Abstract
In some enterobacterial pathogens, but not in Escherichia coli, loss-of-function mutations are a common route to clinically relevant β-lactam antibiotic resistance. We previously constructed an assay system for studying enterobacterial β-lactam resistance mutations using the well-developed genetics of E. coli by integrating enterobacterial ampRC genes into the E. coli chromosome. Like the cells of other enterobacteria, E. coli cells acquire β-lactam resistance by ampD mutation. Here we show that starvation and stress responses provoke ampD β-lactam resistance mutagenesis. When starved on lactose medium, Lac− strains used in mutagenesis studies accumulate ampD β-lactam resistance mutations independent of Lac reversion. DNA double-strand break repair (DSBR) proteins and the SOS and RpoS stress responses are required for this mutagenesis, in agreement with the results obtained for lac reversion in these cells. Surprisingly, the stress-induced ampD mutations require DinB (DNA polymerase IV) and partially require error-prone DNA polymerase V, unlike lac mutagenesis, which requires only DinB. This assay demonstrates that real-world stressors, such as starvation, can induce clinically relevant resistance mutations. Finally, we used the ampD system to observe the true forward-mutation sequence spectrum of DSBR-associated stress-induced mutagenesis, for which previously only frameshift reversions were studied. We found that base substitutions outnumber frameshift mutations, as seen in other experimental systems showing stress-induced mutagenesis. The important evolutionary implication is that not only loss-of-function mutations but also change-of-function mutations can be generated by this mechanism.
Mutation is a primary cause of bacterial resistance to antibiotics. Some mutations promote resistance directly, such as Escherichia coli gyrA and gyrB mutations, which confer quinolone resistance (28). Other mutations ameliorate the otherwise deleterious effects on cell growth conferred by some antibiotic resistance mutations (34). Yet other mutations increase the mutation rate, thereby increasing the likelihood of acquiring a resistance mutation (71). Although antibiotic resistance is a major problem in modern medicine (75) and has been studied intensively, the mechanisms that generate resistance mutations are poorly understood.
Of particular interest in the clinical setting is whether environmental conditions encountered during bacterial infections might promote resistance mutagenesis. For example, variables such as antibiotic exposure can stimulate resistance mutagenesis (1, 10, 60, 64), and various natural environments have been shown to select cells with permanently increased general mutation rates due to mutator mutations (42, 50, 59). Presumably, these mutator mutations are selected because they promote rapid adjustment to changing environments, even though most mutations generated are likely to be deleterious. It is noteworthy that in the studies in which a “high” incidence of mutator mutants was found in commensal and pathogenic bacteria (42, 50, 59), although mutagenesis promotes adaptation, most of the colonizing bacteria were not mutator mutants, indicating that most cells adapted without permanent increases in the mutation rate (66). Environmental stresses have been demonstrated to induce transient, generally mutagenic pathways (for a review, see reference 21). For these reasons, common environmental stressors, such as starvation, have been postulated to stimulate mutagenesis leading to antibiotic resistance (34, 49). In this work we tested this idea directly and showed that starvation stress-induced mutagenesis can indeed induce β-lactam antibiotic resistance.
We studied β-lactam antibiotic resistance using an E. coli model system. β-Lactamases are enzymes that cleave and inactivate β-lactam antibiotics, promoting resistance. Chromosomally encoded AmpC β-lactamases confer β-lactam resistance in many pathogenic and opportunistic bacteria and are ubiquitous in all enterobacteria except the salmonellae, klebsiellae, and some others (33). ampC expression is inducible in all enterobacteria but E. coli and the shigellae (46). In inducible strains, ampC transcription is activated by the AmpR transcriptional activator (3) upon AmpR binding to its allosteric activator molecule, 1-6-anhydromuropeptide (30). AmpD converts the activator molecule to the blocker UDP-N-acetylmuramic acid-pentapeptide, which then binds AmpR and blocks ampC transcription (30). Thus, loss-of-function mutations in ampD lead to constitutive AmpC β-lactamase production and β-lactam resistance (31, 32, 43). ampD missense and nonsense mutations are common in AmpC-mediated β-lactam-resistant clinical isolates (40, 67).
Normally, E. coli lacks ampR and the ampC promoter that it controls (23) and so cannot become resistant via ampD mutation. However, E. coli cells carrying the ampRC genes of other enterobacteria in plasmids (44, 57) or in the chromosome (61) become resistant due to ampD loss-of-function mutations. We constructed a chromosomal model of ampRC-mediated β-lactam resistance to facilitate the use of E. coli genetics to study mutagenesis. This model has the following advantages: (i) the single-copy ampRC locus more closely resembles the situation in clinical isolates and (ii) the chromosomal ampRC system is not affected by mutations that increase the plasmid copy number so that (iii) it is possible to examine effects of mutations in genes affecting DNA repair-recombination and mutation, some of which destabilize plasmid replication (61). In this model, most or all β-lactam resistance mutations are ampD mutations, similar to those seen clinically (61).
We wanted to use the ampRC system as a forward mutational assay in the context of the well-characterized stress-induced mutagenesis in the Lac assay of E. coli (for a review, see reference 21). In this way, the experimental conditions could be replicated, and the results obtained could be directly integrated into the vast amount of data already available for the stress-induced mutagenesis pathway in this strain. Here, we used this E. coli model of enterobacterial β-lactam resistance mutation to address two problems. First, we demonstrated that starvation stress-induced mutagenesis mechanisms can induce β-lactam resistance in this clinically relevant model. Because starvation stress is thought to be a major feature of natural environments encountered by pathogens (14), this indicates that our extensive knowledge of stress-induced mutagenesis mechanisms and environmental conditions that induce them might be relevant to the generation of resistance mutations in nature.
Second, we show that the stress-induced mutagenesis mechanism that can generate a β-lactam resistance mutagenesis is a double-strand break (DSB) repair (DSBR)-associated mutagenesis mechanism, which is a mechanism that has been studied in detail using the E. coli Lac system (for a review, see reference 21), and we used the ampD assay to reveal the true sequence spectrum for DSBR-associated mutagenesis. Stress-induced mutagenesis in the Lac system requires DSBR proteins, the SOS DNA damage response, DinB error-prone DNA polymerase V (Pol IV), and the RpoS stationary-phase and general stress response transcriptional activator, and it results from a switch from high-fidelity DSBR to error-prone DSBR during stress (62). The requirement for an error-prone DNA polymerase and the requirement for stress responses are two features of mutagenesis in the Lac assay which are also found in many other stress-inducible mutagenesis mechanisms in E. coli and other species. DinB DNA polymerase is required for ciprofloxacin-induced resistance mutations in E. coli (10) and stationary-phase mutagenesis in starved cells of both Pseudomonas putida (38, 74) and Bacillus subtilis (70). The SOS response is required for ciprofloxacin-induced resistance mutations (10), as well as mutagenesis in aging colonies (72), in E. coli. RpoS is required for starvation-induced mutagenesis in P. putida (29), for mutagenesis in aging E. coli cells (5), and for stress-induced transposition-mediated deletions in E. coli (22). Another stress response, the competence regulon controlled by the comA and comK gene products, regulates mutagenesis in starved B. subtilis cells (69). Therefore, although several different stress-induced mutagenesis mechanisms seem to occur in nature, the Lac system shares many important features with many of these mechanisms and so is a reasonable general model.
Previously, although base substitution mutations were observed to be the products of other stress-induced mutagenesis mechanisms (10, 72), the mutations that were shown to be generated by the starvation stress-induced DSBR-associated mutagenic mechanism were only frameshift (−1 deletion) mutations. Only mutations that reverted frameshift alleles have been studied previously (7, 15, 16, 65, 76), and the DinB DNA polymerase required makes mostly −1 bp deletions when it is overproduced in vivo (36, 77). The ampD assay selects any loss-of-function mutation, and with this assay we found that stress-induced base substitutions outnumber frameshift mutations. The important evolutionary implication is that not only loss-of-function mutations but also change-of-function mutations can be generated by this stress-induced mutagenesis mechanism, as seen previously for quinolone-induced resistance mutations (10) and mutations in aging colonies (72).
MATERIALS AND METHODS
Strains and plasmids.
The E. coli K-12 strains and plasmids used in this study are shown in Table 1. Phage P1-mediated transduction was performed using standard techniques (54).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| CS85 | ruvC53 eda-51::Tn10 | 68 |
| DM49 | lexA3(Ind−) malB::Tn9 | 45 |
| FC29 | Δ(lac-proAB)XIIIthi ara [F′ proAB+ Δ(lacI-lacZ)] | 8 |
| FC526 | ΔrecG263::Kan | 18 |
| JW2711 | ΔrpoS::FRTKanFRT | 2 |
| N2731 | recG258::Tn10mini-Kan | 47 |
| RW120 | ΔumuDC595::cat | 79 |
| SMR580 | FC40 recB21 argA::Tn10 | 24 |
| SMR593 | FC40 recB21 | 24 |
| SMR4562 | Δ(lac-pro)XIIIthi ara Rifr [F′ α45 lacIqlacI33ΩlacZ] (genotype identical to that of FC40, independent construction) | 52 |
| SMR5201 | Δattλ::ampRC | 61 |
| SMR5222 | SMR4562 Δattλ::ampRC | 61 |
| SMR5225 | SMR5222 Δ(srlR-recA)306::Tn10 | 61 |
| SMR5228 | SMR5222 recB21 argA::Tn10 | SMR5222 × P1(SMR580) |
| SMR5522 | FC29 ΔruvC64::Kan | 7 |
| SMR5578 | SMR5222 ΔrecG263::Kan | 61 |
| SMR5652 | SMR5222 ΔrecG263::Kan Δ(srlR-recA)306::Tn10 | 61 |
| SMR6064 | SMR4562 dinB10 (λxis1 cIts857) [F′ dinB10] | 53 |
| SMR6371 | SMR5222 recB21 | SMR5228 × P1(SMR593) |
| SMR6373 | SMR4562 dinB10 Δattλ::ampRC [F′ dinB10] | SMR6064 × P1(SMR5201) |
| SMR6485 | SMR5222 ruvC53 eda-51::Tn10 | SMR5222 × P1(CS85) |
| SMR6487 | SMR5222 recB21 recG258::Tn10mini-Kan | SMR6371 × P1(N2731) |
| SMR6927 | SMR5222 ΔrecG263::Kan ruvC53 eda-51::Tn10 | SMR5578 × P1(CS85) |
| SMR7055 | SMR5222 ΔrecG263::Kan lexA3(Ind−) malB::Tn9 | SMR5578 × P1(DM49) |
| SMR10316 | SMR5222 ΔumuDC595::cat | SMR5222 × P1(RW120) |
| SMR10317 | SMR5222 ΔrpoS::FRTKanFRT | SMR5222 × P1(JW2711) |
| Plasmids | ||
| pJP2 | pTGV-Light ampRC+ | 61 |
| pJP19 | pACYC184 ampD+ | 61 |
| pKD46 | ori101 repA101(Ts) pBAD-gam-bet-exo AmpraraC+ on plasmid | 13 |
Assay of stress-induced ampD mutations during starvation on lactose with lac strains.
We used a modification of the procedure of Bull et al., who assayed chromosomal reversions of a tet frameshift allele during Lac assay experiments (7). For each strain tested, six independent 5-ml cultures were grown to saturation in minimal M9-glycerol medium (1 × 109 to 2 × 109 cells per ml), washed, and aliquoted in individual plating tubes, and a 20-fold excess of scavenger cells (cells of lac deletion strains that scavenge contaminating carbon sources [FC29 and SMR5522 when ruvC strains were tested]) was added to the “tester” cells. The cells were plated onto plates containing M9 medium with 10 μg/ml vitamin B1 and 0.1% lactose in 2.5 ml of M9 top agar containing vitamin B1 and lactose, and then they were overlaid with a second 2.5-ml layer of M9 top agar containing vitamin B1 and lactose. At least 12 plates were generated for each culture (6 plates with 1× “tester” [ampRC mutation assay] cells and 6 plates with 2× “tester” cells) and then incubated at 37°C (or 30°C for experiments with poorly growing ruv recG strains). Every day or every other day, starting with the first day that cells were plated (day 0), one pair of plates per culture per strain was overlaid with 5 ml of M9 top agar containing 0.12 ml 50% glycerol, 20 μl of 100 mg/ml ampicillin (final concentration, 50 μg/ml), and 10 μl of 20 mg/ml 5-bromo-4-chloro-3-indolyl-β-galactoside (X-Gal) (final concentration, 5 μg/ml). The ampicillin-containing plates were then incubated for an additional 3 days to allow growth of all Ampr cells into colonies. After 3 days, white (Lac−) colonies were counted. These colonies represented the number of cells that had mutated to Ampr at any point in time between inoculation of the original culture and addition of the final ampicillin-glycerol overlay. In early experiments, ampicillin resistance was confirmed by patching colonies onto plates containing Luria-Bertani-Herskowitz medium (1% tryptone, 0.5% NaCl, 0.5% yeast extract, 2 μg/ml thymine, solidified with 1.5% agar) with ampicillin. Ampicillin resistance was attributed to mutation in the ampD gene by complementation with plasmid pJP19 (ampD+) (61). Blue (Lac+) Ampr colonies were avoided, because the resistance mutation could have occurred during growth of the colony once the cell mutated to Lac+. For all stress-induced Ampr mutation experiments, a Lac+ reversion assay (25) was performed in parallel to confirm that the strains tested reverted at expected rates (18, 24, 25, 53). The net cell viability (assayed as described by Harris et al. [25]) monitored during the experiments varied less than 2-fold for all experiments reported.
Sequencing ampD mutations.
On day 5 or later Ampr mutants were obtained as described above and purified on Luria-Bertani-Herskowitz medium containing 100 μg/ml ampicillin. The ampD gene was amplified using primers AmpD no. 1 (5′-GGGTTTTCATGAGAGGCGGCATGTTAAAACTCCAG-3′) and AmpD no. 2 (5′-GGGTTTAAGCTTTCATGTTGT-3′). The PCR products were sequenced (Lone Star Labs, Houston, TX) using primers AmpD no. 3 (5′GCGCGTCTCCGCTCACTGTTT-3′) and AmpD no. 4 (5′-GCATGCCATGCACGTTTATCG-3′).
RESULTS
Accumulation of ampD β-lactam resistance mutations during carbon starvation.
We moved the chromosomal ampRC+ system (61) into E. coli strains used to assay starvation stress-induced reversion of a lac +1 frameshift allele in an F′ episome during starvation (8). When spread on lactose minimal medium, the cells accumulate frameshift reversions in the F′-borne lac gene (16, 65) or a tet gene located either in F′ (15) or in the chromosome (7). This starvation-induced process requires the RpoS stationary-phase starvation and general stress response transcriptional activator (41, 48) and operates by a mechanism distinct from lac reversion in rapidly growing cells (see Introduction and below).
We found that prolonged starvation of the Lac− cells on lactose medium leads to accumulation of mutants resistant to the β-lactam antibiotic ampicillin (Ampr mutants, rec+) (Fig. 1A, C, and E). The mutations are recessive, forward mutations in the chromosomal ampD gene, as shown by complementation with an ampD+ plasmid (see Materials and Methods) and sequencing (see below). The Ampr mutants accumulate over several days of starvation on lactose (Fig. 1A to F). The increase is similar to that observed for reversion of a chromosomal tet frameshift (7), but the frequency of Ampr mutants is about 10-fold higher, perhaps because any loss-of-function mutation in the ∼580-bp ampD gene confers Ampr, whereas only frameshift reversions in a 5-bp window restore the tet+ function. The frequency of Ampr mutants is ∼2-fold lower than that of the F′ lac frameshift revertants in this system, which arise from frameshift mutations in a ∼125-bp region (16, 65).
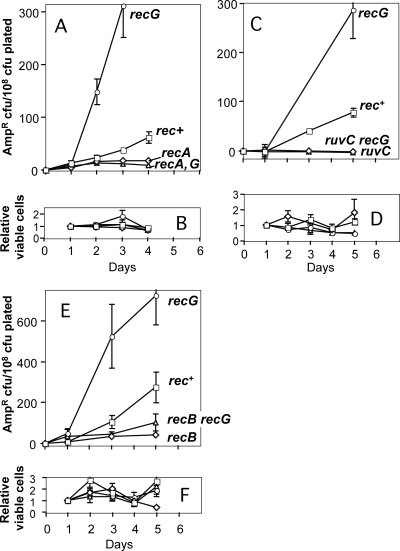
FIG. 1.
DSBR protein-dependent stationary-phase mutation to ampicillin resistance (Ampr) during prolonged starvation. (A, C, and E) Lac assay strains were starved on lactose medium for several days, and replicate plates were removed each day, overlaid with agar containing another carbon source and ampicillin, and reincubated to allow growth of Ampr mutant colonies. The data are the means ± standard errors of the means of six independent cultures per strain assayed in parallel. Each graph is representative of at least three repetitions. The days indicated are the days on which the ampicillin overlay was performed, and the data show the number of Ampr cells that were present on that day. The counts for day 0 (for the generation-dependent mutants present when the cultures were plated) were subtracted from the counts for stress-induced ampD mutants to more clearly show the increases in the numbers of mutants (when present) over time. Negative values were obtained when the average number of mutants from one set of replicate plates overlaid on a day was lower than the number of mutants for the set of plates overlaid on day 0. In all experiments, the rec+ strain (open squares) was SMR5222, and the recG strain (open circles) was SMR5578. (A) Ampr mutants accumulate during prolonged starvation on lactose. RecA is required for, and RecG inhibits, formation of these mutants. Open diamonds, recA strain SMR5225; open triangles, recA recG strain SMR5652. (C) Ampr stationary-phase mutation requires ruvC. Open diamonds, ruvC strain SMR6485; open triangles, ruvC recG strain SMR6927. (E) Ampr stationary-phase mutation requires recB. Open diamonds, recB strain SMR5228; open triangles, recB recG strain SMR6487. (B, D, and F) Daily levels of Lac− viable cells on the plates, normalized to the day 1 count, showing little net growth or death. For each panel the symbols are the same as those for the panel above it.
DSBR proteins in stationary-phase Ampr mutation.
We found that, like stress-induced lac reversion in the F′ (18, 24, 25) and Tet reversion in the chromosome (7), stationary-phase accumulation of Ampr mutants requires homologous recombination (HR) and DSBR proteins RecA, RecBC, and RuvC and is elevated in cells lacking the RecG protein (Fig. 1A to F). recG cells show 10- to 25-fold more Lac+ colonies (18, 25) and 4- to 10-fold more Ampr mutants by day 4 (Fig. 1). The requirement for RecB, which is specific for double-strand ends (DSEs), is the first evidence implicating DSEs in stress-induced chromosomal mutagenesis.
Previously (18, 25) and in this study, we and other workers found that loss of RecG increases mutagenesis via a mechanism that is similar to or the same as the mutagenesis mechanism that operates in Rec+ cells, a mechanism that requires the RecA, RecBC, and Ruv proteins (Fig. 1A to F). The current model for DSBR-associated mutagenesis indicates that invading 3′ ends are the priming site for new DNA synthesis that leads to mutations (for a review, see reference 21). The inhibitory effect of RecG in DSBR-associated mutagenesis is probably due to the unwinding and dissociation of this intermediate that primes DNA synthesis (18, 25), an hypothesis which is further supported by the ability of this protein to perform similar reactions in vitro (51). Because the mutagenesis mechanism operating in recG and wild-type cells is similar, recG cells provided a sensitive way to compare the stress-induced mutagenesis phenotypes of other alleles in these experiments and some of the other experiments described below. recG cells have been used similarly as a sensitizing background in screens for stress-induced mutagenesis-defective mutants (41).
Stress responses in stationary-phase Ampr mutation.
Two stress responses have been implicated in stress-induced mutagenesis in E. coli: the SOS DNA damage response (8, 52) and the RpoS general stress response (41, 48). The SOS regulon is induced when regions of single-stranded DNA that are bound by the RecA protein accumulate. RecA then becomes activated and induces autocleavage of the repressor LexA, leading to an increase in the expression of roughly 40 genes in E. coli (11). lexA mutants unable to undergo autocleavage (Ind−) are therefore impaired in induction of the SOS response (56). RpoS is an alternative sigma factor which accumulates in response to several different stresses, including entry into stationary phase and starvation (27), leading to increased transcription of hundreds of genes.
Figure 2A to D show that both stress responses are required for ampD mutagenesis, as observed previously for stress-induced lac mutagenesis (see the introduction). Additional data presented below confirm by independent means the requirement for the SOS response in ampD mutagenesis in starving cells. These data imply that chromosomal mutagenesis and F′ mutagenesis are triggered by similar environmental signals that stimulate the mutagenic response via RpoS and SOS induction. Because stress responses are required for formation of the ampD mutations, we call these mutations in starving cells “stress-induced” ampD mutations.
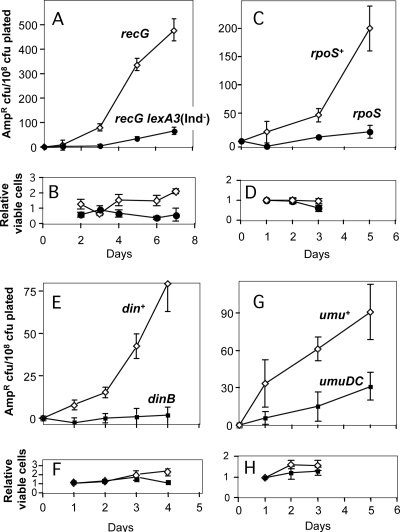
FIG. 2.
Chromosomal Ampr stationary-phase mutation requires stress responses and error-prone DNA polymerases. For an explanation of the experiments and data see the legend to Fig. 1. (A) Induction of the SOS response is required for ampD stress-induced mutagenesis. Open diamonds, recG strain SMR5578; filled circles, recG lexA3 (Ind−) strain SMR7055. The lexA3(Ind−) allele encodes an uncleavable LexA repressor protein that prevents derepression of LexA-controlled SOS genes during an SOS response (45). (C) Ampr stationary-phase mutation requires the general stress response controlled by RpoS. Open diamonds, rpoS+ strain SMR5222; filled circles, rpoS strain SMR10317. (E) DinB is required for Ampr stationary-phase mutagenesis. Open diamonds, din+ strain SMR5222; filled circles, dinB strain SMR6373. (G) The umuDC genes are required for Ampr stationary-phase mutagenesis. Open diamonds, umu+ strain SMR5222; filled squares, ΔumuDC strain SMR10316. (B, D, F, and H) Daily levels of Lac− viable cells on the plates, normalized to the day 1 counts, showing little net growth or death. For each panel the symbols are the same as those for the panel above it.
DinB and Pol V in Ampr mutation.
We found that, as observed for Lac and Tet stress-induced mutagenesis (7, 53), DinB is required for stress-induced ampD mutagenesis (Fig. 2E and F). Despite the requirement for DinB, we found that the data for the sequences of the stress-induced ampD mutations showed that there were more base substitutions than frameshift mutations (Fig. 3; see below). Because the base substitutions observed are not hallmarks of DinB action (see below), we hypothesized that another DNA polymerase(s) might also contribute to chromosomal mutagenesis under stress conditions. The most obvious candidate is Pol V (73), encoded by the umuDC genes. These genes are responsible not only for virtually all SOS-dependent mutagenesis that follows treatment of cells with DNA-damaging agents (35) but also for so-called “SOS untargeted mutagenesis,” which occurs at undamaged sites or sites where there is endogenous DNA damage in SOS-induced cells (78). The umuDC genes are tightly regulated and are virtually not expressed in the absence of an SOS response (58).
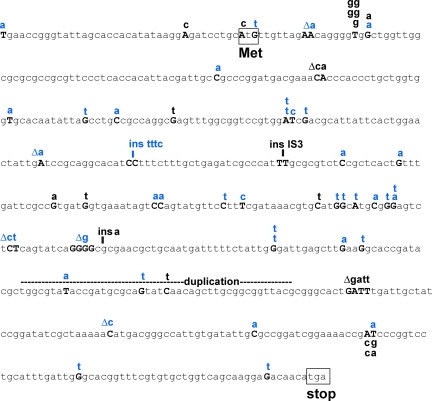
FIG. 3.
Sequences of ampD generation-dependent and stress-induced mutations. The ampD genes of Ampr mutants were sequenced as described previously (61). Above the ampD sequence, generation-dependent mutations determined in a previous study (61) are indicated in black type and stress-induced mutations (selected from overlay plates on day 5 or later) are indicated in blue type. Δ, deletion; ins, insertion.
Figure 2G and H show that a ΔumuDC strain is impaired in stress-induced mutagenesis in the chromosomal ampD locus, showing ∼3-fold less mutagenesis upon starvation than the isogenic umuDC+ strain. This result suggests that there is concerted action of both Pol V and DinB Y family DNA polymerases in stress-induced ampD mutagenesis in E. coli. The results also provide independent confirmation that the SOS response is required for stress-induced ampD mutagenesis, because the umuDC genes are expressed only during an SOS response (for a review, see reference 58).
Base substitutions prevalent in ampD stress-induced mutations.
We sequenced chromosomal ampD stationary-phase mutations in mutants. One potential problem in obtaining stationary-phase ampD mutants is that resistant colonies on plates from later days may also include some early generation-dependent mutants. Plates contain colonies formed by mutant cells that arose at any time between inoculation of the culture and the time when the cells were rescued from starvation and Ampr was selected by overlay with medium containing another carbon source and ampicillin on the lactose plates. To minimize the number of generation-dependent mutation sequences in the data, mutant colonies were chosen from experiments in which there were ≥5- to 7-fold more day 5 or day 7 mutants than day 0 (generation-dependent) mutants. Also, in some experiments preparations were plated using high dilutions so that few, if any, day 0 colonies would appear and the later colonies would be only stationary-phase mutant colonies. Because there were ≥5- to 7-fold more late (day 5) RpoS- and SOS-dependent ampD mutants than early stress response-independent mutants, we estimated that a maximum of ∼20% of our stress-induced ampD mutations could be generation-dependent mutations.
We found 22 mutations in 20 generation-dependent Ampr mutants sequenced and 40 mutations in 38 Ampr mutants obtained at day 5 or later (some mutants had more than one mutation). The ampD generation-dependent mutations included small and large insertions (including insertions of mobile elements), small deletions, and base substitutions (61). Contrary to the expectation that the stress-induced mutations would be mostly −1 deletions in mononucleotide repeats (16, 65), substitutions predominated among the ampD stress-induced mutations (Fig. 3 and Table 2). The proportion of base substitutions resembled that seen in ampD generation-dependent mutations (Table 2) (61), although the kinds of substitutions differed (Table 3 and Fig. 3). Thirty-four of the 40 stress-induced mutations and 17 of the 22 generation-dependent mutations were substitutions (Fig. 3 and Table 2). However, ∼30% (5 of 17) of the generation-dependent substitutions occurred at hot spots (sites showing the same substitution more than once), whereas only 5% (2 of 40) of the stress-induced mutations occurred at hot spots (Fig. 3 and Table 2). The stress-induced mutations also differed from the generation-dependent mutations in containing fewer mobile element insertions and more frameshift mutations in repeated sequences. The two types of mutations included similar numbers of transitions and transversions, but the difference in the types of substitutions observed is striking. In the stress-induced mutations G·C-to-T·A transversions were much more prevalent, comprising more than one-half of the substitutions detected, and more A·T-to-T·A transversions were also observed in these mutations than in the generation-dependent mutations. On the other hand, A·T-to-C·G transversions were observed only for generation-dependent mutations (Fig. 3 and Table 3).
TABLE 2.
Profiles of generation-dependent and stress-induced ampD mutations in sequenced ampD mutants
| Type of mutation | Generation dependenta,b | Stress induceda |
|---|---|---|
| Insertions of insertion element | 1/22 | 0/40 |
| −1 to −4 deletions | 2/22 (0)c | 6/40 (2)c |
| Transitions | 2/22 | 7/40 |
| Transversions | 15/22 | 27/40 |
| Duplications | 1/22 | 0/40 |
| 1-base insertions | 1/22 | 0/40 |
| Substitutions at hot spots/total no. of substitution mutations | 5/17 | 2/34 |
Number of mutations observed/total number of mutations sequenced, unless indicated otherwise.
Data from reference 61.
The numbers in parentheses are the numbers of deletions in repeats.
TABLE 3.
Different base substitutions in generation-dependent and stress-induced ampD mutants
| Type of substitution | No. of substitutions observed/total no. of substitutions
|
|
|---|---|---|
| Generation dependenta | Stress induced | |
| G·C to A·T | 4/17 | 5/34 |
| G·C to T·A | 2/17 | 20/34 |
| A·T to T·A | 1/17 | 7/34 |
| A·T to C·G | 10/17 | 0/34 |
| A·T to G·C | 0/17 | 2/34 |
Data from reference 61.
DISCUSSION
Stress-induced β-lactam resistance mutagenesis.
The data presented here show that starvation stress-induced mutagenesis can promote β-lactam resistance mutation in a clinically relevant model (Fig. 1 and 2). This is important because most antibiotic resistance acquisition and mutation have been modeled using mutation rates determined in nonstress environments, even though many natural environments of pathogens are likely to be stress-inducing environments (14, 49). For example, the stress-inducible mutation process shown here to promote ampD mutagenesis requires induction of the general stress response controlled by RpoS. RpoS is induced during Pseudomonas lung infections and may be a general feature of infection (14). Therefore, the stress-induced mutagenesis mechanisms that occur under RpoS-inducing conditions are likely to provide better models for generation of resistance mutations in situ. Considering that β-lactam antibiotics kill dividing cells, it seems reasonable to conclude that a stationary-phase mutagenesis mechanism may lead to clinically relevant ampD mutations upon antibiotic treatment. For example, in enterobacteria and Pseudomonas aeruginosa, ampD null mutations are found in β-lactam-resistant clinical isolates, although in Pseudomonas other mutations also seem to be required for the β-lactam resistance (40, 67).
Possible error-prone DSBR mechanism of stress-induced ampD mutagenesis.
Like stress-induced mutagenesis at the F′ lac locus (18, 24, 25, 41, 48, 52, 53), accumulation of chromosomal stress-induced ampD mutants during starvation on lactose medium required DNA DSBR and HR proteins RecA, RecB, and RuvC, the ability to induce the SOS DNA damage response, the RpoS stress response activator, and the SOS- and RpoS-inducible error-prone DNA polymerase DinB, and it was stimulated in the absence of RecG (Fig. 1 and 2). This is the first demonstration of a requirement for the DSE-specific RecBC enzyme in chromosomal mutagenesis during starvation stress, and the results imply that DSEs are molecular intermediates in the stress-induced ampD mutagenesis pathway as well. These genetic requirements are reminiscent of two other stress-induced mutagenesis mechanisms: E. coli ciprofloxacin-induced chromosomal ciprofloxacin resistance mutagenesis (10) and Salmonella bile-induced resistance mutagenesis (63). In contrast to stress-induced lac reversion but like ciprofloxacin-induced resistance mutagenesis (10), a significant proportion of the mutagenesis requires the other E. coli error-prone DNA polymerase, Pol V. Because both dinB and umuDC strains show a severe defect in ampD stress-induced mutagenesis, it is likely that these polymerases cooperate in stress-induced mutagenesis in chromosomal genes.
For lac frameshift reversion, the hypothesis that there is an error-prone DSBR mutation mechanism is supported by evidence that DSBs generated by expression of a restriction enzyme in vivo increase mutagenesis nearby >1,000-fold (62). This occurs only in stationary phase or if RpoS is expressed inappropriately in log phase (62). The data indicate that there is a switch from high-fidelity DSBR to error-prone DSBR under stress conditions, mediated by RpoS (62). A similar mechanism of stress-induced switching to error-prone DSBR might also generate the ampD mutations studied here. This mechanism does not produce ampD mutations in rapidly growing cultures of the Lac assay strains, which are RecA and SOS independent and not stimulated by recG mutation (61). The DSEs whose error-prone repair provokes stress-induced mutagenesis might be spontaneous breaks and DSEs generated, for example, by replication mishaps.
Evolutionary significance of stress-induced chromosomal mutation sequences.
The sequences of chromosomal, stress-induced ampD mutations provide the first look at the true sequences of mutations generated by the HR-DSBR-associated stress-induced mutagenesis mechanism, because any loss-of-function mutation in ampD can confer β-lactam resistance. The results are surprising because they show that there were more base substitutions than frameshift mutations (Fig. 3 and Table 2).
Previously, HR-DSBR-associated stress-induced mutagenesis was studied by examining reversion of frameshift alleles, so that only frameshift mutations could be recovered. Stress-induced lac reversions were nearly all −1 bp deletions in small mononucleotide repeats, unlike generation-dependent reversions, which were more heterogeneous, included larger insertions and deletions, and were not confined to mononucleotide repeats (16, 65). Also, the stress-induced mutation sequences in lac (16, 65) and in tet (7) resemble the errors made by DinB error-prone polymerase (36, 77); they are mostly −1 deletions in repeated sequences. Thus, one might have imagined that frameshift mutations would dominate the HR-DSBR-associated stress-induced-mutagenesis mechanism, with substitutions being a minor component. However, the role of Pol V in the type of mutagenesis demonstrated here provides an explanation for the abundance of base substitutions observed in ampD mutants (see below).
The evolutionary consequence of a frameshift-heavy mutation mechanism would be that genes would more often be inactivated and less often have their functions modified. Gene inactivation and reactivation by frameshift mutation in repeated sequences is an important strategy of pathogens that vary expression of surface protein antigens in this way (4), and stress-induced mutagenesis might promote this strategy. However, the results presented here (Fig. 3) imply that at least for chromosomal (and perhaps all) loci mutated during stress, substitutions are the predominant mutations. Therefore, modifications of gene functions, in addition to inactivation and activation, are predicted outcomes of this pathway, as observed in other instances of stress-induced mutagenesis (10, 72). This HR-DSBR stress-induced mutagenesis pathway could therefore contribute far more broadly to bacterial evolution under stress conditions.
Differences between F′ lac reversion and chromosomal ampD mutagenesis during stress.
The involvement of the SOS-inducible Pol V in stress-induced ampD mutagenesis reveals a fundamental difference between the F′ lac gene and ampD. For stress-induced mutagenesis in lac, Pol I (26), Pol II (17), and Pol V (8, 52) are not required. Pol III is difficult to test, because it is essential, but some data suggest that it might compete with Pol IV, decreasing mutagenesis; we suggested that an antimutator Pol III lowered DinB-dependent stress-induced mutagenesis by increased processivity, leading to exclusion of Pol IV from the replisome (53). Thus, there is no compelling evidence that in lac more than one DNA polymerase produces most stress-induced mutagenesis. Additionally, we found that dinB is the only SOS gene required at induced levels for stress-induced mutagenesis in the lac gene (20).
The differences between mutagenesis in lac and mutagenesis in ampD that might account for the different uses of Pol V in mutagenesis are as follows. First, because ampD is a forward mutational target, analysis of ampD allows detection of base substitutions, which are not detectable in the lac reversion assay. Second, ampD is a chromosomal gene, and the DNA polymerases operating in the chromosome might differ somewhat. Third, whereas most of the mutational events in the F′ that occur during stress occur in acts of error-prone DSBR in cis to a DSB (as shown by Ponder et al. [62]), perhaps many chromosomal mutations occur in trans to a DSB (that is, not directly associated with a DSBR event). Different DNA polymerases might be responsible for the mutations generated by the two different mechanisms. This might explain the seemingly different sequence spectra observed for mutations at the two loci.
It is interesting that the following two other biological processes use more than one SOS DNA polymerase in the same mutagenesis pathway: survival in long-term liquid culture, which might include mutagenesis (80), and ciprofloxacin-induced mutation to ciprofloxacin resistance (10), as discussed below.
Mechanistic significance of stress-induced mutation sequences.
Twenty of the 40 ampD stress-induced mutations were G·C-to-T·A transversions. The relatively high number of G·C-to-T·A mutations and the absence of −1 bp deletions in mononucleotide repeats seen here lead us to question whether DinB is the polymerase that is actually responsible for the Ampr mutations, even though DinB is required for the vast majority of the stress-induced mutagenesis in both lac (53) and ampD (Fig. 2E).
First, the error spectrum of purified DinB DNA polymerase includes about 65% −1 bp deletions, mostly deletions in mononucleotide repeats, and there is only a very minor fraction of G-to-T (or C-to-A) changes (37). Second, by measuring mutations in the phage λ cII gene in vivo, Wagner and Nohmi (77) found that although overexpression of dinB from a plasmid increased the numbers of both frameshift and substitution mutations, the number of G-to-T mutations increased less than all the other substitution mutations examined. On the other hand, using the same dinB overexpression plasmid, Kim et al. (36) found that G·C-to-T·A transversions were predominant base substitutions in the lac reversion assay of Cupples and Miller (12), even though −1 bp deletions were still by far the most common type of mutations found. Thus, it is not clear whether G·C-to-T·A transversions could be a major consequence of DinB activity in vivo.
A predominance of G·C-to-T·A and, to a minor extent, A·T-to-T·A transversions is a hallmark of “SOS untargeted mutagenesis,” (i.e., mutagenesis occurring in cells not exposed to DNA-damaging agents but constitutively expressing the SOS response) (55). This is a heavily umuDC-dependent process (6, 9), and the observation that the same type of base substitution predominates in the ampD mutants supports the hypothesis that umuDC has a major role in this stress-induced mutagenesis mechanism. Furthermore, the umuDC-dependent SOS mutator phenotype giving rise to base substitutions is enhanced by DinB (39), particularly for mutagenesis occurring in the lagging strand. Similarly, here we observed base substitutions occurring in a dinB- and umuDC-dependent manner in stressed cells. The sequences of stress-induced Ampr mutants in the umuDC background would, in principle, clarify whether the G·C-to-T·A mutations are really a result of the action of Pol V. However, an experiment to determine these sequences cannot be done, because the generation-dependent mutants outnumber the stress-induced mutants that remain for the umuDC strain.
Our results support a model in which the DNA synthesis that produces the mutations during DSBR-associated stress-induced mutagenesis involves both DinB and Pol V. Previously, Ponder et al. showed that DinB is not required for DSBR but becomes licensed to participate in DSBR and produces DSBR-associated mutations when RpoS is expressed (62). The predominance of Pol V-like mutation sequences in ampD despite a strong DinB dependence of the mutations could be explained by models suggested by others (19), in which one DNA polymerase makes the error and another DNA polymerase makes the initial extension from the mispaired primer terminus. This could account for the base substitutions, whereas the frameshift mutations might be made solely by DinB. This would explain the lack of a requirement for Pol V in stress-induced Lac frameshift reversion (20, 52) and the partial Pol V requirement and sequence signature for stress-induced ampD mutagenesis, which results in mostly base substitutions. This explanation was suggested for DinB-, Pol V-, and Pol II- dependent stress-induced ciprofloxacin resistance mutagenesis induced by ciprofloxacin in E. coli, which produces only base substitutions and is presumed to occur via error-prone DSBR, like Lac stress-induced mutagenesis (10). Apart from the use of Pol V and Pol II, Cirz et al. found requirements identical to those of DSBR-associated stress-induced Lac reversion. Thus, as those authors suggested (10), DSBR-associated stress-induced mutagenesis might involve Pol V and DinB and require both of these enzymes for substitution mutagenesis but only DinB for frameshift mutagenesis.
Acknowledgments
We thank P. J. Hastings, Jesús Blázquez, and an anonymous reviewer for comments on the manuscript.
This work was supported by a Department of Defense Breast Cancer Research Postdoctoral Fellowship (to J.F.P.), by a postdoctoral fellowship from the Pew Latin American Fellows Program (to R.S.G.), and by National Institutes of Health grant GM53158.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Alonso, A., E. Campanario, and J. L. Martinez. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145:2857-2862. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartowsky, E., and S. Normark. 1991. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol. Microbiol. 5:1715-1725. [DOI] [PubMed] [Google Scholar]
- 4.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. [DOI] [PubMed] [Google Scholar]
- 6.Blanco, M., G. Herrera, P. Collado, J. E. Rebollo, and L. M. Botella. 1982. Influence of RecA protein on induced mutagenesis. Biochimie 64:633-636. [DOI] [PubMed] [Google Scholar]
- 7.Bull, H. J., M. J. Lombardo, and S. M. Rosenberg. 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. USA 98:8334-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciesla, Z. 1982. Plasmid pKM101-mediated mutagenesis in Escherichia coli is inducible. Mol. Gen. Genet. 186:298-300. [DOI] [PubMed] [Google Scholar]
- 10.Cirz, R. T., J. K. Chin, D. R. Andes, V. de Crecy-Lagard, W. A. Craig, and F. E. Romesberg. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley, I., P. Marsh, E. M. Wellington, A. W. Smith, and M. R. Brown. 1999. General stress response master regulator rpoS is expressed in human infection: a possible role in chronicity. J. Antimicrob. Chemother. 43:164-165. [DOI] [PubMed] [Google Scholar]
- 15.Foster, P. L. 1997. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 179:1550-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, P. L., and J. M. Trimarchi. 1994. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265:407-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, P. L., G. Gudmundsson, J. M. Trimarchi, H. Cai, and M. F. Goodman. 1995. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:7951-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, P. L., J. M. Trimarchi, and R. A. Maurer. 1996. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics 142:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedberg, E. C., A. R. Lehmann, and R. P. Fuchs. 2005. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell 18:499-505. [DOI] [PubMed] [Google Scholar]
- 20.Galhardo, R. S., R. Do, M. Yamada, E. Friedberg, P. Hastings, T. Nohmi, and S. Rosenberg. 2009. DinB up-regulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182:55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galhardo, R. S., P. J. Hastings, and S. M. Rosenberg. 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42:399-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez-Gomez, J. M., J. Blazquez, F. Baquero, and J. L. Martinez. 1997. H-NS and RpoS regulate emergence of Lac Ara+ mutants of Escherichia coli MCS2. J. Bacteriol. 179:4620-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grundstrom, T., and B. Jaurin. 1982. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 79:1111-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 25.Harris, R. S., K. J. Ross, and S. M. Rosenberg. 1996. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics 142:681-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastings, P. J., A. Slack, J. F. Petrosino, and S. M. Rosenberg. 2004. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2:e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrera, G., V. Aleixandra, A. Urios, and M. Blanco. 1993. Quinolone action in Escherichia coli cells carrying gyrA and gyrB mutations. FEMS Microbiol. Lett. 106:187-191. [DOI] [PubMed] [Google Scholar]
- 29.Ilves, H., R. Horak, and M. Kivisaar. 2001. Involvement of σS in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J. Bacteriol. 183:5445-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs, C., J. M. Frere, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frere. 1995. AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 33.Jacoby, G. A. 2009. AmpC beta-lactamases. Clin. Microbiol. Rev. 22:161-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karunakaran, P., and J. Davies. 2000. Genetic antagonism and hypermutability in Mycobacterium smegmatis. J. Bacteriol. 182:3331-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato, T., and Y. Shinoura. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 156:121-131. [DOI] [PubMed] [Google Scholar]
- 36.Kim, S. R., G. Maenhaut-Michel, M. Yamada, Y. Yamamoto, K. Matsui, T. Sofuni, T. Nohmi, and H. Ohmori. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. USA 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi, S., M. R. Valentine, P. Pham, M. O'Donnell, and M. F. Goodman. 2002. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 277:34198-34207. [DOI] [PubMed] [Google Scholar]
- 38.Koorits, L., R. Tegova, M. Tark, K. Tarassova, A. Tover, and M. Kivisaar. 2007. Study of involvement of ImuB and DnaE2 in stationary-phase mutagenesis in Pseudomonas putida. DNA Repair 6:863-868. [DOI] [PubMed] [Google Scholar]
- 39.Kuban, W., M. Banach-Orlowska, R. M. Schaaper, P. Jonczyk, and I. J. Fijalkowska. 2006. Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J. Bacteriol. 188:7977-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langaee, T. Y., L. Gagnon, and A. Huletsky. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC beta-lactamase expression. Antimicrob. Agents Chemother. 44:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 43.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii beta-lactamase. J. Bacteriol. 169:1923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindquist, S., M. Galleni, F. Lindberg, and S. Normark. 1989. Signalling proteins in enterobacterial AmpC beta-lactamase regulation. Mol. Microbiol. 3:1091-1102. [DOI] [PubMed] [Google Scholar]
- 45.Little, J. W., S. H. Edmiston, L. Z. Pacelli, and D. W. Mount. 1980. Cleavage of the Escherichia coli LexA protein by the RecA protease. Proc. Natl. Acad. Sci. USA 77:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livermore, D. M. 1995. beta-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lloyd, R. G., and C. Buckman. 1991. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J. Bacteriol. 173:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lombardo, M. J., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez, J. L., and F. Baquero. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matic, I., M. Radman, F. Taddei, B. Picard, C. Doit, E. Bingen, E. Denamur, and J. Elion. 1997. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277:1833-1834. [DOI] [PubMed] [Google Scholar]
- 51.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 98:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 54.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Miller, J. H., and K. B. Low. 1984. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell 37:675-682. [DOI] [PubMed] [Google Scholar]
- 56.Mount, D. W., K. B. Low, and S. J. Edmiston. 1972. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutations. J. Bacteriol. 112:886-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicolas, M. H., N. Honore, V. Jarlier, A. Philippon, and S. T. Cole. 1987. Molecular genetic analysis of cephalosporinase production and its role in beta-lactam resistance in clinical isolates of Enterobacter cloacae. Antimicrob. Agents Chemother. 31:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nohmi, T. 2006. Environmental stress and lesion-bypass DNA polymerases. Annu. Rev. Microbiol. 60:231-253. [DOI] [PubMed] [Google Scholar]
- 59.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 60.Perez-Capilla, T., M. R. Baquero, J. M. Gomez-Gomez, A. Ionel, S. Martin, and J. Blazquez. 2005. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J. Bacteriol. 187:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrosino, J. F., A. R. Pendleton, J. H. Weiner, and S. M. Rosenberg. 2002. Chromosomal system for studying AmpC-mediated beta-lactam resistance mutation in Escherichia coli. Antimicrob. Agents Chemother. 46:1535-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponder, R. G., N. C. Fonville, and S. M. Rosenberg. 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19:791-804. [DOI] [PubMed] [Google Scholar]
- 63.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riesenfeld, C., M. Everett, L. J. Piddock, and B. G. Hall. 1997. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob. Agents Chemother. 41:2059-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenberg, S. M., S. Longerich, P. Gee, and R. S. Harris. 1994. Adaptive mutation by deletions in small mononucleotide repeats. Science 265:405-407. [DOI] [PubMed] [Google Scholar]
- 66.Rosenberg, S. M., C. Thulin, and R. S. Harris. 1998. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics 148:1559-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanders, C. C., and W. E. Sanders, Jr. 1992. beta-Lactam resistance in gram-negative bacteria: global trends and clinical impact. Clin. Infect. Dis. 15:824-839. [DOI] [PubMed] [Google Scholar]
- 68.Shurvinton, C. E., R. G. Lloyd, F. E. Benson, and P. V. Attfield. 1984. Genetic analysis and molecular cloning of the Escherichia coli ruv gene. Mol. Gen. Genet. 194:322-329. [DOI] [PubMed] [Google Scholar]
- 69.Sung, H. M., and R. E. Yasbin. 2002. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J. Bacteriol. 184:5641-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sung, H. M., G. Yeamans, C. A. Ross, and R. E. Yasbin. 2003. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J. Bacteriol. 185:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taddei, F., I. Matic, B. Godelle, and M. Radman. 1997. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 5:427-428. [DOI] [PubMed] [Google Scholar]
- 72.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. USA 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tegova, R., A. Tover, K. Tarassova, M. Tark, and M. Kivisaar. 2004. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 186:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tenover, F. C. 2006. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control 34:S3-S10. (Discussion, 34:S64-S73.) [DOI] [PubMed] [Google Scholar]
- 76.Torkelson, J., R. S. Harris, M. J. Lombardo, J. Nagendran, C. Thulin, and S. M. Rosenberg. 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 16:3303-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagner, J., and T. Nohmi. 2000. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182:4587-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Witkin, E. M. 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol. Rev. 40:869-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woodgate, R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]
- 80.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]