Abstract
Disruption of signaling pathways such as those mediated by Shh or Pdgf causes craniofacial disease, including cleft palate. The role that microRNAs play in modulating palatogenesis, however, is completely unknown. We show, in zebrafish, that the microRNA Mirn140 negatively regulates Pdgf signaling during palatal development and we provide a mechanism for how disruption of Pdgf signaling causes palatal clefting. The pdgf-receptor alpha (pdgfra) 3’ UTR contains a Mirn140 binding site functioning in the negative regulation of Pdgfra protein levels in vivo. Both pdgfra mutants and Mirn140-injected embryos share panoply of facial defects including clefting of the crest-derived cartilages that develop in the roof of the larval mouth. Concomitantly, the oral ectoderm beneath where these cartilages develop loses pitx2 and shha expression. Mirn140 modulates Pdgf-mediated attraction of cranial neural crest cells to the oral ectoderm, where crest-derived signals are necessary for oral ectodermal gene expression. Both Mirn140 loss-of-function and pdgfra overexpression alters palatal shape and causes neural crest cells to accumulate around the optic stalk, a source of the ligand Pdgfaa. Conserved molecular genetics and expression patterns of mirn140 and pdgfra suggest that their regulatory interactions are ancient methods of palatogenesis that provide a candidate mechanism for cleft palate.
Cleft palate and other craniofacial diseases are common in humans and have complex cellular and genetic etiologies. In amniotes, the palate serves to separate the nasal and oral cavities and is generated through an intricate series of morphogenic events that include early neural crest cell migration and cell-cell signaling during the formation of facial prominences, as well as later generation and fusion of palatal shelves. While later events involving palatal shelves have not been described in zebrafish, palatal precursors migrate both rostral and caudal to the eye to condense upon the oral ectoderm in amniotes1 as well as zebrafish2,3 and evidence continues to accumulate that the early signaling environment governing palatogenesis is also largely equivalent3–6. For instance, Hh signaling is crucial for palatogenesis in humans and zebrafish3,4,7. Zebrafish and amniotes also share expression patterns of palatogenic genes such as Shh4,8, Fgf84,9,10 and Pdgf receptor alpha (Pdgfra)11–13.
In mouse, the Pdgf family consists of four soluble ligands, Pdgfa, Pdgfb, Pdgfc, and Pdgfd as well as two receptor tyrosine kinases, Pdgfra and Pdgfrb14. Pdgf signaling regulates a myriad of biological processes as demonstrated by analyses of mouse Pdgf ligand and receptor mutants14. Mice null for Pdgfra have a facial clefting phenotype that includes cleft palate12,13. This facial phenotype is fully recapitulated in mice doubly mutant for Pdgfa and Pdgfc 15. Most Pdgfc mutants have cleft palate15 but Pdgfa mutants exhibit either severe phenotypes, dying before palatogenesis, or less severe phenotypes without cleft palate16. The inability to examine the severe phenotypic class for palatal defects precludes a clear understanding of how Pdgfa regulates palatogenesis. Whereas crest require the reception of Pdgf signaling during palatogenesis in mouse13, the palatogenic cell behaviors regulated by Pdgf signaling and the modulation of Pdgf signaling during palatogenesis are unknown.
MicroRNAs (miRNAs) provide a unique mechanism for modulating signaling pathways17–20. Skeletogenic, including palatal, precursors express mirn140 (miR-140) in teleosts21 and amniotes22–24, suggesting that Mirn140 may modulate signaling during palatogenesis across vertebrate species. Despite the function of miRNAs in development and the importance of neural crest in evolution and disease, no miRNA has yet been shown to regulate neural crest development or cellular behaviors.
One important neural crest cell behavior is their migration along highly stereotyped pathways to give rise to a diverse array of differentiated cell types. Across vertebrate species, crest cells at cranial levels migrate in one of three crest streams. Cells in the most anterior, or first, stream will migrate rostrally and caudally around the eye into the first pharyngeal arch and contribute to the jaw and palatal skeleton5. While research in zebrafish and amniotes have uncovered cues that regulate migration of cranial neural crest cells in all crest streams25–27, nothing is known of what cues specifically guide neural crest-derived palatal precursors to the first pharyngeal arch.
Here, we show that Mirn140 attenuates Pdgf-mediated attraction during migration of neural crest-derived palatal precursors. Embryos injected with Mirn140 duplex and pdgfra mutants shared craniofacial phenotypes, including cleft palate and loss of oral ectoderm gene expression, suggesting an interaction between Mirn140 and pdgfra. Binding sites for Mirn140 are conserved in the 3’ untranslated region (UTR) of pdgfra across vertebrate species and Mirn140 interacts with the 3’UTR of the pdgfra transcript to negatively-regulate Pdgfra protein production. Palatal precursors express both mirn140 and pdgfra as they follow a migratory pathway delimited by expression of the ligand Pdgfaa. Attenuation of Pdgf signaling via Mirn140 is critical for rostrally migrating neural crest to migrate beyond the optic stalk, a Pdgfaa source, onward to the oral ectoderm, another Pdgfaa source. Our results demonstrate how delicately orchestrated modulation of Pdgf signaling regulates palatal morphogenesis.
RESULTS
Mirn140 and Pdgf signaling regulate palatal development in zebrafish
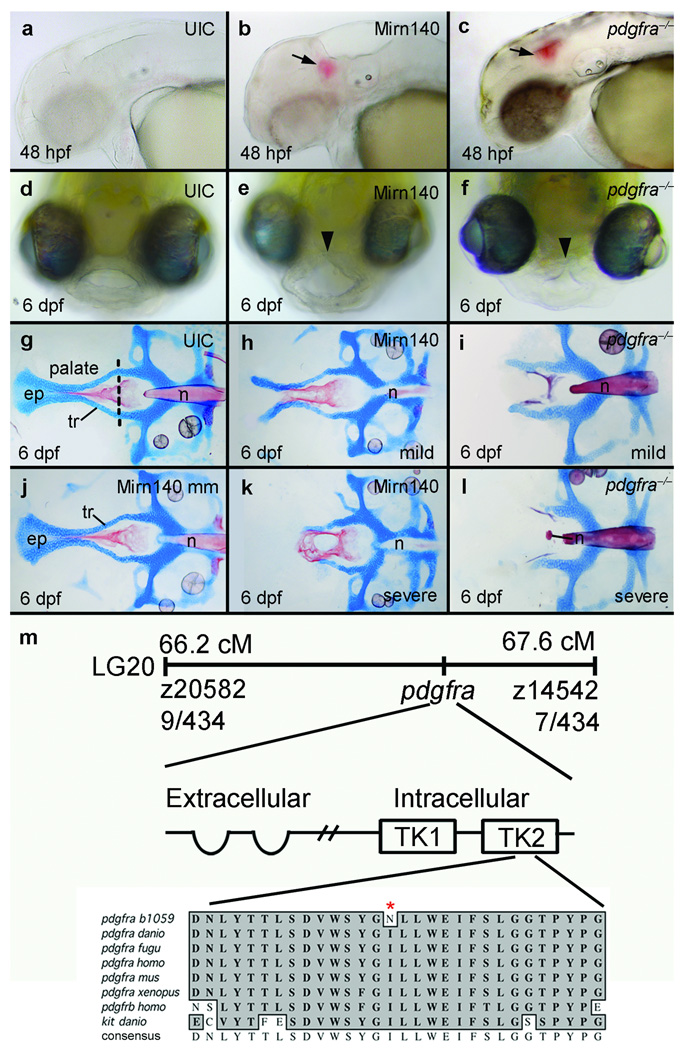
Skeletal precursors across vertebrate species express mirn14021–24 and mirn140 occupies the orthologous intron in wwp2 of zebrafish, human, and other vertebrates (Supplementary Fig. 1), prompting the hypothesis that Mirn140 plays a conserved role in skeletogenesis across vertebrate species. To determine the in vivo role of Mirn140 during zebrafish skeletogenesis, we injected embryos with Mirn140 duplex. By 2 days post-fertilization (dpf), Mirn140 duplex injected embryos had a profound facial phenotype, including cranial hemorrhaging and a hypoplastic roof of the mouth (Fig. 1a,b,d, e). This hypoplasia suggested that the zebrafish skeletal palate was malformed and alcian/alizarin staining confirmed the presence of palatal defects, including cleft palate, in Mirn140 duplex injected embryos (Fig. 1g,h,j,k).
Figure 1.
Overexpression of Mirn140 phenocopies pdgfra mutants. (a-c) Animals injected with Mirn140 duplex had cranial hemorrhaging (arrows) at 48 hpf (b), mimicking the phenotype of zebrafish (c) and mouse pdgfra mutants12,13. (d–f) Frontal views of 6 dpf larvae show that, compared to uninjected controls (UIC) (d), Mirn140 duplex injected (e) and pdgfra mutant (f) animals develop hypoplastic upper lips (arrowheads). (g–l) In Alcian/Alizarin-stained palates of 6 dpf uninjected controls (UIC) (g) or Mirn140 mis-match control injected embryos (j) trabeculae (tr) fuse at the trabecular communis in the midline and extent anteriorly forming the ethmoid plate (ep), but Mirn140 duplex injected (h,k) and pdgfra mutants (i,l) show both mild (h,i) and severe (k,l) phenotypes that include complete clefting of the palatal skeleton. (m) The b1059 allele is a mutation of pdgfrab. 1059 was genetically mapped to linkage group 20 (LG20) between the polymorphic markers z20582 and z14542, with 9 cross-overs and 7 cross-overs, respectively, out of 434 meioses. Sequence analysis of wild-type and b1059 mutant embryos revealed a missense mutation in the second tyrosine kinase (TK) domain of pdgfra. Protein sequence alignment of this region of the second tyrosine kinase domain (amino acids 841–870) of Pdgfra and related receptors shows the non-conservative I855N missense mutation (asterisk) in the second tyrosine kinase domain. This domain is highly conserved in Pdgfra across species as well as across related receptors such as Pdgfrb and Kit. n=notochord.
A clue to the target of Mirn140 during palatogenesis came from the fact that the array of facial defects seen in Mirn140 duplex injected embryos precisely phenocopy those observed in pdgfra b1059 (pdgfra‒/‒) mutants (Fig. 1c,f,i,l), identified in our forward genetic screen for craniofacial mutants (Fig. 1m). The pdgfrab1059 allele is likely hypomorphic since, unlike mouse Pdgfra mutants12 and Mirn140 duplex injected zebrafish embryos (Supplementary Fig. 1), zebrafish pdgfra mutants had normal somites and were typically of similar size to their wild-type siblings (data not shown). The molecular nature of the pdgfrab1059 allele is also consistent with b1059 being hypomorphic. In pdgfrab1059 an I855N missense mutation is present near the activation loop of the second tyrosine kinase domain of the receptor. The sequence of this kinase core is highly conserved in Pdgfra across species and even across other related tyrosine kinase receptors such as Pdgfrb and Kit (Fig. 1m). Therefore the non-conservative hydrophobic to hydrophilic amino acid substitution is likely to attenuate, but not necessarily obliterate, receptor signaling. Pharmacologic inhibition of Pdgf receptor signaling, via Pdgfr inhibitor V, phenocopies the b1059 allele and injection of pdgfra mRNA can rescue the palate of b1059 embryos, providing confirmation that b1059 lesions pdgfra (Supplementary Fig. 2). It is noteworthy that injection of pdgfra mRNA occasionally caused palatal defects in wild-type embryos (see below), suggesting that the overall level of Pdgf signaling must be strictly regulated for proper development of the palatal skeleton.
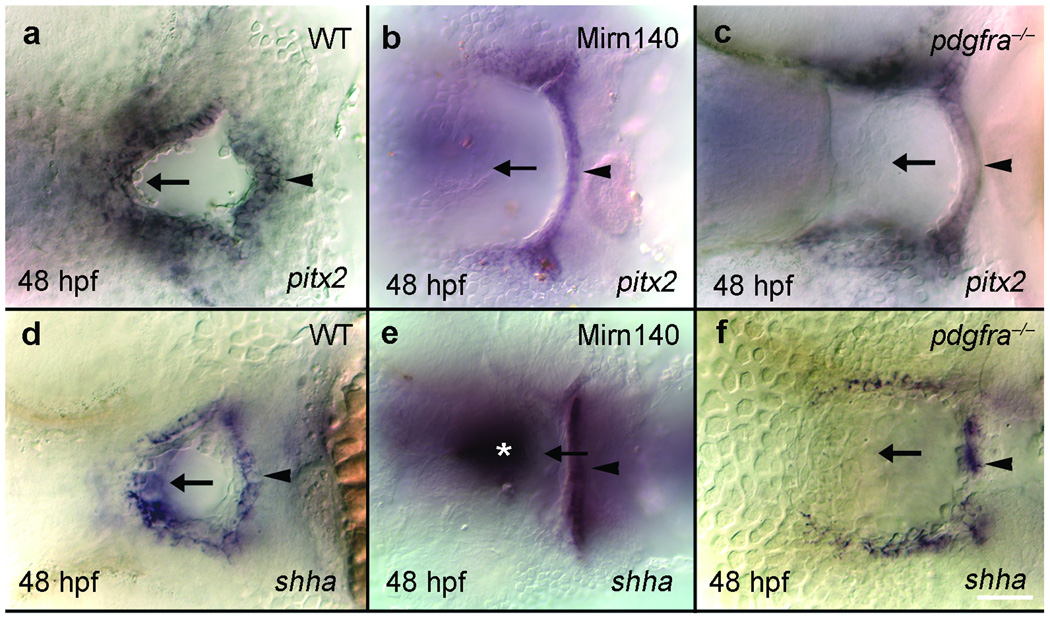
As in amniotes, the zebrafish palatal skeleton rests on the ectodermal roof of the oral cavity. In addition to their skeletal defect, the oral ectoderm of both Mirn140 duplex injected embryos and pdgfra mutant embryos failed to express regulatory genes such as pitx2 (Fig. 2a–c) and shha (formerly called shh; Fig. 2d–f). The oral ectoderm is present, albeit misshapen, and its fate map is not altered as determined by anti-pan-cadherin antibody staining and Kaede photoconversion, respectively (Supplementary Fig 3). The oral ectoderm’s morphological defect is not due to developmental delay because at all time points we’ve examined Mirn140 duplex injected embryos and pdgfra mutants display this defect (see Fig. 1e,f). The mechanism underlying this ectodermal defect is unclear, we do not detect elevated levels of cell death or loss of cell proliferation in the oral ectoderm (data not shown). In sum, the similarity of defects observed in Mirn140 duplex injected embryos and pdgfra mutants suggest a model where Mirn140 modulates Pdgf signaling, coordinating development of the palatal skeleton and the oral ectoderm.
Figure 2.
The oral ectoderm is similarly disrupted in Mirn140 duplex injected embryos and pdgfra mutants. Ventral views, anterior to the left, of oral ectoderm labeled with pitx2 (a–c) and shha (d–f) riboprobe in wild-type (a,d), Mirn140 duplex injected embryos (b,e) and pdgfra mutants (c,f). The roof of the oral ectoderm (arrows), adjacent to the normal location of palatal precursors, expressed neither gene in Mirn140 duplex injected embryos or pdgfra mutants. Loss of gene expression was specific to the roof of the oral ectoderm as the floor of the oral ectoderm expressed both pitx2 and shha (arrowheads). The embryo in e is severely affected and shha staining in the ventral brain is evident (asterisk).
Mirn140 regulates Pdgfra protein levels during palatogenesis
If Mirn140 modulates Pdgf signaling, Mirn140 binding sites should be present in the 3’ UTR of one or more members of the Pdgf signaling pathway and their protein levels should be regulated by Mirn140. Sequence comparisons of the Pdgf signaling family revealed Mirn140 binding sites in the 3’ UTR of pdgfra in all sequenced vertebrate genomes examined (Supplementary Table 1) and injection of Mirn140 duplex lowered Pdgfra protein levels (Fig. 3a). In contrast to pdgfra, the genes encoding zebrafish Pdgf ligands pdgfaa, pdgfab, pdgfba, pdgfbb, and pdgfd do not possess predicted Mirn140 binding sites. Although pdgfc has a Mirn140 binding site, the craniofacial region expresses pdgfc at 30 hpf, after the time we show that Mirn140 has its effects on palatogenesis (see below). An alternative hypothesis, that Mirn140 acts via inhibition of hdac4, as in mouse tissue culture cells24, is unlikely. Zebrafish hdac4 does not have a candidate Mirn140 binding site, nor does the scheduling or quantity of bone in Mirn140 over-expression or knockdown animals change as predicted by the tissue culture studies, suggesting that hdac4 is not a significant in vivo target of Mirn140 in zebrafish embryos. These results suggest that Mirn140 has its effects on palatal morphogenesis through inhibition of Pdgfra.
Figure 3.
Mirn140 regulates Pdgfra levels. (a) Compared to uninjected controls (UIC), Mirn140 duplex injection reduces and mirn140 morpholino injection elevates the level of the endogenous Pdgfra protein, respectively, detected by anti-human PDGFRA antibody. Anti-mouse Actin antibody is used as a loading control. (b) Schematic of the GFP-pdgfra mRNA injected to test for interaction of Mirn140 with the pdgfra 3’UTR, which bears a predicted Mirn140 binding site. (c–f) 27 hpf embryos injected with GFP-pdgfra alone (c) or with GFP-pdgfra and Mirn140-mismatch (Mirn140mm) duplex (d) fluoresce more strongly than animals injected with GFP-pdgfra and Mirn140 duplex (e). Morpholino knockdown of Mirn140 with Dicer-inhibitor MO increased GFP fluorescence above controls (f). (g) Pixel density analysis of GFP fluorescence confirmed results of c–f; Error bars indicate standard deviations. (h–k) Synthetic pdgfra mRNA truncated to remove the Mirn140 binding site (pdgfra*) rescued the cleft palate phenotype of Mirn140 over-expression, in three independent trials. (h) Distribution of palatal phenotypes after injection of Mirn140 duplex alone (n=54) or co-injection of Mirn140 and pdgfra* mRNA (n=117), in percent of animals. Mildly affected fish had near normal trabeculae and lateral ethmoid plate, but lacked the medial ethmoid plate like Fig. 1m; and severely affected fish lacked both the ethmoid plate and trabeculae like Fig. 1n. (i–k) Alizarin/Alcian-stained palates of 6 dpf uninjected control (UIC) (i) and co-injected embryos (j, k).
To directly test whether Mirn140 negatively regulates Pdgfra via the 3’UTR of pdgfra, we fused GFP to the 3’UTR of zebrafish pdgfra (GFP-pdgfra, Fig. 3b). Embryos co-injected with Mirn140 duplex and GFP-pdgfra lost GFP fluorescence (Fig. 3c–e,g), compared to controls. In contrast to GFP-pdgfra, reporter constructs fusing GFP to the 3’UTR of nog3, lacking a Mirn140 binding site, failed to respond to Mirn140 duplex (data not shown). To determine the extent of Pdgfra modulation via Mirn140, we injected embryos with mirn140 morpholino and assayed Pdgfra protein levels and expression of the GFP-pdgfra reporter. Knockdown of endogenous Mirn140 elevated Pdgfra protein levels (Fig. 3a) and GFP fluorescence compared to controls (Fig. 3c,f,g). These experiments show that the 3’UTR of pdgfra is a target of Mirn140.
Because miRNAs have many predicted targets 28, we co-injected Mirn140 duplex along with pdgfra mRNA lacking the Mirn140 binding site (pdgfra*) to test if the effects of Mirn140 on craniofacial development are mediated by Pdgfra. Co-injection of pdgfra* mRNA rescued Mirn140 duplex injected embryos (Fig. 3h–k), indicating that the cleft palate phenotype of Mirn140 duplex injected embryos is primarily due to loss of Pdgf signaling.
Our results show that Mirn140 attenuates Pdgf signaling and that Pdgf signaling is required for palatogenesis, but the cellular events mediated by Pdgf signaling during palatogenesis are unknown. Examining neural crest migration in mouse Pdgfra mutants uncovered no defect13, yet early phenotypic changes in zebrafish postmigratory crest (described below) were suggestive that migration of palatal precursors was abnormal. Therefore, we examined the expression of mirn140 and Pdgf signaling components during crest migration to elucidate how Pdgf signaling is involved in neural crest cell migration.
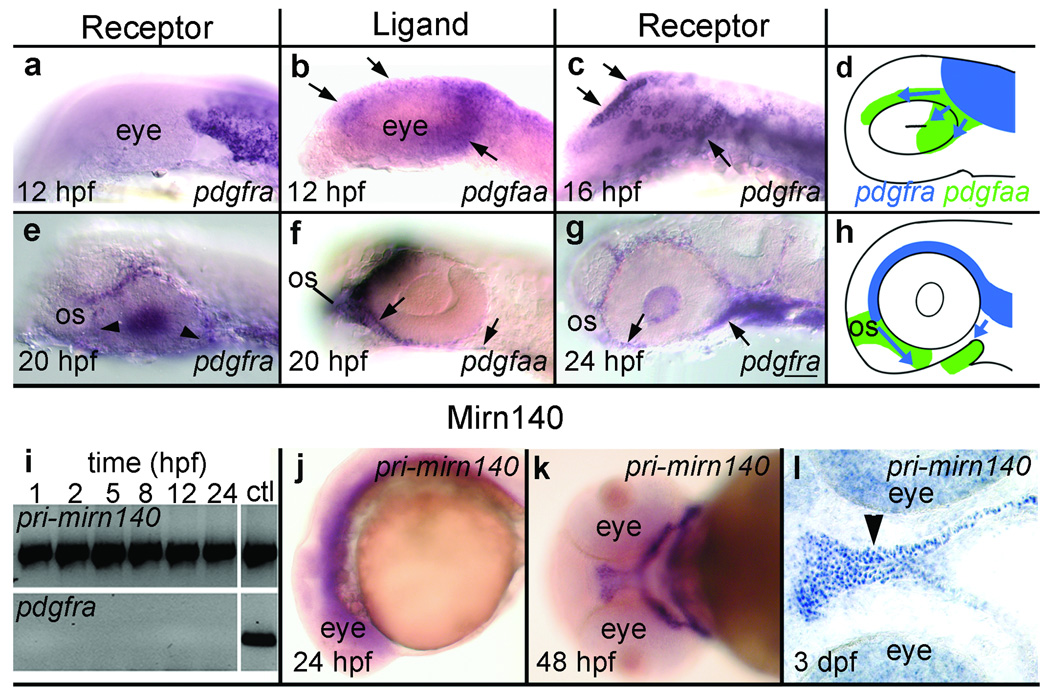
Consistent with previous reports11, we found that most, if not all, cranial neural crest cells express pdgfra. Crest-derived palatal precursors, fate mapped in previous studies3,4, migrate around the eye to reach the oral ectoderm and the subset of these crest cells migrating rostrally to the eye must additionally migrate around the optic stalk (see below). Both rostrally and caudally migrating crest cells express pdgfra throughout migration to the oral ectoderm (Fig. 4a,c,e,g). This expression pattern suggests that Pdgfra mediates migration of palatal precursors to the oral ectoderm.
Figure 4.
mirn140 overlaps with pdgfra during crest cell migration. (a–h) pdgfaa expression predicts the migratory pathway of pdgfra-expressing crest cells. (a–c) Lateral views of pdgfra (a,c) or pdgfaa (b) expression in 12 hpf (a,b) and 16 hpf (c) embryos. Premigratory crest expresses pdgfra (a) while the midbrain rudiment expresses pdgfaa (b). By 16 hpf, the position of pdgfra-expressing crest cells (c) mirrors the 12 hpf distribution of pdgfaa (compare arrows in b,c). (d) Schematic of early crest cell migration (blue arrows) relative to 12 hpf pdgfaa expression (green). (e-g) Lateral views of 20 hpf (e,f) and 24 hpf (g) embryos stained with pdgfra (e,g) or pdgfaa (f) riboprobe. When pdgfra-expressing crest cells have migrated to the optic stalk (os) and are near the oral ectoderm (e, arrowheads) the optic stalk, oral ectoderm, and cells between these two tissues express pdgfaa (f, arrows). By 24 hpf, neural crest cells are condensing on the oral ectoderm (g, arrows). (h) Schematic depicting crest cell migration (blue, arrows) to the oral ectoderm relative to pdgfaa expression (green). (i-l) mirn140 expression overlaps with pdgfra. (i) RT-PCR detects mirn140 from one hpf onward. Controls for genomic contamination utilized primers targeted to intronic sequence and failed to yield PCR product except in PCRs utilizing genomic DNA (ctl) (j-l) mirn140 transcripts detected by pri-mirn140 riboprobe are broadly distributed during crest cell migration (j, lateral view) and become restricted to post-migratory crest cells (k, ventral view), including the palatal skeleton (l, arrowhead, horizontal section).
To determine what ligand Pdgfra utilizes during crest cell migration, we cloned and surveyed the expression of the zebrafish homologues of Pdgfa and Pdgfc, because mice doubly mutant for these genes recapitulate the mouse Pdgfra mutant phenotype12,15. The zebrafish genome (http://www.ensembl.org/Danio_rerio/index.html) contains a single copy of pdgfc and duplicates of pdgfa, we designate these pdgfaa (formerly called pdgf-a29) and pdgfab. We found that only pdgfaa has spatiotemporal expression appropriate for gene encoding a candidate crest cell guidance molecule. Just before migration initiates, the midbrain expresses pdgfaa in a pattern that predicts the location of pdgfra-expressing palatal skeleton precursors four hours later (Fig. 4a–d). Expression is dynamic, and shifts to include the optic stalk and oral ectoderm as rostral crest cells migrate around the eye and optic stalk to reach the oral ectoderm (Fig. 4e–h). Well after palatogenic crest have reached the oral ectoderm, the optic stalk and oral ectoderm maintain expression of pdgfaa (Supplementary Fig. 4) and other Pdgf ligands begin to be expressed in the pharyngeal arches (Supplementary Fig. 4). These results are consistent with Pdgfaa acting as an attractant cue guiding palatal precursors to the oral ectoderm.
Because our reporter constructs demonstrated that Mirn140 functions by negatively regulating Pdgfra levels, we expect to see mirn140 and pdgfra co-expressed. RT-PCR detected mirn140 transcripts as early as 1 hpf and throughout crest cell migration (Fig. 4i) and in situ hybridization demonstrated that these transcripts were distributed broadly as late as 24 hpf (Fig. 4j). Following crest cell migration, transcripts for mirn140 became localized to skeletogenic crest (Fig. 4k,l), similar to the later expression of pdgfra11. This expression profile of mirn140 supports the conclusion that it modulates Pdgfra levels during palatal development, including crest cell migration.
Together, our results suggest the hypothesis that Mirn140, acting via Pdgfra, modulates Pdgf-mediated attraction of palatal precursor cells, which is required for correct migration.
Pdgf signaling is an attractant cue for palatal precursor cells
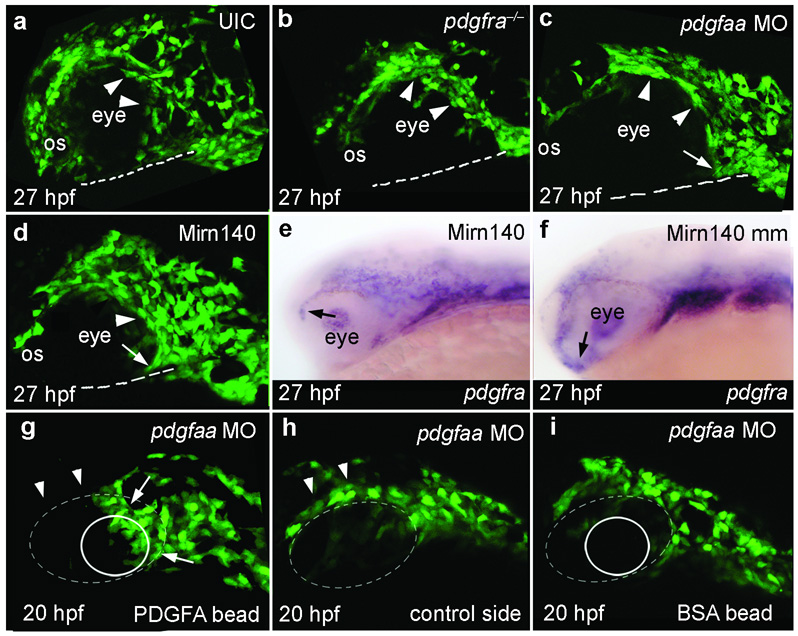
Our hypothesis predicts that palatal precursor migration3,4 will be disrupted in pdgfra mutants. Analyses in sox10:EGFP transgenic embryos show that palatal precursor cells[JP1] normally disperse rapidly rostrally and caudally around the eye to reach the oral ectoderm, where they condense (Supplementary Movie 1). Rostrally migrating cells must migrate around the optic stalk before reaching the oral ectoderm (Fig. 5a, Supplementary Movie 1). In pdgfra mutants, palatal precursors did not disperse but rather stayed tightly grouped together (Fig. 5b). Additionally, rostral crest did not migrate around the optic stalk (Fig. 5b), and most crest cells never reached the oral ectoderm (Supplementary Movie 2). We did not detect elevated levels of cell death in the crest cells or their migratory pathway during crest migration (data not shown), although it remains a possibility that cell death does occur at a later time. Crest cells that reached the oral ectoderm in pdgfra mutants did so via a circuitous route, avoiding a cell-free region normally invaded by crest (Supplementary Fig. 5). In contrast to palatal precursors, most other neural crest cells in pdgfra‒/‒;sox10:EGFP embryos migrated appropriately (Supplementary Fig. 5), similar to findings in mouse13. Hence, a subpopulation of neural crest cells, namely palatal precursors, requires Pdgfra function for proper migration.
Figure 5.
Pdgf signaling, modulated by Mirn140, guides palatal skeleton precursors to the oral ectoderm. (a–d) Lateral views of 27 hpf sox10:EGFP transgenic embryos. (a) Neural crest cells disperse and migrate around the eye and optic stalk (os) to reach the oral ectoderm (dashed line) in uninjected control (UIC) embryos. (b–d) In contrast, most palatal precursor cells do not disperse (arrowheads) or migrate to the oral ectoderm in pdgfra mutants (b), pdgfaa morpholino injected embryos (c), or Mirn140 duplex injected embryos (d). In all three circumstances rostrally migrating crest cells stop migrating at the optic stalk. Caudally migrating crest is less severely effected in pdgfaa morpholino injected embryos and Mirn140 duplex injected embryos than in pdgfra mutants (arrows, c,d, see Supplementary Figure 5). (e–f) Lateral views of 27 hpf embryos stained with pdgfra riboprobe. pdgfra mRNA is similarly present in Mirn duplex (e) and mismatch control (f) injected embryos demonstrating that Mirn140’s effect on migration is not through regulation of pdgfra transcripts. (g–i) Lateral views of pdgfaa morpholino injected (depleting endogenous Pdgfaa), sox10:GFP transgenic embryos implanted with rat recombinant PDGFA (g,h) or BSA (i) loaded beads (circles) just medial to the eye (dashed oval). (g) Crest cells accumulated adjacent to PDGFA beads (arrows, n=8) and fewer crest cells were present above the eye (arrowheads), compared to the control side of the same embryo (h) suggesting that crest cells were rerouted to the Pdgfa bead. (i) Control BSA beads did not attract crest cells.
Our hypothesis also predicts that both pdgfaa loss-of-function and Mirn140 overexpression should result in palatal precursor migration defects as observed in pdgfra mutants. Similar to pdgfra mutants, neural crest cells in pdgfaa morpholino injected or Mirn140 duplex injected, sox10:EGFP transgenic embryos failed to disperse and rostral crest cells stopped migrating at the optic stalk (Fig. 5c,d) resulting in clefting or reduction in the palatal skeleton (Fig. 1h,k; Supplementary Fig. 6). This effect on migration of pdgfra-expressing crest cells (Fig. 5e,f, arrows) in Mirn140 duplex injected embryos was not through loss of pdgfra transcripts, suggesting that Mirn140 attenuates Pdgf signaling by blocking Pdgfra translation. Collectively, we have shown that loss of Pdgf signaling either by blocking the ligand, by mutating the receptor, or by Mirn140 overexpression, results in the failure of palatal precursors to reach the oral ectoderm.
Phenotypes resulting from loss of Pdgf signaling and our expression analyses are consistent with Pdgf signaling being a positive guidance cue for cranial neural crest. To directly test the nature of Pdgf signaling during crest migration, we implanted beads soaked in PDGFA or BSA (bovine serum albumin), as a negative control, into pdgfaa morpholino injected sox10:EGFP transgenic embryos. Neural crest cells accumulated next to PDGFA beads, but not BSA beads (Fig. 5g–i, arrows), demonstrating that PDGFA is a positive cue, probably a chemoattractant as it is in oligodendrocyte migration30, that guides crest to the oral ectoderm.
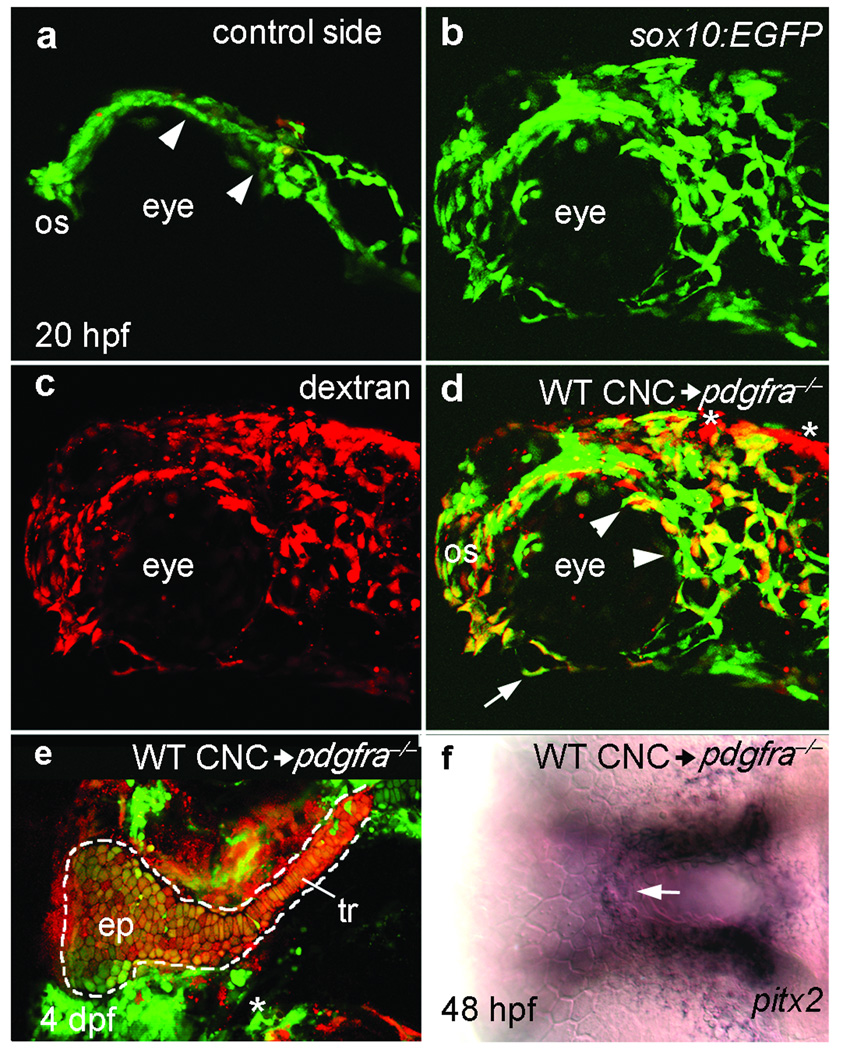
Loss of Pdgf signaling causes defects in both cranial neural crest cell migration and oral ectodermal gene expression, yet we never detected pdgfra transcripts in the oral ectoderm. In mouse, Pdgfra function is required in neural crest for proper palatogenesis13. Additionally, interspecific neural crest transplantation in avian species has shown that the oral ectoderm responds to crest-derived signals31, predicting that loss of oral ectodermal gene expression in pdgfra mutants is secondary to the failure of crest to reach the oral ectoderm. We tested both these predictions by transplanting pdgfra+ crest into pdgfra‒/‒ hosts and assaying neural crest migration and oral ectoderm gene expression. Neural crest cells from pdgfra+;sox10:EGFP donors dispersed and migrated to the oral ectoderm normally in pdgfra‒/‒ hosts (Fig. 6a,b). Not only did transplanted pdgfra+ crest restore the palatal skeleton (Fig. 6e), but it also rescued gene expression (Fig. 6f) in the pdgfra‒/‒ oral ectoderm. These transplants frequently contain contaminant, non-neural crest cells, yet these contaminant cells were typically distant from the oral ectoderm (Fig. 6d, asterisks) and thus unlikely to influence oral ectodermal development. Therefore, a crest migration defect is the primary cause of both the palatal skeleton and oral ectodermal phenotypes observed in embryos lacking functional Pdgf signaling.
Figure 6.
Pdgfra is required in neural crest for neural crest cell migration, palatal skeleton development and proper oral ectoderm specification. (a–d) Lateral views of the control (a) and experimental side (b–d) of a 20 hpf pdgfra mutant embryo that received a neural crest cell transplantation from a pdgfra+;sox10:EGFP embryo (WT CNC). (a) Mutant neural crest cells (green) are not dispersed (arrowheads) and failed to migrate beyond the optic stalk (os) on the side of the embryo that did not receive transplanted crest cells. (b–d) Donor pdgfra+;sox10:EGFP transgenic cells, labeled with Alexa dextran 568 (c), have dispersed, migrated around the optic stalk, and reached the oral ectoderm (arrow in d) in a pdgfra‒/‒ environment (n=21). (d) Merged imaged of b,c. Asterisks mark the location of two small patches of non-neural crest cells in the transplant. (e) Flat mounted palatal skeleton of another pdgfra mutant that received a neural crest cell transplant from a pdgfra+;fli1:EGFP transgenic donor (n=5). pdgfra+ crest-derived cartilage was present unilaterally in the ethmoid plate (ep) and trabeculae (tr) of the palatal skeleton (outlined). The trabecula and a portion of the ethmoid plate on the control side of the mutant embryo host were missing (asterisk). (f) 48 hpf ventral view of the embryo in a-d showing pdgfra+ crest rescues pitx2 expression in the roof of the pdgfra‒/‒ oral ectoderm (arrow, n=8). The contaminant cells, shown in d, are distant from the oral ectoderm and unlikely to influence oral ectodermal gene expression.
Mirn140 is necessary for rostrally migrating neural crest cells to reach the oral ectoderm
We have shown that Mirn140 negatively regulates Pdgf signaling, which is required for neural crest cells to migrate to the oral ectoderm and alter oral ectodermal gene expression. To further probe the normal function of Mirn140, we utilized morpholinos to knock down Mirn140 activity.
Loss of Mirn140 activity resulted in dramatically elevated levels of Pdgfra protein (Fig. 3a) and caused alteration in the shape of the palatal skeleton (Fig. 7a). Co-injection of Mirn140 duplex rescues this palatal phenotype, showing specificity of the morpholino (Fig. 7a). Normally in zebrafish, caudally migrating neural crest cells give rise to the lateral palatal skeleton, while rostrally migrating cells fill in the medial palatal skeleton3. Therefore, shape change after Mirn140 knockdown could be due to changes in the relative contributions of caudal versus rostral crest cells to the palatal skeleton.
Figure 7.
Loss of Mirn140 function alters palatal skeleton morphology and neural crest cell migration. (a–e) At 6dpf the length-to-width ratio (as shown in b–e) was calculated in injected and control embryos. Compared to uninjected controls (UIC, n=9) and mirn140 morpholino (MO) + Mirn140 duplex co-injected embryos (n=13), ratios were significantly larger and smaller in mirn140 morpholino injected embryos (n=8) and Mirn140 duplex injected embryos (n=11), respectively. Levels not connected by the same letter are significantly different at the 0.05% level (Tukey-Kramer HSD; one-way ANOVA: F1,57=186.7, p<0.0001). Error bars indicate standard deviation. (f-h) Compared to controls (f, n=8) fewer rostrally migrating neural crest cells had migrated from the optic stalk (os) to the oral ectoderm (dashed line) in sox10:EGFP transgenic embryos injected with mirn140 morpholino (g, n=10) or pdgfra* mRNA (h, n=10). Neural crest cells did encircle the optic stalk in these embryos, unlike the effects of Pdgf loss-of-function (see Fig. 5). (i–k) The resultant palatal phenotypes in the same embryos imaged in f–h. Compared to controls (i) Palatal morphology was altered in mirn140 morpholino injected embryos (j) but not pdgfa* injected embryos (k).
Rostrally migrating crest cells must pass the optic stalk, a Pdgfaa source, to reach the oral ectoderm. Therefore, elevating Pdgf signaling by injections of either mirn140 morpholino or pdgfra* mRNA, which lacks the Mirn140 binding site, may alter the number of rostral crest cells migrating past the optic stalk. In sox10:EGFP transgenic embryos, we found that many crest cells had migrated past the optic stalk (Fig. 7f, os) to the oral ectoderm by 24 hpf. However, after injection with either mirn140 morpholino or pdgfra* mRNA neural crest cells enveloped the optic stalk, yet few had migrated on to the oral ectoderm (Fig. 7g,h). The palatal skeleton’s morphology was altered in mirn140 morpholino injected embryos (Fig. 7j), but appeared fairly normal in pdgfra* injected embryos (Fig. 7k). This difference could be due to the labile nature of mRNA allowing the embryo to recover by 6 days post injection. We conclude that elevation of Pdgf signaling causes alterations in the shape of the palatal skeleton by reducing the number of rostrally migrating crest cells that reach the oral ectoderm.
Discussion
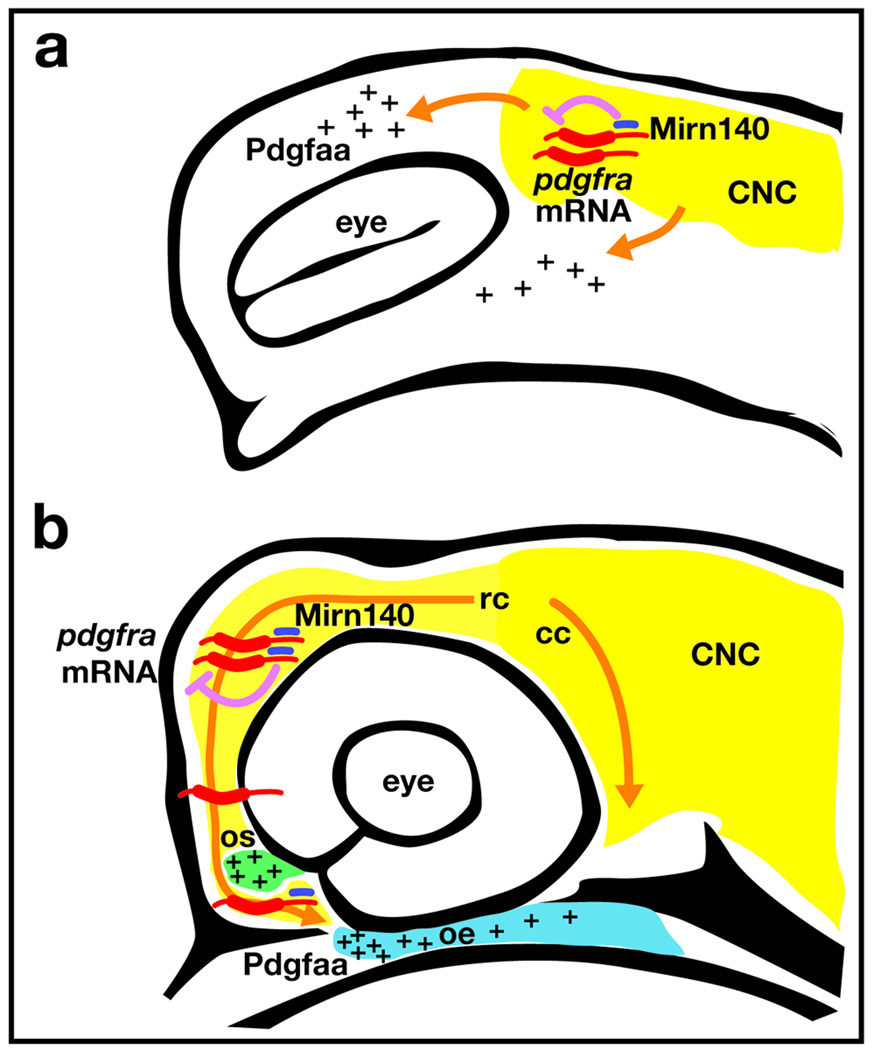
Here we show that precise control of Pdgf signaling is crucial for at least two separate events in crest migration. First, Pdgf signaling is necessary for neural crest cell dispersion (Fig. 8a). Most or all palatal precursors do not disperse if Pdgf signaling levels are low, whether by pdgfra mutation, pdgfaa morpholino injection, or Mirn140 duplex injection. How dispersion may be interrelated with migration remains to be elucidated, but caudally migrating crest cells are initially located very close to the oral ectoderm. Therefore, proper dispersion could play a large part in the migration of caudal cells to the oral ectoderm.
Figure 8.
Model of how Mirn140 modulates Pdgf signaling during palatogenesis. (a) Pdgfra signaling is required for neural crest cell dispersion. Neural crest cells (yellow) express pdgfra (red) and mirn140 (blue) as they disperse into regions of pdgfaa expression (pluses). Mirn140 inhibits Pdgfra production (pink arrow), but the overall level of Pdgf signaling is sufficient to promote crest dispersion along Pdgfaa-positive pathways. (b) When rostrally migrating crest cells reach the optic stalk, stoichiometric differences in pdgfra and Mirn140 levels regulate their final migration. To envelope the optic stalk (green), crest cells require relatively higher levels of Pdgfra. Crest cells that migrate on towards the oral ectoderm (blue) must first decrease the levels of Pdgfra via Mirn140, in order to leave the optic stalk.
Our results show, secondly, that modulated Pdgf signaling is critical for rostrally migrating cells to reach the oral ectoderm. In embryos with reduced Pdgf signaling, rostrally migrating neural crest cells do not migrate around the optic stalk, whereas in embryos with elevated Pdgf signaling, crest cells encircle the optic stalk, but few migrate on to the oral ectoderm. The optic stalk continues to express pdgfaa while palatal precursors migrate on to the oral ectoderm. Since Pdgfaa is an attractant cue for neural crest cells, there would normally be little reason for crest to leave the optic stalk unless Pdgf signaling was attenuated. In addition to the facial skeleton, crest cells normally contribute to structures associated with optic stalk derivatives 32. We propose a model where subtle stoichiometric differences between Mirn140 and pdgfra mRNA levels mediate the choice of neural crest cells to stay at the optic stalk or move on to the oral ectoderm to form the palatal skeleton (Fig. 8b).
Mirn140-mediated attenuation of Pdgf signaling may also play a significant role in the development of tissues outside the craniofacial skeleton. Mirn140 injected embryos have defects in cardiac and somite development as well as body axis elongation (see Supplementary Fig. 1). Mouse Pdgfra mutants share defects in all these systems but, while zebrafish pdgfra mutants do have cardiac defects, somite development and body axis elongation appear fairly normal (data not shown). The similarities in phenotypes between mouse Pdgfra mutants and Mirn140 injected zebrafish suggests conservation of Pdgfra function across vertebrate species, even though the presumptive hypomorphic pdgfrab1059 allele does not cause defects in all these developmental systems. Since, it is possible that Mirn140 regulates multiple signaling pathways, analyses in zebrafish null pdgfra alleles, yet to be identified, will be necessary to determine the full extent of conservation of Pdgfra function.
Our findings suggest that miRNA-mediated modulation of Pdgfra signaling may have evolutionary significance. In amniotes, snout length is associated with the relative contributions of the frontonasal prominence, derived from rostrally migrating cells, and the maxillary prominence, derived from caudally migrating cells, to the facial skeleton33,34. Therefore, the evolution of skull morphology could utilize miRNAs, in particular Mirn140, to tweak the number of crest cells arriving at an individual prominence within the first pharyngeal arch.
Cranial neural crest cells migrate to the pharyngeal arches in three crest streams, with the first stream populating the first pharyngeal arch,2 and yet no guidance cue responsible for the migration of any individual stream has been discovered. We show that loss of Pdgf signaling disrupts the migration of a subpopulation of first stream crest, those destined for the zebrafish palate. As in mouse13, all three crest streams express pdgfra. Hence, it is the expression of pdgfaa, encoding a ligand for Pdgfra, which provides the mechanism for Pdgf signaling to specifically guide palatal precursors. As crest cells migrate pdgfaa expression consistently and specifically predicts the pathway to be taken by palatal precursors. We propose that the restricted expression of pdgfaa attracts only palatal precursors since Pdgf signaling acts at short range14 and, therefore, cranial crest in more posterior streams would not receive Pdgf signaling. The same tissues that express pdgfaa in zebrafish also express Pdgfa in amniotes 35–37, consistent with the possibility that Pdgfa signaling plays similar roles in zebrafish and amniotes. Crest migration defects have not been described in mouse Pdgfra or Pdgfa mutants12,13,16, this could be due to species-specific developmental differences or to improved visualization of crest in zebrafish. It will be of great interest to determine if palatal precursor migratory defects are evident in mouse Pdgfra and Pdgfa mutants, although conditional mutation of Pdgfa would be necessary to overcome the early death of severely effected Pdgfa mutant mice16.
Further exploration of how microRNAs and other factors modulate signaling pathways such as Pdgf during palatogenesis will assuredly continue to provide insights into the cause of, and possible treatments for, human craniofacial disease.
Materials and Methods
Oligonucleotides
All oligonucleotides are listed in Supplementary Table 2.
Zebrafish care and use
The b1059 mutant allele was obtained via ENU mutagenesis39 in an AB background and was out-crossed to a WIK background for genetic mapping. PCR-based microsatellite mapping placed the b1059 allele in a 1.4 cM interval of LG20, between microsatellite markers z14542 (7 cross overs/434 meioses) and z20582 (2 cross overs/434 meioses), which contains 23 known genes including pdgfra. Given the phenotypic similarities of mouse Pdgfra mutants and b1059 mutants, pdgfra was an excellent candidate for the gene lesioned in b1059 mutants. Sequence analysis of pdgfra in wild-type and b1059 embryos revealed an adenosine-to-thymidine nucleotide change resulting in an I855N missense mutation, thus placing a charged residue in the hydrophobic core of the kinase C-lobe in the second tyrosine kinase domain of the receptor. pdgfrab1059 mutants were identified by either phenotype or PCR amplification using dCAPs primers40, forward: 5’ TGTCTCCAAAGGAAGCGTG 3’ and reverse: 5’ ACCGAGAGAGAAGATCTCCCATAACTAG 3’, followed by digestion with SpeI, resulting in a wild-type fragment of 263 bp and a b1059 fragment of 239 bp. Throughout the text we use pdgfra+ to refer to embryos with either pdgfra+/‒ or pdgfra+/+ genotypes.
All embryos were raised and cared for using established protocols39 with IACUC approval. Tg(fli1:EGFP)y1 transgenic embryos express GFP in neural crest cells shortly after the onset of migration, and in the vasculature 41, while Tg(-4.9sox10:EGFP)ba2 transgenic embryos express GFP in neural crest prior to the onset of migration3; here they are called fli1:EGFP and sox10:EGFP, respectively, through the text. We used heterozygous sox10:EGFP transgenics for our analyses, because homozygous embryos can have craniofacial defects3. Embryos were treated with 0.5 µM Pdgfr inhibitor V (Calbiochem) from 10 hpf- 4 dpf and Kaede photoconversion was carried out as described previously4.
Morpholino and RNA injection
Gene Tools (Philomath, OR, USA, http://www.gene-tools.com) supplied morpholino oligonucleotides (MOs) with the sequences: mirn140 MO (mature), 5’-CTACCATAGGGTAAAACCACTG-3’; mirn140 MO (Dicer inhibitor), 5’-GACGTAACCTACCATAGGGTAAAACCACTGA-3’; p53 MO, 5’-GCGCCATTGCTTTGCAAGAATTG-3’ 42 pdgfaa I1E2 (splice inhibitor) 5’ GGAATTGGTGCTTCCTGTTAAAGA 3’ and pdgfaa I2E3 (splice inhibitor) 5’ CCTCCAGCACTTCATTCTCTGCAAC.
We injected one or two-cell stage zebrafish embryos with approximately 3 nl of morpholinos: 0.4 mM pdgfaa, 1.2 mM mature mirn140, or 0.6 mM Dicer inhibitor mirn140. Injecting higher concentrations of pdgfaa morpholino resulted in embryos with disrupted body axes, consistent with pdgfaa playing a role in gastrulation movements43. To control for the effects of nonspecific cell death, we co-injected either pdgfaa morpholino or mirn140 morpholino (Dicer inhibitor, 1.2 mM) with p53 morpholino (0.3 mM)44.
Integrated DNA Technology (Coralville, IA, USA, http://www.idtdna.com) supplied RNA oligonucleotides with the sequences: Mirn140, 5’-CAGUGGUUUUACCCUAUGGUAG-3’; Mirn140 mismatch, 5’-CACACCAAGAACCCUAUGGUAG-3’; Mirn140* (the complementary natural strand), 5’-UACCACAGGGUAGAACCACGGAC-3’. To make 50 µM working stocks, 10 µL of 100 µM Mirn140 or Mirn140 mismatch plus 10 µL of 100 µM Mirn140 * were mixed together, boiled briefly to denature, and then slowly cooled to 4°C and stored at −80°C until injection. Embryos were injected with 3 nl of this 50 µM working stock or a 25 µM dilution of this stock, with identical results. To rescue the cleft palate phenotype in Mirn140 duplex injected embryos (25 uM), we co-injected mirn140 MO (Dicer inhibitor, 1.2 mM).
Reporter constructs
The primers pdgfraUTR-F (5’-TCTGCGTCATCTTGTCACTTTTTCTTCAC-3’) and pdgfraUTR-R (5’-AACACAGCCATTTTCTTCATTTTAGGAC-3’) amplified a 653 bp long fragment from genomic DNA containing the pdgfra 3’ untranslated region (UTR), which was inserted into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA, http://www.invitrogen.com). To fuse GFP with the pdgfra 3’UTR, we used NotI and SpeI enzymes to liberate the 3’UTR region of pdgfra gene from pCR4-TOPO and NotI and XhoI to extract GFP from pEGFP-N3 (Clontech, Mountain View, CA, USA, www.clontech.com). We ligated the fragments into PCRII-TOPO (Invitrogen) between SpeI and XhoI sites, to make GFP-pdgfra. To make mRNA, the vector was linearized with SpeI.
Co-injections were accomplished by double injection, first by injecting 0.6 mM Dicer inhibitor mirn140 MO or 25 µM Mirn140 duplex, and then by injecting 250 ng/µL of the synthetic GFP-pdgfra UTR mRNA. GFP-nog3 UTR construct was made similarly to the GFP-pdgfra UTR with the primers: nog3-F (5’-GAAATAAGCTCCGCACTCATCCTCACAT-3’) and nog3-R (5’-TCCATTCCCCTTATATTTACAGCACACCA-3’), amplifying a 504 bp fragment containing the nog3 3’UTR.
We amplified full-length pdgfra lacking the Mirn140 binding site (pdgfra*) using primers pdgfra-F (5’-TCATGTTCCCGGTGCTGCC-3’) and pdgfra-R (5’-GGGCTCCATAAGACTGAGGTGAAG-3’). The resulting 3418 bp PCR product was ligated into pCR4-TOPO. To make mRNAs, plasmid was linearized with SpeI and about 1 µg of purified linear template was transcribed by T7 RNA polymerase using mMessenger mMachine kit (Ambion, Austin, TX, USA, http://ambion.com) at 37 °C for 2 h. mRNAs were purified with RNA clean up kit (Zymo Research, Orange County, CA, http://www.zymoresearch.com) and diluted to 15 µL with nuclease-free water. We used this truncated mRNA for all of our pdgfra mRNA injections, including rescue of pdgfrab1059 mutants. The truncated pdgfra mRNA only alters the levels of Pdgfra protein produced from the mRNA and therefore does not alter the interpretation of our pdgfrab1059 rescue experiment.
Cloning and in situ hybridization
Primers for generating PCR products containing the full open reading frame of Pdgf family members and mirn140 are: pdgfra (Ensembl Gene ID ENSDARG00000030379) forward 5’ TCATGTTCCCGGTGCTGCC 3’ and reverse 5’ GGGCTCCATAAGACTGAGGTGAAG, pdgfaa (Ensembl Gene ID ENSDARG00000030379) forward 5’ TGGGACACTTTTGGACCACAGG 3’ and reverse 5’ TCGTTTTTCAGGCTGTCGTTG 3’, pdgfab (Ensembl Gene ID ENSDARG00000058424) forward 5’ TGACATTGGAAGGAGATGAGAACC 3’ and reverse 5’ TTATTGAATATCCTTGTTGATCAGTGC 3’ and pdgfc (Genbank accession XM 683962.2) forward 5’ CCAAATGATTCCGTTGCTTCTG 3’ and reverse 5’ GCGTCTTCTCTCTGGGACTGATT 3’. A 902 bp fragment of pri-mirn140 was isolated from 24 hpf embryo cDNA library by the primers primiR140-F (5’-GCAAGTCAAACCCTGTAGCATCCCGTT-3’) and primiR140-R (5’-GCGAGCCGATAGAGCGATTGTTT-3’). mirn140 (dre-mir-140 primary transcript, EU116273) was cloned by RT-PCR and 3’ RACE from 24 hpf zebrafish embryo cDNA. PCR products were cloned in pCR4-TOPO (Invitrogen). ClustalX alignment confirmed our Pdgf orthologue assignments. Capped mRNA was synthesized using the mMessage mMachine kit (Ambion, Austin, TX).
Integrated DNA Technology synthesized mirn140 LNA (Locked Nucleic Acid) probe (5’-CtACcATaGGgTAaAAcCAcTG-3’, lowercase nucleotides represent LNA nts) with 3’ end Digoxigenin-labeling. Probe was diluted to 0.5 µM in hybridization buffer (50% formamide, 2XSSC, 0.3% Tween-20, 0.5 mg/mL baker’s yeast RNA and 0.05 mg/mL heparin). In situ hybridization occurred at 43 °C on whole mounted embryos and frozen sections following protocols provided by Exiqon (Woburn, MA, USA, http://exiqon.com). In situ hybridizations utilizing mirn140 LNA yielded results identical to pri-mirn140 probe (not shown). Conventional in situ hybridization with digoxigenin-labeled RNA probes utilized a protocol similar to that for the LNA probe, but at a hybridization temperature of 68 °C. Cutting circular plasmid with NotI and synthesizing RNA with T3 polymerase generated antisense pdgfra, pdgfaa, and pdgfc riboprobes, whereas cutting with SpeI and synthesizing with T7 polymerase produced pdgfab and pri-mirn140 riboprobe. pitx2 and shh riboprobes have been described4.
Accession numbers for sequences used in this work: pdgfaa, NM_194426; pdgfab, NM_001076757; pdgfb, ESTs DV584985 and EB884207; pdgfc, XM_683962 and EST EH559317; pdgfd, XM_001333193; pdgfra, NM_131459 ENSDART00000011915;
Cartilage staining
Six day postfertilization zebrafish embryos were stained with Alcian Blue and flat mounted45.
Cell transplants and bead implants
For all transplants, donors were injected with 10,000 MW Alexa 568 dextran, labeling all cells. Shield stage transplants were carried out as described elsewhere4.
Affi-gel blue gel (BioRad) was incubated overnight at 4˚ with 10 µg/ml rat recombinant Pdgfaa (R&D Systems) or BSA (Sigma). Individual Affi-gel beads from the gel were inserted into pdgfaa morpholino injected embryos at 12 hpf and implanted embryos were imaged at 20 hpf.
Time-lapse analysis, confocal microscopy, and figure processing
Confocal z-stacks were collected on a Zeiss LSM Pascal. Images were processed in Adobe Photoshop CS and Adobe Illustrator. Recordings, 15 min/frame, and confocal analysis of transgenic embryos were performed according to established protocols4. To quantitate GFP levels, pixels of individual layers were selected by setting Fuzziness level to 170. The histogram function was then used to calculate the number and standard deviations of green pixels for each fish.
Western blotting
Immunoblotting used the Upstate (http://www.upstate.com) protocol with minor modifications. Proteins extracted from thirty 24 hpf fish were separated by SDS polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane using 3% blocking solution. The blot was probed with anti-human PDGFRA antibody (catalog# 07–276, Upstate) and anti-mouse ACTIN antibody (product# A4700, Sigma, St. Louis, MO, USA, http://www.sigmaaldrich.com) using HRP-conjugated secondary antibodies, anti-rabbit (catalog # AP132P) (Chemicon, Temecula, CA, USA, http://www.chemicon.com) and anti-mouse (AP124P) (Chemicon). Protein blots were visualized by ECL Western Blotting Detection System (Catalog # RPN2132, Amersham, Piscataway, NJ, USA. http://www.amersham.com).
Statistical Analysis
JMP version 5.1 software (SAS Institute, Cary, NC) was used for oneway ANOVA.
Supplementary Material
Supplementary Movie 1 Crest cell migration in a pdgfra+ embryo.
Supplementary Movie 2 Crest cell migration in a pdgfra−/− embryo.
Acknowledgements
We thank Ruth BreMiller for help with histology and John Dowd and the University of Oregon Zebrafish Facility for animal care. Morpholinos were provided by John Moulton at Gene Tools as part of Multi-Blocker field testing. This work was supported by the National Center for Research Resources (5R01RR020833 to JHP) and National Institutes of Health (P01 HD22486 to CBK and JHP and 5 K99 DE018088 to JKE); the contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
References
- 1.Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Developmental Biology. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- 2.Trainor PA, Melton KR, Manzanares M. Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. International Journal of Developmental Biology. 2003;47:541–553. [PubMed] [Google Scholar]
- 3.Wada N, et al. Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development. 2005;132:3977–3988. doi: 10.1242/dev.01943. [DOI] [PubMed] [Google Scholar]
- 4.Eberhart JK, Swartz ME, Crump JG, Kimmel CB. Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development. 2006;133:1069–1077. doi: 10.1242/dev.02281. [DOI] [PubMed] [Google Scholar]
- 5.Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Current Opinion in Cell Biology. 2001;13:698–705. doi: 10.1016/s0955-0674(00)00273-8. [DOI] [PubMed] [Google Scholar]
- 6.Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. Journal of Anatomy. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roessler E, et al. Mutations in the human sonic hedgehog gene cause holoprosencephaly. Nature Genetics. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 8.Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- 9.Riley BM, et al. Impaired FGF signaling contributes to cleft lip and palate. Proceedings of the National Academy of Sciences USA. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachler M, Neubüser A. Expression of members of the Fgf family and their receptors during midfacial development. Mechanisms of Development. 2001;100:313–316. doi: 10.1016/s0925-4773(00)00518-9. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Chong SW, Balasubramaniyan NV, Korzh V, Ge R. Platelet-derived growth factor receptor alpha (pdgfr-alpha) gene in zebrafish embryonic development. Mechanisms of Development. 2002;116:227–230. doi: 10.1016/s0925-4773(02)00142-9. [DOI] [PubMed] [Google Scholar]
- 12.Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 13.Tallquist MD, Soriano P. Cell autonomous requirement for PDGFRalpha in populations of cranial and cardiac neural crest cells. Development. 2003;130:507–518. doi: 10.1242/dev.00241. [DOI] [PubMed] [Google Scholar]
- 14.Betsholtz C, Karlsson L, Lindahl P. Developmental roles of platelet-derived growth factors. BioEssays. 2001;23:494–507. doi: 10.1002/bies.1069. [DOI] [PubMed] [Google Scholar]
- 15.Ding H, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nature Genetics. 2004;36:1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- 16.Boström H, et al. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell. 1996;85:863–873. doi: 10.1016/s0092-8674(00)81270-2. [DOI] [PubMed] [Google Scholar]
- 17.Hornstein E, Shomron N. Canalization of development by microRNAs. Nature Genetics. 2006;38 Suppl:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 18.Lee CT, Risom T, Strauss WM. MicroRNAs in mammalian development. Birth Defects Research. Part C, Embryo Today. 2006;78:129–139. doi: 10.1002/bdrc.20072. [DOI] [PubMed] [Google Scholar]
- 19.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local Architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Computational Biology. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song L, Tuan RS. MicroRNAs and cell differentiation in mammalian development. Birth Defects Research. Part C, Embryo Today. 2006;78:140–149. doi: 10.1002/bdrc.20070. [DOI] [PubMed] [Google Scholar]
- 21.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 22.Ason B, et al. Differences in vertebrate microRNA expression. Proceedings of the National Academy of Sciences USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darnell DK, et al. MicroRNA expression during chick embryo development. Developmental Dynamics. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 24.Tuddenham L, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Letters. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 25.Gammill LS, Gonzalez C, Bronner-Fraser M. Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. Developmental Neurobiology. 2007;67:47–56. doi: 10.1002/dneu.20326. [DOI] [PubMed] [Google Scholar]
- 26.McLennan R, Kulesa PM. In vivo analysis reveals a critical role for neuropilin-1 in cranial neural crest cell migration in chick. Developmental Biology. 2007;301:227–239. doi: 10.1016/j.ydbio.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Yu HH, Moens C. Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Developmental Biology. 2005;280:373–378. doi: 10.1016/j.ydbio.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Korzh V, Balasubramaniyan NV, Ekker M, Ge R. Platelet-derived growth factor A (pdgf-a) expression during zebrafish embryonic development. Development Genes and Evolution. 2002;212:298–301. doi: 10.1007/s00427-002-0234-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Vutskits L, Calaora V, Durbec P, Kiss JZ. A role for the polysialic acid-neural cell adhesion molecule in PDGF-induced chemotaxis os oligodendrocyte precursor cells. Journal of Cell Science. 2004;117:93–103. doi: 10.1242/jcs.00827. [DOI] [PubMed] [Google Scholar]
- 31.Schneider RA, Helms JA. The cellular and molecular origins of beak morphology. Science. 2003;299:565–568. doi: 10.1126/science.1077827. [DOI] [PubMed] [Google Scholar]
- 32.Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Developmental Biology. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- 33.Osumi-Yamashita N, Ninomiya Y, Doi H, Eto K. The contribution of both forebrain and midbrain crest cells to the mesenchyme in the frontonasal mass of mouse embryos. Developmental Biology. 1994;164:409–419. doi: 10.1006/dbio.1994.1211. [DOI] [PubMed] [Google Scholar]
- 34.Brugmann SA, et al. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3282–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- 35.Orr-Urtreger A, Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development. 1992;115:1045–1058. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- 36.Ho L, Symes K, Yordan C, Gudas LJ, Mercola M. Localization of PDGF A and PDGFR alpha mRNA in Xenopus embryos suggests signalling from neural ectoderm and pharyngeal endoderm to neural crest cells. Mechanisms of Development. 1994;48:165–174. doi: 10.1016/0925-4773(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 37.Tallquist MD, Weismann KE, Hellström M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 38.Foppiano S, Hu D, Marcucio RS. Signaling by Bone Morphogenetic Proteins directs formation of an ectodermal signaling center that regulates craniofacial development. Developmental Biology. 2007 doi: 10.1016/j.ydbio.2007.09.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westerfield M. The Zebrafish Book; A guide for the laboratory use of zebrafish (Brachydanio rerio) 1993. [Google Scholar]
- 40.Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. The Plant Journal. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 41.Lawson N, Weinstein BM. In vivo imiagining of embryonic vascular development using transgenic zebrafish. Developmental Biology. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 42.Robu ME, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagel M, Tahinci E, Symes K, Winklbauer R. Guidance of mesoderm cell migration in the Xenopus gastrula requires PDGF signaling. Development. 2004;131:2727–2736. doi: 10.1242/dev.01141. [DOI] [PubMed] [Google Scholar]
- 44.Robu ME, et al. p53 activation by knockdown technologies. PLos Genetics. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotechnic and Histochemistry. 2006;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- 46.Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 2004;14:472–477. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1 Crest cell migration in a pdgfra+ embryo.
Supplementary Movie 2 Crest cell migration in a pdgfra−/− embryo.