Abstract
In Staphylococcus, the twin-arginine translocation (Tat) pathway is present only in some species and is composed of TatA and TatC. The tatAC operon is associated with the fepABC operon, which encodes homologs to an iron-binding lipoprotein, an iron-dependent peroxidase (FepB), and a high-affinity iron permease. The FepB protein has a typical twin-arginine (RR) signal peptide. The tat and fep operons constitute an entity that is not present in all staphylococcal species. Our analysis was focused on Staphylococcus aureus and S. carnosus strains. Tat deletion mutants (ΔtatAC) were unable to export active FepB, indicating that this enzyme is a Tat substrate. When the RR signal sequence from FepB was fused to prolipase and protein A, their export became Tat dependent. Since no other protein with a Tat signal could be detected, the fepABC-tatAC genes comprise not only a genetic but also a functional unit. We demonstrated that FepABC drives iron import, and in a mouse kidney abscess model, the bacterial loads of ΔtatAC and Δtat-fep mutants were decreased. For the first time, we show that the Tat pathway in S. aureus is functional and serves to translocate the iron-dependent peroxidase FepB.
The Sec pathway is the major secretion system that exports the majority of extracytosolic proteins in pro- and eukaryotes. Proteins are translocated through this pathway in a more or less unfolded state. A second protein export pathway was identified first in the chloroplast thylakoid membrane (8) and later in several bacteria (4, 14, 35). This pathway has been designated the twin-arginine translocation system (Tat), as the preproteins targeted to this pathway carry a characteristic amino acid motif, including two consecutive arginine residues, which are essential for the recognition by the Tat translocon. The Tat pathway operates independently of the Sec pathway and exports exoproteins across the bacterial cytoplasmic membrane, apparently in a fully folded conformation (3). Many of these proteins are complexed with cofactors.
Studies of several bacterial species, including Escherichia coli (37), Bacillus subtilis (22, 23), Pseudomonas aeruginosa (31), Legionella pneumophila (10, 11), and Mycobacterium smegmatis (29), have demonstrated that they possess a functional Tat export pathway. In E. coli, the TatA, TatB, and TatC proteins have been demonstrated to be essential for Tat-dependent protein translocation (4). However, several bacterial and archaeal species lack a TatB-like protein. For example, the B. subtilis genome encodes three TatA- and two TatC-like proteins. Thus, at least one copy of the TatA homologue and one copy of the TatC homologue are required for a functional Tat pathway. In Bacillus subtilis, several proteins were predicted that could potentially use the Tat pathway, as their signal peptides (SPs) contain RR or KR motifs. However, proteomic analysis revealed that 13 proteins with potential RR/KR SPs were Tat independent, showing that the Tat machinery does not recognize their RR/KR motifs. In fact, only the phosphodiesterase PhoD and the newly identified YwbN protein were shown to be secreted in a strictly Tat-dependent manner (22). YwbN is part of the YwbLMN operon product and is involved in the uptake of free ferric iron (32).
For staphylococci, very little information regarding the function of the Tat system exists. A tatC-deficient mutant showed little difference from the wild type (WT) in its extracellular protein pattern (48). In another study, green fluorescent protein (GFP) was fused with the SP of the E. coli TorA protein and the subcellular localization of the hybrid protein was compared in Staphylococcus carnosus and its tatC mutant (30). The results showed that GFP was secreted in neither the WT nor the tatC mutant but was stuck in the cell wall fraction of the WT. However, no Tat-transported protein has been identified in staphylococci so far.
Here we show that the tat operon encoding TatA and TatC is present in Staphylococcus aureus, S. carnosus, and Staphylococcus haemolyticus but is missing in a number of other staphylococcal species. By comparative analysis of WT and tatAC mutant strains, the functionality of the Tat pathway in S. carnosus and S. aureus was demonstrated, and it was found that the iron-dependent peroxidase (FepB) is translocated by the Tat system. The Tat system can efficiently translocate heterologous proteins such as lipase or protein A when it is fused with the SP of FepB. 55Fe transport experiments demonstrated that FepABC is involved in iron uptake.
MATERIALS AND METHODS
Identification of Tat substrates by ParSeq, a modified TATFIND algorithm.
The Tat SP of peroxidase was identified in staphylococcal genomes by use of the software tool ParSeq, a program that combines the search for motifs with a search for certain structural properties (40). The search pattern used was X1RRX2X3X4, in which the amino acid at position X1 had a hydrophobicity score of <0.26, X2 had to be one of certain residues (ARKDEF), X3 had a hydrophobicity score of >0.77 (positively charged residues were excluded from this position), and X4 was one of certain residues (ILVMF). All hydrophobicity values were described previously (9). In addition, the following three criteria were considered: (i) whether there was an uncharged stretch of at least 13 residues in the 22 residues following the RR; (ii) whether the hydrophobicity value of the first 13 residues of the uncharged region was <8.0; and (iii) since it is known that SPs should be in the beginning of an open reading frame, the twin-arginine motif was expected to be within the first 30 amino acids. All of these constraints were used to construct a search pattern in the form of a regular expression in ParSeq. With this regular expression, we searched the whole genomes of S. aureus and S. carnosus. If the search identified a sequence that matched the above pattern, the corresponding gene was considered to possess a putative Tat signal sequence.
Bacterial strains.
Bacterial strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains used in this studya
| Species and strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. aureus | ||
| SA113 (ATCC 35556) | Derivative of RN1 (NCTC8325), agr, 11-bp deletion in rbsU | 21 |
| RN4220 | NCTC8325 derivative | 21 |
| RN1HG | Derivative of RN1 (NCTC8325), with rsbU repaired | Herbert et al., unpublished data |
| SA113 (ΔtatAC) | tatAC::ermC | This study |
| SA113 (Δtat-fep) | tat-fep::kan | This study |
| S. carnosus | ||
| TM300 | WT | 38 |
| ΔtatAC | tatAC::ermC | This study |
| S. epidermidis ATCC 14990 | WT | 17 |
| S. haemolyticus CCM2737 | WT | 17 |
| S. simulans ATCC 27848 | WT | 24 |
| S. saprophyticus DSM20229 | WT | 39 |
| S. lugdunensis ATCC 43809 | WT | 15 |
| S. gallinarum Tu 3928 | WT | 13 |
| S. muscae DSM 7068 | WT | 20 |
| S. sciuri subsp. lentus DSM 20352 | WT | |
| S. capitis subsp. capitis CCM 2734 | WT | 24 |
| S. hyicus subsp. hyicus NCTC10350 | WT | 12 |
For more detailed information regarding the species characteristics, see reference 16.
Construction of tatAC deletion mutants in S. carnosus and S. aureus.
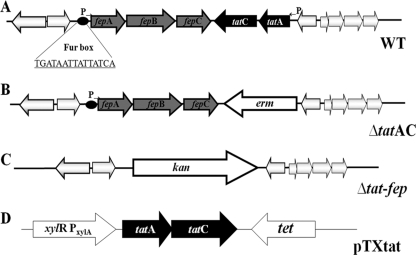
The upstream flanking regions of tatA were amplified using the primer pair SCtat up F (5′-TAACATGAATTCCATTGATATTCATTTC-3′; EcoRI site is underlined) and SCtat up R (5′-TACATTGGTACCAAGTTGATCCAATAG-3′; KpnI site is underlined) for S. carnosus TM300 and the primer pair SAtatC up F (5′-TAACATGAATTCTAATTACACTGTAAATG-3′; EcoRI site is underlined) and SAtatC up R (5′-TACATTGGATCCTAAAATTTTTACTAACCG-3′; BamHI site is underlined) for S. aureus SA113. To amplify the downstream flanking regions of tatC, the primer pairs SCtat down F (5′-TAACATCTGCAGAACAAACACCCGTC-3′; PstI site is underlined) and SCtat down R (5′-TACATTGCTAGCTTCAGATTTGGC-3′; NheI site is underlined) for S. carnosus TM300 and SAtatC downF (5′-TAACATCTGCAGCCTTATACGAATCAATGC-3′; PstI site is underlined) and SAtatC downR (5′-TACATTGATATCAAATTCAAACTAAAGACGG-3′; EcoRV site is underlined) for S. aureus SA113 were used for PCR amplification and cloning into the multiple cloning site of pBT2 (7). The ermB cassette from pEC2 was introduced between the flanking regions. S. carnosus ΔtatAC and S. aureus ΔtatAC mutants were constructed by a standard homologous recombination method (7). The tatAC deletion mutants were identified by their erythromycin-resistant and chloramphenicol-sensitive phenotype and were confirmed by PCR and DNA sequence analyses.
Construction of tat-fep deletion mutant in S. aureus.
A 1-kb fragment resembling the upstream region of tatA and a 1-kb fragment identical to the region upstream of SA0331 were amplified by PCR, using the primers SA ptupF (5′-TATTATGGATCCAAAATTCCACCTATGATACC-3′; BamHI site is underlined), SA ptupR (5′-TAACATCCATGGCCTCACTCATAAGTAG-3′; NcoI site is underlined), Sa ptdwnF (5′-TAACATAGATCTTTTGACACCTCATTATAGAAATTC-3′; BglII site is underlined), and Sa ptdwnR SmaR (5′-TATTATCCCGGGTGGGTCCGTAATTTC-3′; SmaI site is underlined). The resulting PCR products were cloned into the multiple cloning site of pDG782 (19). The Δtat-fep::kan fragment, representing the upstream fragment, the aphAIII cassette, and the downstream fragment, was moved into the plasmid pKOR1 (1) by using Gateway technology (Invitrogen), resulting in plasmid pKOR1-Δtat-fep::kan. Allelic replacement in S. aureus was performed as described previously (1). The Δtat-fep deletion mutants were identified by their kanamycin-resistant and chloramphenicol-sensitive phenotype and were confirmed by PCR and DNA sequence analyses.
Construction of vectors containing Tat SP fused to lipase and protein A.
Protein A and prolipase were used as reporter enzymes. Prolipase was originally derived from Staphylococcus hyicus (18, 26). The prolipase (pro-lip) and protein A (spa) genes were fused with the Tat SP sequence of the S. carnosus-specific iron-dependent peroxidase gene (SC2214). The DNA fragments were inserted in pCX19 in such a way that expression of the target gene was under the control of the xylose-inducible promoter (5, 47). The DNA fragment encoding the peroxidase Tat SP was amplified from S. carnosus chromosomal DNA by use of the primer pair 5′-TACATTGGATCCTAGGAGGTGACGATATGACACAAGACAAGC-3′ (BamHI site is underlined) and 5′-TATCATTCTAGAGAAAGAAAAAATGCCGCCGACTCC-3′ (XbaI site is underlined). The prolipase gene was amplified from pTX15 by PCR (33), using the primers 5′-TACATTTCTAGAAATGATTCGACAACACAAAC-3′ (XbaI site is underlined) and 5′-TATCATGAGCTCTCATTATGCGTTCTTTGTGCTTTC-3′ (SacI site is underlined). The prolipase gene was ligated with the signal sequence of the peroxidase gene and cloned into pDG782 (19). The gene fusion was subsequently recloned into the staphylococcal vector pCX19, using BamHI and PstI, resulting in pCXRR-lip; the corresponding gene was under the control of the xylose-inducible promoter. The vector pCX19 encodes prolipase fused to the Sec SP. The protein A (spa) gene was fused to the Tat signal sequence by amplifying spa from the S. aureus DNA template by PCR using the following primers: TACATTGTCGACGCGCAACACGATGAAGC (SalI site is underlined) and TAACATCCCGGGTTATAGTTCGCGACG (SmaI site is underlined). The PCR fragment was digested and ligated into pCXRR-lip precut with the same restriction enzymes, replacing the lip gene. The resulting plasmid was designated pCXRR-spa.
Lipase and peroxidase activity assays.
Staphylococcal cultures were induced with 0.5% xylose and grown until late stationary phase. Cells were pelleted by centrifugation at 13,000 × g for 5 min, and the culture supernatant was filtered through 0.22-μm-pore-size filters and used for the detection of secreted lipase, peroxidase, and protein A.
Lipase activity was measured spectrophotometrically at 405 nm, using p-nitrophenyl-caprylate as a substrate (45, 47), in stationary-phase culture supernatants as well as in the zymogram gel (26). Briefly, after sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, the gels were washed once in 20% isopropanol for 2 min, followed by three washes with 50 mM Tris-HCl (pH 7.5) for 5 min. Finally, the gels were laid on lipase test medium (0.1% agarose, 1 mM CaCl2, 50 mM Tris-HCl, 1% Tween 20, pH 7.5) and incubated at 37°C until halo bands were visible.
To detect the amount of secreted peroxidase, an Amplex red hydrogen peroxide/peroxidase assay kit (Molecular Probes, Eugene, OR) was used according to the manufacturer's instructions. This assay was performed spectrophotometrically. Absorbance was measured at 560 nm by using a microtiter plate reader (SpectraMAX UV-visible spectrophotometer).
Isolation of anchored and cytoplasmic protein A.
To detect protein A, stationary-phase staphylococcal cultures (50 ml) were pelleted by centrifugation at 13,000 × g for 15 min, and the supernatant was removed (secreted fraction). The cell pellet was resuspended in 5 ml of TRN buffer (50 mM Tris-HCl [pH 7.5], 30% raffinose, 145 mM NaCl) containing 0.1 mg/ml lysostaphin and incubated at 37°C for 30 min. Raffinose was added to prevent cell lysis by lysostaphin treatment. The sample was centrifuged, and the supernatant was removed (cell wall-associated fractions). To obtain cytoplasmic fractions, the cell pellet was resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 145 mM sodium chloride, 0.1 mg of lysostaphin per ml) and subjected to lysis at 37°C for 30 min, followed by centrifugation of the lysate for 30 min at 100,000 × g, and the supernatant was collected (cytoplasmic fraction). Subsequently, an aliquot of each fraction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting analysis using a protein A-specific antibody (Gene Tex, Inc.).
Immunofluorescence microscopy.
To detect cell wall-anchored protein A by immunofluorescence microscopy, S. carnosus and its ΔtatAC deletion mutant harboring the plasmid pCXRR-spa were grown in Trypticase soy broth to mid-log phase, and the culture was centrifuged and washed twice in phosphate-buffered saline (PBS) before incubation (60 min at room temperature) with an Alexa Fluor 594-conjugated anti-rabbit secondary antibody. Following two washes in PBS, the cells were resuspended in 1 ml PBS to an optical density at 600 nm (OD600) of 1, and slides were prepared.
Analysis of exoproteins by 2D gel electrophoresis.
S. aureus and S. carnosus parent strains and their respective tatAC mutant strains were cultured for 12 h and 24 h at 37°C. Cells were pelleted by centrifugation, and the culture supernatants were filtered through 0.22-μm-pore-size filters. Exoproteins were isolated and analyzed by two-dimensional (2D) gel electrophoresis as described earlier (34).
Identification of tatAC genes in Staphylococcus.
The genomic DNAs of 12 different staphylococcal species, S. aureus, S. carnosus, Staphylococcus epidermidis, S. haemolyticus, Staphylococcus saprophyticus, Staphylococcus simulans, Staphylococcus lugdunensis, Staphylococcus gallinarum, Staphylococcus muscae, Staphylococcus lentus, Staphylococcus capitis, and S. hyicus, were used as templates for the identification of the putative tatA and tatC genes, using the primer pair tatAF (5′-GCTTTAATTATTTTTGGTCC-3′) and tatCR (5′-CAGGTGCAATGAATGCCCACAATTG-3′). As described elsewhere (2), broad-range primers SSU-bact-27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and SSU-bact-907r (5′-CCGTCAATTCMTTTRAGTTT-3′) were used to amplify the 16S rRNA gene as a PCR control.
Iron transport assay.
For iron uptake experiments, cells were grown in Chelex-treated M9 minimal medium supplemented with 1% Casamino Acids and 5% minimum essential medium-vitamin solution. The medium was made iron deficient by being supplemented with 200 μM 2,2′-dipyriyl. Cells were harvested when the cultures reached an OD578 of 0.8 to 1 and were washed twice with M9 transport medium (Chelex 100-treated 1× M9 salts, 1 ml of 1 mM MgSO4, 0.1% glucose) at 4°C. Cells were resuspended in M9 transport medium (Chelex 100-treated 1× M9 salts, 100 mM morpholineethanesulfonic acid, pH 6, 1 ml of 1 mM MgSO4, 0.1% glucose) to an OD578 of 1.0 and kept on ice; the cells were incubated with shaking at 37°C for 5 min before the onset of transport. 55Fe reduced with ascorbate (100 μM [5.5 kBq/ml]/100 mM ascorbate) was diluted 100-fold in the cell suspension. Samples (0.7 ml) were taken at the indicated times, filtered on mixed-cellulose GN-6 Metricel membrane filters (Pall), washed twice with 2 ml 0.1 M LiCl solution, dried, and counted with a liquid scintillation counter after addition of a scintillator.
Mouse infection studies.
Female BALB/c mice (16 to 18 g) were purchased from Charles River, Sulzfeld, Germany, housed in polypropylene cages, and received food and water ad libitum.
S. aureus strains were cultured for 18 h in B medium, washed three times with sterile 0.9% NaCl, and suspended in sterile 0.9% NaCl to 7.0 × 107 CFU/100 μl. As a control, selected dilutions were plated on B agar. Mice were inoculated with 100 μl of S. aureus via the tail vein. Control mice were treated with sterile NaCl. For each strain, seven mice were used. Five days after challenge, the animals were sacrificed and kidneys were aseptically harvested and homogenized in 3 ml of 0.9% NaCl, using Dispomix (Bio-Budget Technologies GmbH, Krefeld, Germany). Serial dilutions of the organ homogenates were cultured on mannitol salt-phenol red agar plates for at least 48 h at 37°C. CFU were calculated as CFU/gram of kidney. The statistical significance of bacterial load differences was determined using the Mann-Whitney test.
RESULTS
Presence of the Tat system in Staphylococcus species.
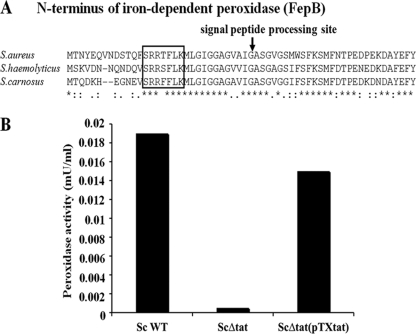
A genome-wide search of S. carnosus for the Tat SP revealed that the iron-dependent peroxidase FepB (Sca2214) contains a typical Tat signal sequence composed of the core sequence SRRFFLK followed by a weakly hydrophobic domain and a possible cleavage site (IGA). FepB revealed 40% similarity to YwbN of Bacillus subtilis, and among the staphylococcal species, it showed 71 to 75% sequence identity. As opposed to the published sequence annotation (25, 46), it turned out that the translational start sites of FepB in the S. aureus N315 (SA0332) and S. haemolyticus (SH0685) sequences begin 38 and 27 codons upstream, respectively (see Fig. 2A). This explains why the Tat SP was apparently not recognized in the corresponding reports. The FepB homologues of other bacterial species from different phyla also have typical Tat signal sequences (Table 2).
FIG. 2.
The iron-dependent peroxidase FepB is a Tat substrate. (A) Multiple amino acid sequence alignment of the N terminus of FepB of S. aureus, S. haemolyticus, and S. carnosus, using Clustal W. Asterisks indicate identical amino acid residues. The twin-arginine motif in the sequence is boxed. The arrow indicates the processing site. (B) Culture supernatants of S. carnosus WT, the S. carnosus Δtat mutant, and its complemented mutant, S. carnosus Δtat(pTX-tat), were tested for the presence of peroxidase by use of an Amplex red peroxidase assay kit.
TABLE 2.
SP sequences of FepB homologs in other bacteria
| Speciesa | SP sequence |
|---|---|
| sau | MTNYEQVNDSTQFSRRTFLKMLGIGGAGVAIGA |
| eco | MQYKDENGVNEPSRRRLLKVIGALALAGSCPVAHA |
| apl | MMAEIRSRRDFLKNTALVGAGLLSAPAFA |
| bma | MLVGAGRSGLTSRSARGAAPRARRHRVGGRAPRWAEANATRRSRGACTRAVA |
| bsu | MTASNEEKWMNKKISRRDMLKLTGIGVAGVAIGASGLGGLA |
| cgl | MVSRRGFLGGAGLIAGASALA |
| eta | MAKKTDEVASASRRRLLKGVGILSGALAVAGGCPAHA |
| kpn | MAQQKPHDVNEPSRRRLLKGIGALGGALAITGGCPVAHA |
| lmo | MTDKKSENQTEKTETKENKGMTRREMLKLSAVAGTGIAVG A |
| msm | MTPAQPSGISRRKLFGAAGVTAAVVGAASAGALAGRASA |
| nmc | MSKKQPAQPTRRTLFKTAIAAGA |
| pst | MKTPDSNPDATNAPVSLQRRRVLMGLGAAGAALAGSSLSGNVLAAA |
| sbo | MQYKDENGVNEPSRRRLLKVIGALALAGSCPVAHA |
| sco | MTDTDSPAPAPSPSRRSLIGWGGAGLALGAAAAAGGAVA |
| sgo | MTNDEKWFEKKMDRREFLKKAGIGGAGLALGVSGASA |
| ssa | MTNDEKWFNKKMDRREFLKKAGIGGAGLALGVSGASA |
| ste | MTDKKFLDQKMDRREFLKKSGIGGAGLALGLSGASA |
| ype | MRDKTGPKFGPYQPDDEAVSPSRRRLILGMGMVSGALVLGGAKTAQA |
| yps | MRDKTGPKFGPYQPDDEAVSPSRRRLILGMGMVSGALVLGGAKTAQA |
sau, Staphylococcus aureus; eco, Escherichia coli; bsu, Bacillus subtilis; ype, Yersinia pestis; sco, Streptomyces coelicolor; kpn, Klebsiella pneumoniae; msm, Mycobacterium smegmatis; lmo, Listeria monocytogenes; cgl, Corynebacterium glutamicum; yps, Yersinia pseudotuberculosis; eta, Erwinia tasmaniensis; sbo, Shigella boydii; sgo, Streptococcus gordonii; ssa, Streptococcus sanguinis; apl, Actinobacillus pleuropneumoniae; pst, Pseudomonas syringae; nmc, Neisseria meningitidis; ste, Streptococcus thermophilus; bma, Burkholderia mallei.
In the genome of S. aureus, tatA (SA0335) and tatC (SA0334), encoding the Tat secretion system, are organized in an operon. Another operon, encoding a lipoprotein (SA0331), an iron-dependent peroxidase (SA0332), and a high-affinity iron transporter (SA0333), is found downstream of tatC. It has been designated the fepABC operon. A region of dyad symmetry resembling a ferric uptake regulatory binding site (Fur box) precedes the lipoprotein gene (fepA). Two REGXE motifs, which have been implicated in iron binding (42), are found in the high-affinity iron transporter protein, at positions 331 to 335 and 449 to 453. The organization of this tat and fep operon cluster is conserved in the genomes of S. aureus, S. carnosus, and S. haemolyticus (Fig. 1A). Homologs of the fep operon are found in several bacteria, including Escherichia coli, Bacillus subtilis, Streptomyces coelicolor, Streptococcus thermophilus, Pseudomonas syringae, Neisseria meningitidis, Yersinia pestis, and Klebsiella pneumoniae.
FIG. 1.
(A) Gene organization of fepABC and tat operons of Staphylococcus. The fepABC operon encodes a lipoprotein, an iron-dependent peroxidase, and a high-affinity iron transporter. Arrows indicate the orientation of the genes, and the circle represents a putative Fur box. (B) Allelic replacement of tatAC operon by an erythromycin resistance cassette in S. aureus and S. carnosus. (C) Allelic replacement of fepABC and tatAC operons by a kanamycin resistance cassette in S. aureus. (D) Relevant part of plasmid pTX-tat, carrying tatAC genes under the control of a xylose-inducible promoter.
Tat deletion mutants of S. carnosus and S. aureus.
To examine the functionality of the Tat pathway in Staphylococcus species, deletion mutants were created in S. carnosus TM300 and S. aureus SA113 by replacing both the tatA and tatC genes with an erythromycin resistance cassette (ermB) (7) (Fig. 1B). The S. aureus Δtat-fep operon was created by replacing the tatAC and fep operons with a kanamycin (kan) resistance cassette (Fig. 1C). The ΔtatAC mutants were complemented with the plasmid pTX-tat, which was constructed by cloning the tat genes under the control of a xylose-inducible promoter (Fig. 1D). The mutant strains from both species are similar to their respective WT strains in terms of growth rate and colony morphology in tryptic soy broth and B medium (data not shown).
FepB, an iron-dependent peroxidase, is a Tat substrate.
To test whether the iron-dependent peroxidase FepB, with a predicted Tat signal sequence, is secreted in a Tat-dependent manner, we examined the presence of peroxidase activity in the culture supernatants of S. carnosus WT, its ΔtatAC deletion mutant, and its complemented mutant, S. carnosus Δtat (pTX-tat). Peroxidase activity was found only in the culture supernatants of S. carnosus WT and its complemented mutant, not in the culture supernatants of the ΔtatAC mutant (Fig. 2B). Similar results were obtained with S. aureus WT and its ΔtatAC mutant.
tat mutants are unable to export lipase fused to the FepB SP.
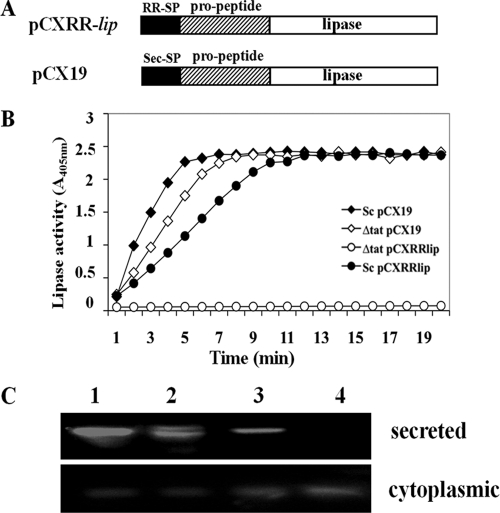
In order to investigate the ability of the Tat system to secrete heterologous proteins in S. carnosus, we fused the Tat SP of FepB to the prolipase (pro-lip) gene (Fig. 3A). The resulting plasmid, pCXRR-lip, was transformed into S. carnosus and S. carnosus ΔtatAC, and their culture supernatants were tested for lipase activity. In contrast to S. carnosus WT(pCXRR-lip), no lipase was detected in the culture supernatants of S. carnosus ΔtatAC(pCXRR-lip) (Fig. 3B). Analysis of the cytoplasmic fractions of the strains revealed the expression of lipase in both S. carnosus WT and the ΔtatAC mutant, suggesting that in the absence of the Tat transport pathway, lipase secretion is blocked and accumulates within the cells of the ΔtatAC mutant (Fig. 3C). This intracellular accumulation of lipase is very likely responsible for the slower growth (data not shown) of S. carnosus ΔtatAC(pCXRR-lip).
FIG. 3.
Expression of lipase fused to RR-SP. (A) Illustration of the primary structures of lipase with the Tat SP and propeptide, as expressed with plasmid pCXRR-lip, and the lipase fused to the Sec SP and propeptide, as expressed with plasmid pCX19. (B) p-Nitrophenyl-caprylate-based lipase assay with culture supernatants of S. carnosus(pCX19) (⧫), S. carnosus ΔtatAC(pCX19) (⋄), S. carnosus(pCXRR-lip) (•), and S. carnosus ΔtatAC(pCXRR-lip) (○). (C) Lipase activity staining (zymogram) of culture supernatants and whole-cell extracts. Lane 1, S. carnosus(pCX19); lane 2, S. carnosus(pCXRR-lip); lane 3, S. carnosus ΔtatAC(pCX19); lane 4, S. carnosus ΔtatAC(pCXRR-lip). Plasmid pCX19 encodes lipase with a Sec SP, while pCXRR-lip encodes lipase with the Tat SP of FepB. The expression of lipase was induced by the addition of 0.5% xylose.
To confirm that the secretion deficiency of lipase in the S. carnosus ΔtatAC mutant was indeed specific for the Tat system, Sec-dependent lipase secretion was also analyzed in this mutant. The secretion efficiencies of the lipase via the Sec machinery in S. carnosus ΔtatAC and its parental S. carnosus strain carrying plasmid pCX19 were comparable, indicating that secretion of the lipase with its Sec SP is not affected in the ΔtatAC mutant (Fig. 3B and C).
The Tat secretion system allows the anchoring of protein A (Spa) to peptidoglycan.
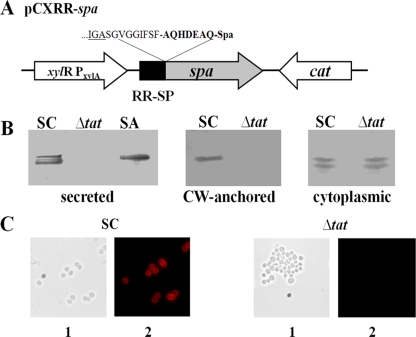
Protein A has a sorting sequence and is covalently anchored to peptidoglycan (41). To find out whether the sortase can recognize a Tat-secreted protein and anchor it to the peptidoglycan, the Tat SP of FepB from S. carnosus was fused with the protein A (spa) gene and transformed via plasmid pCXRR-spa (Fig. 4A) into S. carnosus and S. carnosus ΔtatAC. Protein A expression was monitored by Western blotting. In the culture supernatant and cell wall-associated fractions, protein A was detected only in S. carnosus(pCXRR-spa) and S. aureus (control). However, no protein A was detectable in S. carnosus ΔtatAC(pCXRR-spa) (Fig. 4B). Alexa Fluor 594-conjugated antibody labeling also revealed the anchoring of protein A only in S. carnosus WT(pCXRR-spa), not in S. carnosus ΔtatAC(pCXRR-spa) (Fig. 4C). In the cytoplasmic fraction, protein A was detected in both S. carnosus WT and its ΔtatAC mutant (Fig. 4B), indicating that although protein A is expressed in both strains, it can be neither secreted nor anchored in the ΔtatAC mutant.
FIG. 4.
Expression of pCXRR-spa. (A) Relevant part of plasmid pCXRR-spa encoding protein A under the control of a xylose-inducible promoter. Amino acids at the fusion site are indicated. (B) Western blot analysis of protein A expression in the culture supernatant, cell wall (CW)-anchored, and cytoplasmic fractions of S. carnosus(pCXRR-spa) and S. carnosus ΔtatAC(pCXRR-spa). S. aureus SA113 was used as a control. (C) Immunofluorescence labeling of protein A anchored to the cell wall, using Alexa Fluor 594-conjugated antibody. SC, S. carnosus WT. (1) Phase-contrast image. (2) Protein A visualized by immunofluorescence following antibody labeling.
Exoprotein pattern analysis by 2D gel electrophoresis.
To identify proteins secreted via the Tat pathway, the exoproteins of the S. aureus and S. carnosus parent strains and their respective tatAC mutants were analyzed by 2D gel electrophoresis. However, no marked difference was observed between the parent strains and their ΔtatAC mutants. Although a clear difference in peroxidase activity between S. carnosus WT and its ΔtatAC mutant could be seen, we were unable to identify the peroxidase protein in 2D gels in all four independent assays (data not shown). This result is in accordance with a previous study (48).
Identification of tatAC genes in other staphylococci.
Genome sequence analysis showed that the tatAC operon is present in S. carnosus, S. aureus, and S. haemolyticus and absent in S. epidermidis and S. saprophyticus. PCR analysis with various strains suggested that tatAC genes are also present in S. gallinarum and S. lugdunensis but are absent in quite a number of other staphylococcal species, including S. capitis, S. hyicus, S. lentus, S. muscae, and S. simulans, indicating that the Tat pathway is not a common secretory pathway in staphylococci. Our PCR results were corroborated by the genome sequence databases available for various staphylococcal species (including all sequenced strains of S. aureus and S. epidermidis and sequenced representatives of S. carnosus, S. haemolyticus, and S. saprophyticus).
Role of FepABC in iron transport.
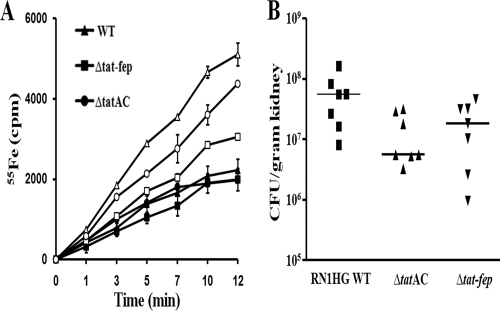
The ability of the fepABC operon to transport iron was investigated using 55Fe uptake assays. S. aureus, S. aureus ΔtatAC, and S. aureus Δtat-fep were grown under iron-sufficient and iron-depleted conditions. Although growth was decreased in iron-deficient medium, no difference in growth rate was observed among the strains (data not shown). Strains grown under iron-sufficient conditions displayed comparable iron uptake rates. A significant decrease in the ability to transport iron was observed in the Δtat-fep mutant compared to the WT. The rate of iron uptake by the ΔtatAC mutant was also decreased compared to that of the WT, although to a lesser degree (Fig. 5A).
FIG. 5.
(A) Iron transport. 55Fe uptake was measured using cells grown under iron-sufficient (closed symbols) and iron-deficient (open symbols) conditions. The strains employed were S. aureus WT (▵), S. aureus ΔtatAC (○), and S. aureus Δtat-fep (□). Values presented are representative of three independent experiments. (B) Role of S. aureus RN1HG TatAC and FepABC in kidney infection. Bacterial loads of parental S. aureus and the tatAC and tat-fep mutant strains in the kidneys of BALB/c mice (n = 7) were determined 5 days after intravenous infection with 7 × 107 CFU. Each dot represents the bacterial load for one animal per gram of kidney.
Comparative virulence in a mouse kidney abscess model.
To assess the role of the tat and fep operons in virulence, the ΔtatAC and Δtat-fep mutations were transduced into strain RN1HG, which is more virulent than strain SA113 (S. Herbert et al., unpublished data) and has functional global regulators, such as agr, sigB, and sarA. Virulence was investigated in a hematogenic kidney abscess model with S. aureus RN1HG WT, RN1HG ΔtatAC, and RN1HG Δtat-fep. Mice were challenged with 7 × 107 CFU via the tail vein, and bacterial loads in kidneys were determined 5 days after infection. Our analysis revealed a significant lower virulence of the ΔtatAC mutant than that of the WT (P = 0.041). The median bacterial load in kidneys for the WT was 1.7 × 107 CFU, compared to 1.7 × 106 CFU for the ΔtatAC mutant. The difference between the WT and Δtat-fep strains was less pronounced, but the bacterial load of the mutant was repeatedly lower than that of the WT (median, 1.7 × 107 versus 5.6 × 106 CFU) (Fig. 5B).
DISCUSSION
The twin-arginine translocation (Tat) pathway exists in many bacteria, archaea, and chloroplasts. In S. aureus, homologs of TatA and TatC have been identified. However, as mentioned in a recent review by Sibbald et al., the functionality of the S. aureus Tat translocase remains to be demonstrated (43). Yamada et al. compared a tatC-deficient mutant with its parent S. aureus strain by 2D gel electrophoresis and found no difference in the amounts of secreted staphylococcal enterotoxins and toxic shock syndrome toxin 1, whose SPs have some features often seen in signal sequences of Tat-dependent proteins, and they finally came to the conclusion that the Tat pathway does not play a major role in the secretion system of S. aureus (48). Here it is shown for the first time that the Tat pathway in S. aureus is functional and serves to translocate the iron-dependent peroxidase FepB.
In the genomes of various staphylococcal species, only one copy each of TatA and TatC could be identified; like the case in Bacillus subtilis, TatA seems to perform the functions of both the TatA and TatB components of E. coli (23). The TatA protein of staphylococci also contains both the phenylalanine residue (F20 in E. coli TatA) which is strictly conserved in the TatA/E proteins and the proline residue (P22 in E. coli TatB) which is strictly conserved in TatB proteins of gram-negative bacteria (see Fig. S1 in the supplemental material). Originally, our search in the S. aureus genome for potential Tat-dependent target proteins with a twin-arginine SP (RR-SP) motif revealed no promising candidates. However, in a search of the S. carnosus genome (36), the RR-SP in the iron-dependent peroxidase FepB was identified. As already mentioned, the RR-SP motif in the iron-dependent peroxidase (FepB) of S. aureus was wrongly annotated and therefore overlooked. Resequencing of the S. aureus fepB gene region indicated that the RR-SP motif was also present in S. aureus FepB.
Apart from FepB, no further protein with an RR-SP motif was discovered in the genome of S. aureus or S. carnosus, even though various search programs were used. Apparently, FepB is very likely the only Tat-dependent protein in staphylococci. As illustrated in Fig. 1A, FepB is encoded in an operon composed of a lipoprotein gene, fepA, the iron-dependent peroxidase gene, fepB, and a high-affinity iron transporter gene, fepC. In the opposite direction are the two TatA- and TatC-encoding genes. The fep-tat genes comprise not only a genetic but also a functional unit, since in the ΔtatAC mutants of S. aureus and S. carnosus the extracellular peroxidase activity is drastically decreased. To see whether other proteins were Tat dependent, we compared the secretomes of S. carnosus WT, S. aureus WT, and their ΔtatAC mutants by 2D gel electrophoresis; however, like Yamada et al. (48), we found no difference in the protein pattern, a further indication that FepB is the only Tat-dependent protein. However, the FepB spot could not be identified, as very likely it was masked by another protein spot. All results speak in favor of the hypothesis that the only function of the Tat system in S. aureus and also in S. carnosus is to translocate FepB. This was a bit surprising, as in many other gram-positive genera, such as Bacillus, Streptomyces, and Mycobacterium, many more proteins appear to be transported by the Tat system.
To our surprise, the analysis of the genomes of various staphylococcal species revealed that the fep-tat gene cluster is not present in all species. It is present in the genomes of all sequenced S. aureus strains as well as in S. carnosus and S. haemolyticus, but it is absent from the genomes of S. epidermidis and S. saprophyticus. By tat-specific PCR analysis, the list of staphylococcal species that possess or lack this gene cluster could be extended (see Results). It is conspicuous that the most aggressive human staphylococcal species, such as S. aureus, S. haemolyticus, and S. lugdunensis, possess the gene cluster, suggesting that it may contribute to aggravating an acute infection. Among those species that lack the gene cluster are S. epidermidis and S. saprophyticus, which are less aggressive and rather play a role in chronic infections. On the other hand, there are two apathogenic species representatives that also contain the fep-tat gene cluster. The natural habitat of S. carnosus is meat (38), and that of S. gallinarum is the chicken crest (13). Presumably, in their particular environments, the iron-dependent peroxidase (FepB) is advantageous. The fact that the fep-tat gene cluster is not a common genus marker raises the question of its origin. Was it once a common part in the genome of the genus and got lost in those species where it was not needed, or was it acquired from other bacteria and kept in those staphylococcal species that benefitted most from the acquisition, such as the aggressive pathogens? There is some indication that at least in S. aureus and S. carnosus the fep-tat cluster might be a relic of a prophage, as immediately upstream of tatA there is a gene (SA0336 and SC2209, respectively) which encodes a protein with similarity to a phage envelope protein present in various staphylococcal phages but also in the phage of Streptococcus mitis SK137 (27). It might be possible that the fep-tat cluster in staphylococci has been spread by phages. On the other hand, in S. haemolyticus the fep-tat cluster is located differently and there is no phage-related gene nearby.
The advantage of the fep-tat cluster could lie in iron uptake and in the external detoxification of H2O2, for example, in oxidative bursts during infection. Indeed, iron uptake under iron limitation was significantly decreased in the fep-tat mutants (Fig. 5A). The clear difference in iron uptake rates was surprising, as S. aureus has various Fe acquisition systems. However, there is only one inorganic Fe transporter (FeoAB), as the others are either siderophore dependent (such as SstABCD, SirABC, and FhuCBG) or transport heme-Fe (IsdDEF and HtsABC) (6). Our Fe transport assay suggests that the FepABC system transports inorganic Fe, with no indication that siderophores like staphyloferrins or aurochelin are involved. If the Fep system is also an inorganic Fe transporter, then its absence explains the observed clear decrease in the iron uptake rate.
To see whether the Fep-Tat system also has an effect in vivo, virulence studies with a mouse hematogenic kidney abscess model were carried out with strain RN1HG, which is an rsbU-repaired 8325 strain and is more virulent than SA113 (Herbert et al., unpublished data). Therefore, the ΔtatAC and Δtat-fep mutations were transduced into RN1HG. Indeed, it turned out that the bacterial load in the kidneys after 5 days was significantly lower for the ΔtatAC and Δtat-fep mutants (Fig. 5B). The ΔtatAC mutant appeared even more attenuated than the Δtat-fep mutant, which might be due to a fitness cost by accumulation of Fep protein in the cytoplasm. These results show that in the presence of the fep-tat cluster, fitness and virulence are increased during infection.
The functionality and specificity of the Tat system in both the S. aureus and S. carnosus backgrounds were also investigated, and it could be demonstrated that FepB is a Tat substrate that is recognized specifically by the Tat system. For the ΔtatAC mutants, the peroxidase activity was drastically decreased in the culture supernatant. The little residual activity seen in the mutants was probably due to some release of the cytoplasmic thioredoxin-dependent thiol peroxidase, which also breaks down H2O2 to water; a corresponding enzyme (SaurTPx) is at least annotated for various staphylococci. It has also been shown that the RR-SP is specific for the Tat system. Lipase fused with the FepB-derived RR-SP can be secreted only if the Tat system is functional, while secretion of lipase with its native SP was not affected in the ΔtatAC mutants. Most of these studies were carried out in the S. carnosus TM300 background, as this strain does not secrete active lipase or protein A that would interfere with monitoring (36). However, the results obtained with S. carnosus were frequently verified with S. aureus and its ΔtatAC mutant, which behaved quite similarly.
It is generally assumed that the Tat secretion system, in contrast to the Sec secretion system, translocates proteins that were completely folded and assembled in the cytoplasm. An interesting question was therefore whether proteins, such as protein A, that are covalently anchored by the sortase to the cell wall (28) can still be anchored when they are translocated by the Tat system. To address this question, the FepB-derived RR-SP was fused with protein A, and we compared its expression in TM300 and its ΔtatAC mutant. As illustrated in Fig. 4B, protein A could only be anchored in the WT, since the ΔtatAC mutant was unable to both translocate protein A to the medium and anchor it to the cell wall, although protein A was expressed in the cytoplasm of the ΔtatAC mutant. It is not known whether protein A is really completely folded during Tat-dependent translocation, but if it is folded it must be folded in such a way that the C-terminal sorting sequence is still accessible to the sortase to tether protein A to the cell wall. The cell wall-bound protein A was further demonstrated by immunofluorescence labeling (Fig. 4C).
The present study shows that the Tat pathway is functional in S. aureus and S. carnosus and that the iron-dependent peroxidase FepB is transported via this pathway. As indicated in Table 2, FepB homologues of other bacterial species from different phyla also have typical Tat signal sequences. Although peroxidase activity was detected in the supernatant of S. carnosus WT and no activity was detected for its ΔtatAC mutant, comparative 2D gel electrophoresis revealed no difference in the protein patterns. It may be that FepB is expressed only at low levels, such that its concentration is below the sensitivity limit of the current 2D gel electrophoresis technique.
Like the accessory Sec system, which exports exclusively one substrate, SraP (44), in S. aureus, the Tat system also appears to have only one substrate, FepB, or it could be that there are more Tat substrates which we failed to detect due to the sensitivity limitations of the techniques used. The most interesting finding was that the proteins routed through the Tat system can also be sortase substrates, and it would be interesting to know more about this mechanism. It would also be of interest to know why FepB should be transported via the Tat pathway.
Supplementary Material
Acknowledgments
We thank Stefan Stevanovic (Institute for Cell Biology, University of Tübingen) for N-terminal sequencing of protein A and Regine Stemmler for technical assistance.
This work was supported by the DFG, graduate college GK685, the SFB766, and the BMBF PathoGenoMik (031U213B).
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. [DOI] [PubMed] [Google Scholar]
- 2.Becker, K., D. Harmsen, A. Mellmann, C. Meier, P. Schumann, G. Peters, and C. von Eiff. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berks, B. C., T. Palmer, and F. Sargent. 2005. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8:174-181. [DOI] [PubMed] [Google Scholar]
- 4.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, R., L. Voggu, U. K. Simon, P. Hentschel, G. Thumm, and F. Götz. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259:260-268. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 7.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 8.Chaddock, A. M., A. Mant, I. Karnauchov, S. Brink, R. G. Herrmann, R. B. Klosgen, and C. Robinson. 1995. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J. 14:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cid, H., M. Bunster, M. Canales, and F. Gazitua. 1992. Hydrophobicity and structural classes in proteins. Protein Eng. 5:373-375. [DOI] [PubMed] [Google Scholar]
- 10.De Buck, E., I. Lebeau, L. Maes, N. Geukens, E. Meyen, L. Van Mellaert, J. Anne, and E. Lammertyn. 2004. A putative twin-arginine translocation pathway in Legionella pneumophila. Biochem. Biophys. Res. Commun. 317:654-661. [DOI] [PubMed] [Google Scholar]
- 11.De Buck, E., L. Maes, E. Meyen, L. Van Mellaert, N. Geukens, J. Anne, and E. Lammertyn. 2005. Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem. Biophys. Res. Commun. 331:1413-1420. [DOI] [PubMed] [Google Scholar]
- 12.Devriese, L. A., V. Hájek, P. Oeding, S. A. Meyer, and K. H. Schleifer. 1978. Staphylococcus hyicus (Sompolinsky 1953) comb. nov. and Staphylococcus hyicus subsp. chromogenes subsp. nov. Int. J. Syst. Bacteriol. 28:482-490. [Google Scholar]
- 13.Devriese, L. A., B. Poutrel, R. Kilpper-Bälz, and K. H. Schleifer. 1983. Staphylococcus gallinarum and Staphylococcus caprae, two new species from animals. Int. J. Syst. Bacteriol. 33:480-486. [Google Scholar]
- 14.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freney, J., Y. Brun, M. Bes, H. Meugneir, F. Grimont, P. A. D. Grimont, C. Nervi, and J. Fleurette. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int. J. Syst. Bacteriol. 38:168-172. [Google Scholar]
- 16.Götz, F., T. Bannerman, and K. H. Schleifer. 2006. The genera Staphylococcus and Macrococcus, p. 5-75. In M. Dworkin (ed.), Procaryotes, vol. 4. Springer, New York, NY. [Google Scholar]
- 17.Götz, F., and K. H. Schleifer. 1975. Purification and properties of a fructose-1,6-diphosphate activated l-lactate dehydrogenase from Staphylococcus epidermidis. Arch. Microbiol. 105:303-312. [DOI] [PubMed] [Google Scholar]
- 18.Götz, F., H. M. Verheij, and R. Rosenstein. 1998. Staphylococcal lipases: molecular characterisation, secretion, and processing. Chem. Phys. Lipids 93:15-25. [DOI] [PubMed] [Google Scholar]
- 19.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 20.Hájek, V., W. Ludwig, K. H. Schleifer, N. Springer, W. Zitzelsberger, R. M. Kroppenstedt, and M. Kocur. 1992. Staphylococcus muscae, a new species isolated from flies. Int. J. Syst. Bacteriol. 42:97-101. [DOI] [PubMed] [Google Scholar]
- 21.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 22.Jongbloed, J. D., H. Antelmann, M. Hecker, R. Nijland, S. Bron, U. Airaksinen, F. Pries, W. J. Quax, J. M. van Dijl, and P. G. Braun. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068-44078. [DOI] [PubMed] [Google Scholar]
- 23.Jongbloed, J. D., U. Grieger, H. Antelmann, M. Hecker, R. Nijland, S. Bron, and J. M. van Dijl. 2004. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54:1319-1325. [DOI] [PubMed] [Google Scholar]
- 24.Kloos, W. E., and K. H. Schleifer. 1975. Isolation and characterization of staphylococci from human skin. II. Description of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int. J. Syst. Bacteriol. 25:62-79. [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.Liebl, W., and F. Götz. 1986. Studies on lipase directed export of Escherichia coli beta-lactamase in Staphylococcus carnosus. Mol. Gen. Genet. 204:166-173. [DOI] [PubMed] [Google Scholar]
- 27.Llull, D., R. Lopez, and E. Garcia. 2006. Skl, a novel choline-binding N-acetylmuramoyl-l-alanine amidase of Streptococcus mitis SK137 containing a CHAP domain. FEBS Lett. 580:1959-1964. [DOI] [PubMed] [Google Scholar]
- 28.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 29.McDonough, J. A., K. E. Hacker, A. R. Flores, M. S. Pavelka, Jr., and M. Braunstein. 2005. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 187:7667-7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meissner, D., A. Vollstedt, J. M. van Dijl, and R. Freudl. 2007. Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different gram-positive bacteria. Appl. Microbiol. Biotechnol. 76:633-642. [DOI] [PubMed] [Google Scholar]
- 31.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peschel, A., B. Ottenwälder, and F. Götz. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279-284. [DOI] [PubMed] [Google Scholar]
- 34.Resch, A., S. Leicht, M. Saric, L. Pasztor, A. Jakob, F. Gotz, and A. Nordheim. 2006. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6:1867-1877. [DOI] [PubMed] [Google Scholar]
- 35.Robinson, C., and A. Bolhuis. 2004. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim. Biophys. Acta 1694:135-147. [DOI] [PubMed] [Google Scholar]
- 36.Rosenstein, R., C. Nerz, L. Biswas, A. Resch, G. Raddatz, S. C. Schuster, and F. Götz. 2009. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 75:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santini, C. L., B. Ize, A. Chanal, M. Muller, G. Giordano, and L. F. Wu. 1998. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleifer, K. H., and U. Fischer. 1982. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153-156. [Google Scholar]
- 39.Schleifer, K. H., and W. E. Kloos. 1975. Isolation and characterization of staphylococci from human skin. I. Amended descriptions of Staphylococcus saprophyticus and descriptions of three new species: Staphylococcus cohnii, Staphylococcus haemolyticus, and Staphylococcus xylosus. Int. J. Syst. Bacteriol. 25:50-61. [Google Scholar]
- 40.Schmollinger, M., I. Fischer, C. Nerz, S. Pinkenburg, F. Gotz, M. Kaufmann, K. J. Lange, R. Reuter, W. Rosenstiel, and A. Zell. 2004. ParSeq: searching motifs with structural and biochemical properties. Bioinformatics 20:1459-1461. [DOI] [PubMed] [Google Scholar]
- 41.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 42.Severance, S., S. Chakraborty, and D. J. Kosman. 2004. The Ftr1p iron permease in the yeast plasma membrane: orientation, topology and structure-function relationships. Biochem. J. 380:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sibbald, M. J., A. K. Ziebandt, S. Engelmann, M. Hecker, A. de Jong, H. J. Harmsen, G. C. Raangs, I. Stokroos, J. P. Arends, J. Y. Dubois, and J. M. van Dijl. 2006. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70:755-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siboo, I. R., D. O. Chaffin, C. E. Rubens, and P. M. Sullam. 2008. Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 190:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauss, A., and F. Götz. 1996. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol. Microbiol. 21:491-500. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wieland, K. P., B. Wieland, and F. Gotz. 1995. A promoter-screening plasmid and xylose-inducible, glucose-repressible expression vectors for Staphylococcus carnosus. Gene 158:91-96. [DOI] [PubMed] [Google Scholar]
- 48.Yamada, K., I. Sanzen, T. Ohkura, A. Okamoto, K. Torii, T. Hasegawa, and M. Ohta. 2007. Analysis of twin-arginine translocation pathway homologue in Staphylococcus aureus. Curr. Microbiol. 55:14-19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.