Abstract
Although peptidoglycan synthesis is one of the best-studied metabolic pathways in bacteria, the mechanism underlying the membrane translocation of lipid II, the undecaprenyl-disaccharide pentapeptide peptidoglycan precursor, remains mysterious. Recently, it was proposed that the essential Escherichia coli mviN gene encodes the lipid II flippase. Bacillus subtilis contains four proteins that are putatively homologous to MviN, including SpoVB, previously reported to be necessary for spore cortex peptidoglycan synthesis during sporulation. MviN complemented the sporulation defect of a ΔspoVB mutation, and SpoVB and another of the B. subtilis homologs, YtgP, complemented the growth defect of an E. coli strain depleted for MviN. Thus, these B. subtilis proteins are likely to be MviN homologs. However, B. subtilis strains lacking these four proteins have no defects in growth, indicating that they likely do not serve as lipid II flippases in this organism.
Peptidoglycan synthesis is vital for cell growth and maintenance of cell shape in both gram-positive and gram-negative bacteria. This polymer of glycan chains that are cross-linked by peptide bridges forms an extracellular shell which provides protection against osmotic stresses as well as a sturdy scaffolding for extracellular appendages. The enzymes responsible for peptidoglycan synthesis are highly conserved in all bacteria with a cell wall. In the cytoplasm, the enzymes MurA to MurE synthesize the soluble MurNAc-pentapeptide starting with UDP-GlcNAc. MraY links this molecule to an isoprenoid chain, forming the membrane-associated lipid I precursor. MurG then adds UDP-GlcNAc to make lipid II, which is subsequently flipped across the cytoplasmic membrane and attached by penicillin-binding proteins via transglycosylation and transpeptidation reactions to the mature peptidoglycan.
While these cytoplasmic and extracellular steps are well characterized, comparatively little is known about the mechanism of membrane translocation. Fluorescently tagged lipid II does not spontaneously flip in protein-free liposomes (31), as would be expected given its large hydrophilic carbohydrate and protein groups. This observation suggests that that flipping is a protein-mediated process, and, consistent with this prediction, fluorescent lipid II molecules were translocated across vesicles made from Escherichia coli membranes. Genetic data have pointed to proteins belonging to the SEDS family as potential lipid II flippases (14). These proteins are highly conserved and contain multiple membrane-spanning domains (generally 10 to 12 transmembrane helices). Since they are in most cases essential for viability, it has been problematic to demonstrate their function. However, depletion or temperature-sensitive mutations result in phenotypes consistent with a block in peptidoglycan synthesis. A nonessential SEDS protein, Bacillus subtilis SpoVE, is necessary for the formation of peptidoglycan during a later step in spore development (13), and point mutations in SpoVE block peptidoglycan synthesis without disturbing protein production or localization (24).
Recently, the integral membrane protein MviN, encoded by an essential E. coli gene, was proposed to be the lipid II flippase (26). Strains carrying a temperature-sensitive mutation in MviN underwent lysis following incubation at the nonpermissive temperature and showed a twofold increase in lipid II accumulation (16). While the operon that includes mviN is essential in the gram-negative bacteria Sinorhizobium meliloti and Burkholderia pseudomallei (20, 25), mviN mutations in Rhizobium tropici, Salmonella enterica serovar Typhimurium, and Bdellovibrio bacteriovorus have not been fully characterized, and therefore the essentiality of MviN in these species remains to be demonstrated (4, 19, 21). Due to the high degree of conservation of other proteins involved in peptidoglycan synthesis between gram-positive and gram-negative bacteria and the essential nature of peptidoglycan synthesis, the protein(s) necessary for flipping of lipid II should also be essential and conserved in a gram-positive organism. We therefore set out to identify and examine the MviN (MurJ) homologs of B. subtilis.
MATERIALS AND METHODS
Standard procedures were used to prepare and handle recombinant DNA and to transform E. coli. Plasmids and details of their construction are described in Table S1 in the supplemental material. Oligonucleotides used are described in Table S2 in the supplemental material. B. subtilis strains were derivatives of PY79 (Table 1) unless otherwise noted (34), and the E. coli strain used was DH5α. The inducible mviN allele was moved to MG1665 from NR1154 via P1 transduction (29). B. subtilis was transformed using competent cells made by the two-step method (12), and antibiotic selection used either macrolides-lincosamides-streptogramin B (MLS) (1 μg/ml erythromycin and 250 μg/ml lincomycin), 5 μg/ml chloramphenicol, 100 μg/ml spectinomycin, or 10 μg/ml tetracycline. Sporulation used either Difco sporulation medium (DSM) (12) or CH medium for growth and A+B medium for resuspension (30). Heat resistance of spores was assayed following sporulation by exhaustion in DSM in 2-ml cultures for 24 h. For induction, 0.5 mM IPTG or 0.5% xylose was added 6 h after starting DSM cultures. Serial dilutions were plated before and after the cells were heated to 80°C 20 min, and the ratio of CFU obtained before and after heat treatment was determined.
TABLE 1.
Bacterial strains
| Strain | Genotype | Source or reference |
|---|---|---|
| B. subtilis | ||
| PY79 | Wild type | Lab stock |
| JDB1097 | spoVBΔ::tet | 9 |
| JDB1395 | spoIIIAG-HΩkan | 3 |
| JDB1752 | ΔspoVE::tet | 24 |
| JDB2327 | ΔytgP::mls | This work |
| JDB2354 | ΔytgP::mls::spc | This work |
| JDB2330 | ΔykvU::mls | This work |
| JDB2355 | ΔykvU::mls::spc | This work |
| JDB2386 | yabM::cm | This work |
| JDB2347 | spoVBΔ::tet ΔytgP::mls | This work |
| JDB2361 | spoVBΔ::tet ΔytgP::mls ΔykvU::mls::spc | This work |
| JDB2398 | spoVBΔ::tet ΔytgP::mls ΔykvU::mls::spc yabM::cm | This work |
| JDB2329 | spoVB-gfp spc | This work |
| JDB2356 | ytgP-gfp spc | This work |
| JDB2358 | ykvU-gfp spc | This work |
| JDB2357 | yabM-gfp spc | This work |
| JDB2416 | spoVB-gfp spc spoIIIAG-HΩkan | This work |
| JDB2418 | spoVB-gfp spc ΔspoVE::tet | This work |
| JDB2377 | ykvU-gfp spc spoIIIAG-HΩkan | This work |
| JDB2378 | ykvU-gfp spc ΔspoVE::tet | This work |
| JDB2402 | amyE::Pspank-uppS lacI spc | This work |
| JDB2403 | amyE::Pspank-uppS-flag lacI spc | This work |
| JDB2404 | spoVBΔ::tet amyE::Pspank-uppS lacI spc | This work |
| JDB2405 | spoVBΔ::tet amyE::Pspank-uppS-flag lacI spc | This work |
| JDB2437 | spoVBΔ::tet amyE::Pxyl-spoVB-gfp xylR spc | This work |
| JDB2504 | spoVBΔ::tet amyE::Pxyl-mviN xylR spc | This work |
| JDB2506 | amyE::Pxyl-mviN xylR spc | This work |
| E. coli | ||
| NR1154 | lysA::kan murJ Ω(−14::bla araC PBAD) | 26 |
| MG1665 | Wild type | Lab stock |
| JDE1118 | murJ Ω(−14::bla araC PBAD) | This work |
| JDE1140 | murJ Ω(−14::bla araC PBAD) pAF374 (Ptac-ytgP-gfp cm) | This work |
| JDE1147 | murJ Ω(−14::bla araC PBAD) pAF375 (Ptac-spoVB-gfp cm) | This work |
| JDE1148 | murJ Ω(−14::bla araC PBAD) pMMB207(Ptac cm) | This work |
MviN homolog deletions.
Strains carrying single ΔytgP and ΔykvU mutations were generated with a modified long flanking homology protocol using PY79 genomic DNA as the template (33). For ytgP, the upstream flank was amplified with AFO606 and AFO607 and the downstream flank was amplified with AFO608 and AFO609. For ykvU, the upstream flank was amplified with AFO610 and AFO611 and the downstream flank was amplified with AFO612 and AFO613. Each of these PCR products was gel purified and used in the second-round reaction with linearized pDG1730, which was used as the source of the erythromycin cassette. For ytgP, AFO606 and AFO609 were added to the second-round reaction mixture, and for ykvU, AFO610 and AFO613 were added to the second-round reaction mixture. Following verification of the PCR product, it was transformed into PY79. MLS-resistant transformants were confirmed to contain an appropriate deletion by assays based on product length, pattern of restriction enzyme digestion, and sequencing of a PCR-amplified allele. The spectinomycin-resistant allele of ykvU (JDB2355) was generated by transformation of JDB2330 with pEr:Sp, selecting for spectinomycin resistance, and screening for MLS resistance.
The yabM disruption strain was generated by transformation of PY79 with pAF344 (Cmr). Transformants were selected for chloramphenicol resistance and tested for sensitivity to spectinomycin (indicating a double-crossover event). Confirmation of the yabM deletion used assays based on product length, pattern of restriction enzyme digestion, and sequencing of a PCR amplified allele.
A strain carrying mutations in all four MviN homologs was constructed by transforming JDB1097 (spoVBΔ::tet) with genomic DNA from JDB2327 (ΔytgP::mls), selecting for MLS resistance, and screening for tetracycline resistance. This strain, JDB2347 (spoVBΔ::tet ΔytgP::mls), was then transformed with genomic DNA from JDB2355 (ΔykvU::mls::spc), selected for spectinomycin resistance, and screened for tetracycline resistance and MLS resistance. The resulting strain, JDB2361 (spoVBΔ::tet ΔytgP::mls ΔykvU::mls::spc), was transformed with genomic DNA from JDB2386 (ΔyabM::cm) and selected for chloramphenicol resistance (and screened for MLS, tetracycline, and spectinomycin resistance) to construct the quadruple mutant strain JDB2361. Strains used are listed in Table 1 and were verified by PCR for individual deletions.
All original mutations made were transformed into PY79. Transformation efficiencies were indistinguishable from those of a control genomic DNA (amyE::cm). The transformation efficiency for construction of a double mutant strain (ΔytgP::mls ΔyabM::cm) was also indistinguishable from that for construction of a control strain (ΔytgP::mls amyE::cm).
Fluorescence microscopy.
Samples (100 μl) of cells were taken at designated times during vegetative growth in CH medium or following resuspension in A+B medium. To each sample, 0.5 ml of FM4-64 (Invitrogen; 100 μg/ml) was added just before the cells were collected by centrifugation. The pellet was resuspended in 10 ml phosphate-buffered saline and added to a poly-l-lysine pretreated coverslip. All microscopy was performed on a Nikon Eclipse 90i with a 100× objective using phase contrast and captured by a Hamamatsu Orca-ER camera using Nikon Elements BR software. Exposure for fluorescein isothiocyanate or tetramethyl rhodamine isothiocyanate images was 400 ms. ImageJ (National Institutes of Health [NIH]) was used for analysis.
RESULTS
Identification of B. subtilis MviN homologs using BLAST and topology analysis.
BLAST analysis demonstrated that E. coli MviN is conserved among gram-negative bacteria; however, a similar analysis in gram-positive bacteria was not reported (26). Since E. coli and B. subtilis both utilize m-diaminopimelic acid-containing lipid II, the proteins involved in the synthesis, translocation, and polymerization of this molecule are likely to be highly conserved. This is true for enzymes in the pathway leading to lipid II synthesis (MurA to MurG and MraY) and to its polymerization (E. coli PBP2/3 and B. subtilis PBP2B/3) that all have e values of >10−20 (Table 2). Using BLASTp and the sequence of E. coli MviN as the query, the B. subtilis genome yielded YtgP as the top hit (e = 10−7 [Table 2]). Using the sequence of YtgP as the query against the E. coli genome yielded MviN as the top hit (e = 8 × 10−10). Thus, while YtgP and MviN were not nearly as highly conserved as any other proteins in the peptidoglycan pathway, the reciprocity of the BLASTp searches indicated that YtgP was the most likely candidate for an MviN homolog in B. subtilis. We further analyzed the presence of MviN homologs in the genomes of 11 gram-positive organisms, including representatives from the Bacillus, Streptococcus, and Staphylococcus species. YtgP homologs were the top hits, with BLASTp scores similar to those seen with B. subtilis (data not shown).
TABLE 2.
BLAST results for MviN and peptidoglycan biosynthetic homologsa
| Query | Subtilist best hit | e value |
|---|---|---|
| E. coli proteins | ||
| MviN | YtgP | 9.00E-07 |
| MraY | MraY | 4.00E-59 |
| FtsW | SpoVE | 1.00E-60 |
| MurG | MurG | 4.00E-41 |
| PBP1a (MrcA) | PonA (PBP1) | 4.00E-53 |
| B. subtilis YtgP | YtgP | 0.0 |
| YabM | 1.00E-38 | |
| SpoVB | 5.00E-29 | |
| YkvU | 1.00E-15 |
BLAST results utilizing Subtilist (http://genolist.pasteur.fr/SubtiList/) and Colibri (http://genolist.pasteur.fr/Colibri/) databases. The top portion shows e values of best hits from BLAST analysis of MviN and conserved proteins involved in peptidoglycan synthesis from E. coli against B. subtilis genome (Subtilist). The bottom portion shows e values of MviN best hit from B. subtilis, YtgP, against B. subtilis genome (Subtilist).
To explore the possibility that YtgP was not the sole MviN homolog in B. subtilis, the YtgP sequence was used as a BLASTp query against the B. subtilis genome. This analysis resulted in the identification of SpoVB, YkvU, and YabM, which were more homologous to each other and to YtgP than to MviN (Fig. 1; Table 2). Consistent with the proposed role of MviN as the lipid II flippase, strains lacking SpoVB are defective in the synthesis of spore cortex peptidoglycan and accumulate peptidoglycan precursors (32).
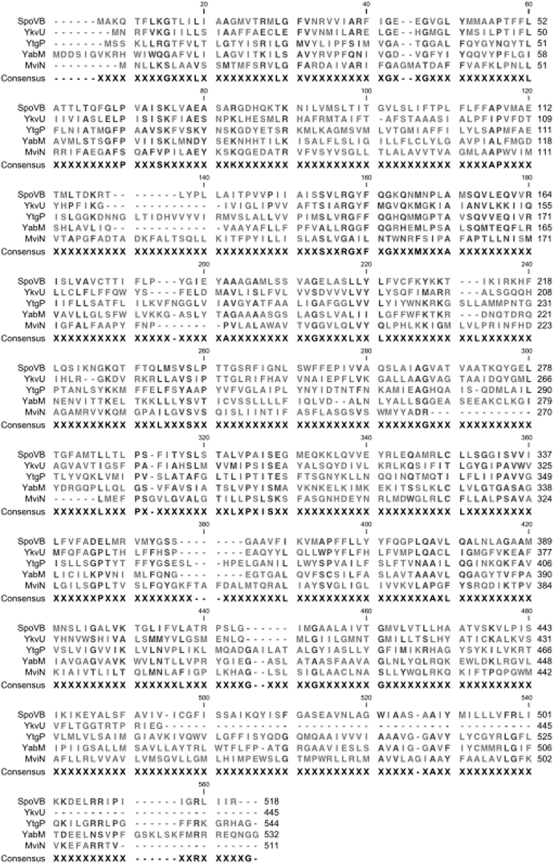
FIG. 1.
Alignment of B. subtilis MviN homologs. A ClustalW alignment of E. coli MviN and B. subtilis SpoVD, YtgP, YabM, and YkvU is shown. The alignment was generated after a BLASTp query of the B. subtilis genome with the E. coli MviN sequence followed by a second BLASTp query using YtgP, the highest-scoring sequence.
While YtgP was the best hit, two additional proteins, YvbV and YtxB, had BLASTp scores greater than those of SpoVB, YkvU, and YabM when MviN was used as the query. Reciprocal BLASTp analysis of YvbV or YtxB against the E. coli genome did not return MviN as a top hit, as did YtgP. YvbV did not return MviN as a significant hit, and YtxB returned MviN as the fifth significant hit (e = 0.042). Both YvbV and YtxB are not predicted to be essential in B. subtilis (17). Since MviN was predicted to contain 14 transmembrane helices by TMHMM (18), true MviN homologs would be expected to have a similar number. YvbV and YtxB contain fewer than 10 predicted transmembrane helices. YvbV and YtxB were therefore excluded from further analysis. We focused on YtgP, SpoVB, and YabM, each with 14 predicted transmembrane helices, and on YkvU, with 12 predicted transmembrane helices. Also, as predicted for MviN, all four proteins have N and C termini predicted to be located in the cytoplasm.
Generation of deletions in genes encoding putative MviN homologs.
Since lipid II translocation is necessary for peptidoglycan biosynthesis, the protein(s) responsible for this process are assumed to be essential. While E. coli MviN is essential (26), the genes encoding the most homologous B. subtilis protein, YtgP, or any of the other putative homologs were not identified during a genome-wide screen of essential B. subtilis genes (17). However, this analysis provided only a limited characterization of these genes and would not identify a gene that has a paralog providing a redundant function. The only B. subtilis MviN homolog that has been closely examined to date is SpoVB, and although a strain carrying a spoVBΔ::tet mutation is blocked in stage V of sporulation and results in the accumulation of peptidoglycan precursors, this strain exhibits no vegetative growth defects (23). However, if SpoVB functioned only during sporulation, one or more of the identified MviN homologs may be necessary for vegetative growth. As a precedent, the SpoVD and SpoVE sporulation specific proteins necessary for peptidoglycan synthesis of the spore cortex have no vegetative phenotypes (5, 13), but their homologs (FtsW/PBP2B and RodA/PBP3) function during growth and are essential.
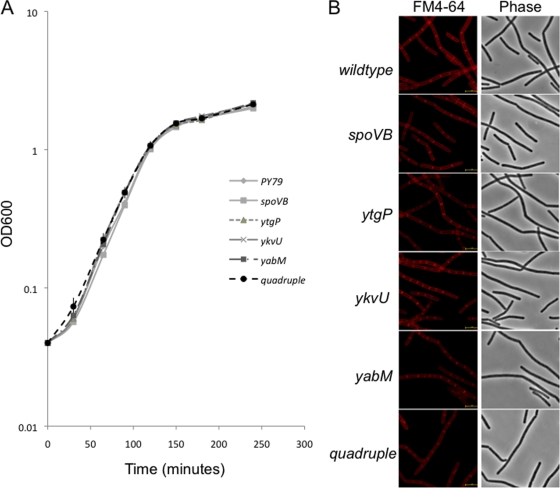
We examined whether the three additional candidate MviN homologs were essential either singly or in combination by generating chromosomal deletions of their genes. Insertions in ykvU have been reported but not a full deletion (1), suggesting that ykvU was not essential. Thus, for ykvU and ytgP, the ribosome binding site and the entire coding region were replaced with an erythromycin resistance gene (33). These strains (and their isogenic relatives that had a spectinomycin resistance gene inserted in place of the erythromycin resistance gene) had normal growth rates (Fig. 2A) and cell morphology (Fig. 2B). Since yabM coding sequences partially overlap with the gene directly downstream, yabN, the last 300 bp of yabM was maintained. Again, this strain exhibited no growth defect in log phase (Fig. 2A), no change in cell morphology (Fig. 2B), and no sporulation defect (see Table 4).
FIG. 2.
Growth of B. subtilis strains carrying mutations in putative MviN homologs. (A) Strains carrying the single mutation ΔytgP::mls (JDB2327), ΔykvU::mls (JDB2330), ΔyabM::cm (JDB2386), or spoVBΔ::tet (JDB1097) or all four mutations (JDB2361) were grown in LB at 37°C following dilution to an optical density at 600 nm (OD600) of 0.04 from an overnight culture grown in LB. OD600 was measured at a series of time points for each of the strains. (B) B. subtilis strains carrying the mutations in single MviN homologs or mutations in all four homologs were grown in LB to an OD600 of 0.9. After collection, cells were resuspended in phosphate-buffered saline with 1 mg/ml FM4-64 and imaged. Bar, 5 mm.
TABLE 4.
Sporulation of B. subtilis strains carrying an inducible MviN allele
| Strain | Genotype | CFUa
|
% Sporulation (±SD) | |
|---|---|---|---|---|
| Preheat | Postheat | |||
| PY79 | Wild type | 5.5 × 108 | 5.0 × 108 | 91 ± 5.7 |
| JDB2327 | ΔytgP::mls | 3.5 × 108 | 3.3 × 108 | 90 ± 2.6 |
| JDB2330 | ΔykvU::mls | 4.7 × 108 | 4.5 × 108 | 89 ± 4.7 |
| JDB2386 | yabM::cm | 3.7 × 108 | 3.5 × 108 | 91 ± 2.7 |
| JDB1097 | spoVBΔ::tet | 1.6 × 108 | 0 | 0 |
| JDB2329 | spoVB-gfp spc | 4.8 × 108 | 4.4 × 108 | 90 ± 2.4 |
| JDB2437 | spoVBΔ::tet amyE::Psweet-spoVB-gfp spc | 3.3 × 108 | 3.0 × 108 | 91 ± 4.9 |
| JDB2506 | amyE::Psweet-mviN spc | 3.7 × 108 | 3.5 × 108 | 92 ± 4.4 |
| JDB2504 | spoVBΔ::tet amyE::Psweet-mviN spc | 3.9 × 108 | 2.4 × 108 | 60 ± 5.8 |
Strains were sporulated in DSM for 24h at 37°C. Cultures were diluted, and CFU were counted before and after exposure to 80°C for 20 min.
While none of the four single mutations produced vegetative growth defects that would be expected for a putative lipid II flippase (Fig. 2), it is possible that they could compensate for the loss of each other. A similar redundancy is seen with the B. subtilis class A PBPs, where single mutations lead to very mild growth defects (7, 22). To investigate this possible redundancy, we generated a quadruple mutant strain (JDB2361) that contained null mutations in ytgP, ykvU, yabM, and spoVB. Transformation efficiencies for the movement of each single mutant allele for the construction of the quadruple mutant strain did not vary significantly from that of control allele markers (Table 3). This strain showed a stage V sporulation defect, consistent with the presence of the spoVB mutation; however, no growth defect or change in cell morphology was seen, as would be expected if the homologs acted redundantly (Fig. 2). Thus, the viability of the quadruple mutant suggests that none of the MviN homologs acts as the putative lipid II flippase.
TABLE 3.
Transformation efficiencies for construction of mutant strainsa
| Donor and/or selected mutation | Recipient | CFU/μg genomic DNA |
|---|---|---|
| amyE::cm | PY79 | 415 |
| JDB1097 (ΔspoVB::tet) | PY79 | 435 |
| JDB2327 (ΔytgP::mls) | PY79 | 489 |
| JDB2330 (ΔykvU::mls) | PY79 | 420 |
| JDB2386 (yabM::cm) | PY79 | 438 |
| sacA::mls | JDB1097 | 280 |
| JDB2327 (ΔytgP::mls) | JDB1097 | 227 |
| amyE::spc | JDB2347 | 354 |
| JDB2355 (ΔykvU::mls:spc) | JDB2347 | 380 |
| amyE::cm | JDB2361 | 335 |
| JDB2386 (yabM::cm) | JDB2361 | 322 |
Competent cells were transformed with 1 μg of donor genomic DNA and selected for the marker listed. No single MviN homolog mutation differed significantly from the control donor genomic DNA. Introduction of the yabM::cm allele into the triple mutant strain carrying the ytgP::mls allele resulted in DNA that did not differ significantly from the control donor DNA. The similar transformation efficiencies obtained indicated that second-site suppressors were probably not generated.
Cellular localization of MviN homologs.
Given the lack of an observable phenotype of strains carrying mutations in the MviN homologs, GFP fusions were generated to further examine their physiological roles. We investigated whether any of the putative homologs were recruited to sites of peptidoglycan synthesis by constructing C-terminal GFP fusions based on the predicted location of both N and C termini of all four homologs inside the cytoplasm. When these fusions were placed at their endogenous loci, the resulting strains did not show any defect in growth or sporulation, including the strain expressing SpoVB-GFP, indicating that this fusion was fully functional for spore cortex formation (Table 4). Since the other three homologs did not have any phenotype when mutated, it was not possible to assess whether the GFP fusions complemented for function.
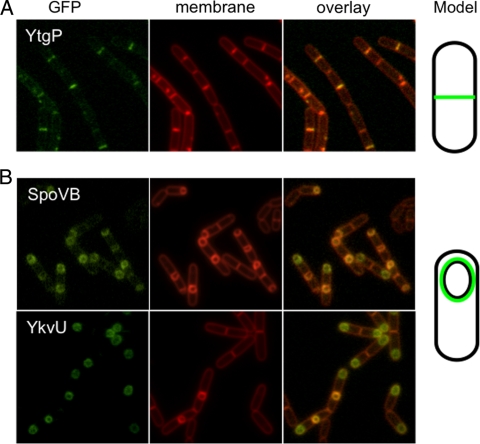
All fusions with the exception of YabM-GFP produced detectable GFP signals. SpoVB-GFP was seen only during sporulation (Fig. 3B), consistent with the sporulation defect observed in a spoVBΔ::tet mutant (23) and with the observation that it is under σE control (32). Recruitment to the outer forespore membrane was scored by taking the ratio of average maximum forespore fluorescence to average maximum mother cell fluorescence (24). SpoVB-GFP is likely at least partially recruited to the outer forespore membrane, although the ratio obtained (2.50 ± 0.57) was lower than that reported for sporulation proteins such as SpoVE (24). YtgP-GFP was observed at the septum during vegetative growth (Fig. 3A), suggesting that it has a role in division. This localization was not unexpected despite a lack of a phenotype for ΔytgP, as E. coli MviN was implicated in growth and septation. YkvU-GFP accumulated at the outer membrane of an early engulfing forespore (Fig. 3B) as previously described (10), and no vegetative signal was detected. While YkvU-GFP was recruited to the outer forespore membrane (3.62 ± 0.64), it does not seem to play a vital role in spore cortex formation, since a strain carrying a ΔykvU mutation produced wild-type levels of heat-resistant spores (Table 4).
FIG. 3.
Localization of GFP fusions. (A) A strain expressing YtgP-GFP (JDB2356) was grown in CH growth medium, cells were collected at an OD600 of 0.7, and the membranes were stained with FM4-64. (B) Strains expressing SpoVB-GFP (JDB2329) and YkvU-GFP (JDB2358) were sporulated by resuspension in A+B medium, cells were collected at T3 of sporulation, and membranes were stained with FM4-64.
MviN homologs require additional sporulation proteins for localization.
Since the targeting of proteins to the outer forespore membrane during sporulation is the subject of much interest (3), we investigated whether other proteins were necessary for the recruitment of YkvU and SpoVB. For example, SpoVD, like SpoVB, is required for the spore cortex synthesis, and its localization to this membrane is dependent on SpoVE (A. Fay and J. Dworkin, unpublished data). However, both YkvU-GFP and SpoVB-GFP were recruited appropriately in the absence of SpoVE. We examined their localization in a ΔspoIIIAH background, since SpoIIIAH is necessary for the proper recruitment of SpoIVFA to the forespore (3, 8). SpoVB-GFP was recruited to the outer forespore membrane in a SpoIIIAH-dependent manner, since the ratio was significantly lower in a spoIIIAH mutant strain (P < 0.001) than in the wild type (Fig. 4A). Furthermore, SpoVE-GFP was recruited to the outer forespore membrane in a spoVBΔ::tet background (data not shown), suggesting that these proteins localize independently of each other and that defects in spore cortex formation are not due merely to a localization effect on each other. YkvU-GFP followed a similar pattern, in which it failed to be recruited in the absence of SpoIIIAH, with a ratio of 2.09 ± 0.38 (P < 0.001), but was recruited in the absence of SpoVE, with a ratio of 3.71 ± 0.54 (Fig. 4B).
FIG. 4.
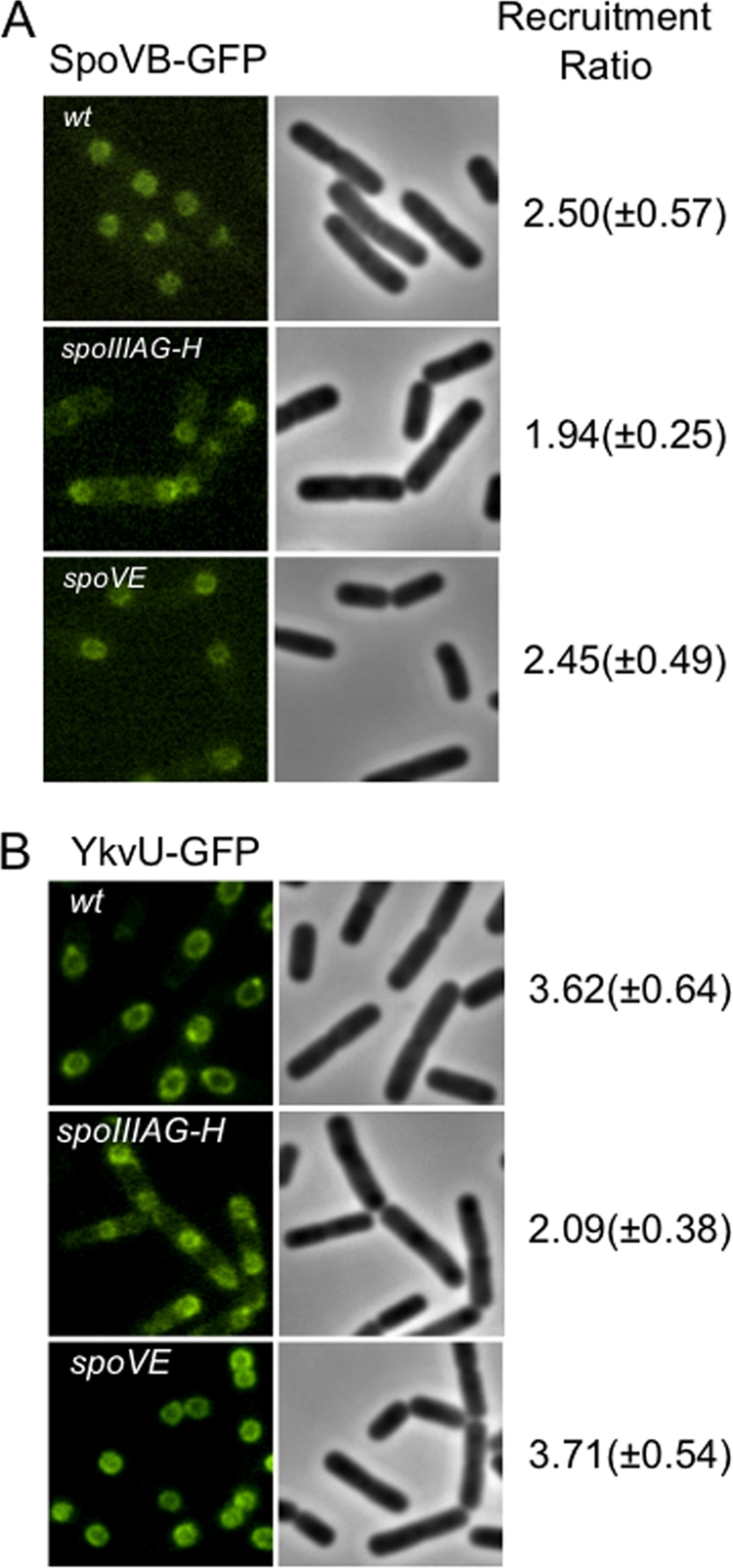
SpoVB-GFP and YkvU-GFP targeting requires additional sporulation-specific proteins. (A) SpoVB-GFP-expressing strains JDB2329 (wt), JDB2416 (spoIIIAG-H::kan), and JDB2418 (spoVE::tet) were sporulated by resuspension, and images were taken at 3 h. (B) YkvU-GFP-expressing strains JDB2358 (wt), JDB2377 (spoIIIAG-H::kan), and JDB2378 (spoVE::tet) were sporulated by resuspension, and images were taken at T3. Recruitment ratios are the ratios of the average maximum forespore fluorescence to the average maximum mother cell fluorescence.
A spoVBΔ::tet mutant is not rescued by overexpression of UppS.
A plasmid-borne copy of uppS, encoding the undecaprenyl pyrophosphate synthase, rescues the growth defect at the nonpermissive temperature of an E. coli strain carrying an mviN temperature-sensitive mutation (16). This mutation lies in the mviN promoter region and likely leads to a decrease in MviN levels. Since the undecaprenyl chain is used to form a number of metabolic intermediates, including lipid I (the immediate precursor of lipid II), this result suggests that MviN is involved in lipid II synthesis. We therefore investigated whether a spoVBΔ::tet mutation could be similarly rescued by constructing a merodiploid strain that carried an additional, inducible copy of uppS. However, the sporulation defect of the spoVBΔ::tet background was not rescued by overexpression of UppS or UppS-FLAG (data not shown). We confirmed that UppS was overexpressed by introducing a FLAG tag into the construct (data not shown). While the inability of UppS to rescue the spoVBΔ::tet mutation may indicate that SpoVB and MviN are not true homologs, this may be due to unrelated reasons. The lack of a detectable phenotype of mutations in any of the other candidate MviN homologs excludes examining their possible suppression by UppS.
MviN complements SpoVB function during sporulation.
SpoVB is essential for spore formation, since a strain carrying a spoVB deletion does not produce any heat-resistant spores (23) (Table 4). This phenotype is complemented by expression of a SpoVB-GFP fusion (Table 4). We demonstrated that MviN (MurJ) was a homolog of SpoVB by placing MviN under inducible (Psweet) control in a ΔspoVB strain. In the absence of inducer, no spores were observed in this strain, similar to what was observed with the ΔspoVB parental strain (Table 4). However, addition of inducer resulted in spore production by this strain that was only slightly reduced compared to that of the wild type (Table 4). This complementation indicates that MviN (MurJ) and SpoVB are homologs.
YtgP or SpoVB complements the growth defect of an E. coli MviN depletion strain.
An E. coli strain carrying MviN under inducible (PBAD) control is dependent on arabinose for growth (16, 26). To determine whether the growth defect in the absence of arabinose could be complemented by a B. subtilis homolog, we introduced into this strain a plasmid containing a C-terminal GFP fusion of one of these homologs, YtgP, under the control of Ptac. This strain was now able to grow in the absence of arabinose, unlike the parent strain, indicating that YtgP complemented this growth defect (Fig. 5). We also introduced into the MviN depletion strain a plasmid containing a C-terminal GFP fusion to a second homolog, SpoVB, under Ptac control. As with YtgP, this strain was able to grow in the absence of arabinose, unlike the parent strain, indicating that SpoVB also complemented this growth defect (Fig. 5). Lethality was noted when YtgP-GFP or SpoVB-GFP expression was induced in parental strain MG1665 and the MviN depletion strain; however, low levels of leaky expression were not. All complementation analysis was therefore performed without addition of the inducer IPTG to avoid this toxicity (data not shown). Thus, two of the B. subtilis homologs identified by BLAST (Fig. 1; Table 2) complemented the lethality resulting from MviN depletion, even at the low levels of induction resulting from read-through transcription. Taken together, these genetic complementation experiments demonstrate that the B. subtilis proteins we identified by bioinformatics are likely to be true MviN homologs.
FIG. 5.
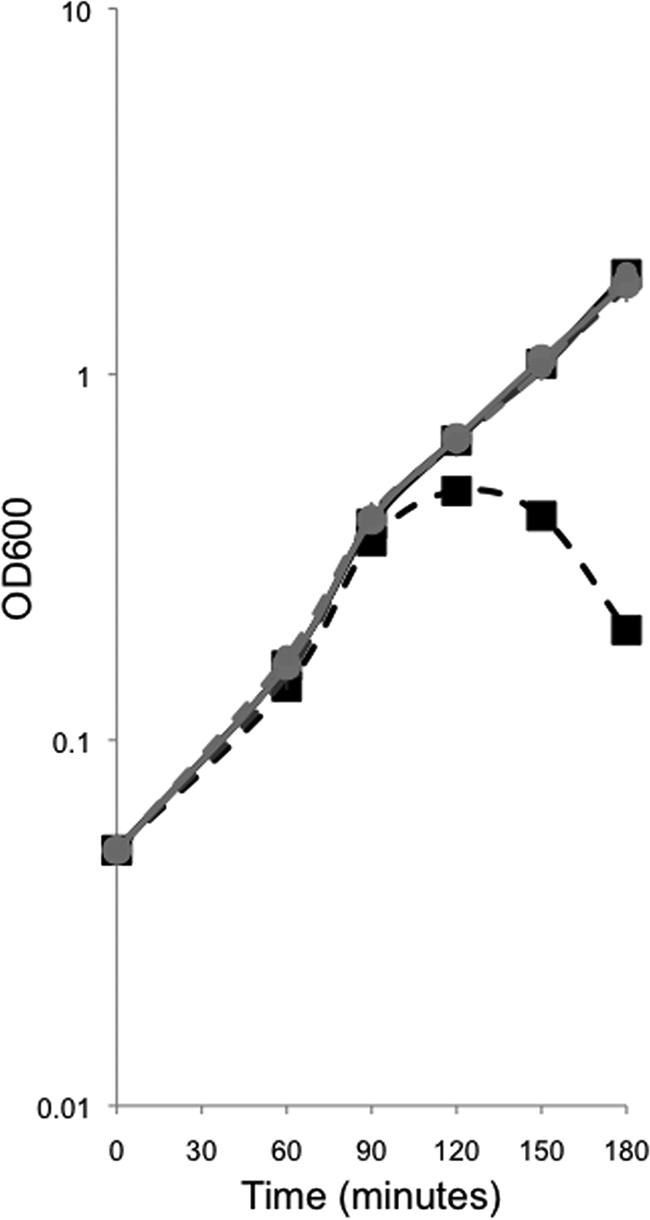
SpoVB-GFP and YtgP-GFP rescue the mviN depletion mutant. JDE1148 (pMMB207) (▪), JDE1140 (pAF374) (+), and JDE1147 (pAF375) (•) were depleted for mviN by growth in LB in the absence (dashed) or presence (solid) of the inducer arabinose. The presence of pMMB207 did not affect mviN depletion, as the strain required arabinose for growth and IPTG had no effect. Leaky expression of YtgP-GFP or SpoVB-GFP rescued the mviN mutation, as seen by growth without arabinose.
DISCUSSION
Very recently, it was proposed that the essential MviN protein of E. coli is the lipid II flippase in this organism (26). Consistent with this hypothesis, an mviN temperature-sensitive mutation results in the accumulation of peptidoglycan precursors and cell lysis, similarly to depletion of MviN (16). If MviN is the lipid II flippase, then MviN homologs in organisms that also utilize lipid II are likely to be essential as well. We examined this possibility in B. subtilis, a gram-positive organism that, like E. coli, uses diaminopimelic acid-containing lipid II, by identifying putative MviN homologs using BLASTp-based analyses. Four proteins showed significant homology either to MviN or to each other and were predicted to contain a number of transmembrane sequences similar to that in MviN (Fig. 1). We confirmed that MviN complemented the sporulation defect of a B. subtilis strain carrying a null mutation in SpoVB, one of these homologs (Table 4). In addition, two of these homologs, YtgP and SpoVB, complemented the growth defect of an E. coli strain carrying an inducible allele of MviN (Fig. 5). Thus, these four proteins are likely to be real MviN (MurJ) homologs. Recently, an essential YtgP homolog from Streptococcus pyogenes, Spy_0390, was shown to complement MviN depletion strains of E. coli (27), although its physiological role in S. pyogenes is not well understood.
Strains carrying mutations in each or all of these homologs did not exhibit any shape or growth rate defects (Fig. 2). This result is inconsistent with the expected requirement for lipid II flipping for growth and suggests that other proteins mediate this function. It is formally possible, however, that an additional unidentified gene that lacks significant sequence homology to MviN could be functionally redundant for lipid II flipping.
If these MviN homologs are not lipid II flippases, what role do they play in B. subtilis physiology? SpoVB is necessary for spore cortex formation, and a ΔspoVB mutation results in the accumulation of peptidoglycan precursors (32). Further, the localization of SpoVB-GFP to the outer forespore membrane (Fig. 3) is consistent with a role in spore peptidoglycan synthesis. However, this localization does not depend on SpoVE, another protein essential for spore peptidoglycan synthesis (Fig. 4). This lack of dependence is different from that observed with another protein, SpoVD, that is also essential for spore peptidoglycan synthesis, suggesting that SpoVB and SpoVE/SpoVD are present in different complexes and therefore may mediate different functions.
The recruitment of YtgP-GFP to septa (Fig. 3) indicates a possible role in division, although it is not essential and vegetative cells lacking YtgP divide and grow normally (Fig. 2). Mutagenesis of Listeria monocytogenes identified YtgP as a virulence factor in a mouse model (2). This particular screen identified several other genes involved in peptidoglycan modifications, suggesting that YtgP may play a role in such processes. YkvU localizes to the outer forespore membrane (Fig. 3), and its dependence on SpoIIIAH for this targeting (Fig. 4) suggests that it may play a role in late spore formation and/or germination (1, 15), although it is not required for the production of heat-resistant spores. Our failure to observe any specific localization of YabM-GFP and the lack of any observable phenotype associated with a ΔyabM mutation make it difficult to identify a physiological role for this protein.
Our data do not rule out the possibility that these MviN homologs play an accessory but nonessential role in lipid II translocation. Recently, it was suggested that the yeast protein Rft1, a member of the family that includes MviN, may not act as a flippase directly but rather may facilitate glycolipid flipping (11), since in vitro flippase activity appeared to result from a protein or proteins other than Rft1 (28). By analogy, E. coli MviN and the B. subtilis MviN homologs we have characterized would have a role in peptidoglycan synthesis that is essential in E. coli but that is not essential in B. subtilis.
Alternatively, MviN could be involved in a cell wall modification that utilizes the isoprenoid chain that is also a component of lipid II. Thus, cell lysis would result from an indirect defect in the recycling of the isoprenoid chain and not from a direct defect in peptidoglycan synthesis. A similar scenario occurs in the teichoic acid synthesis pathway in B. subtilis, which was long thought to be essential. However, strains lacking TagO, the first enzyme in the pathway, are viable, and mutations of enzymes later in the pathway are probably lethal due to their sequestration of the isoprenoid chain (6). It is therefore possible that the suppression of the mviN temperature-sensitive mutant by uppS, the gene encoding the undecaprenyl pyrophosphate synthetase, is similarly the result of sequestration of the isoprenoid chain component of lipid II. This suggestion is reinforced by the ability of either YtgP or SpoVB to complement the growth defect of a strain carrying an inducible allele of MviN but only in the absence of induction. This lethality suggests that the overexpression of YtgP or SpoVB depletes the cell of an essential substrate, such as the isoprenoid chain. Regardless of the biochemical activity of these proteins, the observation that they are not required for growth indicates that they are not likely to mediate the essential process of lipid II flipping.
Supplementary Material
Acknowledgments
We thank David Popham for generously communicating results prior to publication. We thank Natasha Ruiz (Princeton) for the kind gift of E. coli strain NR1154. We also thank members of our laboratory for helpful suggestions and Howard Shuman for comments on the manuscript.
This work was funded by NIH grant R01GM081368-01 and by an Irma T. Hirschl Scholar award to J.D. A.J.F. was supported by NIH training grant T32AI07161-29.
Footnotes
Published ahead of print on 7 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aertsen, A., I. Van Opstal, S. C. Vanmuysen, E. Y. Wuytack, and C. W. Michiels. 2005. Screening for Bacillus subtilis mutants deficient in pressure induced spore germination: identification of ykvU as a novel germination gene. FEMS Microbiol. Lett. 243:385-391. [DOI] [PubMed] [Google Scholar]
- 2.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaylock, B., X. Jiang, A. Rubio, C. P. Moran, Jr., and K. Pogliano. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 18:2916-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carsiotis, M., B. A. Stocker, D. L. Weinstein, and A. D. O'Brien. 1989. A Salmonella typhimurium virulence gene linked to flg. Infect. Immun. 57:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 6.D'Elia, M. A., K. E. Millar, T. J. Beveridge, and E. D. Brown. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 188:8313-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doan, T., K. A. Marquis, and D. Z. Rudner. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55:1767-1781. [DOI] [PubMed] [Google Scholar]
- 9.Eichenberger, P., P. Fawcett, and R. Losick. 2001. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol. 42:1147-1162. [DOI] [PubMed] [Google Scholar]
- 10.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 11.Frank, C. G., S. Sanyal, J. S. Rush, C. J. Waechter, and A. K. Menon. 2008. Does Rft1 flip an N-glycan lipid precursor? Nature 454:E3-4, E4-5. [DOI] [PubMed] [Google Scholar]
- 12.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. Wiley, New York, NY.
- 13.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 14.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 15.Imamura, D., K. Kobayashi, J. Sekiguchi, N. Ogasawara, M. Takeuchi, and T. Sato. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 186:5450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, A., Y. Murata, H. Takahashi, N. Tsuji, S. Fujisaki, and J. Kato. 2008. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J. Bacteriol. 190:7298-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 19.Lambert, C., M. C. Smith, and R. E. Sockett. 2003. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5:127-132. [DOI] [PubMed] [Google Scholar]
- 20.Ling, J. M., R. A. Moore, M. G. Surette, and D. E. Woods. 2006. The mviN homolog in Burkholderia pseudomallei is essential for viability and virulence. Can. J. Microbiol. 52:831-842. [DOI] [PubMed] [Google Scholar]
- 21.O'Connell, K. P., S. J. Raffel, B. J. Saville, and J. Handelsman. 1998. Mutants of Rhizobium tropici strain CIAT899 that do not induce chlorosis in plants. Microbiology 144(Pt. 9):2607-2617. [DOI] [PubMed] [Google Scholar]
- 22.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popham, D. L., and P. Stragier. 1991. Cloning, characterization, and expression of the spoVB gene of Bacillus subtilis. J. Bacteriol. 173:7942-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Real, G., A. Fay, A. Eldar, S. M. Pinto, A. O. Henriques, and J. Dworkin. 2008. Determinants for the subcellular localization and function of a nonessential SEDS protein. J. Bacteriol. 190:363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudnick, P. A., T. Arcondeguy, C. K. Kennedy, and D. Kahn. 2001. glnD and mviN are genes of an essential operon in Sinorhizobium meliloti. J. Bacteriol. 183:2682-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz, N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. USA 105:15553-15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz, N. 2009. Streptococcus pyogenes YtgP (Spy_0390) complements Escherichia coli strains depleted of the putative peptidoglycan flippase MurJ. Antimicrob. Agents Chemother. 53:3604-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanyal, S., C. G. Frank, and A. K. Menon. 2008. Distinct flippases translocate glycerophospholipids and oligosaccharide diphosphate dolichols across the endoplasmic reticulum. Biochemistry 47:7937-7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dam, V., R. Sijbrandi, M. Kol, E. Swiezewska, B. de Kruijff, and E. Breukink. 2007. Transmembrane transport of peptidoglycan precursors across model and bacterial membranes. Mol. Microbiol. 64:1105-1114. [DOI] [PubMed] [Google Scholar]
- 32.Vasudevan, P., A. Weaver, E. D. Reichert, S. D. Linnstaedt, and D. L. Popham. 2007. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol. Microbiol. 65:1582-1594. [DOI] [PubMed] [Google Scholar]
- 33.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 34.Youngman, P., J. B. Perkins, and R. Losick. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet. 195:424-433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.