Abstract
Hemagglutinin esterases (HEs), closely related envelope glycoproteins in influenza C and corona- and toroviruses, mediate reversible attachment to O-acetylated sialic acids (Sias). They do so by acting both as lectins and as receptor-destroying enzymes, functions exerted by separate protein domains. HE divergence was accompanied by changes in quaternary structure and in receptor and substrate specificity. The selective forces underlying HE diversity and the molecular basis for Sia specificity are poorly understood. Here we present crystal structures of porcine and bovine torovirus HEs in complex with receptor analogs. Torovirus HEs form homodimers with sialate-O-acetylesterase domains almost identical to corresponding domains in orthomyxo- and coronavirus HEs, but with unique lectin sites. Structure-guided biochemical analysis of the esterase domains revealed that a functionally, but not structurally conserved arginine–Sia carboxylate interaction is critical for the binding and positioning of glycosidically bound Sias in the catalytic pocket. Although essential for efficient de-O-acetylation of Sias, this interaction is not required for catalysis nor does it affect substrate specificity. In fact, the distinct preference of the porcine torovirus enzyme for 9-mono- over 7,9-di-O-acetylated Sias can be explained from a single-residue difference with HEs of more promiscuous specificity. Apparently, esterase and lectin pockets coevolved; also the porcine torovirus HE receptor-binding site seems to have been designed to use 9-mono- and exclude di-O-acetylated Sias, possibly as an adaptation to replication in swine. Our findings shed light on HE evolution and provide fundamental insight into mechanisms of substrate binding, substrate recognition, and receptor selection in this important class of virion proteins.
Keywords: glycobiology, influenza, nidovirus, sialate-O-acetylesterase, X-ray crystallography
To initiate infection, viruses must find and attach to suitable host cells. This process involves the specific recognition of cell surface determinants by dedicated receptor-binding proteins on the virion. Ideally, the receptors would occur exclusively on cells that support viral replication, but this is not always the case. Sialic acids (Sias), which serve as receptors/attachment factors for a multitude of mammalian and avian viruses, are common terminal residues of carbohydrate chains on glycoproteins and glycolipids (1). Consequently, they are found on many different cell types and even on non-cell-associated glycoconjugates. To escape from irreversible attachment to such “decoy” receptors, some viruses produce envelope glycoproteins with receptor-destroying enzyme (RDE) activity. For example, toro- and group 2a coronaviruses (both members of the family Coronaviridae, order Nidovirales; collectively referred to as “nidoviruses” throughout) and influenza C virus (family Orthomyxoviridae) attach to O-acetylated Sias and encode hemagglutinin–esterase proteins (HEs) with sialate-O-acetylesterase RDE activity to reverse off-target attachment (2–6).
The HEs of toro- and coronaviruses are ≈65-kDa class I glycoproteins that are dispensable for propagation in cultured cells but strictly maintained in field strains (for a review, see ref. 3). They share 30% sequence identity and are equally identical to subunit 1 of the influenza C virus hemagglutinin–esterase fusion (HEF) protein. While toro- and coronaviruses are of common ancestry, they apparently acquired their HE genes through horizontal gene transfer on separate occasions after the torovirus–coronavirus split (3). Structural and bioinformatic evidence suggests that the nidovirus HEs originated from trimeric HEF-like fusion proteins (7–9). In the course of events, the coronavirus (CoV) HE was converted into a disulfide-bonded homodimer and remnants of the fusion domain were adapted to establish new intermonomeric contacts (9). The quaternary structure of torovirus (ToV) HEs is not known; whether it is trimeric like HEF, also transformed into a dimer, or adopted another oligomeric structure altogether is an open question.
The toro- and coronaviruses that acquired HE each diverged to give rise to several new (sub)species in various new hosts. Concurrently, their HE proteins evolved to yield several distinct types with 30–50% intertypic variation (10, 11). HE divergence was accompanied by extensive changes in the architecture of the receptor-binding domains and in RDE substrate preference (3, 9). While most CoV HEs are specific for 9-O-acetylated Sias (i.e., Sias carrying O-acetyl groups at carbon atom C9 in the glycerol side chain), one branch of murine CoV HEs instead targets 4-O-acetylated Sias (i.e., with the O-acetyl attached to C4 in the pyranose ring) (11–13). The ToV HEs identified so far all display sialate-9-O-acetylesterase activity, yet whereas those of bovine toroviruses (BToVs) will cleave both 7,9-di-O- and 9-mono-O-acetylated sialic acids, porcine torovirus (PToV) HEs have a strong preference for the latter substrate (ref. 11 and see below).
Here we present and compare the crystal structures of two torovirus HE proteins, namely those of PToV strain Markelo and of BToV strain Breda. The data show that ToV HEs form dimers that in overall structure closely resemble those of CoV HEs with esterase domains highly similar to those of HEF and bovine coronavirus (BCoV) HE, but with unique functional receptor-binding sites. Guided by the crystal structures, we biochemically probed the ToV HE esterase domain. Our data shed new light on HE evolution and provide fundamental insight into the mechanisms of esterase substrate binding and substrate selection.
Results and Discussion
Overall Structure of Torovirus HE.
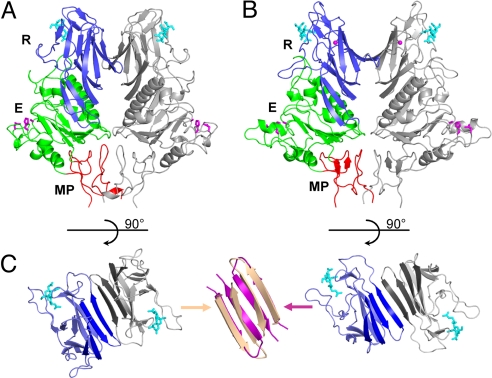
We determined the crystal structures of soluble HE ectodomains of PToV strain Markelo (residues 24–393) and BToV strain Breda (residues 15–392) to resolutions of 2.1 and 1.8 Å, respectively (Table 1). The PToV and BToV HE monomers share 67.7% sequence identity and display a root mean square difference (rmsd) on main chain Cα atoms of 0.87 Å. The data reveal that ToV HEs form symmetrical dimers (Fig. 1A) remarkably similar to those of CoV HE (Fig. 1B). There are no indications from the crystal packing that ToV HE dimers associate to form larger oligomers. We therefore assume that the ToV HE spikes as they occur in virions (6) are homodimeric.
Table 1.
Data collection and refinement statistics
| BToV HE | BToV HE with ligand | PToV HE S46A | PToV HE S46A with ligand | |
|---|---|---|---|---|
| Space group | P212121 | P212121 | C2 | C2 |
| Cell dimension a, b, c (Å), α, β, γ (o) | 67.8, 113.4, 273.5 90.0, 90.0, 90.0 | 67.6, 112.9, 273.0 90.0, 90.0, 90.0 | 156.3, 104.5, 97.5 90.0, 95.8, 90.0 | 156.0, 103.7, 97.1 90.0, 96.0, 90.0 |
| Resolution (Å)* | 50–1.8 (1.89–1.80) | 50–2.0 (2.11–2.00) | 58–2.1 (2.21–2.10) | 48–2.8 (2.94–2.79) |
| Completeness (%) | 95.3 (86.3) | 99.5 (98.6) | 100 (100) | 94.3 (95.3) |
| No. unique reflections | 186,978 | 141,552 | 90,955 | 36,147 |
| Multiplicity | 5.9 (5.8) | 3.7 (3.7) | 3.8 (3.8) | 2.7 (2.6) |
| Rmerge (%) | 7.2 (48.5) | 7.5 (34.0) | 11.6 (72.1) | 8.6 (48.7) |
| I/σ | 14.4 (3.6) | 12.0 (3.8) | 8.4 (2.2) | 10.5 (2.0) |
| Rfactor/Rfree (%) | 18.9/21.8 | 17.8/20.9 | 17.1/20.4 | 19.3/24.3 |
| rmsd bond lengths (Å) | 0.012 | 0.013 | 0.015 | 0.008 |
| rmsd bond angles (°) | 1.4 | 1.4 | 1.6 | 1.2 |
| No. protein atoms | 11,419 | 11,419 | 8,463 | 8,463 |
| No. glycan units | 44 | 31 | 16 | 16 |
| No. Waters | 1,276 | 1,026 | 518 | 11 |
*Values in parentheses refer to the highest-resolution shell of data.
Fig. 1.
Overall structure of ToV HE and comparison to CoV HE. (A) Ribbon representation of the PToV HE dimer. One HE monomer is colored gray, and the other is colored by domain: lectin domain (R: 143–284, blue) with bound Neu4,5,9Ac3α2Me (cyan sticks), esterase domain (E: 38–142 and 285–350, green) with Ser-His-Asp active site triad (magenta sticks), and membrane-proximal domain (MP: 25–37 and 351–386, red). The overall structure of the BToV HE dimer is very similar. (B) Ribbon representation of the BCoV HE dimer colored as in A. (C) Top views of the R domains in the PToV (Left) and the BCoV HE dimer (Right). A continuous 8-stranded β-sheet (emphasized by darker coloring) is formed across the dimer interface in BCoV HE, but not in PToV HE. Superposition (Center) of β-strands at the interface shows that strands in PToV HE (wheat) are more twisted than in BCoV HE (magenta), which prevents formation of a continuous β-sheet.
ToV HE monomers, like those of CoV HE, consist of 3 domains: a central esterase (E) domain displaying an α/β hydrolase fold with a Ser-His-Asp catalytic triad, a receptor-binding jelly-roll lectin (R) domain, and a small membrane-proximal (MP) domain (Fig. 1). The presence of the MP domain, structurally homologous to the F2 segments in the fusion domains of influenza C virus HEF and influenza A virus hemagglutinin (HA) (14, 15), indicates that ToV HE, like CoV HE, originated from an HEF-like fusion protein (9).
The overall arrangement of the ToV HE dimer is very similar to that of BCoV HE. Yet, structure-based sequence alignments [supporting information (SI) Table S1] show that the dimeric ToV and CoV HEs and trimeric influenza C virus HEF1 are essentially “equidistant” with rmsds on Cα atoms of the monomers ranging from 1.6 to 1.8 Å and sequence identities between 30 and 36%. Most conservation is in the E domains (rmsds 1.1–1.4 Å), whereas, as was already evident from comparative sequence analyses (11), the R domains are most variable (rmsds 2.0–2.2 Å). Upon closer inspection of ToV and CoV HE dimers, several differences become apparent, particularly in monomer–monomer interactions. In both types of protein, the dimer interfaces are made up of two contact regions (CRs), CR1 involving the R domains and CR2 predominantly involving the MP domains with a small additional contribution of the E domains in the ToV HEs (Table 2). The buried surface in CR2 of CoV HE is much smaller than in its ToV homolog, but this appears to be compensated by a disulfide bond, absent in ToV HEs, that links monomers near the membrane C-terminal from the MP domains. The β-sheet of the R domain that contributes to CR1 in the nidovirus HEs is also present in HEF, but remains unpaired in the trimer (9, 14). In ToV HEs, this β-sheet is more twisted than in CoV HE (Fig. 1C) and whereas in the CoV HE dimer two R domain β-sheets interact to form a continuous intermolecular β-sheet across the dimer interface, those of the ToV monomers are oriented at angles. This has consequences both for the CR1 contact area, which is significantly smaller in ToV HE, and for the relative position of R domains, which in ToV HE are shifted along the β-strands (Fig. 1C).
Table 2.
Surface area buried in dimer interface (Å2)
| Protein | Total | CR1 | CR2 |
|---|---|---|---|
| PToV HE | 2,563 | 686 | 1,877 |
| BToV HE | 2,044 | 661 | 1,383* |
| BCoV HE | 2,075 | 910 | 1,165 |
*Reduction of the buried surface area in CR2 by ∼500 Å2 with respect to PToV HE is caused by Glu365, a residue that is unique to BToV HE (see Fig. S4).
From their quaternary structure (dimers rather than trimers) it would be tempting to assume that the ToV and CoV HEs are more closely related to each other than to influenza C virus HEF1. However, the current data do not permit definitive conclusions about the course of HE evolution and the direction of gene flow. Trimer-to-dimer transition may have occurred only once and a gene for an ancestral dimeric HE may have found its way into the CoV and ToV genomes (or may even have been passed on from one nidovirus to the other). However, the differences between CoV and ToV HE structure, in particular those in monomer–monomer interaction, still allow for scenarios in which these proteins arose independently from trimeric HEF-like ancestors to transform into dimers through convergent evolution.
The Receptor-Binding Site.
To study ToV HE–Sia interactions, we soaked crystals of an esterase-deficient PToV HE Ser46Ala mutant with 10 mM of the synthetic Sia analog Neu4,5,9Ac3α2Me (Fig. S1) and solved the structure of the complex to 2.8-Å resolution (Table 1). Clear ligand density was visible in the receptor-binding (lectin) domains, but not in the esterase catalytic pockets (Fig. S2 A and B). Crystals of BToV HE mutant Ser37Ala soaked likewise did not bind Neu4,5,9Ac3α2Me for reasons unknown. The binding of ligand may have been hampered by the presence of an O-acetyl group at C4 or the ligand concentration may have been too low. When crystals of wild-type BToV HE were soaked at a higher concentration (30 mM) of the nonhydrolyzable Sia analog methyl 5-N-acetyl-9-S-acetyl-α-9-thioneuraminoside (Neu5N,9SAc2α2Me), a complex formed, the structure of which was solved to 2.0-Å resolution. Again, the Sia analog was found exclusively in the receptor-binding site (Fig. S2 C and D).
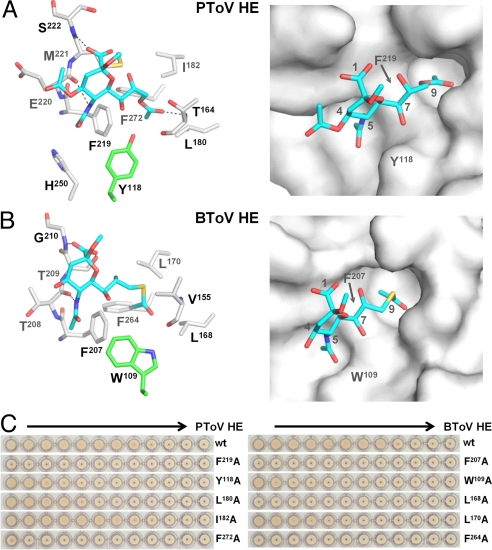
The receptor-binding sites of PToV and BToV HE are similar in overall architecture and mode of ligand binding. They are formed by residues from 4 segments of the R domain together with a single E domain residue, Tyr118 in PToV HE and Trp109 in BToV HE, located at the tip of a long loop (Fig. 2 A and B). Prominent protein–receptor interactions are provided by an internally disulfide-linked β-hairpin (residues 218–225 in PToV HE and 206–213 in BToV HE; Fig. S3A). At the base of this hairpin is a Phe residue (Phe219 and Phe207 in PToV and BToV HE, respectively) that is conserved in all ToV HEs (10) and in most CoV HEs (3) and becomes sandwiched between the Sia 5-N- and 9-O-acetyl groups when ligand is bound (Fig. 2 A and B; Fig. S3). In the ToV HEs and BCoV HE alike this interaction is critical, as in all cases substitution of the Phe residue by Ala abrogates lectin activity (Fig. 2C) (9). Additional β-hairpin–receptor interactions in the ToV HEs are provided by hydrogen bonding from main chain atoms (PToV HE, Glu220 and Ser222; BToV HE, Thr208 and Gly210) to the Sia 5-N-acetyl and carboxyl groups. In BToV HE, the Sia carboxyl group receives an additional hydrogen bond from the side chain of Thr209. A hydrophobic patch composed of Phe219, His250, and Tyr118 in PToV HE and of Trp109 and Phe207 in BToV HE further accommodates the Sia 5-N-acetyl group.
Fig. 2.
Receptor-binding sites of PToV and BToV HE. (A) Stick (Left) and surface (Right) representations of the PToV receptor-binding site (carbon, gray) bound to receptor analogs Neu4,5,9Ac3α2Me (carbon, cyan; nitrogen, blue; oxygen, red). Hydrogen bonds are shown as black dashed lines. (B) Stick (Left) and surface (Right) representations of the BToV receptor-binding site (carbon, gray) bound to receptor analogs Neu5,9Ac2α2Me (carbon, cyan; nitrogen, blue; oxygen, red; sulfur, yellow). (C) Effect of Ala substitutions on receptor binding of PToV and BToV HE. The relative binding affinity of wild-type PToV (Left) and BToV HE (Right) and their derivatives was assessed by hemagglutination assay with rat erythrocytes and 2-fold serial dilutions of each of the HE-Fc chimeras (5,000 to 2 ng of PToV and 10,000 to 5 ng of BToV HE per well; arrows indicate the direction from low to high dilutions). Of note, the PToV HE Phe272Ala and BToV HE Phe264Ala mutants were poorly secreted, indicative of protein misfolding, and hence were not analyzed. All other HE mutants were expressed to levels comparable to those of the wild-type HEs.
The 9-O-acetyl group, i.e., the one Sia modification essential to receptor binding, docks into a conserved hydrophobic pocket formed by Phe219, Ile182, Leu180, and Phe272 in PToV HE and by Phe207, Leu170, Leu168, and Phe264 in BToV HE (Fig. 2 A and B). The importance of each of these residues was confirmed by mutational analysis; their individual substitution by Ala reduced receptor binding of BToV and PToV HE, in most cases to levels below detection (Fig. 2C).
The architecture of the ToV HE lectin site is essentially different from those of influenza C HEF and BCoV HE (Fig. S3). Although the ToV and CoV lectin sites do share the β-hairpin and its phenylalanine, an element noticeably absent in influenza C HEF, they are distinct in all other respects: (i) The degree of divergence in the HE and HEF R domains is illustrated by extensive differences in disulfide-bonding pattern (3). Saliently, the disulfide bridge that stabilizes the ToV β-hairpin is absent in CoV HEs; it involves one cysteine residue conserved only in HEF (Cys213 and Cys225 in PToV and BToV HE, respectively) and one unique to ToV HEs (PToV HE Cys206 and BToV HE Cys218). (ii) The binding site of BCoV HE is organized by a potassium-binding loop that orients two adjacent leucines to interact with the Sia 9-O-acetyl group (9). While residues that coordinate the metal ion are conserved, with rare exception, in CoV HEs (3, 9), they are absent from ToV HEs. (iii) Finally and perhaps most strikingly, although BCoV HE and the ToV HEs employ a common strategy to specifically bind 9-O-acetylated Sias by fitting the critical 9-O-acetyl group into a hydrophobic pocket, the residues from which these pockets are composed are not conserved and even derived from different segments of the respective proteins.
As the orthomyxo- and nidovirus HEs diverged, the influenza C HEF receptor-binding site seems to have changed least. Despite the considerable evolutionary distance between influenza C HEF and influenza A HA, the Sia-binding pockets of these proteins are still very much alike in architecture (8, 9). The relative plasticity of the nidovirus HE receptor-binding domains may be attributed to evolutionary flexibility conferred by functional redundancy between HE and the companioning spike protein S (9). From another perspective, however, the fact that nidovirus HEs with new functional receptor-binding sites did evolve on more than one occasion reinforces the view that HE-mediated attachment to Sias provides a strong selective advantage to both the toro- and group 2a coronaviruses during natural infection.
The Catalytic Site: Arg–Sia Carboxylate Interaction, a Conserved Strategy for HE Substrate Binding.
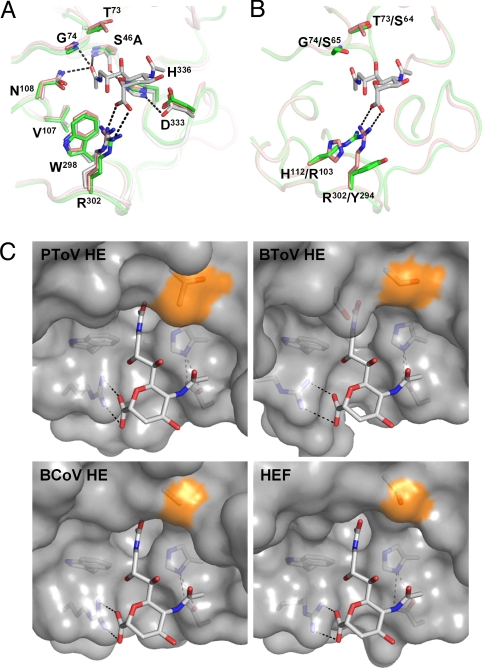
The esterase active site of PToV HE closely resembles those of BCoV HE and HEF (Fig. 3A). BToV-Breda HE, however, displays a number of intriguing amino acid differences that set it apart from most other nidovirus HEs (Fig. 3B; discussed further below). Although we did not succeed in obtaining ToV HE structures with a substrate analog in the catalytic pocket, the similarity to influenza C HEF justifies the use of a low-resolution HEF-inhibitor structure (14) to assess the potential role of individual amino acid residues in substrate binding. In influenza C HEF, the side chain of Asn117 and the NH groups of Gly85 and Ser57 create an oxyanion hole to stabilize the tetrahedral intermediate formed after nucleophilic attack by Ser57. In PToV HE, the residues composing the oxyanion hole are conserved (Asn108, Ser46, and Gly74), but in BToV HE, the Gly residue (Gly74) is replaced by Ser. Mutagenesis of PToV and BToV HE showed that this substitution and its back mutation do not affect enzyme activity toward natural and synthetic substrates nor do they affect substrate preference.
Fig. 3.
Structure of the PToV and BToV HE esterase sites and comparison to BCoV HE and HEF. (A) Superposition of PToV HE (orange), BCoV HE (green) and HEF (gray). Substrate analog 9-N-acetyl sialic acid (Neu5,9NAc2α2Me) present in the structure of HEF is shown in stick representation (carbon, gray; nitrogen, blue; oxygen, red). Hydrogen bonds in the catalytic triad and oxyanion hole of HEF are indicated by dashed black lines. Residue labeling refers to PToV HE0 that carries an active-site Ser46Ala mutation. (B) Superposition of the active sites of BToV HE (green) and PToV HE (orange). As shown, swapping of the active-site arginine between positions 294 and 103 in BToV HE would preserve hydrogen bonding to the sialoside carboxylate; note that optimal bidentate hydrogen bonding between Arg103 and the Sia carboxylate would require a ≈50° rotation and a 1.5-Å shift of the substrate's pyranose ring. Hydrogen bonds are indicated as in B. Residue labeling refers to PToV HE/BToV HE. (C) Surface representation of the PToV HE, BToV HE, BCoV HE, and HEF active sites with surface patches contributed by Thr73, Ser64, Ala74, and Ser84 in the respective proteins colored in orange. Note that the presence of Thr in PToV HE reduces the amount of free space available for substituents at the Sia C7 position, which, as shown by enzymatic analysis (Fig. 4), fully determines PToV HE substrate preference for 9-mono-O-acetylated Sias.
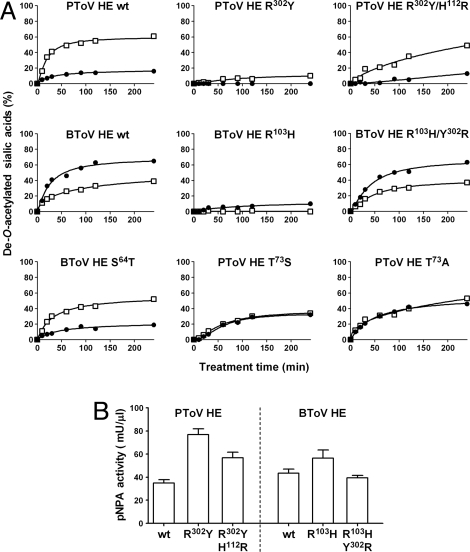
The structure of influenza C HEF revealed that in particular the Sia 9-O-acetate and carboxylate groups are involved in major contacts with the protein (8, 9) and we recently proposed that these groups in correct spatial arrangements are required for strong substrate interaction (16). Especially, HEF Arg322 would aid in binding and orientation of Sias by engaging into a double-hydrogen bond interaction with the Sia carboxylate group. Remarkably, however, while this Arg is conserved in PToV HE (Arg302) and, in fact, in most nidovirus HEs including those of BToV type III strains, it is substituted for Tyr (Tyr294) in BToV-Breda HE and in the HEs of type II BToV strains (for a description of torovirus genotypes, see Fig. S5 and ref. 10). The Tyr294 side chain is directed away from the active site and is thus unlikely to interact with the substrate. When introduced in PToV HE, the Arg302Tyr substitution severely reduced enzymatic activity toward glycosidically bound 9-O-acetylated Sias (Fig. 4A). This result showed that in the context of PToV HE the Arg residue is essential for efficient cleavage of natural substrates, but at the same time it raised the question why this substitution is tolerated in BToV HE. Interestingly, in BToV HE an arginine is found at position 103 (His112 in PToV HE), the side chain of which adopts an extended conformation and points toward the position that would be occupied by the sialoside carboxylate. This prompted us to ask whether the role of an Arg head group in substrate binding might still be essential and preserved in BToV-Breda HE, i.e., whether loss of Arg at the original location may have been compensated by ectopic introduction of a new one (Fig. 3B; Movie S1). Indeed, substitution of Arg103 by His in BToV HE strongly reduced its enzymatic activity toward both 7,9-di- and 9-mono-O-acetylated Sias. This defect, however, could be suppressed by a Tyr302Arg substitution; correspondingly, PToV HE with Arg302Tyr mutation regained enzymatic activity upon replacement of His112 by Arg (Fig. 4A). Importantly, the Arg residues are not essential for general folding of the esterase domain or for the catalytic process. PToV HE Arg302Tyr and BToV HE Arg103His mutants still cleaved p-nitrophenylacetate (pNPA) and, if anything, displayed enhanced activity toward this small synthetic substrate (Fig. 4B). Also of note, the location of the Arg residue did not affect substrate specificity; mutant proteins PToV HE Arg302Tyr/His112Arg and BToV HE Arg103His/Tyr302Arg were identical to their parental proteins with respect to their preference for 9-mono- and 7,9-di-O-acetylated Sias, respectively (Fig. 4A). Our observations provide experimental evidence that the bidentate Arg–Sia carboxylate interaction is of overriding importance for efficient cleavage of glycosidically bound (α-anomeric) Sias, apparently by fixing the Sia pyranose ring in an orientation that would bring the O-acetyl moiety on the C9 of the glycerol side chain in close proximity to the esterase active site nucleophile.
Fig. 4.
Molecular basis of HE substrate specificity. (A) Enzymatic activity of BToV and PToV HE and their derivatives toward O-acetylated Sias of bovine submaxillary mucin (BSM) as determined by GC-MS. The extent of enzymatic de-O-acetylation (expressed in percentages on the y axis) was calculated by integrating mass-chromatogram surfaces of peaks corresponding to Neu5,9Ac2 and Neu5,7,9Ac3 in BSM samples that had been either mock treated or HE treated for different amounts of time (x axis). Open squares, Neu5,9Ac2; closed circles, Neu5,7,9Ac3. (B) Esterase activity toward the synthetic substrate pNPA.
The Catalytic Site: Substrate Selectivity of PToV HE Determined by a Single Active-Site Residue.
The close similarity between the esterase pockets of the various HEs is remarkable given the differences in substrate preference. BToV HE, BCoV HE, and influenza C HEF readily accept both mono- and di-O-acetylated Sias as substrates. In contrast, PToV HEs display a strong preference for 9-mono-O-acetylated Sia (Fig. 4A) (10). Scrutiny of the PToV HE esterase site revealed a subtle yet conspicuous difference from the other HEs in the region proximal to the predicted position of C7 of bound Sia (Fig. 3C; Movie S2). In PToV HE, Thr73 replaces the slightly smaller orthologous residues Ala74, Ser64, and Ser84 in BCoV HE, BToV HE, and HEF, respectively. Biochemical analysis showed that these single-residue differences, modest as they might seem, greatly affect substrate specificity. Substitution of PToV HE Thr73 by either Ala or Ser resulted in a loss of substrate preference and yielded enzymes that now accept di- and mono-O-acetylated Sia substrates apparently with equal efficiency (Fig. 4A). Conversely, the reversed substitution Ser64Thr in BToV HE greatly diminished enzymatic activity toward 7,9-di-O-acetylated Sias and significantly enhanced cleavage of 9-mono-O-acetylated Sia. Thr73 is conserved in all known PToV HEs including those of PToV type II strains (Fig. S5) (11). Our findings thus provide a general molecular explanation for substrate selection in PToV HEs.
In the case of influenza A viruses, the ligand specificity of the viral lectin (HA) and the substrate specificity of the receptor-destroying enzyme, neuraminidase (NA) closely match, apparently as a result of HA–NA coevolution during host adaptation (17). Conceivably, a similar relationship between lectin and RDE preference might exist in the HE proteins. Comparison of the PToV and BToV HE lectin pockets suggests that in both cases the Sia glycerol side chain atom C8 and the C8 hydroxyl group are buried. This finding supports the notion that any major modification at this position would hamper binding by either HE. The lectin pockets do, however, differ, particularly with respect to the area near Sia glycerol side chain atom C7. Whereas in BToV HE there appears to be ample room to accommodate modifications at this position, in PToV HE the side chains of Val166 and Tyr118 would, in principle, obstruct binding of 7-O-acetylated ligands. Our combined observations suggest that porcine toroviruses have evolved to use 9-mono-O-acetylated Sias and to exclude di-O-acetylated receptors possibly as an adaptation to replication in swine. Viral Sia usage and the consequences thereof for host specificity, cell tropism, and pathogenesis merit further investigation.
Materials and Methods
Expression and Purification of ToV HE.
Codon-optimized genes encoding the HE ectodomains of PToV strain Markelo and BToV strain Breda (residues: PToV, 24–393; BToV, 15–392) (10) were cloned in expression plasmid S1-Ig as described (9). To allow expression of an enzymatically inactive HE-Fc chimera (HE0-Fc), the esterase catalytic residue (PToV Ser46; BToV Ser37) was substituted by Ala by site-directed mutagenesis, using the QuikChange XL II kit (Stratagene). Ala substitutions of selected residues in the HE receptor-binding site were made likewise. HE-Fc chimeric proteins were expressed in HEK293 GnT-I(−) cells (18) and purified on protein A beads, cleaved by thrombin to remove the Fc fusion, and concentrated to 10–15 mg/mL for crystallization as described.
Crystallization.
Crystallization conditions were screened as described (9). Crystals of BToV HE with space group P212121 grew to a final size of up to 0.30 × 0.25 × 0.25 mm within 1 or 2 weeks from 0.2 M sodium malonate, 0.1 M Bis-Tris propane, pH 8.5, 25% (wt/vol) PEG 3350 at 18 °C. For data collection, crystals were flash frozen in liquid nitrogen, using reservoir solution containing 12.5% (wt/vol) glycerol as the cryoprotectant. To determine the BToV HE structure in complex with ligand, crystals were soaked with 30 mM of the nonhydrolyzable sialic acid analog Neu5N,9SAc2α2Me (19) in cryoprotectant solution for ≈0.5 h.
PToV HE crystals with space group C2 were obtained from 0.2 M potassium acetate, 18% (wt/vol) PEG 3350 at 18 °C and grew to a final size of up to 0.35 × 0.1 × 0.05 mm. For data collection the fragile crystals were transferred to reservoir solution containing 17.5% (wt/vol) glycerol and flash frozen in liquid nitrogen. For data collection of receptor–analog complexes, crystals of PToV HE0 were soaked in cryoprotectant solution containing 10 mM methyl 5-N-acetyl-4,9-di-O-acetyl-α-neuraminoside (Neu4,5,9Ac3α2Me) (20) for ≈2 min.
Data Collection and Structure Determination.
Diffraction data (Table 1) were collected at ESRF stations ID14–3 (BToV HE native, PToV HE ligand soaked), ID23–1 (BToV HE ligand soaked), and ID23–2 (PToV native). Diffraction data were processed using XDS (21) and scaled using SCALA from the CCP4 suite (22). Molecular replacement was performed using PHASER with BCoV HE as template (PDB ID: 3CL4). Molecular replacement of ToV HE ligand complexes was performed using the refined PToV or BToV HE structures. Models were built by hand using Coot (23). Refinement was carried out using REFMAC (CCP4 package). Water molecules were added using ARP/WARP (24). Molecular graphics were generated with PYMOL (http://pymol.sourceforge.net/).
Hemagglutination Assay.
Hemagglutination assay was performed in V-shaped 96-well plates (Greiner Bio-One). Twofold serial dilutions of each of the HE-Fc chimeras (PToV, 10 μg; BToV, 20 μg) were made in 50 μL PBS supplemented with 0.1% BSA. Rat blood was washed with PBS and 50 μL 0.5% rat blood suspension was added to each well and incubated for 2 h on ice.
Enzyme Analysis.
Substrate preference of wild-type and mutant ToV HEs was determined by enzymatic de-O-acetylation of bovine submaxillary mucin (BSM) essentially as described previously (11). Briefly, BSM (1 mg/mL in PBS, pH 6.5) was treated with 0.5 μg/mL HE-Fc for 10–240 min at 37 °C or mock treated. Sialic acid analysis by gas–liquid chromatography–electron impact mass spectrometry was performed as described (25), using a Fisons Instruments GC8060/MD800 system (Interscience) and an AT-1 column (30 m × 0.25 mm, Alltech). Chromatograms were analyzed and quantitated with MassLab software and graphs were made with GraphPad.
Supplementary Material
Acknowledgments.
We acknowledge the European Synchrotron Radiation Facility for providing beamline facilities and the beamline scientists at ID14–3, ID23–1, and ID23–2 for their help with data collection. We thank P. Rottier and J. de Groot-Mijnes for advice and for reading this manuscript and P. Reeves and M. Farzan for sharing HEK293S GnT-I(−) cells and expression plasmids, respectively. This work was supported by an ECHO grant from the Council for Chemical Sciences of the Netherlands Organization for Scientific Research and by a grant from the Mizutani Foundation for Glycoscience (to R.J.d.G.). The Australian Research Council is acknowledged for its generous support and for the award of a Federation Fellowship (to M.v.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3I26 (BToV Breda HE), 3I27 (BToV Breda HE complexed with Neu5N,9SAc2α2Me), 3I1K (PToV Markelo HE), and 3I1L (PToV Breda HE complexed with Neu4,5,9Ac3α2Me)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0904266106/DCSupplemental.
References
- 1.Angata T, Varki A. Chemical diversity in the sialic acids and related α-keto acids: An evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 2.Herrler G, Klenk HD. Structure and function of the HEF glycoprotein of influenza C virus. Adv Virus Res. 1991;40:213–234. doi: 10.1016/S0065-3527(08)60280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Groot RJ. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlasak R, Luytjes W, Leider J, Spaan W, Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelissen LA, et al. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luytjes W, Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ. Sequence of mouse hepatitis virus A59 mRNA 2: Indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenthal PB, et al. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Q, Langereis MA, van Vliet al., Huizinga EG, de Groot RJ. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci USA. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits SL, et al. Phylogenetic and evolutionary relationships among torovirus field variants: Evidence for multiple intertypic recombination events. J Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits SL, et al. Nidovirus sialate-O-acetylesterases: Evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J Biol Chem. 2005;280:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klausegger A, et al. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regl G, et al. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J Virol. 1999;73:4721–4727. doi: 10.1128/jvi.73.6.4721-4727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenthal PB, et al. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 16.Mayr J, et al. Influenza C virus and bovine coronavirus esterase reveal a similar catalytic mechanism: new insights for drug discovery. Glycoconj J. 2008;25:393–399. doi: 10.1007/s10719-007-9094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 18.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isecke R, Brossmer R. Synthesis of N-acetyl-9-S-acetyl-9-thioneuraminic acid, N-acetyl-9-thioneuraminic acid, and their methyl α-glycosides. Carbohydr Res. 1995;274:303–311. [Google Scholar]
- 20.Haverkamp J, Schauer R, Wember M, Kamerling JP, Vliegenthart JFG. Synthesis of 9-O-acetyl- and 4,9-di-O-acetyl derivatives of the methyl ester of N-acetyl-β-D-neuraminic acid methylglycoside. Their use as models in periodate oxidation studies. Hoppe Seylers Z Physiol Chem. 1975;356:1575–1583. doi: 10.1515/bchm2.1975.356.2.1575. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 22.The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 25.Kamerling JP, Vliegenthart JFG. In: Mass Spectrometry, Clinical Biochemistry - Principles, Methods, Applications. Lawson AM, editor. Vol 1. Berlin: Walter de Gruyter; 1989. pp. 176–263. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.