Abstract
A longitudinal study was conducted to investigate the nature of Escherichia coli O157:H7 colonization of feedlot cattle over the final 100 to 110 days of finishing. Rectal fecal grab samples were collected from an initial sample population of 788 steers every 20 to 22 days and microbiologically analyzed to detect E. coli O157:H7. The identities of presumptive colonies were confirmed using a multiplex PCR assay that screened for gene fragments unique to E. coli O157:H7 (rfbE and fliCh7) and other key virulence genes (eae, stx1, and stx2). Animals were classified as having persistent shedding (PS), transient shedding (TS), or nonshedding (NS) status if they consecutively shed the same E. coli O157:H7 genotype (based on the multiplex PCR profile), exhibited variable E. coli O157 shedding, or never shed morphologically typical E. coli O157, respectively. Overall, 1.0% and 1.4% of steers were classified as PS and NS animals, respectively. Characterization of 132 E. coli O157:H7 isolates from PS and TS animals by pulsed-field gel electrophoresis (PFGE) typing yielded 32 unique PFGE types. One predominant PFGE type accounted for 53% of all isolates characterized and persisted in cattle throughout the study. Isolates belonging to this predominant and persistent PFGE type demonstrated an enhanced (P < 0.0001) ability to adhere to Caco-2 human intestinal epithelial cells compared to isolates belonging to less common PFGE types but exhibited equal virulence expression. Interestingly, the attachment efficacy decreased as the genetic divergence from the predominant and persistent subtype increased. Our data support the hypothesis that certain E. coli O157:H7 strains persist in feedlot cattle, which may be partially explained by an enhanced ability to colonize the intestinal epithelium.
Escherichia coli serotype O157:H7 was first linked to human illness in the early 1980s, when it was determined to cause severe abdominal pain with initially watery diarrhea that progressed to grossly bloody diarrhea accompanied by little or no fever (42). Initially, E. coli O157:H7 can cause nonbloody diarrhea through attachment to, and subsequent destruction of, intestinal microvilli (24). In addition to microvillus damage, serious health complications can arise due to the ability of E. coli O157:H7 to produce Shiga toxins (Stx1 and Stx2). Shiga toxins are very potent cytotoxins that are absorbed into the intestinal microvasculature and initiate apoptosis of vascular epithelium, resulting in hemorrhagic colitis (41). Persistent uptake of these toxins may lead to more severe manifestations of disease, such as hemolytic-uremic syndrome, which may ultimately result in kidney failure (24). Most recent estimates have identified E. coli O157:H7 as the cause of at least 70,000 cases of food-borne illness annually in the United States, and in 4% of cases life-threatening hemolytic-uremic syndrome develops (37). Epidemiological studies have implicated the consumption of meat, dairy products, produce, and water contaminated by animal feces, as well as person-to-person contact and direct contact with farm animals or their environment, as routes of E. coli O157:H7 transmission leading to human illness (36).
It is generally accepted that cattle and other animals are the major reservoir of E. coli O157:H7, but it is still not clear if animals are colonized for prolonged periods with E. coli O157:H7 or if they transiently shed this organism following repeated exposure to it through ingestion of contaminated feedstuffs or water or through exposure to other contaminated environmental sources. Based on results of numerous epidemiological studies (4, 6, 21, 30, 32), the prevalence of E. coli O157:H7 in feedlot cattle is highly variable and can range from less than 1% to 80%. Several other studies (7, 8, 23) have found evidence of persistent E. coli O157:H7 colonization in individual cattle, supporting the hypothesis that at least some animals are susceptible to persistent E. coli O157:H7 colonization. Multiple experimental inoculation studies (15, 23, 39, 46) showed that E. coli O157:H7 persists in the bovine gastrointestinal (GI) tract for at least 14 days up to 140 days postinfection. Studies have implicated the lower GI tract and specifically the recto-anal junction (RAJ) as the major location of E. coli O157:H7 colonization and proliferation (9, 12, 23, 39); however, this organism also can be found throughout the bovine GI tract (7, 8, 31, 40, 54).
It stands to reason that if the E. coli O157:H7 prevalence in cattle presented for harvest were reduced, there would be a decrease in the probability of beef product contamination, if good manufacturing procedures were used. Although there is consensus concerning the importance of preharvest pathogen mitigation and its role in minimizing entry of E. coli O157:H7 into harvest facilities, there is disagreement about the significance of “supershedders” (animals that excrete large quantities of a pathogen for various amounts of time) for E. coli O157:H7 transmission dynamics at the preharvest level (12, 34, 35, 39). Utilizing statistical modeling, researchers have estimated that, on average, the prevalence of “supershedders” in a population is 4% and that these animals excrete 50 times more E. coli O157:H7 than other animals colonized by this organism (34). Additionally, the same researchers suggested that approximately 80% of E. coli O157:H7 transmission is generated by a few “supershedders” (35).
Research by our group discovered a unique association between E. coli O157:H7 prevalence in pen floor fecal pats and carcass contamination by this pathogen (57). When the prevalence in fecal pats from a pen floor exceeded 20%, carcasses of animals from the pen had E. coli O157:H7 prevalence values of 14.3, 2.9, and 0.7% before evisceration, after evisceration, and after final intervention, respectively. However, when the prevalence in pen floor fecal pats was less than 20%, the preeviscerated carcass prevalence value was 6.3%, and there was no detectable E. coli O157:H7 contamination of carcass samples after evisceration and after final intervention (57). Thus, we hypothesize that animals which persistently excrete normal levels of E. coli O157:H7 over prolonged periods (persistent shedders [PS]) rather than animals that periodically shed abnormally high levels (supershedders) are the most significant source of E. coli O157:H7 contamination in the food continuum. Although previous studies suggested that cattle may be persistently colonized by E. coli O157:H7 and shed this organism in their feces for prolonged periods, molecular subtyping data are required to further investigate whether cattle are persistently colonized by the same strain (i.e., molecular subtype) or if they are repeatedly exposed to different strains through contaminated feedstuffs, water, or other environmental sources. Thus, the objectives of this study were to determine if naturally colonized feedlot cattle persistently shed E. coli O157:H7, using combined cultural microbiological analyses, molecular subtyping approaches, and in vitro virulence phenotype assays to probe the factors (agent, host, environment, or a combination of these factors) that contribute to the complex ecology of E. coli O157:H7 persistence at the preharvest level.
MATERIALS AND METHODS
Study design.
Holstein steers from different calf farms in Wisconsin (n = 788) consuming a high-concentrate finishing ration at a commercial feedlot in eastern Kansas (all research protocols were reviewed and approved by the Colorado State University Animal Care and Use Committee [approval 05-233A-01]) that had never been exposed to directly fed antimicrobials (e.g., Bovamine) were enrolled in the study. The steers were housed in five pens; three of the pens shared fence lines, and the two remaining pens were independently located in other areas of the feedlot. All animals were fed the same finishing diet during the sampling period. Animals were permanently removed from the study population following treatment for any clinical illness.
Rectal fecal grab samples were collected every 20 to 22 days during the final 100 to 110 days (June to October) of the finishing period. Every animal was sampled during the first two collection periods to establish its E. coli O157:H7 shedding status along with the prevalence of shedding in the population of feedlot cattle studied (Table 1). Animals whose E. coli O157:H7 shedding status varied during the first two sample collections were excluded from further sample collection as the focus of this study was to further investigate the PS status. The four remaining sample collections focused on identification of animals that were either consecutively E. coli O157:H7 positive or consecutively E. coli O157 negative as determined using the results for each previous sample collection. Additionally, a random subset of animals whose E. coli O157:H7 shedding status varied was included during each of the final four sample collections to determine if these animals reverted back to their original shedding patterns.
TABLE 1.
Distribution of E. coli O157 virulence genes among animals during the first two antemortem collection periods
| Genotypea | Collection 1b
|
Collection 2b
|
||
|---|---|---|---|---|
| No. of animals shedding | % of animals | No. of animals shedding | % of animals | |
| rfb fliCh7eae stx1stx2 | 314 | 39.8 | 265 | 33.6 |
| rfb stx1 | 32 | 4.1 | 156 | 19.8 |
| rfb stx1stx2 | 3 | 0.4 | 0 | 0.0 |
| rfb eae stx1stx2 | 9 | 1.1 | 42 | 5.3 |
| rfb fliCh7stx1stx2 | 1 | 0.1 | 0 | 0.0 |
| Total | 359 | 45.5 | 463 | 58.7 |
rfb encodes the O157 antigen, fliCh7 encodes the H7 antigen, eae encodes intimin, stx1 encodes Shiga toxin 1, and stx2 encodes Shiga toxin 2.
All 788 animals were sampled during collections 1 and 2.
At every predetermined collection period, steers were processed through conventional processing facilities, where rectal fecal grab samples were collected from each targeted animal. Feces were transferred to a sterile Whirl-Pak bag (Nasco, Modesto, CA) and subsequently placed in a cooler with ice packs. Following sampling, the fecal samples were transported to the Pathogen Reduction Laboratory of the Center for Meat Safety & Quality at Colorado State University (Fort Collins, CO), where they were stored at 4°C until a microbiological analysis was performed (within 48 h after collection).
Analysis of fecal E. coli O157:H7.
Fecal samples (10 g) were enriched using procedures outlined by Barkocy-Gallagher et al. (5). Following incubation, fecal slurries were stored at 4°C until they were subjected to immunomagnetic bead separation. Immunomagnetic bead separation was performed as described by Barkocy-Gallagher et al. (3), and ultimately 50 μl of each sample was plated onto Rainbow agar (Biolog Inc., Hayward, CA) supplemented with 10 mg/liter of novobiocin (Sigma-Aldrich, St. Louis, MO) and 0.8 mg/liter of potassium tellurite (Sigma), as well as sorbitol MacConkey agar (Becton, Dickinson and Company, Sparks, MD) supplemented with 20 mg/liter of novobiocin and 2.5 mg/liter of potassium tellurite (mSMAC). Rainbow plates were incubated for 24 ± 2 h at 37°C, and mSMAC plates were incubated for 36 ± 2 h at 37°C. After incubation, up to three colonies displaying E. coli O157:H7 morphology were selected for each medium and initially screened for the O157 antigen using the RIM E. coli O157:H7 latex agglutination test (Remel, Lenexa, KS). All agglutination-positive colonies were cultured in 5 ml of tryptic soy broth for 24 ± 2 h at 37°C, streaked onto mSMAC, and incubated for 36 ± 2 h at 37°C to determine purity.
Presumptive colonies were confirmed to be E. coli O157:H7 colonies by a multiplex PCR assay performed using a 96-well plate format and 25-μl reaction mixtures that included a primer master mixture containing forward and reverse primers at concentrations specified previously (25) to amplify rfbE (which encodes the O157 antigen), fliCh7 (which encodes the H7 antigen), eae (which encodes intimin), stx1 (which encodes Shiga toxin 1), and stx2 (which encodes Shiga toxin 2), GoTaq Green master mixture (Promega, Madison, WI), nuclease-free water, and a DNA template. The PCR cycling conditions outlined by Hu et al. (25) were used. PCR products were separated by electrophoresis in 2% agarose gels, stained with ethidium bromide, and visualized with UV illumination. Multiplex PCR profiles were assigned based on the presence of each targeted gene. Isolates in which amplicons corresponding to rfbE, fliCh7, and at least one stx gene were detected were designated E. coli O157:H7 isolates. For the purposes of this study, we chose to follow up with animals that consecutively shed the most common E. coli O157:H7 genotype (indicated by the presence of all five genes, as determined by multiplex PCR [Table 1]). Up to three E. coli O157:H7 isolates from each positive fecal sample were stored in 15% glycerol at −80°C for subsequent further characterization.
Enumeration of E. coli O157:H7 PS animals.
A five-tube most-probable-number MPN assay (19) was used to enumerate E. coli O157:H7 in fecal samples collected from PS animals just before slaughter to determine whether these animals excreted elevated quantities of E. coli O157:H7 in their feces. A 10-g aliquot of each fecal sample was combined with 90 ml of Butterfield's phosphate buffer (BPB) (Becton) and pummeled in a stomacher for 2 min. Three 1:10 serial dilutions were prepared from each BPB sample. From each of a sample's three serial BPB dilutions, 1 ml was removed and added to five different tubes of lauryl tryptose broth (LTB) (Becton), resulting in a total of 15 tubes for each sample. The inoculated LTB tubes were incubated for 24 h at 37°C. After incubation, the contents of all turbid LTB tubes were streaked on mSMAC and appropriately incubated, and the identities of morphologically typical colonies were confirmed by performing multiplex PCR as previously described.
GI tissue and content samples.
All animals identified as PS (n = 8) and nonshedders (NS) (n = 11), as well as a subsample of animals identified as transient shedders (TS) (n = 18), were harvested at a commercial facility in the upper Midwest. The entire GI tract (esophagus, reticulum, rumen, omasum, abomasum, gall bladder, small intestine, large intestine, colon, and bung) was collected from each animal and transported to a vacant area of the facility to allow sample collection. Additionally, the liver of each animal was examined for the presence of abscesses. Samples of tissue and contents were collected aseptically from the reticulum, rumen, omasum, abomasum, duodenum (proximal to the anterior side of the first loop), ileocecal valve, distal colon (∼60 cm proximal to the anus), RAJ, and two mesenteric lymph nodes (at a position ∼30 cm proximal to the anterior root of the mesentery and the ileal-cecal colic node) (only tissue samples were collected from lymph nodes) for microbiological analysis. Tissue samples were washed with sterile phosphate-buffered saline (PBS) with 0.05% Tween 20 (Sigma) to remove visible organic matter before they were placed into a Whirl-Pak bag (Nasco). All microbial samples were placed in ice pack-filled coolers and shipped to the Pathogen Reduction Laboratory of the Center for Meat Safety & Quality at Colorado State University (Fort Collins, CO).
GI tissue and content samples were analyzed microbiologically as described above. Ten-gram aliquots of the epithelial layer of each tissue were aseptically removed and placed in a Whirl-Pak bag containing 90 ml of phosphate-buffered tryptic soy broth (5). Lymph nodes were first aseptically trimmed to remove excess adipose tissue, and a 10-g aliquot of each tissue sample was placed in a Whirl-Pak bag containing 90 ml of phosphate-buffered tryptic soy broth. All tissue samples were pummeled (IUL Instruments, Barcelona, Spain) for 2 min and processed as described above. Additionally, pH measurements were obtained by preparing an additional 1:10 dilution of each GI content sample with distilled water, pummeling (IUL Instruments) the preparation for 2 min, and submerging a glass pH electrode (Denver Instruments, Arvada, CO) into the preparation.
GI tissue samples from selected PS (n = 3) and NS (n = 3) animals were fixed in 4% paraformaldehyde and shipped to the Colorado State University Pathology Diagnostic Laboratory, where they were embedded in paraffin using an automated tissue processor, 5-μm sections were cut with a microtome, and the sections were stained with hematoxylin and eosin. Histopathological evaluation and characterization were performed by a trained pathologist who was blinded with respect to sample identification information.
PFGE.
Pulsed-field gel electrophoresis (PFGE) typing of E. coli O157:H7 isolates was performed using the standardized Centers for Disease Control and Prevention PulseNet protocol (10). At least one isolate from all six sample collections for each PS animal and representative isolates from TS animals were selected for characterization by PFGE. All isolates were previously confirmed to be E. coli O157:H7 isolates using the eae, stx1, and stx2 genes before PFGE analysis. Briefly, isolates were grown on tryptic soy agar (Becton) plates and incubated at 37°C for 18 h. Bacterial cultures were imbedded in 1% agarose (SeaKem Gold Agarose; Cambrex BioScience Rockland, Inc., Rockland, ME), lysed, washed, and digested with XbaI overnight at 37°C. Restricted agarose plugs were then placed into 1% agarose gels and electrophoresed with a CHEF Mapper XA (Bio-Rad Laboratories, Hercules, CA) for 21 h with switch times of 2.16 s to 54.17 s. XbaI-digested Salmonella enterica serovar Braenderup (H9812) DNA was used as a reference size standard (26). Agarose gels were stained with ethidium bromide, and the resulting images were captured with a FOTO/Analyst Investigator system (FOTODYNE, Inc., Hartland, WI). PFGE patterns then were analyzed and compared using the Applied Maths Bionumerics software package (v3.5; Applied Maths, Saint-Matins-Latem, Belgium). Similarity clustering analyses were performed with Bionumerics software using the unweighted-pair group matching algorithm and the Dice correlation coefficient (26).
Cell attachment assay.
The attachment efficiencies of E. coli O157:H7 isolates representing the predominant PFGE subtype were compared to those of the E. coli O157:H7 isolates representing the more genetically diverse PFGE subtypes by performing attachment assays using the Caco-2 human intestinal epithelial cell line. Up to 10 antemortem fecal isolates representing each of the following categories were selected for characterization by this Caco-2 attachment assay: (i) the dominant PFGE subtype, (ii) subtypes closely related to the dominant subtype, (iii) subtypes possibly related to the dominant subtype, and (iv) subtypes distantly related to the dominant subtype (based on the criteria defined previously [53]). Caco-2 cells were seeded into 24-well flat-bottom plates (Corning Inc., Corning, NY) at a density of 1 × 105 cells/well in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) containing 20% heat-inactivated fetal bovine serum (Gibco) without antibiotics and grown to confluence (approximately 72 h). E. coli O157:H7 overnight cultures were prepared by inoculating a colony isolated in a single well into a 10-ml tube containing brain heart infusion (BHI) (Becton) broth and incubating the tube at 37°C for 12 to 18 h without shaking. Overnight E. coli O157:H7 cultures (1 ml) were pelleted by centrifugation (11,337 × g, 5 min) and reconstituted in 1 ml of PBS. Confluent Caco-2 monolayers were infected with approximately 2 × 107 E. coli O157:H7 cells/well. After infection for 3 h at 37°C, nonadherent bacteria were removed by washing preparations three times with PBS. Caco-2 cells were lysed by adding 0.5 ml of ice-cold sterile ultrapure water and vigorous pipetting, followed by vortexing of the cell suspension. Adherent E. coli O157:H7 cells, along with the cells in overnight E. coli O157:H7 cultures, were enumerated by spread plating appropriate serial dilutions onto BHI medium plates, in duplicate. The BHI medium plates were incubated at 37°C for 24 h, and the resultant CFU were enumerated. The attachment efficiency of each E. coli O157:H7 isolate was expressed as a percentage based on the initial inoculum that was recovered as adherent E. coli O157:H7 cells. The attachment efficiency of each isolate was measured in duplicate wells in at least three independent experiments.
Statistical analysis.
The GI content pH data were analyzed with PROC MIXED of SAS (version 9.3; SAS Institute, Cary, NC). Analysis of variance techniques were employed to determine if there were differences (P < 0.05) among main effects, including GI tissue location and shedding status, as well as all appropriate interactions.
The proportion of the study population expected to be classified as PS or NS animals by chance alone was calculated by multiplying the prevalence estimates of animals shedding and not shedding E. coli O157:H7 with the five-gene multiplex PCR profile at each of the six time points. These joint probabilities represented the expected frequency in each category, which was compared to the observed number in each category by using chi-square goodness-of-fit tests. Within-table independence (i.e., independence of the expected and observed outcomes) was determined using a P value of <0.05.
Chi-square analysis was utilized to detect differences in subtype frequency between shedding status groups using the PROC FREQ procedure of SAS. Initially, data were collapsed to form three subtype categories: the dominant subtype, subtypes that differed from the dominant subtype by 1 to 3 bands, and subtypes that differed from the dominant subtype by 4 to >7 bands before analysis. The estimated probability of shedding a given PFGE subtype during antemortem collection was analyzed using a repeated-measures generalized estimating equations marginal logistic model with PROC GLIMMIX of SAS with the empirical difference in standard errors. Differences between predicted probabilities were considered significant at P values of <0.05.
The difference between the log-transformed value for cells inoculated into a well and the log-transformed counts for adherent cells recovered in the well was used as the dependent variable to compare Caco-2 attachment data. Analysis of variance techniques were employed to determine if there was a difference between the attachment abilities of different PFGE subtype categories, as previously described. Differences in attachment efficiency were analyzed using PROC MIXED of SAS with least-squares means generated for each PFGE subtype category. Ultimately, least-squares means were separated using pairwise t tests incorporating a Tukey adjustment, and significant inferences were noted when differences between means were detected at the P < 0.05 level.
RESULTS
E. coli O157:H7 carriage in feedlot steers.
All animals enrolled in this study were sampled during the first two sample collections, which allowed us to determine the overall prevalence of E. coli O157:H7 in the study population. Presumptive E. coli O157:H7 isolates were characterized by using a five-gene multiplex PCR that detects gene fragments unique to serotype O157:H7 along with genes encoding three key virulence determinants (eae, stx1, and stx2). Overall, 45.5 and 58.7% of the study population shed E. coli isolates belonging to serotype O157 and carrying at least one stx gene during collections 1 and 2, respectively (Table 1). Over the first two collection periods, animals shed E. coli O157 isolates with five virulence genotypes as determined by their multiplex PCR profiles (Table 1). During the first two collection periods, E. coli O157:H7 isolates possessing the eae, stx1, and stx2 genes (five-gene multiplex PCR profile) were shed most frequently, as 39.8% and 33.6% of animals shed isolates with this genotype during the first and second collections, respectively (Table 1).
Since the purpose of this study was to determine if cattle become persistently colonized by E. coli O157:H7, which would imply that the same E. coli O157:H7 strain is able to persist in the GI tract of a given animal over time, our strategy for the remaining four sample collections was to target animals that consistently shed the predominant genotype (isolates with the five-gene multiplex PCR profile). As a result, the prevalence of less common genotypes could not be determined after the first two sample collections, and we focused on monitoring the persistence of the dominant genotype throughout the study (Table 2). Specifically, PS animals were defined as animals that shed an E. coli O157:H7 isolate carrying eae, stx1, and stx2 (five-gene-positive multiplex genotype) over the six collection periods. Animals that intermittently shed an E. coli O157:H7 isolate with a genotype that included all three virulence factors were classified as TS animals, while animals that never shed morphologically typical E. coli O157 (i.e., colonies that were not able to rapidly ferment sorbitol) during the 110-day sample collection period were classified as NS animals. Overall, based on these criteria, 8 of 788 animals (1.0%) were classified as PS animals, while 11 of 788 animals (1.4%) never shed a detectable amount of morphologically typical E. coli O157 and were thus classified as NS animals. Chi-square goodness-of-fit tests revealed the independence of the observed and expected frequency of PS status (P < 0.001), while the number of animals classified as having NS status was similar to that expected by chance alone (P = 0.23). The remaining 769 animals were classified as TS animals since they shed an E. coli O157 isolate carrying at least one stx gene at least once during this study. The distribution of both PS and NS animals was balanced for the five pens, and each pen contained at least one animal with each E. coli O157:H7 shedding status. These results demonstrated that small subpopulations of cattle in a feedlot population appear to be persistently colonized by E. coli O157:H7 for extended periods of time during the final 100 to 110 days of finishing.
TABLE 2.
Prevalence of E. coli O157:H7 in feedlot cattle during the final phase of finishing
| Collection | No. of animals sampled | No. of positive samplesa |
|---|---|---|
| 1 | 788 | 314 |
| 2 | 788 | 265 |
| 3 | 476 | 206 |
| 4 | 197 | 39 |
| 5 | 100 | 28 |
| 6 | 35 | 15 |
Positive samples were samples that contained E. coli O157:H7 with the eae, stx1, and stx2 virulence genes. This E. coli O157:H7 genotype was targeted as it was the most common genotype identified during the first two sample collections and the purpose of this study was to gain insight into persistent shedding of E. coli O157:H7.
Feces from the eight animals classified as PS animals that were collected during the final antemortem sampling were examined using the five-tube MPN methodology. Only one PS animal (PS-7) shed E. coli O157:H7 at levels (46 MPN/g) detectable by our method. The remaining seven PS animals shed E. coli O157:H7 at levels below the detectable limit, 1.8 MPN/g (data not shown). Our results indicate that an animal that becomes persistently colonized by E. coli O157:H7 does not necessarily shed high levels of the organism in its feces.
GI tissue and GI content analysis.
E. coli O157:H7 was detected in tissue and content samples collected from both upper and lower sites in the GI tracts of PS and TS animals (Table 3). Although the differences were not significant, more lower GI tissue and content samples from PS animals than from TS animals tested positive for E. coli O157:H7. An upper GI tissue sample (omasum) and an anterior root lymph node tissue sample from only one PS animal tested positive for E. coli O157:H7. Two upper GI tissue samples (reticulum and omasum) from two different TS animals tested positive for E. coli O157:H7. No gall bladder samples were positive for E. coli O157:H7 regardless of the previous shedding status (Table 3).
TABLE 3.
Distribution of postmortem GI tissues and contents positive for E. coli O157:H7 among PS and TS animals
| Location | Site | No. of positive samples
|
|||
|---|---|---|---|---|---|
| PS (n = 8)
|
TS (n = 18)
|
||||
| Tissue | Contents | Tissue | Contents | ||
| Upper GI | Reticulum | 0 | 0 | 1 | 0 |
| tract | Omasum | 1 | 0 | 1 | 0 |
| Rumen | 0 | 0 | 0 | 0 | |
| Abomasum | 0 | 0 | 0 | 0 | |
| Duodenum | 0 | 0 | 0 | 0 | |
| Lower GI | Ileal-cecal junction | 1 | 1 | 1 | 0 |
| tract | Colon | 0 | 1 | 0 | 0 |
| RAJ | 1 | 1 | 1 | 2 | |
| Ileal-cecal colic node | 0 | 0 | 0 | 0 | |
| Anterior root node | 1 | 0 | 0 | 0 | |
| Gall bladder | 0 | 0 | 0 | 0 | |
Histology and pathology.
Histological analyses of tissue samples collected from PS and TS animals in this study did not reveal any notable differences between tissues of PS and NS animals. As expected, all tissue samples from PS and NS animals were normal or had minor lesions commonly found in GI tissues characteristic of fed cattle. In addition, the livers examined from animals representing the PS, TS, and NS groups had no visible surface lesions (data not shown). Thus, there do not appear to be significant physiological differences between animals that become persistently colonized by E. coli O157:H7 and animals that are not colonized by this organism.
Molecular characterization.
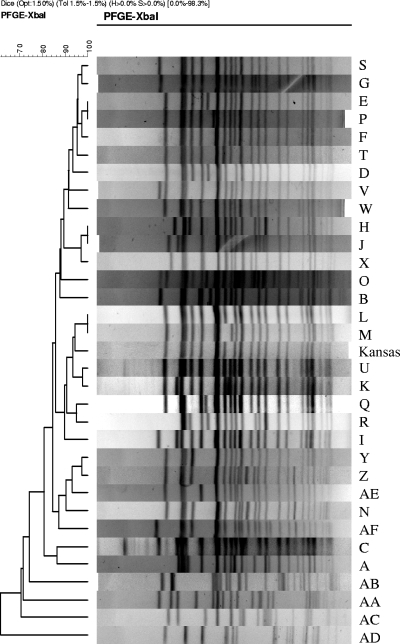
At least one fecal isolate was selected to represent each antemortem collection period for each of the eight PS animals along with all postmortem isolates (for a total of 82 isolates), and a random representative set of fecal isolates was selected for 16 TS animals along with all postmortem isolates (for a total of 50 isolates), resulting in a set of 132 isolates that were characterized by PFGE typing. The 132 E. coli O157:H7 isolates analyzed by PFGE typing were classified into 32 different subtypes (Fig. 1; Table 4). A single, predominant PFGE subtype (subtype F) (Fig. 1; Table 4) accounted for 53% of the 132 isolates characterized. A chi-square test of independence showed that subtype F was distributed similarly in the PS and TS animal populations (P > 0.05), and this subtype persisted throughout the study (Fig. 2). Twenty-two of the 32 unique PFGE subtypes were found in PS animals, and 17 of the 32 unique PFGE subtypes were found exclusively in PS animals. Interestingly, only five PFGE subtypes (subtypes E, F, H, I, and N) overlapped in the PS and TS animal populations (Table 4). All eight PS animals shed subtype F during the first sample collection, at least twice over the entire collection period, and in at least two consecutive sample collection periods (Table 5). One PS animal (PS-4) shed subtype F over the entire collection period, while two other PS animals (PS-1 and PS-7) shed subtype F consecutively over the first four sample collections (Table 5). During fecal sample collection, PS animals shed 15 PFGE subtypes in addition to subtype F, and 10 of these subtypes differed from subtype F by three or fewer bands (Table 4). Tenover et al. (53) concluded that strains with differences of three or fewer bands compared to a reference strain (subtype F was used as the reference strain) were closely related. The remaining five subtypes differed from subtype F by four or more bands and thus were not considered closely related to PFGE subtype F according to the criteria of Tenover et al. (53). The postmortem tissue and GI content sample collection yielded an additional 10 unique PFGE types for PS and TS animals that were not present during antemortem fecal sample collections, and 4 of these postmortem PFGE subtypes were closely related to the dominant antemortem subtype, subtype F.
FIG. 1.
Partial dendrogram and representative banding patterns for the 32 unique PFGE banding patterns, including the banding pattern for the E. coli O157:H7 isolate (Kansas) obtained in a previous study (11) conducted 2 years before the current study. The Kansas isolate was determined to have the same virulence genotype as the other E. coli O157:H7 isolates utilizing multiplex PCR before the PFGE analysis. The letters on the right correspond to the PFGE subtypes shown in Table 4.
TABLE 4.
PFGE characterization of E. coli O157:H7 isolates from PS and TS animals collected ante- and postmortem
| PFGE subtype | No. of band differences from subtype F | PS
|
TS
|
||
|---|---|---|---|---|---|
| No. of isolates collected antemortem (no. of isolates collected postmortem) | No. of animals | No. of isolates collected antemortem (no. of isolates collected postmortem) | No. of animals | ||
| A | 3 | 1 (0) | 1 | 0 (0) | 0 |
| B | 4 | 0 (0) | 0 | 0 (1) | 1 |
| C | 4 | 1 (0) | 1 | 0 (0) | 0 |
| D | 2 | 0 (3) | 1 | 0 (0) | 0 |
| E | 1 | 0 (2) | 1 | 1 (0) | 1 |
| F | 40 (0) | 8 | 24 (6) | 11 | |
| G | 3 | 0 (5) | 2 | 0 (0) | 0 |
| H | 3 | 2 (0) | 2 | 3 (0) | 3 |
| I | 2 | 8 (0) | 3 | 2 (0) | 1 |
| J | 4 | 0 (4) | 2 | 0 (0) | 0 |
| K | 1 | 1 (0) | 1 | 0 (0) | 0 |
| L | 1 | 1 (0) | 1 | 0 (0) | 0 |
| M | 1 | 0 (0) | 0 | 2 (0) | 1 |
| N | 6 | 1 (0) | 1 | 1 (0) | 1 |
| O | 4 | 0 (0) | 0 | 0 (1) | 1 |
| P | 1 | 3 (0) | 1 | 0 (0) | 0 |
| Q | 2 | 0 (0) | 0 | 1 (0) | 1 |
| R | 3 | 1 (0) | 1 | 0 (0) | 0 |
| S | 2 | 0 (0) | 0 | 3 (0) | 1 |
| T | 1 | 1 (0) | 1 | 0 (0) | 0 |
| U | 1 | 1 (0) | 1 | 0 (0) | 0 |
| V | 2 | 0 (0) | 0 | 1 (0) | 1 |
| W | 1 | 1 (0) | 1 | 0 (0) | 0 |
| X | 3 | 0 (0) | 0 | 0 (1) | 1 |
| Y | 1 | 0 (0) | 0 | 1 (0) | 1 |
| Z | 4 | 1 (0) | 1 | 0 (0) | 0 |
| AA | >7 | 2 (0) | 1 | 0 (0) | 0 |
| AB | >7 | 1 (0) | 1 | 0 (0) | 0 |
| AC | >7 | 0 (1) | 1 | 0 (0) | 0 |
| AD | >7 | 0 (0) | 0 | 0 (1) | 1 |
| AE | 3 | 0 (1) | 1 | 0 (0) | 0 |
| AF | >7 | 0 (0) | 0 | 0 (1) | 1 |
| Total | 66 (16) | 39 (11) | |||
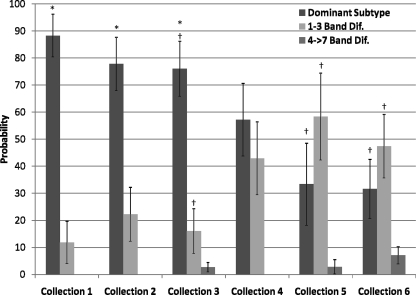
FIG. 2.
Estimated probabilities of encountering the three PFGE subtype categories during antemortem sampling. The probability of encountering an E. coli O157:H7 isolate belonging to the dominant subtype, a subtype differing from the dominant subtype by 1 to 3 bands, or a subtype differing from the dominant subtype by 4 to >7 bands is indicated on the y axis, while the collections are indicated on the x axis. Isolates possibly related (4- to 6-band difference) to the dominant subtype and isolates divergent (>7-band difference) from the dominant subtype were collapsed to form the 4- to >7-band difference subtype category due to low numbers. The low frequency of the 4- to >7-band difference subtype category prevented inclusion of this category in the analysis, which prevented calculation of errors. The error bars indicate the standard error calculated for each estimated probability. For each independent collection, the estimated probability of encountering the dominant subtype was greater (P < 0.05) than the probability of encountering strains that differed by one to three bands (indicated by an asterisk). Also, for an individual collection, a dagger indicates that the probability of encountering the dominant subtype and subtypes with 1 to 3 band differences was greater (P < 0.05) than the probability of encountering subtypes with 4- to >7-band differences. Dif., differences.
TABLE 5.
Distribution of PFGE subtypes for each PS animal during the antemortem collection period
| Animal | PFGE subtype(s)
|
|||||
|---|---|---|---|---|---|---|
| Collection 1 | Collection 2 | Collection 3 | Collection 4 | Collection 5 | Collection 6 | |
| PS-1 | F | F | F | F, I | AB | I |
| PS-2 | F | F, I | F | I | I | I |
| PS-3 | F | F | C | T, K | Z | N |
| PS-4 | F | F | F, U | F | F | F |
| PS-5 | F | F | F | H | I | A |
| PS-6 | F | F | F | L | R | H |
| PS-7 | F | F, W | F | F, A | P | F |
| PS-8 | F | F | F | A | F | AA |
The ability of the predominant E. coli O157:H7 subtype to persist in the GI tracts of feedlot cattle was indicated by the percentage of animals that shed this subtype in their feces throughout the study. More specifically, the estimated probabilities of finding subtype F were 89.1, 77.0, 78.5, 59.3, 36.0, and 30.2% for sample collections 1, 2, 3, 4, 5, and 6, respectively (Fig. 2). Although there was a decrease (P < 0.05) in the probability of shedding the dominant subtype with time, the dominant subtype was still detected and actually accounted for 31.6% (6/19) of the isolates collected during the final antemortem collection. Interestingly, our results also suggest that the dominant E. coli O157:H7 molecular subtype (subtype F) underwent microevolutionary changes during the study, as shown by the emergence of molecular subtypes that were closely related to the dominant subtype and the decline in the presence of the dominant subtype as the study progressed (Fig. 2). These results indicate that a dominant E. coli O157:H7 strain (subtype F) and other closely related strains persisted in the population of feedlot cattle over a 100- to 110-day period.
In an effort to further investigate the ability of E. coli O157:H7 to persist in the feedlot environment, we molecularly characterized an additional E. coli O157:H7 isolate from a previous study performed by Childs et al. (11) that was collected from the environment of the same feedlot that was used in the current study more than 2 years before our cattle arrived. This E. coli O157:H7 isolate (designated the Kansas isolate) was analyzed first with multiplex PCR to determine its genotype. After it was determined that the Kansas isolate had the same genotype as the persistent E. coli O157:H7 strain obtained in the current study (based on a five-gene multiplex PCR profile), the Kansas isolate was characterized by PFGE typing and compared to isolates obtained in the current study (Fig. 1). The Kansas isolate differed by only a single band from the dominant subtype and exhibited 86% similarity (Fig. 1), supporting the hypothesis that there is long-term persistence of closely related E. coli O157:H7 strains in the feedlot environment.
Cell attachment.
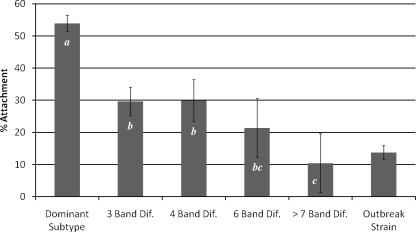
The abilities of E. coli O157:H7 isolates belonging to the dominant PFGE subtype (subtype F), closely related PFGE subtypes (<3-band difference from subtype F), possibly related subtypes (between 4- and 6-band difference from subtype F), and divergent subtypes (>7-band difference from subtype F) to adhere to the Caco-2 human intestinal epithelial cell line were compared. The attachment efficiency was expressed as the percentage of the initial inoculum that adhered to host cells, and the mean attachment efficiencies for the strain categories described above ranged from 10.7 to 53.9% (Fig. 3). E. coli O157:H7 isolates belonging to the persistent PFGE subtype (subtype F) demonstrated an enhanced (P < 0.05) ability to adhere to human intestinal epithelial cells compared to isolates belonging to closely related, possibly related, and genetically divergent subtypes. A relationship was observed between genetic diversity and attachment efficacy; as the genetic difference from subtype F increased (based on the number of band differences), the ability to attach to Caco-2 cells decreased (Fig. 3). As a reference, an E. coli O157:H7 isolate obtained from a food sample associated with an outbreak of human illness (ATCC 43895) with the same genotype (five-gene multiplex PCR profile) was included in all cell attachment assays. The attachment efficiencies of the E. coli O157:H7 isolates representing the dominant subtype were more than threefold greater than that of the outbreak-associated isolate (Fig. 3). The results of the Caco-2 attachment assays suggest that E. coli O157:H7 subtypes that persist in cattle have an enhanced ability to adhere to human intestinal epithelial cells.
FIG. 3.
Caco-2 attachment efficacies for E. coli O157:H7 isolates representing genetically diverse PFGE subtypes and an E. coli O157:H7 isolate known to have caused an outbreak of food-borne illness, which was included as a reference. The data are expressed as the percentage of adherent E. coli O157:H7 cells recovered from each well. The PFGE subtype or subtype category is indicated on the x axis, while the percent attachment is indicated on the y axis. The bars indicate the least-squares means for attachment efficacies obtained for at least two strains for each subtype category (except for the 6- and >7-band bars, where only one E. coli O157:H7 strain was assayed for each category) for three independent experiments. The error bars indicate the standard errors calculated for each least-squares mean. Bars that do not contain the same letter are significantly different (P < 0.05). The outbreak strain was not included in the analysis, but data for this strain are included for reference. Dif., differences.
DISCUSSION
To date, there has not been an extensive investigation of the molecular ecology of E. coli O157:H7 persistence and shedding in naturally colonized feedlot cattle. Our results demonstrate that closely related E. coli O157:H7 strains may persist in the feedlot ecosystem (i.e., cattle and the feedlot environment) for extended periods, which may be explained in part by an enhanced ability of these persistent strains to adhere to intestinal epithelial cells. Additionally, we demonstrated that most (97.6%) feedlot steers shed Shiga toxin-encoding E. coli O157 during the final 100 to 110 days of the feeding period. Molecular characterization of E. coli O157:H7 isolates revealed that a predominant E. coli O157:H7 strain persisted throughout the study and that this persistent strain diversified during the study. Further phenotypic characterization of isolates belonging to the persistent subtype and closely related and more genetically divergent PFGE subtypes using a cell culture attachment assay revealed an increased attachment efficiency of the persistent E. coli O157:H7 strain found in the feedlot population. Our results illustrate that certain E. coli O157:H7 strains may persist in cattle populations and that, ultimately, these strains may represent an increased risk to human health due to the increased likelihood that they could enter the human food supply and due to their subsequent enhanced ability to attach to human intestinal epithelial cells.
Most feedlot cattle appear to shed Shiga toxin-encoding E. coli O157 at some point during the final phase of finishing.
During collections 1 and 2, we detected an E. coli O157 isolate carrying at least one Shiga toxin-encoding gene in 45.5 and 58.7% of all animals, respectively. Before now, the point prevalence of Shiga toxin-encoding E. coli O157, a more indicative estimate of true risk (e.g., a Shiga toxin-encoding E. coli O157 would elicit a regulatory response if it was transferred to a carcass [20]), was reported to range from 0.3 to 19.7% in feedlot cattle (28). More importantly, we observed that 97.6% of steers shed a Shiga toxin-encoding E. coli O157 isolate at least once during the final 100 to 110 days of the feeding period, a level that had been reported previously. During the first two sample collection periods, 39.8 and 33.6% of the animals shed an E. coli O157:H7 isolate with the same genotype (i.e., carrying the eae, stx1, and stx2 genes), which are higher percentages than those previously reported (12, 18, 27, 45, 50, 55, 56).
Small subpopulations of feedlot cattle appear to become persistently colonized by E. coli O157:H7.
Few studies have evaluated the persistence of E. coli O157:H7 fecal shedding in large populations of feedlot cattle naturally colonized by this human pathogen. We found that 1% of feedlot steers persistently shed an E. coli O157:H7 isolate with the same genotype (i.e., a strain carrying eae, stx1, and stx2) during the final 100 days of the feeding period, a level that is significantly greater than expected by chance. Intensive fecal sampling (multiple samples collected each day) that was conducted with two different cohorts of 6- to 11-month-old dairy calves revealed two animals in each cohort (14 and 12.5% of the animals) that persistently shed E. coli O157:H7 for 4 and 15 days (43). Observation of E. coli O157:H7 fecal shedding in feedlot cattle determined that a small number of animals (n = 8) shed the organism for a maximum of 4.5 weeks, about three-quarters of the time that we found E. coli O157:H7 persistence to last, and the remaining animals in the sample population shed this bacterium for 2.5 weeks on average (30). For dairy cattle, the duration of E. coli O157:H7 shedding was estimated to be approximately 1 month (6). These previous studies of naturally colonized populations were limited by the lack of molecular subtyping to characterize isolates in order to determine if animals were persistently colonized by the same E. coli O157:H7 strain or if they were continuously exposed to, and subsequently shed, genetically diverse strains. Experimental inoculation of calves with E. coli O157:H7 established that shedding occurred for periods between 14 and 140 days long (7, 15, 39, 44). Alternatively, experimental inoculation of cattle that were >0.5 year old increased the minimum number of days of persistent fecal shedding to 29 days but reduced the maximum shedding time to only 98 days (15, 23, 46). Assessment of previously published data supported the hypothesis that there is a general trend of increased E. coli O157:H7 shedding duration for neonates and calves less than 1 year old. As the animal age increases, persistent shedding decreases and is increasingly intermittent.
It is plausible that the eight animals that we found to be colonized by the same E. coli O157:H7 strain throughout this study continued to be reexposed to the organism via animal-to-animal transmission or a contaminated pen environment, which resulted in the persistent E. coli O157:H7 shedding status. Animal-to-animal transmission was previously observed when uninoculated young calves (10 weeks old) that commingled with calves inoculated with E. coli O157:H7 began shedding the organism (7). However, animal-to-animal transmission of E. coli O157:H7 was found to be very inefficient and unlikely for a group of 5- to 8-month-old calves (46), further substantiating our conclusion that the animals were persistently colonized with E. coli O157:H7, particularly since the PS animals were fed and maintained in different pens. Additionally, it appears that observations from one time point to the next are not independent with regard to positive animals. Furthermore, previous literature suggests that while the microbiological assays are highly sensitive, the sampling methodology is quite insensitive (17a). Thus, the true PS population was likely underestimated and the true NS population overestimated; if so, there is even more true dependency from time point to time point than that observed herein.
E. coli O157:H7 may show specificity for colonization of the lower GI tract, and there do not appear to be notable histopathological differences between PS and NS animals.
We did not obtain conclusive evidence regarding the preferential site of E. coli O157:H7 colonization due to the limited prevalence of this organism in the GI tract tissue and content samples collected postmortem, which was presumably a result of transportation stress (2) and extended lairage (38) at the plant. A comparable situation was reported for a population of sheep inoculated with E. coli O157:H7 that shed detectable amounts of the organism in their feces, but the organism could not be detected in any GI tract tissue or content samples following necropsy (23). Despite the limited E. coli O157:H7 prevalence, we found that at least one ileal-cecal sample and one RAJ tissue sample were positive for E. coli O157:H7 both in animals identified as PS animals and in animals identified as TS animals. We also obtained three positive tissue samples from the fore-stomach, the earliest GI site believed to be a site of E. coli O157:H7 propagation (8). Recent research has concluded that the colon (23, 52), specifically an area designated the RAJ, is the principal site of E. coli O157:H7 colonization (33, 39, 46), and this colonization site is particularly important for E. coli O157:H7 excretion in feces (12). In cattle naturally colonized by E. coli O157, tissue samples obtained 1 cm from the RAJ contained larger quantities of E. coli O157 than samples obtained 15 cm from the RAJ (33). A relationship between increased E. coli O157:H7 concentration and areas closest to the RAJ was observed with tissue samples obtained from experimentally infected animals, naturally colonized animals, and calves exposed to infected animals (39). It was concluded that animals are more likely to become consistent long-term shedders when they are infected with E. coli O157:H7 directly at the RAJ rather than through oral inoculation (46). While our findings do not confirm the preferential site of E. coli O157:H7 colonization in cattle, they do corroborate the conclusion that the lower GI tract is the preferred site of colonization.
Histopathological evaluation of the all of the GI tracts obtained from PS, TS, and NS animals did not reveal any gross abnormalities or discernible lesions. More specifically, no attaching and effacing (A/E) lesions, which are characteristic of E. coli O157:H7 colonization, were identified in PS animal GI tissue samples. All animals enrolled in the study remained healthy throughout the entire time of sample collection, and cattle colonized with E. coli O157:H7 do not generally have clinical symptoms (15). There does, however, appear to be an association between animal age and susceptibility to clinical symptoms for animals exposed to very high levels of E. coli O157:H7. Experimental infection of young calves (<12 h old or 30 to 36 h old) with 10 logs of E. coli O157:H7 resulted in severe diarrhea and A/E lesions throughout the lower GI tract (16). In contrast, 1-day-old calves remained clinically normal after inoculation with 8 logs of E. coli O157:H7 (44), an inoculation level believed to encourage A/E lesion development (17). Once animals reach 3 weeks of age, their susceptibility to E. coli O157:H7 infection (10 logs) appears to diminish; E. coli O157:H7 infection results in slight increases in the body temperature and watery diarrhea, but these symptoms last for only a couple of days following inoculation and no A/E lesions are formed (8, 15). Still, infection of 3- to 4-month-old calves with 10 logs of E. coli O157:H7 caused watery diarrhea (17) and A/E lesion formation (17, 52) and in one case resulted in translocation of the bacteria to the gall bladder, where they were able to produce A/E lesions (52). The general consensus that E. coli O157:H7 colonization is not associated with clinical symptoms in animals that are at least 1 year old (15, 23) is further validated by the lack of pathological symptoms observed for the animals enrolled in our study. Furthermore, histopathological comparisons between PS and NS animals support the conclusion that host-associated factors do not appear to be as significant as E. coli O157:H7's ability to orchestrate persistent colonization in cattle more than 1 year old.
A single E. coli O157:H7 strain may persist in a population of feedlot cattle.
We observed that a single predominant PFGE subtype accounted for 53% of all isolates characterized; furthermore, 87% of the isolates belonged to the predominant PFGE subtype or to PFGE subtypes that were closely related to the predominant subtype (only one to three bands were different [53]). Our data contribute to the growing body of evidence indicating that E. coli O157:H7 persists in cattle populations. More specifically, along with several previous studies (6, 43, 45, 48, 51), our study supports the conclusion that a given population of feedlot cattle appears to be colonized by a single predominant strain and a few closely related strains. The predominant PFGE subtype in our study was disseminated into each animal pen, was found on each sample collection date, and was shed by each PS animal on at least two consecutive sample collection dates. Similarly, in another study, all 54 E. coli O157 isolates obtained from two different dairies belonged to the same PFGE subtype (43). Analysis of the genetic diversity of E. coli O157:H7 collected from four dairies located in a 30-km area in southern Alberta also demonstrated that there was a highly clonal E. coli O157:H7 population, as three dominant subtypes, which were detected at every dairy, accounted for a majority of the isolates characterized (50). It can be argued that dairy farm environments have an increased likelihood of sustaining highly clonal E. coli O157:H7 populations because of reduced animal turnover compared to commercial feedlots, where new animals presumably are the main source of new E. coli O157:H7 subtypes. However, feedlots also appear to maintain highly related populations of E. coli O157:H7, even with their increased animal turnover rates. For example, PFGE analyses of 103 and 230 E. coli O157:H7 isolates obtained from two different commercial feedlots revealed that isolates clustered with 80% similarity (45) and that 60% of the isolates belonged to four closely related subtypes (32), respectively. Additionally, indistinguishable E. coli O157:H7 subtypes were recovered from two feedlots that were approximately 100 km apart and did not share any common source of animals that entered them (56). Previously, we demonstrated that transit to, or holding at, the processing plant can introduce E. coli O157:H7 isolates with diverse PFGE subtypes (11). We obtained similar results, as 10 unique E. coli O157:H7 subtypes that were never observed during antemortem sampling were obtained from either GI tract tissue or GI content samples.
The persistence of predominant E. coli O157:H7 subtypes in beef cattle feedlots was characterized and was determined to last for several years (32). Strains of E. coli O157:H7 persist in the environment (1) and can potentially be rapidly disseminated throughout a cattle population (45). We compared the PFGE banding pattern of the dominant PFGE subtype observed in the current study to that of an E. coli O157:H7 strain (with the same genotype) obtained from an environmental sample (11) from the same feedlot 2 years before the arrival of our cattle and determined that the two subtypes were highly related, with only one band difference and only 14% divergence from the dominant subtype (Fig. 1). Our findings provide further evidence that certain E. coli O157:H7 strains likely persist in the feedlot environment and subsequently colonize exposed animals.
E. coli O157:H7 strains that persist in cattle populations demonstrate an enhanced ability to adhere to intestinal epithelial cells.
Based on our observations that a predominant PFGE subtype and other closely related PFGE subtypes accounted for the majority of E. coli O157:H7 isolates obtained from the population of feedlot cattle studied here and that these subtypes persisted in these cattle throughout the study, we hypothesized that these strains may represent an “ecotype” that adapted to colonize and persist in the GI tract. We explored this hypothesis by characterization of E. coli O157:H7 isolates representing the predominant and persistent PFGE subtype (subtype F), along with isolates representing PFGE subtypes that were closely related, possibly related, and distantly related to subtype F. Our investigation showed that E. coli O157:H7 isolates that represent the predominant PFGE subtype demonstrate an enhanced (P < 0.05) ability to adhere to the Caco-2 human intestinal epithelial cell line. Interestingly, as genetic diversity of the predominant PFGE subtype increases, attachment efficacy decreases. E. coli O157:H7 isolates representing the predominant subtype demonstrated an ability to attach to Caco-2 cells that was greater than that of a reference E. coli O157:H7 isolate from an outbreak of human illness, substantiating the human-pathogenic potential of E. coli O157:H7 strains that persist in feedlot cattle.
We characterized the attachment efficacies of E. coli O157:H7 isolates using a human intestinal epithelial cell line because of the lack of an immortal bovine intestinal epithelial cell line. Although our initial objectives were to elucidate the nature of E. coli O157:H7 colonization and persistence in the bovine GI tract, we discovered that E. coli O157:H7 strains that persist in feedlot cattle appear to have an accentuated ability to adhere to human intestinal epithelial cells, which is essential for disease manifestation. E. coli O157:H7 depends on intimin and the translocated intimin receptor (Tir) for intimate adherence to a host cell (29). The role of intimin and its importance in bacterial adherence have been investigated and validated using bovine models (14, 22, 47) and appear to be no different in human cell lines (13). We do not discount the significance of intimin in bacterial attachment, but taking into consideration the fact that all of the E. coli O157:H7 isolates screened during the attachment assay contained the gene responsible for intimin production, our results provide evidence that there are other influential mechanisms that are responsible for attachment efficacy. Further work is required to elucidate the molecular mechanisms responsible for the disparate attachment efficacies of diverse E. coli O157:H7 strains that encode the same virulence determinants.
Conclusions.
We provide compelling evidence that in a population of healthy feedlot cattle, a small subpopulation of animals appears to become persistently colonized by closely related E. coli O157:H7 strains. We found no physiological differences between animals that we classified as PS, TS, and NS based on our observations of animal health status and postmortem histopathology. In addition, PS and TS animals appeared to become colonized by a single predominant E. coli O157:H7 molecular subtype along with other closely related molecular types, supporting the hypothesis that new genotypes emerged. Finally, our findings provide evidence that cattle may be more likely to be colonized by E. coli O157:H7 molecular subtypes that demonstrate accentuated human-pathogenic potential, as shown by the enhanced ability of persistent strains to adhere to human intestinal epithelial cells. Additionally, it stands to reason that there is an increased likelihood that these E. coli O157:H7 subtypes are transferred through the production continuum and subsequently into the human population because of their increased prevalence in feedlot cattle. Our results highlight the importance of preharvest food safety interventions to reduce the load of E. coli O157:H7 that enters the human food supply and support the conclusion that such efforts should be targeted at strains that persist in cattle populations which seem to represent the greatest risk to human health. Further research is needed to elucidate the underlying pathogen factors associated with persistent colonization of healthy cattle by E. coli O157:H7, including work to further probe molecular mechanisms associated with enhanced adhesion to intestinal cells and to develop mitigation strategies to control E. coli O157:H7 in feedlot populations with the ultimate goal of reducing the risk of human infection.
Acknowledgments
This work was supported by the Beef Checkoff and by the Colorado State University Agricultural Experiment Station.
We are indebted to M. Wiedmann and Y. Soyer of Cornell University for their expertise and assistance with PFGE typing and PFGE pattern cluster analysis.
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Bach, S. J., L. J. Selinger, K. Stanford, and T. A. McAllister. 2005. Effect of supplementing corn- or barley-based feedlot diets with canola oil on faecal shedding of Escherichia coli O157:H7 by steers. J. Appl. Microbiol. 98:464-475. [DOI] [PubMed] [Google Scholar]
- 2.Barham, A. R., B. L. Barham, A. K. Johnson, D. M. Allen, J. R. Blanton, J. R., and M. F. Miller. 2002. Effects of the transportation of beef cattle from the feedyard to the packing plant on prevalence levels of Escherichia coli O157 and Salmonella spp. J. Food Prot. 65:280-283. [DOI] [PubMed] [Google Scholar]
- 3.Barkocy-Gallagher, G. A., E. D. Berry, M. Rivera-Betancourt, T. M. Arthur, X. Nou, and M. Koohmaraie. 2002. Development of methods for the recovery of Escherichia coli O157 and Salmonella from beef carcass sponge samples and bovine fecal and hide samples. J. Food Prot. 65:1527-1534. [DOI] [PubMed] [Google Scholar]
- 4.Barkocy-Gallagher, G. A., T. M. Arthur, M. Rivera-Betancourt, X. Nou, S. D. Shackelford, T. M. Wheeler, and M. Koohmaraie. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J. Food Prot. 66:1978-1986. [DOI] [PubMed] [Google Scholar]
- 5.Barkocy-Gallagher, G. A., K. K. Edwards, X. Nou, J. M. Bosilevac, T. M. Arthur, S. D. Shackelford, and M. Koohmaraie. 2005. Methods for recovering Escherichia coli O157:H7 from cattle fecal, hide, and carcass samples: sensitivity and improvements. J. Food Prot. 68:2264-2268. [DOI] [PubMed] [Google Scholar]
- 6.Besser, T. E., D. D. Hancock, L. C. Pritchett, E. M. McRae, D. H. Rice, and P. I. Tarr. 1997. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 175:726-729. [DOI] [PubMed] [Google Scholar]
- 7.Besser, T. E., B. L. Richards, D. H. Rice, and D. D. Hancock. 2001. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol. Infect. 127:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, C. A., B. G. Harmon, T. Zhao, and M. P. Doyle. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 63:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchko, S. J., R. A. Holley, W. O. Olsen, V. P. J. Gannon, and D. M. Veira. 2000. The effects of different grain diets on fecal shedding of Escherichia coli O157:H7 by steers. J. Food Prot. 63:1467-1474. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2001. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7 by pulsed-field gel electrophoresis (PFGE). Centers for Disease Control and Prevention, Atlanta, GA.
- 11.Childs, K. D., C. A. Simpson, W. Warren-Serna, G. Bellinger, B. Centrella, R. A. Bowling, J. Ruby, J. Stefanek, D. J. Vote, T. Choat, J. A. Scanga, J. N. Sofos, G. C. Smith, and K. E. Belk. 2006. Molecular characterization of Escherichia coli O157:H7 hide contamination routes: feedlot to harvest. J. Food Prot. 69:1240-1247. [DOI] [PubMed] [Google Scholar]
- 12.Cobbold, R. N., D. D. Hancock, D. H. Rice, J. Berg, R. Stilborn, C. J. Hovde, and T. E. Besser. 2007. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl. Environ. Microbiol. 73:1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cookson, A. L., and M. J. Woodward. 2003. The role of intimin in the adherence of enterohemorrhagic Escherichia coli (EHEC) O157:H7 to HEp-2 tissue culture cells and to bovine gut explants tissues. Int. J. Med. Microbiol. 292:547-553. [DOI] [PubMed] [Google Scholar]
- 14.Cornick, N. A., S. L. Booher, and H. W. Moon. 2002. Intimin facilitates colonization by Escherichia coli O157:H7 in adult ruminants. Infect. Immun. 70:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cray, Jr., W. C., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean-Nystrom, E. A., B. T. Bosworth, W. C. Cray, Jr., and H. W. Moon. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect. Immun. 65:1842-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean-Nystrom, E. A., B. T. Bosworth, and H. W. Moon. 1999. Pathogenesis of Escherichia coli O157:H7 in weaned calves. Adv. Exp. Med. Biol. 473:173-177. [DOI] [PubMed] [Google Scholar]
- 17a.Echeverry, A., G. H. Loneragan, B. A. Wagner, and M. M. Brashears. 2005. Effect of intensity of fecal pat sampling on estimates of E. coli O157 prevalence. Am. J. Vet. Res. 66:2023-2027. [DOI] [PubMed] [Google Scholar]
- 18.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcass of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Food and Drug Administration, United States Department of Health and Human Services. 2001. Bacteriological analytical manual. http://vm.cfsan.fda.gov/∼ebam/bam-toc.html. Accessed 20 September 2006.
- 20.Food Safety Inspection Service, United States Department of Agriculture. 2008. Detection, isolation and identification of Escherichia coli O157:H7 from meat products. http://www.fsis.usda.gov/PDF/MLG_5_04.pdf. Accessed 13 May 2008.
- 21.Galland, J. C., D. R. Hyatt, S. S. Crupper, and D. W. Acheson. 2001. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 67:1619-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girard, F., F. Dziva, P. van Diemen, A. D. Phillips, M. P. Stevens, and G. Frankel. 2007. Adherence of enterohemorrhagic Escherichia coli O157, O26, and O111 strains to bovine intestinal explants ex vivo. Appl. Environ. Microbiol. 73:3084-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. J. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyles, C. L. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45-E62. [DOI] [PubMed] [Google Scholar]
- 25.Hu, Y., Q. Zhang, and J. C. Meitzler. 1999. Rapid and sensitive detection of Escherichia coli O157:H7 in bovine faeces by a multiplex PCR. J. Appl. Microbiol. 87:867-876. [DOI] [PubMed] [Google Scholar]
- 26.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussein, H. S. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85(Suppl. E):E63-E72. [DOI] [PubMed] [Google Scholar]
- 28.Hussein, H. S., and L. M. Bollinger. 2005. Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J. Food Prot. 68:2224-2241. [DOI] [PubMed] [Google Scholar]
- 29.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 30.Khaitsa, M. L., D. R. Smith, J. A. Stoner, A. M. Parkhurst, S. Hinkley, T. J. Klopfenstein, and R. A. Moxley. 2003. Incidence, duration, and prevalence of Escherichia coli O157:H7 fecal shedding by feedlot cattle during the finishing period. J. Food Prot. 66:1972-1977. [DOI] [PubMed] [Google Scholar]
- 31.Laven, R. A., A. Ashmore, and C. S. Stewart. 2003. Escherichia coli in the rumen and colon of slaughter cattle, with particular reference to E. coli O157. Vet. J. 165:78-83. [DOI] [PubMed] [Google Scholar]
- 32.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stillborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low, J. C., I. J. McKendrick, C. McKechnie, D. Fenlon, S. N. Naylor, C. Currie, D. G. E. Smith, L. Allison, and D. L. Gally. 2005. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. J. Appl. Microbiol. 71:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews, L., I. J. McKendrick, H. Ternent, G. J. Gunn, B. Synge, and M. E. J. Woolhouse. 2006. Super-shedding cattle and the transmission dynamics of Escherichia coli O157. Epidemiol. Infect. 134:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews, L., J. C. Low, D. L. Gally, M. C. Pearce, D. J. Mellor, J. A. P. Heesterbeek, M. Chase-Topping, S. W. Naylor, D. J. Shaw, S. W. J. Reid, G. J. Gunn, and M. E. J. Woolhouse. 2006. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. USA 103:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mead, P. S., and P. M. Griffin. 1998. Escherichia coli O157:H7. Lancet 352:1207-1212. [DOI] [PubMed] [Google Scholar]
- 37.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illnesses and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minihan, D., M. O'Mahony, P. Whyte, and J. D. Collins. 2003. An investigation on the effect of transport and lairage on the fecal shedding prevalence of Escherichia coli O157 in cattle. J. Vet. Med. 50:378-382. [DOI] [PubMed] [Google Scholar]
- 39.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, G. E. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterhemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen, M. A., W. C. Cray, T. A. Casey, and S. C. Whipp. 1993. Rumen contents as a reservoir of enterohemorrhagic Escherichia coli serotype O157:H7. FEMS Microbiol. Lett. 114:79-84. [DOI] [PubMed] [Google Scholar]
- 41.Ray, P. E., and X. H. Lui. 2001. Pathogenesis of Shiga toxin-induced hemolytic uremic syndrome. Pediatr. Nephrol. 16:823-839. [DOI] [PubMed] [Google Scholar]
- 42.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 43.Robinson, S. E., E. J. Wright, C. A. Hart, M. Bennett, and N. P. French. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 97:1045-1053. [DOI] [PubMed] [Google Scholar]
- 44.Sanderson, M. W., T. E. Besser, J. M. Gay, C. C. Gay, and D. D. Hancock. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet. Microbiol. 69:199-205. [DOI] [PubMed] [Google Scholar]
- 45.Scott, L., P. McGee, D. Minihan, J. J. Sheridan, B. Earley, and N. Leonard. 2006. The characterization of E. coli O157:H7 isolates from cattle faeces and feedlot environment using PFGE. Vet. Microbiol. 114:331-336. [DOI] [PubMed] [Google Scholar]
- 46.Sheng, H., M. A. Davis, H. J. Knecht, and C. J. Hovde. 2004. Rectal administration of Escherichia coli O157:H7: novel model for colonization of ruminants. Appl. Environ. Microbiol. 70:4588-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheng, H., J. Y. Lim, H. J. Knecht, J. Li, and C. J. Hovde. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74:4685-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reference deleted.
- 50.Stanford, K., D. Croy, S. J. Bach, G. L. Williams, H. Zahiroddini, and T. A. McAllister. 2005a. Ecology of Escherichia coli O157:H7 in commercial dairies in southern Alberta. J. Dairy Sci. 88:4441-4451. [DOI] [PubMed] [Google Scholar]
- 51.Stanford, K., S. J. Bach, T. H. Marx, S. Jones, J. R. Hansen, G. L. Wallins, H. Zahiroddini, and T. A. McAllister. 2005b. Monitoring Escherichia coli O157:H7 in inoculated and naturally colonized feedlot cattle and their environment. J. Food Prot. 68:26-33. [DOI] [PubMed] [Google Scholar]
- 52.Stoffregen, W. C., J. F. L. Pohlenz, and E. A. Dean-Nystrom. 2004. Escherichia coli O157:H7 in the gall bladders of experimentally infected calves. J. Vet. Diagn. Investig. 16:79-84. [DOI] [PubMed] [Google Scholar]
- 53.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelson, B. A. Murray, D. A. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tkalcic, S., C. A. Brown, B. G. Harmon, A. V. Jain, E. P. Mueller, A. Parks, K. L. Jacobsen, S. A. Martin, T. Zhao, and M. P. Doyle. 2000. Effects of diet on rumen proliferation and fecal shedding of Escherichia coli O157:H7 in calves. J. Food Prot. 63:1630-1636. [DOI] [PubMed] [Google Scholar]
- 55.Van Donkersgoed, J., T. Graham, and V. Gannon. 1999. The prevalence of verotoxins, Escherichia coli O157:H7, and Salmonella in the feces and rumen of cattle at processing. Can. Vet. J. 40:332-338. [PMC free article] [PubMed] [Google Scholar]
- 56.Van Donkersgoed, J., J. Berg, A. Potter, D. Hancock, T. Besser, D. Rice, J. LeJeune, and S. Klashinsky. 2001. Environmental sources and transmission of Escherichia coli O157 in feedlot cattle. Can. Vet. J. 42:714-720. [PMC free article] [PubMed] [Google Scholar]
- 57.Woerner, D. R., J. R. Ransom, J. N. Sofos, G. A. Dewell, G. C. Smith, M. D. Salman, and K. E. Belk. 2006. Determining the prevalence of Escherichia coli O157 in cattle and beef from the feedlot to the cooler. 2006. J. Food Prot. 69:2824-2827. [DOI] [PubMed] [Google Scholar]