes, several proteins engage with newly transcribed RNA as it is extruded from the RNA polymerase. For example, in S. cerevisiae, the THO/TREX complex binds transcripts in the nucleus (33) and in chicks and HeLa cells, nascent RNA interacts with ASF/SF2 (34). Such early interactions with the newly exiting mRNAs are important to prevent the RNA from forming RNA/DNA hybrids with the partially unwound DNA left behind during transcription by the RNA polymerase. The DNA strand not forming a hybrid would be in single strand form and prone to breakage, with potential deleterious effects.
In a different in vivo situation, formation of R-loops followed by RNase H cleavage could generate RNA primers that might be used in DNA replication/repair, such as in E. coli, where RNase HI is important for creating RNA primers in colE1 replication. Likewise, reverse transcription of retroviral RNAs such as HIV-AIDS requires generation of RNA primers by the RT type 1 RNase H to prime replication from the PPT (polypurine tract) (see Champoux and Schultz). Alternatively, these potential primers could be eliminated to prevent damage to DNA. In E. coli, the absence of RNase HI permits DNA replication to initiate from sites others than the normal replication initiation at oriC (35). Involvement in Okazaki fragment processing is unlikely, because RNase H1 cannot remove the last ribonucleotide attached to the DNA. Recent models suggest that Fen1 and/or Dna2 are responsible for RNA primer removal from Okazaki fragments and do not require any participation of RNase H (36).
Sequence preferences
The co-crystal structures of type 1 RNases H tell us a lot about the details for recognition and cleavage but provide only clues as to why some sites are preferred over others. Interactions of the RNA strand with the protein occur through four 2′-OH residues on consecutive ribonucleotides explaining the failure of this class of RNase H to cleave anything shorter than a four ribonucleotide stretch in a DNA duplex (Figure 3). The DNA has to be distorted to fit in the phosphate-binding pocket, and the sequence of the DNA and/or the proximity to an end may influence its malleability resulting in a variety of sequence preferences related to the ease with which the DNA bends. A requirement for flexibility of the DNA strand has been demonstrated by Lima et al. (18) when they showed that chemical modifications of a single deoxyribonucleotide that result in an A-form sugar pucker prevent cleavage of the opposing ribonucleotide and also of two ribonucleotides downstream. These studies have been extended in a more systematic way by increasing, one nucleotide at the time, various modifications on the DNA strand (18;19).
Most of the studies on cleavage site preferences have used 5′- or 3′-labeled RNAs annealed to complementary DNA oligonucleotides. When only a small fraction of the starting substrate is hydrolyzed, it is possible to see one or a few RNA products arising from site-preference cleavages. However, when a preferred site is near the labeled end of the RNA, that cleavage may obscure other sites that are also readily cleaved. Cleavage site preferences are also generally examined in a single RNA/DNA hybrid and therefore do not address the issue of what might be the best (or better) site when two hybrids are examined simultaneously.
Further work with emphasis of the role and function of RNase H1 in antisense therapy can be seen in the references to Lima et. al (15;16) and references therein.
RNases H2
Composition of Eukaryotic RNase H2
For many years following its discovery in 1969, the composition of RNase H2 was difficult to determine, due in part to its low abundance that made its purification problematic. With the completion of the genome sequence of S. cerevisiae, it was straight forward to find the homologue of E. coli RNase HII, a protein that had 25% identity (45% when allowing for similarities) and all the residues known to be important for catalysis. Frank and Wintersberger's group gave the name RNase H35 to this protein based on its molecular mass (37). We also examined this S. cerevisiae RNase HII homolog and discovered that it lacked RNase H activity (27). We obtained cDNA clones from human, mouse, and C. elegans that would express the putative homologous RNase H protein and found that none of them, when expressed in E. coli, exhibited RNase H activity. To determine whether these proteins could restore RNase H2 activity in an S. cerevisiae strain deleted for the gene encoding the yeast protein, we expressed the counterpart cDNAs and found that only the yeast clone could complement its own deficiency. These results were the first indication that S. cerevisiae RNase H2 might be composed of more than one subunit.
From the heroic efforts by Büsen and Frank to purify endogenous calf thymus RNase H2 (at that time called RNase H1) (38), it was concluded that the enzyme has several subunits. In fact, Frank et al. (see GenBank AF312034 –noted as small subunit of human RNase H1 – using the older terminology) found that their most purified calf thymus RNase H2 contained an additional protein, which they identified as related to AYP1 and were then able to examine the equivalent human subunit. This protein was later found to be the RNASEH2B subunit (6).
By expressing the tagged yeast protein homologous to E. coli RNase HII, we were able to purify an active enzyme from S. cerevisiae and identified two co-purifying proteins (39;39). Finding three proteins was surprising. We examined S. cerevisiae deletion strains for each of the genes encoding the three proteins, and found no RNase H2 activity. Moreover, expression of the three yeast proteins together in E. coli showed that they are necessary and sufficient to form a stable active RNase H2. Another surprise came when we searched for proteins with sequences similar to the two additional S. cerevisiae RNase H2 subunits. We found these proteins only in closely related fungi, not in human or S. pombe (39).
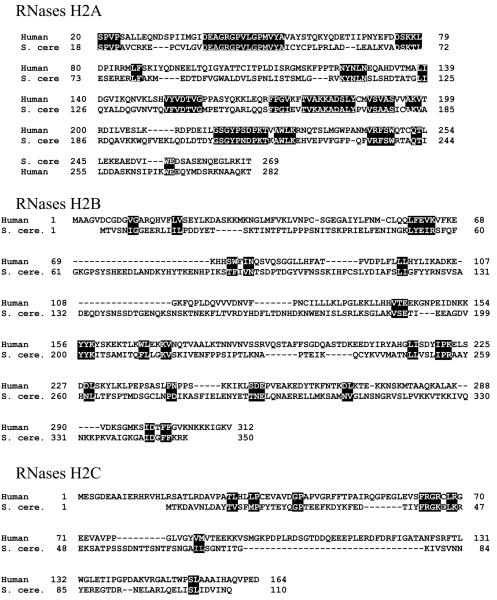
Taking a similar tagging approach, we identified two proteins co-purifying with the human tagged RNASEH2A (by homology to S. cerevisiae RNH201 – or RNase H2A) subunit (40). These same three proteins were identified by Crow et al. (6) (41) as subunits of human RNase H2 through some elegant work on Aicardi-Goutières Syndrome (AGS). Using human genetics they demonstrated that the neurological defects and other symptoms of AGS could be attributed to any of several mutations in each of the three subunits of human RNase H2, thereby identifying the three components of human RNase H2. Thus, RNase H2 seems to have evolved rather extensively and rapidly from a monomeric enzyme in prokaryotes to a heterotrimer in eukayotes, with the S. cerevisiae type B/C subunits being distantly related to most others, as it is shown in the alignment in Figure 4 (6). Such disparity in amino acid sequence explains the failure of human RNase H2A to substitute in vivo its yeast counterpart. When the three subunits of S. cerevisiae or human RNase H2 are expressed in E. coli, only the heterotrimeric complex is active, but heterodimers of RNase H2B and H2C could be obtained in a soluble form.
Figure 4.
(A) Comparison of amino acid sequences of human and S. cerevisiae RNase H2 subunits. Alignments are based on the consensus alignment of RNase H2 subunits from Crow et al. Reverse letters highlight homologies between the subunits. For RNases H2B and RNases H2C, a more generous highlighting is used and even then it is clear there is very little conservation between the human and yeast B and C subunits. (B) Schematic representation of the three components of human RNase H2. On top of each subunit are shown AGS-related mutations, and on the bottom the amino acids involved in catalysis, in RNASEH2A, and the PIP (PCNA Interacting Peptide) site of RNASEH2B.
Bacterially expressed human RNase H2 has very similar biochemical properties with that isolated from HeLa cells expressing a tagged RNASEH2A (40). The wild type protein has the ability to degrade poly-rA/poly-dT in a processive manner. Prokaryotic RNase HII act distributively, therefore the ability of the eukaryotic RNase H2 to cleave processively must be attributed to the contribution of the RNase H2B and H2C components. We also found that the RNase H2B subunit interacts with PCNA (proliferating cell nuclear antigen) via a PCNA-interacting peptide (PIP) at its C-terminus (Fig 4). PCNA is a protein essential for eukaryotic DNA replication/repair, as it is responsible for bringing to the replication fork the elongating polymerase and other factors involved in Okazaki fragment processing (42). In doing so, PCNA might affect the activities of the interacting proteins. However, we observed neither stimulation nor inhibition of RNase H2 activity in the presence of a wide concentration of PCNA. Similarly, inactivation or deletion of the PIP sequence on the RNase H2B subunit did not alter the activity of RNase H2, including processivity. Interaction with PCNA clearly points to a role for RNase H2 in replication/repair processes.
Crow et al. reported that the G37S (RNASEH2A subunit) mutant protein lost most of its RNase H activity (6). This conserved Glycine is near the catalytic center as seen in the three dimensional structure of several prokaryotic RNases HII (43). Based on these structures, Crow et al. suggested the G37S substitution altered the region near the catalytic center thereby decreasing the activity of the protein (6). Rohman et al. made the equivalent of G37S (G42S) in Thermococcus kodakareansis RNase HII and observed a significant loss of RNase H activity. Substitution of G42 by several other amino acids led to the conclusion that the presence of bulkier side chains on the amino acid replacing G42 limits access to the nearby catalytic aspartic acid residue (43), confirming the hypothesis of Crow et al.
AGS individuals also are known with mutations in the RNASEH2B and RNASEH2C subunits of RNase H2, with the A177T mutation in the RNASEH2B protein being the one most commonly observed (6;41). We expressed six different human RNases H2, each with a change found in AGS patients (Fig. 4B). In agreement with the data reported by Crow et al., the G37S mutant protein was defective in RNase H activity. Moreover, the G37S enzyme was less processive than the wild type enzyme. Proteins with RNase H2B K162T, A177T, V185 and RNase H2C K143I all exhibited enzymatic activity and processivity very similar to the wild type protein. However, the protein with the RNase H2C R69W substitution, while having processivity similar to wild type RNase H2, showed a clear decrease in RNase H2 specific activity. These findings are surprising because it was expected that all mutations would lead to loss of function. Some of the mutations may affect stability, interaction with some other cellular components or some other in vivo properties.
The exact role(s) of the “extra” subunits of eukaryotic RNase H2 remains unclear but in light of our data we propose that the RNase H2B and RNase H2C proteins provide (i) a platform for assembly of the complete enzyme (RNase H2B and RNase H2C from human and S. cerevisiae can form, stable, soluble heterodimers), (ii) processivity, and (iii) interaction with PCNA (suggesting a role in DNA replication and repair) and other cellular proteins (40).
Mutations in TREX1, a single-stranded DNA exonuclease also cause AGS (44). A recent paper reported that Trex1−/− mice accumulate short single-stranded DNAs that induce interferon α (45). In the absence of TREX1 activity, an increase in DNAs derived from endogenous retroviral sequences was seen. This observation led the authors to suggest a connection between reverse transcriptase, RNase H2 and TREX1 (45;46). According to this model, mRNAs that are reverse transcribed into DNA forming hybrids could be degraded by RNase H2 (the RNA strand) and TREX1 (the remaining ssDNA).
Substrates for RNase H2
Eder and Walder (47) were the first to show that RNase H2 differs from RNase H1 by recognizing and cleaving a single ribonucleotide embedded in a DNA duplex (Figure 3), and suggested that the role of RNase H2 is to cleave at sites in DNA where DNA polymerase(s) had mistakenly incorporated a ribo- rather than a deoxyribo-nucleotide and was thus a repair enzyme. Moreover, they found that mismatches at either the deoxynucleotide 5′ to the single ribonucleotide or at the RNA/DNA site in a DNA7-RNA1-DNA5/DNA13 substrate were cleaved. Bambara's group (48), using model in vitro substrates, later showed that RNase H2 could hydrolyze an Okazaki fragment RNA primer leaving a single ribonucleotide of the primer attached to the newly synthesized DNA and described this activity as a junction RNase H. Murante et al. also examined mismatched RNA/DNA hybrid substrates and obtained results similar to Eder and Walder. In addition, using non-canonical hybrids they obtained cleavages at sites where the RNA was in single stranded form, the significance of which remains to be determined.
S. cerevisiae RNase H2 cleaves a DNA12-RNA4-DNA12/DNA hybrid at two sites within the RNA segment leaving a ribonucleotide residue attached to each end of the DNA (Figure 3). D155A or D183A mutations in the catalytic center of RNase H2A retained partial activity and showed preferential cleavage near the junction at the 3′-end of the four ribonucleotides (39). Because of the absence of a co-crystal structure of RNase H2 with a hybrid, it is not possible to infer sites of cleavage preferences within a substrate, but at least in vitro RNase H2 showed some site specificity for cleavage a model substrate designed by Pileur et al. (49) to distinguish between RNase H1 and RNase H2 hydrolysis (Figure 3). RNase H1 and RNase H2 show clear differences in hybrid hydrolysis (Figure 3), indicating they might have distinct in vivo substrates. More detailed analysis of substrate specificity for these two types of enzymes may aid in elucidating the so far elusive in vivo functions of RNase H1 and RNase H2.
Perspectives
Probably the most outstanding questions about RNases H are what are their in vivo substrates and in which processes these enzymes are involved. From their activities, we can assume they recognize almost any RNA/DNA hybrid and they may have multiple roles, similarly to E. coli RNase HI and the RNase H of reverse transcriptases, at times hydrolyzing illicit hybrids to prevent genome instability, while under other circumstances they could generate specific primers to initiate DNA replication. Elucidating the role of RNase H1 in mtDNA replication would be crucial to understanding the mechanism by which mitochondria duplicates their DNAs, a quite controversial topic. The nuclear form of RNase may also have an important function that so far has been masked by the critical role of the mitochondrial isoform. Therefore, finding the substrates of the nuclear RNase H1 should be a major focus of interest for future studies. How the connection domain brings together and coordinates the HBD and RNase H domain of eukaryotic RNases H1 would aid our understanding of how this type of RNases H functions. Finally, determining how mutations in any of the three subunits of RNase H2 lead to AGS would help to elucidate the processes in which RNase H2 is normally involved, as well as revealing which nucleic acids accumulate when RNase H2 is deficient, and how these nucleic acids induce AGS.
Acknowledgements
This work was supported by the intramural program of the National Institutes of Health, NICHD. We thank Marcin Nowotny and Hyongi Chon for help with figures.
Reference List
- 1.Stein H, Hausen P. Enzyme from Calf Thymus Degrading the RNA Moiety of DNA-RNA Hybrids: Effect on DNA-Dependent RNA Polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 2.Büsen W, Frank P. Bovine RNases H. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. INSERM; Paris: 1998. pp. 113–146. [Google Scholar]
- 3.Itaya M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc. Natl. Acad. Sci.U.S.A. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouch RJ, Arudchandran A, Cerritelli SM. RNase H1 of Saccharomyces cerevisiae: methods and nomenclature. Methods Enzymol. 2001;341:395–413. doi: 10.1016/s0076-6879(01)41166-9. 395-413. [DOI] [PubMed] [Google Scholar]
- 5.Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase I-III and Mg2+-dependent RNase HIII from Bacillus subtilis: Classification of RNases H into three families. Biochemistry. 1999;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 6.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat.Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 7.Cerritelli SM, Crouch RJ. The non-RNase H domain of Saccharomyces cerevisiae RNase H1 binds double-stranded RNA: magnesium modulates the switch between double-stranded RNA binding and RNase H activity. RNA. 1995;1:246–259. [PMC free article] [PubMed] [Google Scholar]
- 8.Cerritelli SM, Crouch RJ. Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics. 1998;53:300–307. doi: 10.1006/geno.1998.5497. [DOI] [PubMed] [Google Scholar]
- 9.Cerritelli SM, Frolova EG, Feng CG, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol.Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 10.Gaidamakov SA, Gorshkova II, Schuck P, Steinbach PJ, Yamada H, Crouch RJ, Cerritelli SM. Eukaryotic RNases H1 act processively by interactions through the duplex RNA-binding domain. Nucleic Acids Res. 2005;33:2166–2175. doi: 10.1093/nar/gki510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowotny M, Cerritelli SM, Ghirlando R, Gaidamakov SA, Crouch RJ, Yang W. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008;27:1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochiwa H, Tomita M, Kanai A. Evolution of ribonuclease H genes in prokaryotes to avoid inheritance of redundant genes. BMC Evol.Biol. 2007;7:128. doi: 10.1186/1471-2148-7-128. 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Cerritelli SM, Fedoroff OY, Reid BR, Crouch RJ. A common 40 amino acid motif in eukaryotic RNases H1 and caulimovirus ORF VI proteins binds to duplex RNAs. Nucleic Acids Res. 1998;26:1834–1840. doi: 10.1093/nar/26.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans SP, Bycroft M. NMR structure of the N-terminal domain of Saccharomyces cerevisiae RNase HI reveals a fold with a strong resemblance to the N-terminal domain of ribosomal protein L9. J. Mol. Biol. 1999;291:661–669. doi: 10.1006/jmbi.1999.2971. [DOI] [PubMed] [Google Scholar]
- 16.Gaidamakov SA, Gorshkova II, Schuck P, Steinbach PJ, Yamada H, Crouch RJ, Cerritelli SM. Eukaryotic RNases H1 act processively by interactions through the duplex RNA-binding domain. Nucleic Acids Res. 2005;33:2166–2175. doi: 10.1093/nar/gki510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shivaprasad PV, Rajeswaran R, Blevins T, Schoelz J, Meins F, Jr., Hohn T, Pooggin MM. The CaMV transactivator/viroplasmin interferes with RDR6-dependent trans-acting and secondary siRNA pathways in Arabidopsis. Nucl.Acids Res. 2008;36:5896–5909. doi: 10.1093/nar/gkn590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima WF, Rose JB, Nichols JG, Wu H, Migawa MT, Wyrzykiewicz TK, Siwkowski AM, Crooke ST. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol.Pharmacol. 2007;71:83–91. doi: 10.1124/mol.106.025015. [DOI] [PubMed] [Google Scholar]
- 19.Lima WF, Rose JB, Nichols JG, Wu H, Migawa MT, Wyrzykiewicz TK, Vasquez G, Swayze EE, Crooke ST. The positional influence of the helical geometry of the heteroduplex substrate on human RNase H1 catalysis. Mol.Pharmacol. 2007;71:73–82. doi: 10.1124/mol.106.025429. [DOI] [PubMed] [Google Scholar]
- 20.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol.Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol.Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 24.Kanaya S. Enzymatic activity and protein stability of E. coli ribonuclease HI. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. INSERM; Paris: 1998. pp. 1–38. [Google Scholar]
- 25.Morikawa M, Katayanagi K. Crystal structures of RNase H from prokaryotes. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. INSERM; Paris: 1998. pp. 181–194. [Google Scholar]
- 26.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol.Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 27.Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 28.Williamson D. The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 2002;3:475–481. doi: 10.1038/nrg814. [DOI] [PubMed] [Google Scholar]
- 29.Arudchandran A, Cerritelli SM, Bowen NJ, Chen X, Krause MW, Crouch RJ. Multiple ribonuclease H-encoding genes in the Caenorhabditis elegans genome contrasts with the two typical ribonuclease H-encoding genes in the human genome. Mol.Biol.Evol. 2002;19:1910–1919. doi: 10.1093/oxfordjournals.molbev.a004015. [DOI] [PubMed] [Google Scholar]
- 30.Kochiwa H, Itaya M, Tomita M, Kanai A. Stage-specific expression of Caenorhabditis elegans ribonuclease H1 enzymes with different substrate specificities and bivalent cation requirements. FEBS J. 2006;273:420–429. doi: 10.1111/j.1742-4658.2005.05082.x. [DOI] [PubMed] [Google Scholar]
- 31.Luna R, Gaillard H, González-Aguilera C, Aguilera A. (8 A.D.) Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 32.Broccoli S, Rallu F, Sanscartier P, Cerritelli SM, Crouch RJ, Drolet M. Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation. Mol.Microbiol. 2004;52:1769–1779. doi: 10.1111/j.1365-2958.2004.04092.x. [DOI] [PubMed] [Google Scholar]
- 33.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol.Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Kogoma T, Foster PL. Physiological functions of E. coli RNase HI. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. INSERM; Paris: 1998. pp. 39–66. [Google Scholar]
- 36.Rossi ML, Purohit V, Brandt PD, Bambara RA. Lagging Strand Replication Proteins in Genome Stability and DNA Repair. Chem.Rev. 2006;106:453–473. doi: 10.1021/cr040497l. [DOI] [PubMed] [Google Scholar]
- 37.Frank P, Braunshofer-Reiter C, Wintersberger U. Yeast RNase H(35) is the counterpart of the mammalian RNase HI, and is evolutionarily related to prokaryotic RNase HII. FEBS Lett. 1998;421:23–26. doi: 10.1016/s0014-5793(97)01528-7. [DOI] [PubMed] [Google Scholar]
- 38.Frank P, Braunshofer-Reiter C, Wintersberger U, Grimm R, Busen W. Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc.Natl.Acad.Sci.U.S A. 1998;95:12872–12877. doi: 10.1073/pnas.95.22.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong HS, Backlund PS, Chen HC, Karavanov AA, Crouch RJ. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004;32:407–414. doi: 10.1093/nar/gkh209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn913. doi:10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Baxter P, Benko WS, Bergmann C, Bertini E, Biancheri R, Blair EM, Blau N, Bonthron DT, Briggs T, Brueton LA, Brunner HG, Burke CJ, Carr IM, Carvalho DR, Chandler KE, Christen HJ, Corry PC, Cowan FM, Cox H, D'Arrigo S, Dean J, De Laet C, De Praeter C, Dery C, Ferrie CD, Flintoff K, Frints SG, Garcia-Cazorla A, Gener B, Goizet C, Goutieres F, Green AJ, Guet A, Hamel BC, Hayward BE, Heiberg A, Hennekam RC, Husson M, Jackson AP, Jayatunga R, Jiang YH, Kant SG, Kao A, King MD, Kingston HM, Klepper J, van der Knaap MS, Kornberg AJ, Kotzot D, Kratzer W, Lacombe D, Lagae L, Landrieu PG, Lanzi G, Leitch A, Lim MJ, Livingston JH, Lourenco CM, Lyall EG, Lynch SA, Lyons MJ, Marom D, McClure JP, McWilliam R, Melancon SB, Mewasingh LD, Moutard ML, Nischal KK, Ostergaard JR, Prendiville J, Rasmussen M, Rogers RC, Roland D, Rosser EM, Rostasy K, Roubertie A, Sanchis A, Schiffmann R, Scholl-Burgi S, Seal S, Shalev SA, Corcoles CS, Sinha GP, Soler D, Spiegel R, Stephenson JB, Tacke U, Tan TY, Till M, Tolmie JL, Tomlin P, Vagnarelli F, Valente EM, Van Coster RN, Van der AN, Vanderver A, Vles JS, Voit T, Wassmer E, Weschke B, Whiteford ML, Willemsen MA, Zankl A, Zuberi SM, Orcesi S, Fazzi E, Lebon P, Crow YJ. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am.J.Hum.Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majka J, Burgers PMJ. The PCNA-RFC Families of DNA Clamps and Clamp Loaders. In: Kivie M, editor. Prog. in Nucleic Acid Res. and Mol. Biol. Academic Press (Elsevier); N.Y.: 2004. pp. 227–260. [DOI] [PubMed] [Google Scholar]
- 43.Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S. Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. FEBS J. 2008;275:4836–4849. doi: 10.1111/j.1742-4658.2008.06622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat.Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 45.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 Prevents Cell-Intrinsic Initiation of Autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhoj VG, Chen ZJ. Linking Retroelements to Autoimmunity. Cell. 2008;134:569–571. doi: 10.1016/j.cell.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Eder PS, Walder RY, Walder JA. Substrate specificity of Human RNase H1 and its Role in Excision Repair of Ribose Residues Misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 48.Murante RS, Henricksen LA, Bambara RA. Junction ribonuclease: an activity in Okazaki fragment processing. Proc.Natl.Acad.Sci.U.S A. 1998;95:2244–2249. doi: 10.1073/pnas.95.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pileur F, Toulme JJ, Cazenave C. Eukaryotic ribonucleases HI and HII generate characteristic hydrolytic patterns on DNA-RNA hybrids: further evidence that mitochondrial RNase H is an RNase HII. Nucleic Acids Res. 2000;28:3674–3683. doi: 10.1093/nar/28.18.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]