Abstract
Cytosine-5 methylation within CpG dinucleotides is a potentially important mechanism of epigenetic influence on human traits and disease. In addition to influences of age and gender, genetic control of DNA methylation levels has recently been described. We used whole blood genomic DNA in a twin set (23 MZ twin-pairs and 23 DZ twin-pairs, N = 92) as well as healthy controls (N = 96) to investigate heritability and relationship with age and gender of selected DNA methylation profiles using readily commercially available GoldenGate bead array technology. Despite the inability to detect meaningful methylation differences in the majority of CpG loci due to tissue type and locus selection issues, we found replicable significant associations of DNA methylation with age and gender. We identified associations of genetically heritable single nucleotide polymorphisms with large differences in DNA methylation levels near the polymorphism (cis effects) as well as associations with much smaller differences in DNA methylation levels elsewhere in the human genome (trans effects). Our results demonstrate the feasibility of array-based approaches in studies of DNA methylation and highlight the vast differences between individual loci. The identification of CpG loci of which DNA methylation levels are under genetic control or are related to age or gender will facilitate further studies into the role of DNA methylation and disease.
Introduction
Epigenetic factors have recently been found to play an important role in developmental plasticity [1]–[3]. The molecular mechanisms that underlie the epigenetic modifications of phenotype are increasingly well understood. Coordinated changes in the methylation of cytosine-5 at CpG dinucleotides in the promoter regions of specific genes, changes in chromatin structure through histone acetylation and methylation and post-transcriptional control by microRNAs [4] have been shown to alter gene transcription levels. Specifically, methylation of CpG-rich promoter areas (CpG islands) has been identified as an important mechanism in the dynamic regulation of gene transcription of mammalian genes [5], [6]. DNA methylation is established early in embryogenesis [7] and is the prime mechanism of X-chromosomal inactivation as well as of imprinting effects [8]. Differential DNA methylation can be age-related [9], [10], and is involved in a number of disorders including human cancers [11]. As DNA methylation plays an important role in the mechanism of epigenetic memory [12] it is of primary interest for the study of epigenetic influences on human traits and diseases.
Studies involving monozygotic and dizygotic twins have been used to investigate the influence of DNA methylation on phenotypic variation. A study using monozygotic twins discordant for caudal duplication anomaly revealed significant differences in DNA methylation patterns of the disease associated gene AXIN1 (NC_000016) in peripheral blood [13]. In monozygotic twins discordant for low birth weight, differential DNA methylation was observed in the COMT gene region (NC_000022), suggesting that DNA methylation may provide a mechanism through which birth weight affects the risk for schizophrenia [14]. Several studies have since tried to find differences in DNA methylation in twins discordant for schizophrenia with mixed results [15]–[18]. Analysis of the heritability of DNA methylation in the human IGF2/H19 locus (NC_000011) in twins revealed substantial heritability and cis genetic associations with single nucleotide polymorphisms (SNPs) [19]. The first systematic evidence for familial clustering of DNA methylation presented longitudinal DNA methylation changes as well as genetic influences [20]. Further evidence that DNA methylation patterns are heritable came from a recent twin study using white blood cells and buccal tissue although this study suggested that the molecular mechanisms of heritability may not be restricted to DNA sequence differences [21].
Based on these recent findings a picture is emerging in which DNA methylation levels at specific loci are to some degree under genetic control [19], [21]–[23] and subject to changes over time. Previous studies observed that patterns of epigenetic modification diverge in twins as they become older and suggest that this could be due to small defects in transmitting epigenetic information through successive cell divisions, or maintaining it in differentiated cells [24], [25]. This process, termed “epigenetic drift”, is associated with the aging process. In addition to the reported associations of DNA methylation levels with age and genotype, there is evidence that methylation levels are related to gender. There are several studies that found higher global DNA methylation levels in males [22]. Studies on gender-associated differences in DNA methylation at specific loci have yielded contrasting results [26].
To investigate the heritability of DNA methylation as well as the effects of age and gender, we performed a pilot study of DNA methylation in healthy twin and control subjects using the Illumina GoldenGate Methylation assay. In this assay 1,505 CpG sites from more than 800 genes in the human genome are interrogated simultaneously. The aim of the study was two-fold. First, we were interested to see whether it was feasible to successfully measure DNA methylation levels in blood using commercially available arrays. Second, we determined whether previously reported effects of age, gender and genotype on DNA methylation levels were restricted to specific loci and whether the effects sizes were large enough to be detected.
Results
DNA Methylation analysis
We generated DNA methylation data of MZ twins (N = 43), DZ twins (N = 43) and 96 healthy singletons using peripheral blood DNA samples and the GoldenGate Methylation assay. First, we used BeadStudio software to obtain intensity measures (the mean cy3+cy5 value) for each sample. Using a threshold intensity value of 4,000, we eliminated eight subjects from five pairs (including replicate pairs) in the twin set from further analysis. We next excluded eight probes that had a detection p-value>0.05 in more than 5 % of the hybridizations. The failure rate in the remaining 1,497 probes was <0.01 %. We repeated the above steps for the 96 healthy control subjects included in the second set. In the latter series, 11 of the 1,505 DNA methylation probes and one subject failed quality control and were removed from further analysis.
We performed two-dimensional unsupervised hierarchical clustering of autosomal loci in the filtered data set (Heatmap and histogram shown in Supplementary Figures S1 and S2). These analyses showed a bimodal distribution of DNA methylation [27]. A large proportion of the probes showed no or very little variation in DNA methylation levels and were either fully methylated or unmethylated. Since Illumina arrays can only reliably estimate DNA methylation differences greater or equal to 17 percent only probes with a sufficient range were included for further analysis (See methods). From the 411 probes with normal distribution in both sets as well as in the combined set, 131 failed the range criterion leaving 280 high quality probes in 250 genes for further study of heritability, age and gender effects.
Age-related DNA methylation
We investigated the association between DNA methylation levels of the 280 high-quality probes with age. We observed a strong correlation between the two experiments with an overall Spearman correlation for t-values of 0.77 (95% CI: 0.71–0.81). The combined analyses identified a total of 58 probes that were significantly related to age (21.5 percent). Six loci also showed a significant correlation with age in both samples series independently (Table 1). The genes represented by these probes were activin A receptor type I (ACVR1) (NM_001105.2), interleukin 6 (IL6) (NM_000600.1), caspase recruitment domain-containing protein 15 (CARD15) (NM_022162.1), platelet-derived growth factor receptor alpha (PDGFRA) (NM_006206.3), nuclear factor kappa-B subunit 1 (NFKB1) (NM_003998.2), and ETS-domain protein (ELK) (NM_005229.2). The associations with age of all 251 DNA methylation probes are listed in Supplementary Table S1.
Table 1. DNA methylation probes with significant effects of age and gender (listed are genes for which significance was detected in both series independently).
| Range | Twins | Singletons | ||||||||||||
| Probe | min | max | B.x | Chi2.x | p.x | p.fdr.x | B.y | Chi2.y | p.y | p.fdr.y | Symbol | GenBank | Chr | Locus |
| Age | ||||||||||||||
| ACVR1_E328_R | 0.78 | 0.99 | −0.403 | 11.361 | 7.50E-04 | 3.88E-02 | −0.648 | 55.757 | 8.20E-14 | 2.30E-11 | ACVR1 | NM_001105.2 | 2 | 158402708 |
| IL6_E168_F | 0.00 | 0.32 | 0.476 | 18.525 | 1.68E-05 | 3.86E-03 | 0.443 | 37.288 | 1.02E-09 | 9.51E-08 | IL6 | NM_000600.1 | 7 | 22733513 |
| CARD15_P302_R | 0.17 | 0.61 | −0.390 | 11.309 | 7.71E-04 | 3.88E-02 | −0.440 | 33.265 | 8.04E-09 | 5.63E-07 | CARD15 | NM_022162.1 | 16 | 49288249 |
| PDGFRA_E125_F | 0.25 | 0.98 | 0.471 | 17.580 | 2.75E-05 | 3.86E-03 | 0.412 | 30.935 | 2.67E-08 | 1.29E-06 | PDGFRA | NM_006206.3 | 4 | 54790329 |
| NFKB1_P496_F | 0.08 | 0.50 | −0.422 | 15.125 | 1.01E-04 | 9.39E-03 | −0.361 | 30.863 | 2.77E-08 | 1.29E-06 | NFKB1 | NM_003998.2 | 4 | 103641022 |
| ELK3_P514_F | 0.01 | 0.28 | 0.385 | 11.172 | 8.30E-04 | 3.88E-02 | 0.380 | 28.501 | 9.37E-08 | 3.75E-06 | ELK3 | NM_005230.2 | 12 | 95111824 |
| Gender | ||||||||||||||
| TDGF1_P428_R | 0.22 | 0.64 | 0.791 | 10.777 | 1.03E-03 | 2.88E-02 | 1.050 | 41.315 | 1.30E-10 | 1.81E-08 | TDGF1 | NM_003212.1 | 3 | 46593789 |
| EVI2A_E420_F | 0.27 | 0.97 | 0.816 | 12.815 | 3.44E-04 | 1.60E-02 | 0.796 | 30.494 | 3.35E-08 | 2.34E-06 | EVI2A | NM_001003927.1 | 17 | 26672423 |
| NDN_E131_R | 0.54 | 0.87 | 1.028 | 23.063 | 1.57E-06 | 4.39E-04 | 0.487 | 26.130 | 3.19E-07 | 1.79E-05 | NDN | NM_002487.2 | 15 | 21483412 |
| PLAGL1_E68_R | 0.63 | 0.91 | 0.883 | 17.177 | 3.41E-05 | 4.77E-03 | 0.628 | 24.507 | 7.40E-07 | 3.46E-05 | PLAGL1 | NM_002656.2 | 6 | 144371178 |
| DNMT2_P199_F | 0.75 | 0.96 | 0.858 | 13.209 | 2.79E-04 | 1.56E-02 | 0.686 | 23.858 | 1.04E-06 | 4.15E-05 | DNMT2 | NM_004412.3 | 10 | 17283886 |
| ERN1_P809_R | 0.18 | 0.58 | −0.868 | 13.505 | 2.38E-04 | 1.56E-02 | −0.468 | 22.692 | 1.90E-06 | 6.66E-05 | ERN1 | NM_001433.2 | 17 | 59562017 |
| PTHR1_P258_F | 0.63 | 0.86 | 0.720 | 9.892 | 1.66E-03 | 3.87E-02 | 0.611 | 17.822 | 2.43E-05 | 6.17E-04 | PTHR1 | NM_000316.2 | 3 | 46893982 |
| XPC_P226_R | 0.75 | 0.99 | 0.904 | 14.454 | 1.44E-04 | 1.34E-02 | 0.553 | 20.202 | 6.97E-06 | 1.95E-04 | XPC | NM_004628.3 | 3 | 14195369 |
| PYCARD_P150_F | 0.18 | 0.48 | 0.756 | 11.670 | 6.35E-04 | 2.22E-02 | 0.263 | 10.500 | 1.19E-03 | 1.19E-02 | PYCARD | NM_145182.1 | 16 | 31121902 |
| SGCE_P250_R | 0.33 | 0.97 | 0.803 | 11.878 | 5.68E-04 | 2.22E-02 | 0.279 | 9.005 | 2.69E-03 | 1.98E-02 | SGCE | NM_003919.1 | 7 | 94123664 |
Gender-related DNA methylation
We next investigated the relationship of DNA methylation levels with gender. As expected, non-parametric analysis with Mann-Whitney U test showed that all X-chromosomal probes on the bead array were significantly more methylated in females. However, all but two X-linked probes failed the stringent normality test. For this reason we restricted the analysis of gender effects to the autosomal loci only. A total of 56 of the autosomal probes (20%) showed a significant effect of gender in the combined samples (see Supplementary Table S2 for complete list). The DNA methylation status at ten loci was also significantly associated with gender in the twin and singleton samples independently (Table 1). Although highly significant, the mean difference in DNA methylation level even at the most significant locus (TDGF1_P248_R, NM_003212.1) in the combined group was modest (0.75, sd 0.04 in females compared to 0.79, sd 0.04 in males. t = 4.5734, df = 169.39, p = 9.23e–06). Similar to the age-specific findings, we observed fairly good overall correlation of the gender effects on DNA methylation levels in the two sample sets at 0.69 (95% CI: 0.62–0.74).
Heritability
Third, the twin sample was used to obtain heritability estimates of the observed high-quality DNA methylation profiles. In total, 96 of the 431 CpG sites (23%) yielded a significant heritability (Table 2). The most significant heritable DNA methylation levels included sites in AXL receptor tyrosine kinase isoform (AXL) (NM_001699.3), glutathione S-transferase M1 isoform 2 (GSTM1) (NM_000561.2), T-cell lymphoma invasion and metastasis 1 (TIAM1) (NM_003253.2) and TEK tyrosine kinase (TEK) (NM_000459.2) among others (See supplementary table S3 for full heritability result).
Table 2. Heritability of probes (top 25 only).
| Probe | Her | Chi2 | p-value | Symbol | GenBank | Chr | Locus |
| AXL_P223_R | 0.94 | 54.390 | 1.64E-13 | AXL | NM_021913.2 | 19 | 46416440 |
| GSTM1_P363_F | 0.90 | 36.833 | 1.29E-09 | GSTM1 | NM_146421.1 | 1 | 110031602 |
| TIAM1_P188_R | 0.76 | 23.136 | 1.51E-06 | TIAM1 | NM_003253.1 | 21 | 31853349 |
| CHGA_P243_F | 0.87 | 20.119 | 7.28E-06 | CHGA | NM_001275.2 | 14 | 92459002 |
| TEK_P479_R | 0.72 | 19.197 | 1.18E-05 | TEK | NM_000459.1 | 9 | 27098962 |
| HPN_P823_F | 0.76 | 19.065 | 1.26E-05 | HPN | NM_182983.1 | 19 | 40222427 |
| CREBBP_P712_R | 0.73 | 17.853 | 2.39E-05 | CREBBP | NM_004380.1 | 16 | 3871424 |
| JAK3_P1075_R | 0.67 | 16.326 | 5.33E-05 | JAK3 | NM_000215.2 | 19 | 17820875 |
| NPR2_P618_F | 0.64 | 16.126 | 5.93E-05 | NPR2 | NM_003995.3 | 9 | 35781788 |
| CCL3_E53_R | 0.77 | 15.769 | 7.16E-05 | CCL3 | NM_002983.1 | 17 | 31441547 |
| NPR2_P1093_F | 0.57 | 15.696 | 7.44E-05 | NPR2 | NM_003995.3 | 9 | 35781313 |
| HRASLS_P353_R | 0.66 | 15.472 | 8.37E-05 | HRASLS | NM_020386.2 | 3 | 194441259 |
| MET_E333_F | 0.64 | 15.122 | 1.01E-04 | MET | NM_000245.2 | 7 | 116100028 |
| HOXA5_P1324_F | 0.68 | 15.054 | 1.04E-04 | HOXA5 | NM_019102.2 | 7 | 27151136 |
| TSP50_P137_F | 0.73 | 14.607 | 1.32E-04 | TSP50 | NM_013270.2 | 3 | 46734505 |
| FZD9_E458_F | 0.66 | 14.545 | 1.37E-04 | FZD9 | NM_003508.2 | 7 | 72486503 |
| CRK_P721_F | 0.60 | 14.229 | 1.62E-04 | CRK | NM_005206.3 | 17 | 1307015 |
| HHIP_P578_R | 0.63 | 13.141 | 2.89E-04 | HHIP | NM_022475.1 | 4 | 145786045 |
| IL16_P226_F | 0.59 | 12.836 | 3.40E-04 | IL16 | NM_004513.3 | 15 | 79262029 |
| ACVR1_E328_R | 0.64 | 12.729 | 3.60E-04 | ACVR1 | NM_001105.2 | 2 | 158402708 |
| DNAJC15_E26_R | 0.67 | 12.536 | 3.99E-04 | DNAJC15 | NM_013238.2 | 13 | 42495388 |
| ABCC2_E16_R | 0.58 | 12.367 | 4.37E-04 | ABCC2 | NM_000392.1 | 10 | 101532577 |
| AXIN1_P995_R | 0.61 | 12.179 | 4.83E-04 | AXIN1 | NM_003502.2 | 16 | 343460 |
| DDR1_P332_R | 0.67 | 12.085 | 5.08E-04 | DDR1 | NM_001954.3 | 6 | 30959508 |
| LIG3_P622_R | 0.57 | 11.919 | 5.56E-04 | LIG3 | NM_013975.2 | 17 | 30331029 |
Linkage Disequilibrium mapping
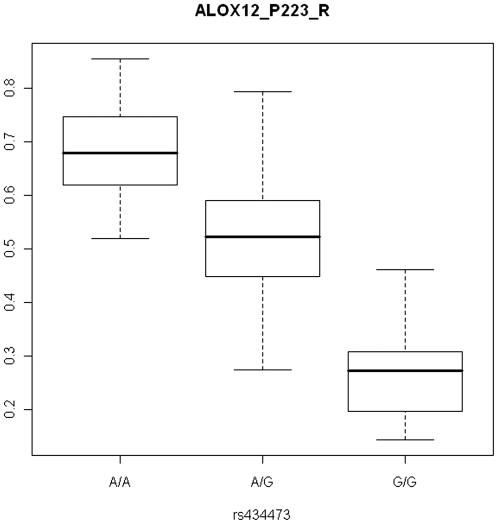
Based on the high heritability estimate of DNA methylation profiles of multiple CpG sites, we performed association mapping using DNA methylation levels as quantitative traits. For 91 subjects of the second set, sample genotype data was available (Illumina 550K BeadChip), with a total of 437,968 SNPs genome-wide after quality control filtering. Genomic inflation factor as a measure of population stratification ranged from 0.96 to 1.05, the mean chi-square statistic ranged between 0.98 and 1.03 indicating an absence of population stratification. We observed significant associations between DNA methylation at 11 different CpG loci representing 35 cis and 5 trans effects after stringent multiple testing correction and confirmation by permutation testing (Supplementary Table S4 shows all 106 methylation probes that showed significant associations after FDR correction for multiple testing, uncorrected for the 512 phenotypes tested). It is noticeable that all significant trans effects are observed with common variants located within genes, and not positioned in regions with known micro RNA's (http://microrna.sanger.ac.uk/). We here present genetic associations of three DNA methylation loci with nine SNPs that constituted a mean difference in DNA methylation level larger or equal than 0.17 to rule out the possibility of technical artifact and survived corrections for multiple testing both for the number of SNPs as well as the 512 phenotypes employed (Table 3). An example plot of DNA methylation differences of ALOX12 (NM_000697.1), the gene locus with the most significant association of DNA methylation with genotype, is shown in Figure 1. The heritability of these CpG loci as estimated in the twin data by means of non-parametric analysis (polychoric correlations) was significant for ALOX12_E85_R (heritability: 0.48, 95% CI: 0.24–0.61) and KRAS_E82_F (heritability: 0.9 95% CI: 0.79–1), but non-significant for ALOX12_P223_R (heritability: 0.03, 95% CI: 0–0.3) or MET_E333_F (heritability: 0.07 95% CI:0–0.42). Associations of DNA methylation level of two X- chromosomal CpG loci (GRPR (Xp22.2, NM_005314.2), and PCTK1 (Xp11.3, NM_033018.2)) with the SNPs rs12743401, rs3881953, rs12734338 rendered non-significant after including gender as a covariate. The association signals are most likely from a homolog area on the Y chromosome as the association of these SNPs with male gender (p<1E-8) confirms.
Table 3. Genetic associations with DNA methylation levels.
| Probe | CHR probe | SNP | CHR | Base | MAF | AA Mean (sd) | AB Mean (sd) | BB Mean (sd) | BETA | SE | R2 | T | p | FDR |
| ALOX12_E85_R | 17 | rs434473 | 17 | A/G | 0.49 | 0.649 (0.133) | 0.493 (0.142) | 0.303 (0.151) | −0.173 | 0.023 | 0.395 | −7.629 | 2.49E-11 | 6.37E-03 |
| ALOX12_E85_R | 17 | rs1126667 | 17 | A/G | 0.50 | 0.315 (0.156) | 0.492 (0.143) | 0.649 (0.133) | −0.167 | 0.023 | 0.380 | −7.383 | 7.86E-11 | 2.01E-02 |
| ALOX12_P223_R | 17 | rs434473 | 17 | A/G | 0.49 | 0.686 (0.093) | 0.523 (0.120) | 0.277 (0.112) | −0.204 | 0.018 | 0.583 | −11.160 | 1.33E-18 | 3.40E-10 |
| ALOX12_P223_R | 17 | rs1126667 | 17 | A/G | 0.50 | 0.291 (0.127) | 0.522 (0.121) | 0.686 (0.093) | −0.198 | 0.019 | 0.560 | −10.650 | 1.50E-17 | 3.82E-09 |
| ALOX12_P223_R | 17 | rs2292350 | 17 | A/G | 0.38 | 0.693 (0.081) | 0.558 (0.112) | 0.372 (0.180) | 0.168 | 0.022 | 0.391 | 7.554 | 3.53E-11 | 9.03E-03 |
| MET_E333_F | 7 | rs38840 | 7 | C/T | 0.20 | 0.835 (0.004) | 0.701 (0.062) | 0.575 (0.084) | 0.127 | 0.015 | 0.452 | 8.567 | 2.95E-13 | 7.54E-05 |
| MET_E333_F | 7 | rs1002399 | 7 | C/T | 0.21 | 0.835 (0.004) | 0.694 (0.067) | 0.575 (0.086) | 0.121 | 0.015 | 0.417 | 7.976 | 4.87E-12 | 1.25E-03 |
| KRAS_E82_F | 12 | rs1566753 | 6/12 | G/T | 0.37 | 0.606 (0.067) | 0.518 (0.065) | 0.433 (0.074) | −0.087 | 0.010 | 0.461 | −8.719 | 1.44E-13 | 3.67E-05 |
| KRAS_E82_F | 12 | rs4516942 | 6/12 | A/G | 0.37 | 0.606 (0.067) | 0.518 (0.065) | 0.433 (0.074) | −0.087 | 0.010 | 0.461 | −8.719 | 1.44E-13 | 3.67E-05 |
Figure 1. Sample plot of DNA methylation levels at probe ALOX12_P223_R by genotype of SNP rs434473.
Sequencing
To investigate whether sequence variation at the probe target regions may affect hybridization and cause spurious association findings, we performed a targeted sequencing effort of 15 gene regions. All subjects of the second set were included for sequencing EVI2A (NM_001003927.1), which showed high heritability and strong associations with DNA methylation. We used a subset of subjects representing the upper and lower quartile of DNA methylation values and homozygosity of the associated alleles for four additional gene regions (ALOX12_P223_R, TDG_E129_F, CTSD_P726_F, TMPRSS4_P552_F). We observed one novel sequence variation within the EVI2A_E420_F probe region in one subject. The remaining six loci did not show any genetic variation. (Detailed results of sequence data are available upon request.)
Discussion
This study surveys inter-individual DNA methylation in relation to genotype, age and gender effects simultaneously. Using genomic DNA extracted from peripheral blood, we analyzed DNA methylation patterns of a large number of CpG sites from more than 800 genes spread throughout the genome in 23 monozygotic and 23 dyzygotic healthy twin-pairs and 96 healthy singletons. We demonstrate that the application of readily available array based technology can provide meaningful results that open the opportunity of incorporating epigenetic information in genetic studies.
Despite the fact that in the majority (86 percent) of CpG sites interrogated by this panel, no substantial DNA methylation variation was present due to tissue type and CpG selection, we did observe replicable, independent, significant effects of age and gender on DNA methylation status for some of the remaining sites. Furthermore, for a number of DNA methylation loci in the twin data, we observed high heritability values for their DNA methylation levels. Genome-wide mapping using DNA methylation as a quantitative trait in 91 healthy singletons with available SNP data yielded highly significant associations. Particularly some of the cis effects constituted large effect sizes that are likely to be biologically relevant and survived strict correction for multiple testing. Other associations, although statistically significant, constituted smaller effect sizes and included trans effects of genetic variation on DNA methylation levels as well. Additional sequence analysis did not identify DNA variation within the targeted CpG probe regions that could have lead to spurious association findings [28]. Our results in the twin data as well as in the singletons demonstrate the role of genetic variation regulating epigenetic signals.
The strongest effects of genetic regulation act locally (cis), within regions that are in linkage disequilibrium with the CpG sites assayed on the array. However, we also observed a number of significant genetic associations with DNA methylation status elsewhere in the genome on other chromosomes (i.e. trans effects) albeit with small effect sizes and with unknown biological relevance. Given the fact that trans effects were already observed in a relatively small sample size of 91 subjects with limited power in a small subset of CpG methylation sites, it is likely that larger studies will identify more trans effects and uncover new genetic regulation of epigenetic profiles. Further non-parametric analysis of association signals may implicate additional genetic control of methylation levels. It is noticeable that all significant trans effects are observed with common variants located within genes instead of gene deserts, suggesting biologically relevant interactions. However caution is warranted as some trans effects were identified as artifacts. For instance the genetic associations in trans from the X-chromsomal CpG sites were due to homologs of the associated SNPs on the Y-chromosome and the SNPs (rs1566753, rs4516942) listed at chromosome 6 but have homologs at chromosome 12 and therefore most likely constitute cis effects rather trans effects on methylation levels of the KRAS locus (NM_033360.2) on chromosome 12. However, another trans effect such as from rs3134269 in the Solute Carrier Family 25, member 32 gene (SLC25A32; NC_000008.10) (involved in folate metabolism) that is associated with DNA methylation levels at TSC2 (tuberous sclerosis 2, NM_001077183.1) constitute a difference of 0.24 percent, well above detection sensitivity (Supplementary Table S5). We are not aware of any studies reporting a direct role of this gene in DNA methylation, but our results suggest that further studies into the molecular mechanisms of these trans effects are warranted.
Recent studies have begun to reveal the complex underlying control mechanisms of DNA methylation but studies of the effect of sequence variant on DNA methylation levels have yielded contrasting results [21], [29], [30]. Our data provides evidence for cis-acting DNA sequences that exert local epigenetic control possibly extending over a cluster of nearby genes and suggest the presence of trans-acting factors not limited to DNA methyltransferases [31]. The SNPs associated with methylation levels in our study were not positioned in genomic regions with microRNAs and we therefore are unable to corroborate recent studies suggesting that microRNAs are associated with DNA methylation levels [32], [33].
In addition to genetic control of DNA methylation levels, we found independent associations of DNA methylation with age and gender. Gender effects for methylation loci on the X chromosome are largely due to the X-inactivation dosage compensation mechanism in females [34]. However, we observed that several of variable autosomal CpG loci on the array are differentially methylated in females as well. We are not the first to report gender specific DNA methylation differences suggesting that autosomal gender dependent DNA methylation is a recurrent phenomenon. Although the absolute differences in methylation level between male and female participants was low, nevertheless they constitute effect sizes similar to previous findings from other groups (Cohen's d = 1) [26]. Our study is also in agreement with previous reports that DNA methylation is age related although this may be restricted to mitotic cells [35] and specific loci [19]. It is noteworthy that about half of the probes that showed significant association to age and gender are non-CpG island probes. The ratio of CpG island in the samples that showed approximate normal distribution and sufficient range (173 of 280) was significant lower than in the original set (1044 of 1505) (Chi-square test: Χ2 = 5.9, df = 1, p = 0.015). These results support recent reports that changes in DNA methylation do not predominantly occur in promoter regions or CpG islands [36].
The DNA methylation loci with high heritability or with significant associations with age or gender identified in our study present some genes that have previously been implicated in disease. For example, the PTHR1 (parathyroid hormone receptor 1, NM_000316.2) gene that was found differentially methylated in females is associated with bone mineral density that shows substantial difference between males and females. Sequence variation of the IL6 gene (NM_000600.1) is related to metabolic syndrome in older males. Finally the AXIN1 locus (NM_003502.2) of which DNA methylation levels in peripheral blood has previously been associated with caudal duplication anomaly showed high heritability in our sample (heritability: 0.61, Χ2 = 12.18, p<0.001). Further studies into the phenotypic consequences of such methylation differences seem warranted.
We realize that these findings are based on a relatively small sample size with limited statistical power. In addition, the Illumina GoldenGate Cancer Panel probes represent only a minute fraction (less than 0.005 percent) of all CpGs in the human genome. On estimate there are 32 million CpG dinucleotides constituting approximately 1 one percent of the genome [37], [38]. The selection of CpG probes predominantly in promoter regions of cancer related genes may not be representative for the vast number of DNA methylation sites throughout the genome and are known to be commonly unmethylated in blood [6], [27]. A recent study of the human B cell methylome suggest that DNA methylation at different regions in the genome (whether in the promoter, intragenic or near the 3′ end) exert different effects on gene transcription [39]. Selection of probes with substantial range and proximal normal distribution as employed in this study focuses on the strongest and biologically most relevant signals. Further non-parametric analysis may tease out additional relevant signals but is beyond the scope of this paper. Technological array-based advances will enable us to study a much larger number of targeted CpGs, while high-throughput, next-generation sequencing studies will provide genome-wide DNA methylation information. Such new developments may also overcome the current limitations in detection sensitivity of the Illumina GoldenGate panel applied in this study. The arrays can detect DNA methylation levels as low as 2.5 percent, but are generally more accurate detecting higher levels of methylation (above 10 percent). Bibikova and colleagues suggested that a beta value difference of ≥0.17 is needed in analyzing GoldenGate methylation data [40]. Whereas the sensitivity to detect differences greater than this difference is excellent, caution is warranted for smaller differences. The mean differences between the genotypes presented in Table 4 are all above this detection threshold as are the ranges in DNA methylation levels that were significantly associated with age or gender. However the mean differences between the male and female groups were smaller than 17 percent and should therefore be interpreted with caution.
Table 4. Sample description.
| Twins | Singletons | All | ||
| Monozygotic | Dizygotic | |||
| N | 46 (23 pairs) | 46 (23 pairs) | 96 | 188 |
| Mean Age (SD; range) | 39.5 (11.1; 20.8–57.8) | 39.6 (10.0; 18.8–53.9) | 34.2 (16.0; 19.6–66.2) | 36.8 (13.3; 18.8–66.2) |
| Male (%) | 20 (43%) | 18 (39%) | 48 (50%) | 86 (46%) |
A remaining challenge will be the study of tissue-specificity of epigenetic control. We do not know what the meaning of differential DNA methylation in blood is for other tissues. Although several studies suggest an important role of DNA methylation in the specialization of tissues [41] other studies suggest that the role of DNA methylation in tissue-specific gene expression is limited to tissue-specific differentially methylated regions (T-DMRs) [42], [43]. Our data are in agreement with those of Kaminsky et al who reported that the intraclass correlation between tissue type was significantly higher compared to intra-individual differences [21]. Nevertheless, further studies that include several tissue types preferably from single subjects are necessary [25]. In absence of information on the exact types of lymphocytes that were included in our blood samples this remains a potential source of bias in this study too.
DNA methylation information can potentially be applied to the study of environmental influences, genetic mapping studies and the development of biomarkers. Further studies may unveil novel mechanisms by which the environment exerts influence on the susceptibility to diseases and may also be important for genetic linkage and association studies. It is possible that DNA methylation of disease-associated alleles may affect the relative risk of carriers to develop the trait, cause phenotypic heterogeneity or may explain some of the phenotypic differences in monozygous twins [44]. For this reason, incorporating DNA methylation data with association and linkage analyses may provide new insights for correlating genetic markers and disease, and thus increase the power of linkage and association studies. This pilot provides a first glimpse of the possibilities and hurdles of large-scale DNA methylation profiling.
Methods
Ethics statement
Written informed consent from all participants was obtained. This study was approved by the Medical Ethical Committee of the UMC Utrecht, The Netherlands.
Subjects
The twin sample included 23 healthy monochorionic or dichorionic monozygotic twin pairs (46 subjects) and 23 healthy dizygotic twin pairs (an additional 46 subjects) recruited via advertisements in local newspapers and media. Inclusion criteria included good general health and at least three Dutch grandparents. Zygosity of twin pairs was investigated previously by the Illumina DNA Test Panel consisting of 360 validated single nucleotide polymorphisms (SNPs). General Health was established using a general medical checklist; all subjects were free of medication. An independent second sample set consisted of 96 healthy controls, matched for age and gender to the original twin series. For this second sample set, genome-wide SNP data (Illumina 550K Infinium array) was available. The second sample was used to validate the age and gender effects observed in the twin sample and to perform genetic mapping using DNA methylation as quantitative traits. The mean age and gender information for both sample sets are listed in Table 4.
Laboratory analysis
Genomic DNA extracted from peripheral blood was used for this study. Experiments of the twin data and singletons were performed independently. Genomic DNA samples (0.5 µg) from both sets were bisulfite converted using the EZ-96 DNA methylation kit (ZYMO Research, Orange, CA, USA) according to the manufacturer's protocol in two batches, first the twins and secondly the singletons. The twins and co-twins were randomly distributed across plates. DNA methylation profiles were obtained using the commercially available Illumina GoldenGate DNA methylation platform (DNA Methylation Cancer Panel I) as recommended by the manufacturer . With this assay, the DNA methylation levels of up to 1,505 CpG sites covering 807 gene regions are interrogated concurrently. The probe IDs are listed in Supplemental Table S5 and online by the manufacturer (www.illumina.com/pages.ilmnID193).
Statistical Analysis
We used Illumina BeadStudio software for normalization and initial quality control. BeadStudio uses a background normalization that subtracts a background value derived by averaging the signals of built-in negative control bead types. The DNA Methylation status for each probe is presented as a Beta value, the ratio of signal from the methylated probe relative to the sum of both methylated and unmethylated probes. Beta values can range from 0 (unmethylated) to 1 (fully methylated). We tested for approximate normal distribution by means of a Lilliefors test for normality at the 0.1 level after FDR correction for multiple testing. Non-normal distributed probes were excluded from subsequent analysis. To ensure biological relevance of the methylation differences, we also excluded those probes that had a range smaller than 0.17 in the methylation levels. For further analysis, we used Z-scores of DNA methylation levels and age in a variance component analysis. Analyses of age and gender effects were conducted for the twins and singletons, both separately and jointly. The effects of age and gender on the level of methylation were tested in Mx [45]. The covariance of the methylation levels in MZ and DZ twin-pairs was estimated to take into account the statistical dependency of the data. Two parameters were used to model the variances and covariances in MZ and DZ twins, representing an additive genetic factor (A), and a unique environmental factor (E). Variances in both MZ and DZ twins are explained by additive genetic variation and unique environmental variation, and are therefore estimated as A+E. The covariance in MZ twins is expected to equal A, as MZ twins share their genetic material. The covariance in DZ twins is expected to equal 0.5*A, as DZ twins share 50% of their genetic material. The heritability was estimated as the additive genetic variance (A) divided by the total amount of variance (A+E). Heritability Chi-square statistic and probability were calculated for all the probes that showed approximate normal distribution and sufficient range in the twins set. We used Mann-Whitney non-parametric testing to test for differences in DNA methylation of X-chromosomal probes between males and females, chi-square test for testing the proportion of CpG islands before and after filtering. Heritability and standard error non-normal distributed methylation levels were estimated based on an ACE model using Falconer's formulae [46]. Under the ACE model the correlation between the DZ twins is expected to be equal or higher than half of the correlation between the MZ twins.
Normalization steps were conducted in BeadStudio, variance component analysis was conducted using Mx [42] all other analyses were done using the R package for statistical computing [47].
Association analysis
We generated SNP genotype data for 91 samples in the singletons using the Illumina HumanHapmapv1.1 550K platform at UCLA. Associations with the 512 DNA methylation levels that were normally distributed in this sample as quantitative phenotypes, were calculated using PLINK [48]. Provided the small sample size, we included only SNPs with minor allele frequency (MAF) at ≥0.1 and a genotyping success rate of >90%. Markers deviating from expected Hardy-Weinberg equilibrium at p<0.001 using an exact test [49] were also excluded. Population stratification was assessed using genomic inflation factor (based on median chi-squared) and the mean chi-square statistic. We applied FDR correction at the 0.05 level [50] for multiple testing by the number of SNPs and the number of phenotypes tested. Adaptive permutation testing (n = 107) was performed for the associated SNPs to confirm our findings. These analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org). Association signals that were obtained from X-chromosomal loci were re-calculated using gender as covariate in a linear regression model.
Sequencing
To investigate whether DNA sequence variation at probe regions affected the hybridization signals, we examined several candidate loci by standard sequencing. Selection of CpG sites for sequencing was based on the presence of highly significant association findings. All subjects from the second set were sequenced for PCR amplicons for the EVI2A_E420 locus; eight selected subjects were used to screen for sequence variation in the probe regions of ALOX12_P223_R, TDG_E129_F, CTSD_P726_F, TMPRSS4_P552_F. The latter subjects were selected by two criteria: DNA methylation levels in the upper or lower quintile, and homozygosity for the associated SNPs. The probes were selected from a range of strength of association signals based on the availability of suitable primers. Primer sequences are available upon request.
Supporting Information
Full age results
(0.12 MB XLS)
Full gender results
(0.13 MB DOC)
Full heritability result
(0.10 MB DOC)
Full mapping results
(0.06 MB XLS)
Probe IDs
(1.09 MB XLS)
Heatmap of twin sample
(1.28 MB TIF)
Heatmap of singleton sample
(1.28 MB TIF)
Acknowledgments
The authors wish to thank the subjects for their participation in this study. We further thank Dr. Peter W. Laird for his support. Drs. Chiara Sabatti, Ye (Jimmie) Chun, Eleazar Eskin and Caroline van Baal are thanked for helpful discussion on the statistical approaches, Juliaan Meijboom for his help with the analysis and Drs. Ronald Vonk and Astrid van der Schot for collection of the twin samples. Some of the analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: RAO gratefully acknowledges support of NIMH grant RO1 MG078075. MB is supported by a NARSAD young investigator grant and RAO is the 2008 NARSAD Adele Selengut Investigator. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc Natl Acad Sci U S A. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Metivier R, Gallais R, Tiffoche C, Le PC, Jurkowska RZ, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 6.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 7.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4 Spec No:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 8.Shemer R, Birger Y, Dean WL, Reik W, Riggs AD, et al. Dynamic methylation adjustment and counting as part of imprinting mechanisms. Proc Natl Acad Sci U S A. 1996;93:6371–6376. doi: 10.1073/pnas.93.13.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopisetty G, Ramachandran K, Singal R. DNA methylation and apoptosis. Mol Immunol. 2006;43:1729–1740. doi: 10.1016/j.molimm.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, et al. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- 11.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 12.Feng YQ, Desprat R, Fu H, Olivier E, Lin CM, et al. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2006;2:e65. doi: 10.1371/journal.pgen.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oates NA, van Vliet J, Duffy DL, Kroes HY, Martin NG, et al. Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet. 2006;79:155–162. doi: 10.1086/505031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mill J, Dempster E, Caspi A, Williams B, Moffitt T, et al. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet. 2006;141:421–425. doi: 10.1002/ajmg.b.30316. [DOI] [PubMed] [Google Scholar]
- 15.Zhang AP, Yu J, Liu JX, Zhang HY, Du YY, et al. The DNA methylation profile within the 5′-regulatory region of DRD2 in discordant sib pairs with schizophrenia. Schizophr Res. 2007;90:97–103. doi: 10.1016/j.schres.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Murphy BC, O'Reilly RL, Singh SM. DNA methylation and mRNA expression of SYN III, a candidate gene for schizophrenia. BMC Med Genet. 2008;9:115. doi: 10.1186/1471-2350-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor CM, Akbarian S. DNA methylation changes in schizophrenia and bipolar disorder. Epigenetics. 2008;3:55–58. doi: 10.4161/epi.3.2.5938. [DOI] [PubMed] [Google Scholar]
- 18.Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, et al. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29:169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- 19.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 20.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 22.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 23.Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci U S A. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin GM. Epigenetic drift in aging identical twins. Proc Natl Acad Sci U S A. 2005;102:10413–10414. doi: 10.1073/pnas.0504743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Maarri O, Becker T, Junen J, Manzoor SS, az-Lacava A, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 27.Rakyan VK, Hildmann T, Novik KL, Lewin J, Tost J, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberts R, Terpstra P, Li Y, Breitling R, Nap JP, et al. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murrell A, Heeson S, Cooper WN, Douglas E, Apostolidou S, et al. An association between variants in the IGF2 gene and Beckwith-Wiedemann syndrome: interaction between genotype and epigenotype. Hum Mol Genet. 2004;13:247–255. doi: 10.1093/hmg/ddh013. [DOI] [PubMed] [Google Scholar]
- 31.Birger Y, Shemer R, Perk J, Razin A. The imprinting box of the mouse Igf2r gene. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- 32.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 33.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, et al. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu MW, Siegmund KD, Eckstam CL, Kim JY, Yang AS, et al. Lack of increases in methylation at three CpG-rich genomic loci in non-mitotic adult tissues during aging. BMC Med Genet. 2007;8:50. doi: 10.1186/1471-2350-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagase H, Ghosh S. Epigenetics: differential DNA methylation in mammalian somatic tissues. FEBS J. 2008;275:1617–1623. doi: 10.1111/j.1742-4658.2008.06330.x. [DOI] [PubMed] [Google Scholar]
- 42.Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, et al. Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics. 2007;89:326–337. doi: 10.1016/j.ygeno.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haque FN, Gottesman II, Wong AH. Not really identical: epigenetic differences in monozygotic twins and implications for twin studies in psychiatry. Am J Med Genet C Semin Med Genet. 2009;151C:136–141. doi: 10.1002/ajmg.c.30206. [DOI] [PubMed] [Google Scholar]
- 45.Neale M, Boker SM, Xie G, Maes HH. Richmond, VA: Department of Psychiatry; 2003. Mx: Statistical Modeling. [Google Scholar]
- 46.Falconer DS, Mackay TFC. Harlow, Essex, UK: Longmans Green; 1996. Introduction to Quantitative Genetics. [Google Scholar]
- 47.R Development Core Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing. 2005.
- 48.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. PLINK: A Tool Set for Whole Genome Association and Population Based Linkage Analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;171:783–790. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full age results
(0.12 MB XLS)
Full gender results
(0.13 MB DOC)
Full heritability result
(0.10 MB DOC)
Full mapping results
(0.06 MB XLS)
Probe IDs
(1.09 MB XLS)
Heatmap of twin sample
(1.28 MB TIF)
Heatmap of singleton sample
(1.28 MB TIF)