Abstract
Growth phase-dependent gene regulation has recently been demonstrated to occur in Bordetella pertussis, with many transcripts, including known virulence factors, significantly decreasing during the transition from logarithmic to stationary-phase growth. Given that B. pertussis is thought to have derived from a Bordetella bronchiseptica-like ancestor, we hypothesized that growth phase-dependent gene regulation would also occur in B. bronchiseptica. Microarray analysis revealed and quantitative real-time PCR (qRT-PCR) confirmed that growth phase-dependent gene regulation occurs in B. bronchiseptica, resulting in prominent temporal shifts in global gene expression. Two virulence phenotypes associated with these gene expression changes were tested. We found that growth-dependent increases in expression of some type III secretion system (TTSS) genes led to a growth phase-dependent increase in a TTSS-dependent function, cytotoxicity. Although the transcription of genes encoding adhesins previously shown to mediate adherence was decreased in late-log and stationary phases, we found that the adherence of B. bronchiseptica did not decrease in these later phases of growth. Microarray analysis revealed and qRT-PCR confirmed that growth phase-dependent gene regulation occurred in both Bvg+ and Bvg− phase-locked mutants, indicating that growth phase-dependent gene regulation in B. bronchiseptica can function independently from the BvgAS regulatory system.
Bordetellae are gram-negative bacterial respiratory pathogens. Bordetella pertussis and Bordetella parapertussishu, the causative agents of whooping cough, are human-adapted variants of Bordetella bronchiseptica, which naturally infects a broad range of mammals, causing a range of diseases from lethal pneumonia to asymptomatic chronic infection (12, 33). Comparative genome sequencing analysis of these three Bordetella strains has revealed a loss or inactivation of a large number of genes in B. pertussis and B. parapertussishu, compared to B. bronchiseptica, indicating that host restriction seems to be primarily due to the loss of genes or gene function, as opposed to acquisition of foreign DNA (32). These results have led to the proposal that B. bronchiseptica is the closest evolutionary ancestor of these strains (32, 33).
A universal underlying pathogenic mechanism is shared among these Bordetella strains in that the majority of virulence gene expression is regulated by a two-component sensory transduction system encoded by the bvg locus. This locus comprises a histidine kinase sensor protein, BvgS, and a DNA-binding response-regulator protein, BvgA. In response to environmental cues, BvgAS controls expression of a spectrum of phenotypic phases transitioning between a virulent (Bvg+) phase and a nonvirulent (Bvg−) phase, a process referred to as phenotypic modulation. During the virulent Bvg+ phase, the BvgAS system is fully active and many of the known virulence factors are expressed, such as filamentous hemagglutinin (FHA), pertactin, fimbriae, adenylate cyclase-hemolysin toxin, and dermonecrotic toxin (DNT), as well as a type III secretion system (TTSS) (5). Conversely, BvgAS is inactive during the Bvg− phase, resulting in the maximal expression of motility loci, virulence-repressed genes (vrg genes), and genes required for the production of urease (2, 3, 26). Previous studies involving phase-locked and ectopic expression mutants demonstrated that the Bvg+ phase promotes respiratory tract colonization by B. pertussis and B. bronchiseptica (1, 6, 7, 23, 27), while the Bvg− phase of B. bronchiseptica promotes survival under conditions of nutrient deprivation (6, 7).
Despite the close genetic relatedness and sharing a key pathogenic mechanism involving the BvgAS regulatory system, B. pertussis and B. bronchiseptica differ in some interesting yet fundamental attributes of bacterial pathogens such as host range, pathologies, and persistence. Recently, growth phase-dependent gene expression changes have been reported to occur in B. pertussis, with many transcripts, including known virulence factors, significantly decreasing during the transition from logarithmic phase to stationary phase (28). Given that B. pertussis is thought to have derived from a B. bronchiseptica-like ancestor (32, 33), we hypothesized that growth phase-dependent gene regulation in B. bronchiseptica would occur in a manner analogous to that in B. pertussis and influence virulence factor expression and virulence-associated phenotypes. Additionally, the data arising from this comparative analysis may advance our understanding of the molecular basis for the differences between these species. Using microarray analysis and quantitative real-time PCR (qRT-PCR), we demonstrate that growth phase-dependent gene regulation occurs in B. bronchiseptica and results in large shifts in global gene expression during growth. Growth phase-dependent changes in two virulence phenotypes associated with these gene expression changes were tested. We found that the growth-dependent increase in expression of some TTSS genes led to a growth-dependent increase in a TTSS-dependent function, cytotoxicity. Additionally, while genes encoding adhesins previously shown to mediate adherence were decreased in late log and stationary phases, in contrast to B. pertussis (28), we found that B. bronchiseptica adherence did not decrease in these later phases of growth. It has been previously shown that a Bvg+ phase-locked B. pertussis mutant failed to exhibit growth-dependent gene regulation, indicating that a BvgAS system capable of modulating is required for growth-dependent gene regulation (28). Thus, to broadly evaluate the role of the BvgAS regulatory system in B. bronchiseptica growth phase-dependent gene regulation, the transcriptional profiles of both Bvg+ and Bvg− phase-locked mutants were assessed during growth. Microarray analysis revealed and qRT-PCR confirmed growth phase-dependent global shifts in gene expression occurring in both phase-locked mutants. Therefore, in contrast to B. pertussis, growth phase-dependent gene regulation in B. bronchiseptica can function independently from the BvgAS regulatory system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. bronchiseptica strains RB50, RB53 (a Bvg+ phase-locked derivative of RB50), RB54 (a Bvg− phase-locked derivative of RB50), and RB50ΔbscN (an isogenic mutant lacking the putative ATPase required for the function of the TTSS apparatus) have been previously described (6, 43). B. bronchiseptica strains RB63 (ΔfimBCD), RBX9 (ΔfhaB), and SP5 (Δprn) lacking single adhesins have been characterized previously (8, 10, 24). All B. bronchiseptica strains were cultured on Bordet-Gengou (BG) agar (Difco, Sparks, MD) containing 10% defibrinated sheep's blood for determination of colony morphology and hemolytic activity or in Stainer-Scholte (SS) broth (39) supplemented with 40 μg/ml streptomycin. Beta-hemolysis on BG agar was verified following growth in liquid cultures to ensure that bacteria remained Bvg+ and were not spontaneous Bvg− mutants. For all growth phase time course experiments, a single colony was inoculated in SS broth supplemented with 40 μg/ml streptomycin at 37°C with shaking. To ensure similar inocula, bacteria were then subcultured at a starting optical density at 600 nm (OD600) of 0.02 into a 250-ml flask containing 50 ml SS broth and grown at 37°C with shaking at 275 rpm. This was repeated twice, resulting in three biological replicates for each strain.
Measurement of growth by optical density, colony counts, and DNA concentration.
Growth was monitored by removing culture samples at 6-h intervals and measuring the OD600 and determining the number of CFU of B. bronchiseptica per ml of culture by plating dilutions on BG plates containing 40 μg/ml streptomycin. Additionally, growth was monitored by measuring the DNA concentration by absolute quantification of genomic DNA (gDNA).
Absolute quantification of gDNA.
B. bronchiseptica RB50 gDNA was purified using the High Pure PCR template preparation kit (Roche Applied Science, Indianapolis, IN). A 53-bp qPCR amplicon of the 16S rRNA gene was amplified using the 16S rRNA forward and reverse primers (see data set S1 in the supplemental material) and cloned into the pCR-XL-TOPO vector (Invitrogen, Carlsbad, CA). The resulting plasmid, pSMS75-16SFrag, was confirmed by sequencing and quantified by UV spectrophotometry. A qPCR amplicon standard set was created by serial 10-fold dilution of pSMS75-16SFrag. In this set, 2 μl of each standard contained 3 × 102, 3 × 103, 3 × 104, 3 × 105, 3 × 106, or 3 × 107 copies of the 16S rRNA qPCR amplicon or target sequence. Growth phase samples were prepared by collecting 200-μl aliquots, in triplicate, from RB50, RB53, and RB54 cultures at 6-hour intervals. Samples were heated to 95°C for 10 min and immediately frozen at −20°C (36). For use in the absolute qPCR assay, the samples were diluted 1:100 in H2O and incubated with 0.25 μg RNase A (Invitrogen, Carlsbad, CA) for 30 min at 37°C. Absolute qPCR was performed in 26-μl reaction volumes containing 1× SYBR green PCR master mix (Applied Biosystems, Foster City, CA), 0.4 mM each of 16S rRNA forward and reverse primers (see data set S1 in the supplemental material), and 2 μl of each growth phase sample. Real-time PCRs were run in 96-well plates on an ABI 7300 real-time PCR system using the following cycling conditions: activation stages of 2 min at 50°C and 10 min at 95°C; 40 cycles of 15 s at 95°C and 1 min at 60°C; and a dissociation stage of 15 s at 95°C, 30 s at 60°C, and 15 s at 95°C. Reactions for all growth phase samples for each strain and 2 μl of each qPCR amplicon standard, with the copy numbers specified above as the template, were run in triplicate on each plate. Creation of a standard curve relating the cycle threshold (CT) values of the standards to target sequence copy number was performed using the SDS v1.3.1 software (Applied Biosystems, Foster City, CA). This standard curve was then used to determine the 16S rRNA target sequence copy number in growth phase samples based on their CT values and reported as the number of genomic 16S rRNA target copies present or genome equivalents (geq) per ml of culture. RB50 genomic data showing that a single B. bronchiseptica genome contains three copies of the 16S rRNA gene were accounted for in this calculation (32).
RNA isolation, preparation of labeled cDNA, and microarray analysis.
Samples were collected, at indicated time points, from three cultures of RB50, RB53, or RB54. Total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA), treated with RNase-free DNase I (Invitrogen, Carlsbad, CA), and purified using RNeasy columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. A two-color hybridization format was used, and dye swap experiments were performed. For each reaction, 5 μg of cDNA representing three distinct phases of growth, mid-logarithmic phase (15 h), late log phase (24 h), and late stationary phase (48 h), was fluorescently labeled and directly compared with other reaction mixtures as previously described (4, 29). The two differentially labeled reaction mixtures to be compared were combined and hybridized to a B. bronchiseptica strain RB50-specific long-oligonucleotide microarray (29). Slides were then scanned using a GenePix 4000B microarray scanner and analyzed with GenePix Pro software (Axon Instruments, Union City, CA). Spots were assessed visually to identify those of low quality, and arrays were normalized so that the median of the ratio across each array was equal to 1.0. Spots of low quality were identified and were filtered out prior to analysis. Ratio data from the two biological replicates were compiled and normalized based on the total Cy3 percent intensity and Cy5 percent intensity to eliminate slide-to-slide variation. Gene expression data were then normalized to 16S rRNA. The statistical significance of the gene expression changes observed was assessed by using the significant analysis of microarrays program (41). A one-class unpaired significant analysis of microarrays using a false-discovery rate of 0.001% was performed. Hierarchical clustering of microarray data using Euclidean distance metrics and average linkage clustering was performed using MeV software from TIGR (35).
qRT-PCR.
DNase-treated total RNA (1 μg) from each biological replicate was reverse transcribed using 300 ng of random oligonucleotide hexamers and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The resulting cDNA was diluted 1:100, and 1 μl of this dilution was used in qPCR mixtures containing 300 nM primers and 2× SYBR green PCR master mix (Applied Biosystems, Foster City, CA) using an Applied Biosystems 7300 real-time PCR detection system (Applied Biosystems, Foster City, CA). All primers were designed using Primer Express software (Applied Biosystems, Foster City, CA) and are available in the supplemental material (see data sets S1, S2, and S3). To confirm the lack of DNA contamination, reactions without reverse transcriptase were performed. Dissociation curve analysis was performed for verification of product homogeneity. Threshold fluorescence was established within the geometric phase of exponential amplification, and the CT value for each reaction was determined. The CT values from all three biological replicates for each strain were compiled, and the 16S rRNA amplicon was used as an internal control for data normalization. Change in transcript level was determined using the relative quantitative method (ΔΔCT) (19).
Cytotoxicity assays.
Cytotoxicity assays were carried out as previously described (25, 43). Briefly, J774 macrophages were cultured in Dulbecco modified Eagle medium (DMEM) broth supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 1% nonessential amino acids, and 1% sodium pyruvate to 85% confluence at 37°C with 5% CO2. Warmed RPMI medium lacking phenol red, with 5% FBS, 1% l-glutamine, 1% nonessential amino acids, and 1% sodium pyruvate was then used to replace the DMEM. Bacterial infections using the indicated bacterial strains were prepared from cultures collected in mid-log phase (15 h), late log phase, and stationary phase (48 h). Bacteria were washed with phosphate-buffered saline and adjusted to a multiplicity of infection (MOI) of 10. Bacterial suspensions were then centrifuged onto the macrophage cells at 250 × g for 5 min and incubated at 37°C with 5% CO2 for 4 hours. The cell culture supernatants were collected, and percent lactate dehydrogenase (LDH) release was analyzed using the Cytotox96 kit (Promega, Madison, WI) according to the manufacturer's instructions. Results were analyzed for significance using the Student t test, and a P value of less than 0.05 was considered significant.
Bacterial adhesion assays.
Rat lung epithelial (L2) cells were grown to ∼80% confluence in a 24-well plate using 50/50 DMEM-F-12 medium supplemented with 10% FBS and then inoculated with bacterial suspensions prepared from cultures collected in mid-log phase (15 h), late log phase, and stationary phase (48 h) diluted to 2.1 × 108 CFU/ml, resulting in an MOI of 100, using growth medium. Dilutions of bacterial inocula were plated on BG plates containing 40 μg/ml streptomycin to determine CFU counts and verify the MOI. Adhesion assays were carried out in triplicate and performed by removing growth medium from L2 cells and then adding either medium alone or 1 ml of each bacterial inoculum to L2 cells. Plates were centrifuged for 5 min at 250 × g and incubated at 37°C with 5% CO2 for 40 min. Wells were then washed four times with 1 ml of the growth medium to remove nonadherent bacteria. L2 cells were then trypsinized using 0.5 ml of 0.125% trypsin and incubated for 10 min at 37°C. The total volume of each well was then brought up to 1 ml with growth medium and homogenized by pipetting. Dilutions were plated on BG plates containing 40 μg/ml streptomycin to determine CFU counts, which were used to calculate the proportion of adherent bacteria, expressed as a percentage of the original inoculum. Results were analyzed for significance using the Student t test, and a P value of less than 0.05 was considered significant.
Microarray data accession number.
All microarray data are available in the supplemental material and have been deposited in Array Express under accession number E-MEXP-2015.
RESULTS
Growth phase-dependent gene regulation occurs in B. bronchiseptica.
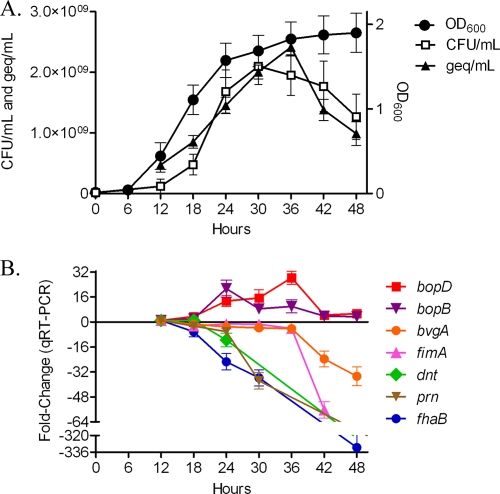
To characterize the different stages of growth for B. bronchiseptica, growth of strain RB50 was monitored by optical density, CFU counts, and DNA concentrations over time (Fig. 1A). A decrease in both recoverable CFU and DNA concentrations was observed during late stationary phase for B. bronchiseptica RB50 (Fig. 1A). Given that virulence factor gene expression in B. pertussis decreases in a growth phase-dependent manner (28), we began evaluating temporal virulence gene expression changes in B. bronchiseptica, by monitoring the expression of seven genes encoding BvgAS-regulated virulence factors (15, 20, 34, 42, 43) at 6-hour intervals starting at early log phase (12 h) and continuing until stationary phase (48 h) using qRT-PCR. All transcripts exhibited growth phase-dependent gene expression changes, albeit in a divergent manner. Transcription of the TTSS effector genes bopB and bopD increased in a growth-dependent manner, with maximal expression of bopB occurring during late log phase (24 h) and maximal expression of bopD occurring during stationary phase (36 h) (Fig. 1B). bvgA expression began a threefold decrease during late log phase (24 h) and continued a fourfold decrease during stationary phase (30 h to 36 h), followed by a substantial 23-fold decrease (42 h) and then a 34-fold decrease (48 h) during late stationary phase (Fig. 1B). Transcription of fimA remained relatively consistent until late stationary phase (36 h), at which time expression severely decreased (Fig. 1B). Lastly, gene expression for fhaB and dnt decreased throughout both log and stationary phases of growth (Fig. 1B). These data suggest that multiple subsets of B. bronchiseptica virulence factors are differentially regulated by growth phase.
FIG. 1.
Growth curves and changes in virulence gene expression of B. bronchiseptica strain RB50. (A) DNA concentrations (geq/ml) and CFU counts (CFU/ml) during growth of RB50 are plotted along the left y axis; optical densities (OD600) are plotted along the right y axis; and time, given in hours, is plotted along the x axis. All data points represent averages obtained from triplicate cultures. (B) Change in gene expression (y axis) occurring at 6-h intervals (x axis), starting at early log phase (12 h) and continuing until late stationary phase (48 h), determined by using qRT-PCR. Data shown are averages obtained from triplicate cultures. The error bars represent standard deviations.
Next, we sought to determine if there are larger sets of genes differentially expressed under different growth phase conditions and to identify novel subsets of similarly regulated genes, implicating involvement in growth phase-dependent functions. Microarray analysis was performed to determine if the growth-dependent gene expression changes were specific to these subsets of B. bronchiseptica virulence factors or occurred on a global scale. For this analysis, cDNAs representing three distinct phases of growth, mid-log phase (15 h), late log phase (24 h), and late stationary phase (48 h), were directly compared. These time points were chosen based on the temporal virulence gene expression changes (Fig. 1B) in an effort to evaluate the maximal changes in gene transcription occurring during these distinct phases of growth.
Evaluating the transcriptional changes specifically occurring during mid-log- to late-log-phase growth revealed a prominent shift in gene expression, with the majority of genes decreasing in transcript levels. There were 1,641 genes found to be significantly regulated by growth, with an overwhelming majority of them (1,614 genes) being downregulated during late-log-phase growth (Fig. 2A; see also data set S1 in the supplemental material). These downregulated genes include 249 metabolism-related transcripts, 234 hypothetical or predicted/probable genes, 111 biosynthesis-related transcripts, 101 transcriptional regulators, 109 periplasmic/exported or lipoprotein genes, and 108 cell envelope genes including members of the lipopolysaccharide (LPS) and O-antigen loci (see data set S1 in the supplemental material). Of the 27 genes found to be significantly upregulated, nine are predicted to be involved in the TTSS and eight within the bsc locus and bopC, a TTSS effector gene located outside the bsc locus. Additionally, four heat shock and chaperone genes (dnaJ, dnaK, groEL, and groES) were also significantly upregulated (see data set S1 in the supplemental material).
FIG. 2.
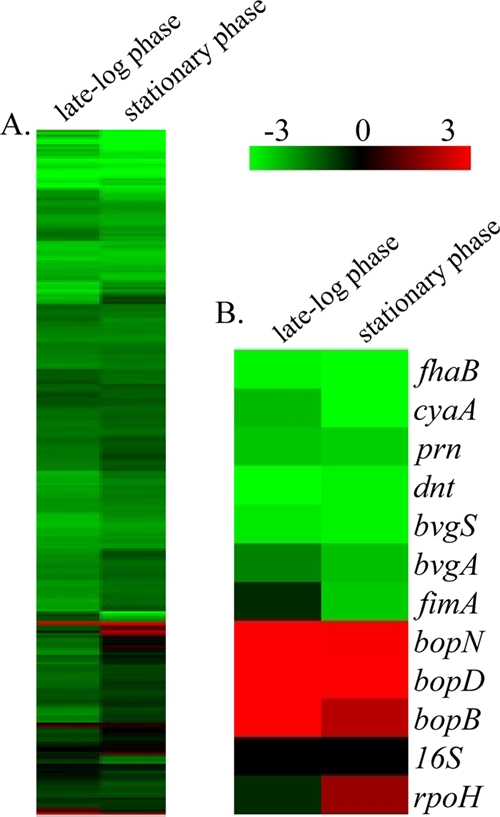
Hierarchical clustering of the transcriptional response of B. bronchiseptica strain RB50 during late-log-phase and stationary-phase growth. (A) Expression profiles representing global transcriptional changes. (B) Expression profiles representing transcriptional changes of genes positively regulated by BvgAS, along with 16S RNA and rpoH. Transcriptional changes identified by comparing cDNA from mid-log phase (15 h) to that from late log phase (24 h) are represented under “late-log phase,” and the transcriptional changes identified by comparing cDNA from mid-log phase (15 h) to that from late stationary phase (48 h) are represented under “stationary phase.” Data are mean centered for each array element and averaged from three biological replicates. All expression profiles of genes are in rows and are represented using the color scale at top, with gray indicating missing data.
In focusing on the transcriptional changes occurring during mid-log to stationary phases of growth, 1,596 transcripts were identified as significantly regulated by growth during this time (Fig. 2A). There were 1,538 genes identified as significantly downregulated, and they included 214 hypothetical or predicted/probable genes, 213 metabolism-related transcripts, 194 transport genes, 111 periplasmic/exported or lipoprotein genes, 90 biosynthesis-related transcripts, 112 cell envelope genes including members of the LPS and O-antigen loci, and 88 transcriptional regulators (see data set S1 in the supplemental material). Fifty-eight genes were identified as being significantly upregulated during stationary phase, including six periplasmic/exported or lipoprotein genes, six transcriptional regulators (including rpoH), five metabolism-related genes, and five TTSS-related transcripts (see data set S1 in the supplemental material).
Traditionally, the paradigm for the study of bacteria is growth in suspension at mid-log phase, which is likely due to the fact that this point reflects an abundance of nutrients, minimal amounts of harmful by-products, and an optimal growth rate. Thus, using cDNA from mid-log phase (15 h) as a reference allowed for the identification of growth phase transcriptional changes occurring specifically during stationary phase or transcriptional changes occurring throughout both log and stationary phases. Of the 1,538 significantly downregulated transcripts identified during stationary phase, 945 were significantly downregulated during log phase as well, indicating that downregulation of these genes began in log-phase growth. Examples of such transcripts include fhaB, dnt, and prn, encoding well-characterized virulence factors (Fig. 1B and 2B; see also data set S1 in the supplemental material). Similarly, of the 58 genes identified as being significantly upregulated during stationary phase, 19 were also significantly upregulated during log phase, indicating that upregulation of these genes initiated during log-phase growth. Examples of these transcripts include the TTSS-secreted effectors bopB and bopD (Fig. 1B and 2B; see also data set S1 in the supplemental material). Focusing on gene expression changes occurring specifically during stationary phase, we identified two small gene clusters. Gene expression for the first cluster of transcripts, referred to as the fimA cluster (see data set S1 in the supplemental material), remained relatively consistent until late stationary phase (36 h), at which time expression significantly decreased. This cluster includes fimA (Fig. 1B and 2B; see also data set S1 in the supplemental material), as well as 17 metabolism-related genes, 14 transport-related genes, 12 periplasmic/exported or lipoprotein genes, and nine TTSS-related transcripts (see data set S1 in the supplemental material). The second gene cluster, referred to as the rpoH cluster, includes 62 genes that significantly increased in expression specifically during stationary-phase growth. Along with rpoH (Fig. 2B; see also data set S1 in the supplemental material), four other transcriptional regulators, 11 hypothetical or predicted/probable genes, 10 transport-related genes, seven metabolism-related transcripts, and seven periplasmic/exported or lipoprotein genes were also identified within this cluster (see data set S1 in the supplemental material). To provide an independent assessment of microarray expression measurements, genes were selected to reflect the full spectrum of changes occurring during both log and stationary phases of growth and analyzed by qRT-PCR. These results provided data consistent with the quantitative measures by microarray analysis (r = 0.96 and r = 0.92), thus providing independent verification of the microarray results (see data set S1 in the supplemental material). Combined, these data demonstrate a pattern of growth phase-dependent gene regulation in B. bronchiseptica.
B. bronchiseptica TTSS-mediated cytotoxicity increases in a growth-dependent manner.
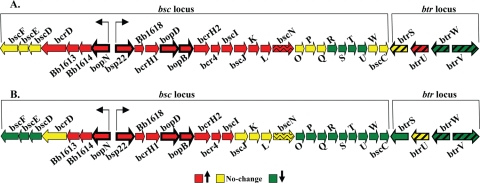
Genes encoding the components of the Bordetella TTSS, including the secretion apparatus, secreted effector proteins, and putative chaperones, are located within the bsc locus (25). Directly adjacent to this locus is the btr locus, which contains genes encoding the regulatory elements for TTSS (25). Specifically, btrS is required for expression of TTSS genes, btrU and btrW are required for secretion, and btrV is required for translation and/or stability of TTSS proteins (17, 25). Microarray analysis revealed growth-dependent transcriptional changes occurring within both loci during both late-log-phase (Fig. 3A) and stationary-phase (Fig. 3B) growth; however, these changes in gene transcription were not similar in that the expression of some genes did not change while the expression for others either increased or decreased in a growth-dependent manner. For example, expression of bopN, bsp22, bopD, and bopB, which encode secreted effector proteins, increased during late log phase (Fig. 3A). Conversely, the expression of other genes within the bsc locus (bscR, bscS, bscT, and bscU) decreased during late-log-phase growth, while the expression of others did not change (bscF, bscE, bscD, bscO, bscP, bscQ, bscW, and bscC) (Fig. 3A). Genes within the btr locus also exhibited expression patterns that differed in late log and stationary phases. Transcription of btrU increased, btrS gene expression did not change, and the expression of btrW and btrV decreased during late-log-phase growth (Fig. 3A). Similarly, differential transcriptional patterns were detected for genes located in both the bsc and btr loci during stationary-phase growth as well (Fig. 3B). Expression of the secreted effectors bopN, bsp22, bopD, and bopB increased, while expression of other TTSS genes either decreased (bscF, bscE, bscD, btrW, and bscV) or remained unchanged (bcrD, bscN, and btrU) during stationary phase (Fig. 3B). These growth-dependent gene expression changes occurring within both loci obtained by microarray analysis were also confirmed by qRT-PCR (see data set S1 in the supplemental material).
FIG. 3.
Gene expression changes occurring in the bsc and btr loci from B. bronchiseptica strain RB50 during late-log-phase (A) and stationary-phase (B) growth. Data are mean centered for each array element and averaged from three biological replicates. All expression profiles are represented using the color scale at bottom. Genes encoding known effector proteins (bopN, bsp22, bopD, and bopB) are shown as bold arrows. The bscN gene, which encodes the known ATPase, is shown by an arrow with wavy lines. Regulatory elements btrS, btrU, btrW, and btrV are shown by arrows with slanted bold solid lines.
Given that gene expression of several members of the TTSS locus, including bopN, bsp22, bopD, and bopB, which encode secreted effector proteins, increased in a growth-dependent manner, we hypothesized that the biological function of the TTSS would increase in a growth-dependent manner as well. A well-characterized function of the Bordetella TTSS is killing of mammalian cells (11, 25, 43). Therefore, to test whether TTSS-mediated cytotoxicity increased in a growth-dependent manner, we measured the release of LDH from macrophages infected with B. bronchiseptica cultures grown to different growth phases. Macrophages treated with medium alone released 2.8% cytoplasmic LDH (Fig. 4). Upon infection with the same dose of bacteria, macrophages infected with RB50 from cultures collected in mid-log phase (15 h), late log phase (24 h), and stationary phase (48 h) released 51%, 88%, and 95% of their LDH, respectively (Fig. 4). This indicates that the cytotoxicity caused by RB50 from late-log-phase (24 h) and stationary-phase (48 h) cultures was greater than the cytotoxicity caused by RB50 from a mid-log-phase (15 h) culture (P < 0.001) (Fig. 4). To confirm that the cytotoxicity induced by RB50 from cultures collected from different growth phases was caused by the TTSS, a previously characterized TTSS-null mutant, RB50ΔbscN (43), was used in the J774 macrophage assay. LDH release caused by infection with RB50ΔbscN was at or below detectable levels for cultures collected in all phases of growth, indicating that the TTSS from RB50 caused the cytotoxicity toward macrophages (Fig. 4). Combined, these data show that the growth phase-dependent increase in expression of TTSS genes correlates with the growth phase-dependent increase in cytotoxicity, a TTSS-dependent function.
FIG. 4.
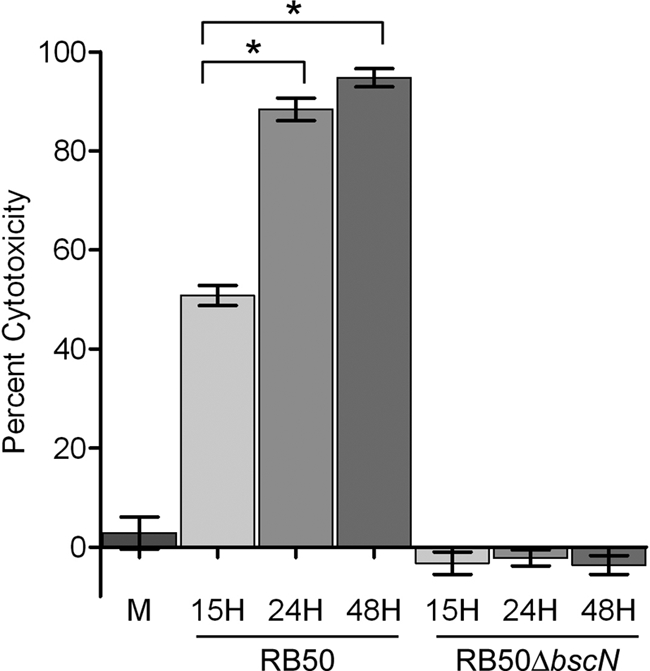
Growth phase-dependent changes in B. bronchiseptica TTSS-mediated cytotoxicity as measured by percent LDH release. J774 macrophages treated with medium alone (M) or infected with RB50 or RB50ΔbscN from cultures collected in mid-log phase (15 h), late log phase (24 h), and stationary phase (48 h) for 4 h at an MOI of 10. The error bars represent standard deviations. An asterisk indicates that the P value is <0.001.
B. bronchiseptica adherence does not decrease in a growth-dependent manner.
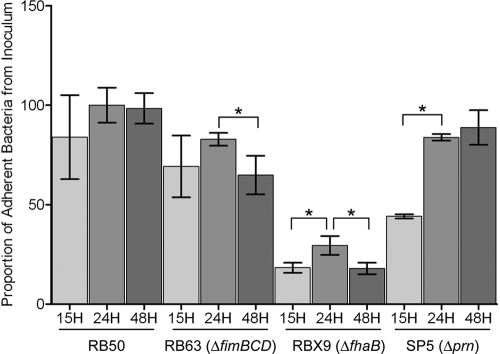
With the exception of TTSS-related genes, expression of most of the characterized virulence genes was decreased in late log and stationary phases relative to mid-log phase. Most notably, this included expression of genes encoding known adhesins, such as fhaB, prn, and fimABCD (8, 10, 24, 30). Similarly, genes encoding known adhesins in B. pertussis have recently been shown to decrease in a growth-dependent manner, along with a growth-dependent decrease in adherence (28). Therefore, we hypothesized that in vitro adherence of B. bronchiseptica would decrease as well. To test this, we compared cultures of B. bronchiseptica RB50, along with previously characterized B. bronchiseptica mutants lacking single adhesins in FHA, pertactin, or fimbriae (8, 10, 24), collected in mid-log phase (15 h), late log phase (24 h), and stationary phase (48 h), for the ability to adhere to a rat epithelial (L2) cell line in a standard adherence assay. We found no statistical difference in the adherence of RB50 to epithelial cells between bacteria grown to mid-log phase (15 h), late log phase (24 h), or stationary phase (48 h) (Fig. 5). In comparing the adherence levels of a B. bronchiseptica fimbria-null mutant, RB63 (ΔfimBCD) (24), we found no statistical difference in the adherence of RB63 to epithelial cells between bacteria grown to mid-log phase (15 h) and late log phase (24 h); however, significantly fewer stationary-phase (48-h) bacteria than bacteria grown to late log phase (24 h) adhered to epithelial cells (P < 0.05) (Fig. 5). Using an FHA-null mutant, RBX9 (ΔfhaB) (8), we found that the adherence of bacteria grown to mid-log phase (15 h) was significantly less than that of bacteria grown to late log phase (24 h) (P < 0.05) and significantly fewer stationary-phase (48-h) bacteria than bacteria grown to late log phase (24 h) adhered to epithelial cells (P < 0.05) (Fig. 5). When we compared the adherence levels of a B. bronchiseptica pertactin-null mutant, SP5 (Δprn) (10), we found that the adherence of bacteria grown to mid-log phase (15 h) was significantly less than that of bacteria grown to late log phase (24 h) (P < 0.05); however, we found no statistical difference in the adherence of SP5 (Δprn) to epithelial cells between bacteria grown to late log phase (24 h) and bacteria grown to stationary phase (48 h) (Fig. 5). In summary, FHA, pertactin, and fimbriae all contribute to B. bronchiseptica adherence, with FHA providing the greatest contribution of these adhesins; however, the contribution of each adhesin differs in a growth-dependent manner. Taken together, these data indicate that despite the growth phase-dependent decrease in expression of most known virulence genes and adhesins, and in contrast to B. pertussis Tohama I (28), B. bronchiseptica RB50 adherence does not decrease in a growth phase-dependent manner.
FIG. 5.
Growth phase-dependent changes in B. bronchiseptica adherence to L2 epithelial cells. L2 cells were incubated with RB50, RB63 (ΔfimBCD), RBX9 (ΔfhaB), or SP5 (Δprn) from cultures collected in mid-log phase (15 h), late log phase (24 h), and stationary phase (48 h) for 40 min at an MOI of 100. Adherence is expressed as the proportion of adherent bacteria to that in the original inoculum. The error bars represent standard deviations. An asterisk indicates that the P value is <0.05.
Growth phase-dependent gene regulation occurs in a Bvg+ phase-locked B. bronchiseptica mutant.
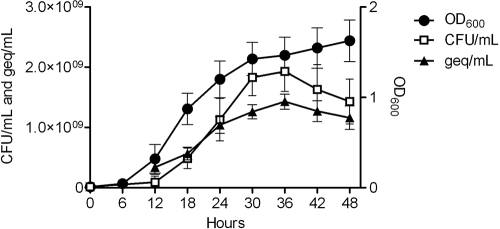
It has recently been shown that Tohama I-Sc3, a Bvg+ phase-locked B. pertussis mutant, failed to exhibit growth-dependent gene regulation, indicating that a BvgAS system capable of modulating is required for growth-dependent gene regulation. Given that BvgAS is the major virulence regulator in Bordetella and many of the classical virulence genes exhibited growth phase-dependent transcriptional changes and the requirement of a modulatable BvgAS system in B. pertussis, we sought to determine if, similar to B. pertussis, the global shifts in gene expression measured in RB50 would require a modulatable BvgAS system. To begin, we first monitored the growth of RB53, a Bvg+ phase-locked derivative of RB50, by optical density, CFU counts, and DNA concentrations over time (Fig. 6). While all growth measurements were generally lower for RB53 than for RB50, similar temporal growth patterns were observed in that both strains exhibited a decrease in recoverable CFU and DNA concentrations during late stationary phase (Fig. 1A and 6). Next, microarray analysis was performed to determine if growth-dependent changes in gene expression occurred on a global scale in RB53 by directly comparing cDNAs representing three distinct phases of growth, mid-log phase (15 h), late log phase (24 h), and late stationary phase (48 h), in the same manner used to evaluate its parental strain, RB50. This analysis revealed prominent growth phase-dependent global shifts in gene expression occurring in RB53, similar to the global transcriptional patterns seen in RB50 (Fig. 7A).
FIG. 6.
Growth curves for B. bronchiseptica strain RB53, a Bvg+ phase-locked mutant. Optical densities (OD600), DNA concentrations (geq/ml), and CFU counts (CFU/ml) during growth of RB53 are plotted along the y axis, and time, given in hours, is plotted along the x axis. All data points represent averages obtained from triplicate cultures, and error bars represent standard deviations.
FIG. 7.
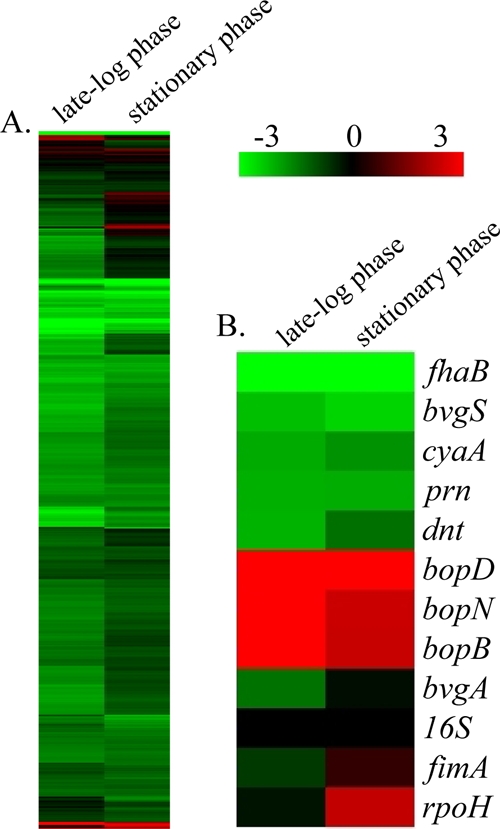
Hierarchical clustering of the transcriptional response of B. bronchiseptica strain RB53, a Bvg+ phase-locked mutant, during late-log-phase and stationary-phase growth. (A) Expression profiles representing global transcriptional changes. (B) Expression profiles representing transcriptional changes of genes positively regulated by BvgAS, along with 16S RNA and rpoH. Transcriptional changes identified by comparing cDNA from mid-log phase (15 h) to that from late log phase (24 h) are represented under “late-log phase,” and the transcriptional changes identified by comparing cDNA from mid-log phase (15 h) to that from late stationary phase (48 h) are represented under “stationary phase.” Data are mean centered for each array element and averaged from three biological replicates. All expression profiles of genes are in rows and are represented using the color scale at top, with gray indicating missing data.
The transcriptional changes occurring during mid-log to late log phases of growth in RB53 comprise 1,545 genes, with the majority of them (1,497 genes) being significantly downregulated (Fig. 7A). These downregulated genes include 225 metabolism-related transcripts, 208 hypothetical or predicted/probable genes, 163 transport-related genes, 103 periplasmic/exported or lipoprotein genes, 123 biosynthesis-related transcripts, 104 cell envelope genes including members of the LPS and O-antigen loci, and 85 transcriptional regulators. Additionally, 25 Bvg-regulated virulence factor genes, including cyaA, dnt, tcfA, brkAB, fhaB, fimN, fimX, fim2, prn, and BB0110, were also significantly downregulated during log-phase growth (Fig. 7B; see also data set S2 in the supplemental material). Forty-eight genes were found to be significantly upregulated, including 10 hypothetical or predicted/probable genes and 10 TTSS genes including bopD, bopN, and bopB (Fig. 7B; see also data set S2 in the supplemental material). Five heat shock and chaperone genes (dnaJ, dnaK, groEL, groES, and htpG) were significantly upregulated as well (see data set S2 in the supplemental material).
Concentrating on the transcriptional changes occurring in RB53 during mid-log to late stationary phases of growth, 1,547 genes were found to be significantly regulated by growth during this time. There were 1,485 genes identified as being significantly downregulated, and they included 221 metabolism-related transcripts, 203 hypothetical or predicted/probable genes, 197 transport-related genes, 100 periplasmic/exported or lipoprotein genes, 105 cell envelope genes including members of the LPS and O-antigen loci, 96 biosynthesis genes, and 72 transcriptional regulators. Conversely, 62 genes were found to be significantly upregulated in RB53 during stationary phase, including 16 hypothetical or predicted/probable genes, six TTSS genes, six transcriptional regulators including rpoH, four periplasmic/exported or lipoprotein genes, and four heat shock and chaperone genes (Fig. 7B; see also data set S2 in the supplemental material). To provide an independent assessment of microarray expression measurements, genes were chosen to reflect the full spectrum of changes occurring during both log and stationary phases of growth and analyzed by qRT-PCR. Once again, the data were consistent with the quantitative measures by microarray analysis (r = 0.98 and r = 0.93) (see data set S2 in the supplemental material). Together, these data suggest that growth phase-dependent gene regulation occurs in B. bronchiseptica when the BvgAS system is constitutively active and results in decreased expression of transcripts that are positively regulated by BvgAS. Additionally, these data indicate that growth phase-dependent gene regulation in B. bronchiseptica can function independently from the BvgAS regulatory system.
Growth phase-dependent gene regulation occurs in a Bvg− phase-locked B. bronchiseptica mutant.
Next, we tested whether the growth-dependent transcriptional changes identified in both RB50 and RB53 were dependent on a functional BvgAS system by evaluating temporal gene expression changes in RB54, a Bvg− phase-locked derivative of RB50. As before, we began these studies by first monitoring the growth of RB54 by optical density, CFU counts, and DNA concentrations over time (Fig. 8A). Temporal growth patterns observed in RB54 were comparable to the temporal growth patterns of RB50 and RB53 (Fig. 1A, 6, and 8A). However, all growth measurements were higher in RB54 and exhibited a modest decrease in recoverable CFU and DNA concentrations during late stationary phase, compared to those in RB50 and RB53 (Fig. 1A, 6, and 8A). Perhaps the best-characterized phenotype of the Bvg− phase is motility and the expression of flagellar synthesis genes, which are negatively regulated by BvgAS and maximally expressed when the BvgAS system is inactive (3). To initially evaluate temporal gene expression changes in RB54, expression of five genes from the motility locus and bvgR was monitored at 6-hour intervals starting at early log phase (12 h) and continuing until late stationary phase (48 h) by qRT-PCR. All transcripts exhibited a growth phase-dependent decrease in expression. Transcription of fliA began a fivefold decrease during late log phase (24 h) that continued through late stationary phase (48 h) (Fig. 8B). Expression of flhD, flhC, and bvgR decreased between 5- and 12-fold during early log phase (12 to 18 h) and continued through late stationary phase (48 h) (Fig. 8B). Transcripts flgB and cheZ exhibited a substantial decrease of 99-fold and 28-fold, respectively, in expression beginning in early log phase (12 to 18 h) and continuing through late stationary phase (48 h) as well (Fig. 8B). These data suggest that this subset of negatively regulated BvgAS genes undergo a growth phase-dependent decrease in expression.
FIG. 8.
Growth curves and changes in gene expression of genes that are negatively regulated by BvgAS and maximally expressed when the BvgAS system is inactive for B. bronchiseptica strain RB54, a Bvg− phase-locked mutant. (A) Optical densities (OD600), DNA concentrations (geq/ml), and CFU counts (CFU/ml) during growth of RB50 are plotted along the y axis, and time, given in hours, is plotted along the x axis. All data points represent averages obtained from triplicate cultures. (B) Change in gene expression (y axis) occurring at 6-h intervals (x axis) starting at early logarithmic phase (12 h) and continuing until late stationary phase (48 h) determined by using qRT-PCR. Data shown are averages obtained from triplicate cultures. The error bars represent standard deviations.
In the same manner used to evaluate RB50 and RB53, microarray analysis was performed on strain RB54 to determine if growth-dependent changes in gene expression occurred on a global scale in the absence of a functional BvgAS system. Similar to the global transcriptional patterns seen in both RB50 and RB53, this analysis revealed prominent growth phase-dependent global shifts in gene expression occurring during mid-log to late log phase in RB54 (Fig. 9A). RB54 was found to differentially express 1,729 genes during this phase of growth, with the majority of them (1,652 genes) being significantly downregulated (Fig. 9A; see also data set S3 in the supplemental material). These transcripts include 235 hypothetical or predicted/probable genes, 226 metabolism-related genes, 174 transport genes, 117 transcriptional regulators, 113 biosynthesis genes, 106 periplasmic/exported or lipoprotein genes, 115 cell envelope genes including members of the LPS and O-antigen loci, and 43 chemotaxis and motility-related transcripts. Seventy-six genes were found to be significantly upregulated during log-phase growth in RB54, including 15 transport-related genes, 12 hypothetical or predicted/probable genes, 11 metabolism-related transcripts, 10 periplasmic/exported or lipoprotein genes, eight transcriptional regulators, and three heat shock and chaperone genes (Fig. 9A; see also data set S3 in the supplemental material).
FIG. 9.
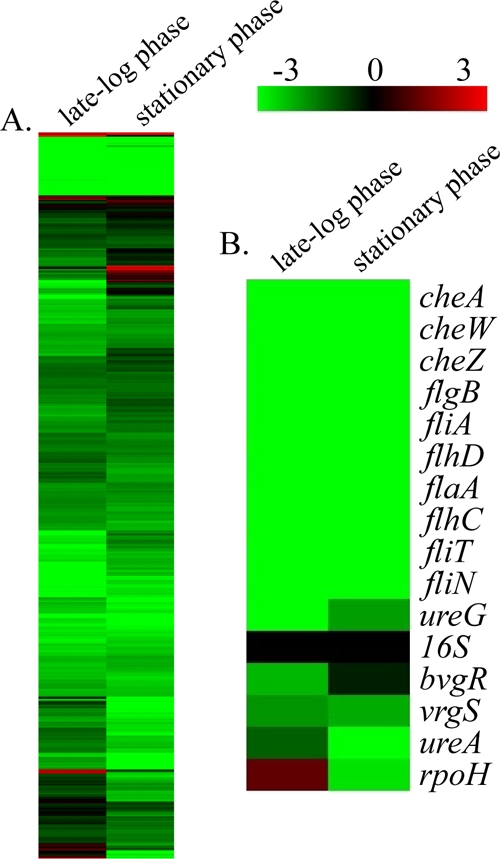
Hierarchical clustering of the transcriptional response of B. bronchiseptica strain RB54, a Bvg− phase-locked mutant during late-log-phase and stationary-phase growth. (A) Expression profiles representing global transcriptional changes. (B) Expression profiles representing transcriptional changes of genes negatively regulated by BvgAS, along with 16S RNA and rpoH. Transcriptional changes identified by comparing cDNA from mid-log phase (15 h) to that from late log phase (24 h) are represented under “late-log phase,” and the transcriptional changes identified by comparing cDNA from mid-log phase (15 h) to that from late stationary phase (48 h) are represented under “stationary phase.” Data are mean centered for each array element and averaged from three biological replicates. All expression profiles of genes are in rows and are represented using the color scale at top, with gray indicating missing data.
Focusing on the transcriptional changes occurring in RB54 during mid-log to late stationary phases of growth, 1,557 genes were found to be differentially expressed, with the majority of them, 1,484 genes, being significantly downregulated (Fig. 9A; see also data set S3 in the supplemental material). These downregulated genes include 236 metabolism-related genes, 148 transport-related genes, 111 periplasmic/exported or lipoprotein genes, 110 biosynthesis-related transcripts, 111 cell envelope genes including members of the LPS and O-antigen loci, and 86 transcriptional regulators. In contrast, 73 transcripts were found to be significantly upregulated during stationary-phase growth, including 23 transcriptional regulators, 13 metabolism-related genes, 12 hypothetical or predicted/probable genes, six transport-related genes, and five periplasmic/exported or lipoprotein genes (Fig. 9A; see also data set S3 in the supplemental material).
Many of the genes significantly downregulated during both late log and stationary phases of growth have been characterized as being negatively regulated by BvgAS and maximally expressed during the Bvg− phase, when BvgAS is inactive. For B. bronchiseptica, these genes include motility loci, virulence-repressed genes (vrg genes), and genes required for the production of urease (Fig. 9B) (2, 3, 26). In both RB50 and RB53, rpoH expression significantly increased during stationary-phase growth. However, in RB54 rpoH expression increased twofold during log phase and then subsequently decreased sixfold during stationary phase (Fig. 9B). qRT-PCR was once again performed to provide an independent assessment of microarray expression measurements for selected genes, representing the full spectrum of changes occurring during both late log and stationary phases of growth, and provided data consistent with the microarray results (r = 0.98 and r = 0.92) (see data set S3 in the supplemental material). Combined, these data suggest that growth phase-dependent gene regulation occurs in RB54, a Bvg− phase-locked B. bronchiseptica mutant, and further suggest that a functional BvgAS system is not necessary for growth phase-dependent gene regulation.
DISCUSSION
Microarray analysis was used to evaluate B. bronchiseptica global gene expression changes occurring in distinct phases of growth, revealing prominent shifts in the global transcriptional program during both late log and stationary phases of growth. Analogous growth phase-dependent gene expression has recently been demonstrated in B. pertussis (28). Based on data arising from the recent completion of the comparative sequencing of three different Bordetella strains, B. pertussis and B. parapertussishu are thought to have derived from a B. bronchiseptica-like ancestor (32). The detailed transcriptome data presented here suggest that growth-dependent gene regulation may be a regulatory mechanism shared by all bordetellae.
The global transcriptional patterns in all three B. bronchiseptica strains (RB50, RB53, and RB54) were largely comprised of decreases in the transcription levels for the majority of genes identified as significantly regulated by growth phase. The functional categories most overrepresented by genes exhibiting decreased expression were translation, metabolism and biosynthesis-related transcripts, cell envelope, and periplasmic/exported or lipoprotein genes. Nutrient limitation is the major signal regulating entry into stationary phase. For example, in Escherichia coli nutrient limitation leads to decreased expression of genes involved in translation, metabolism, and flagellum/chemotaxis and the upregulation of genes involved in amino acid biosynthesis or acquisition (9). Outside of the lack of induction of genes involved in amino acid biosynthesis, this global response is similar to that of all three B. bronchiseptica strains, suggesting that nutrient or specifically amino acid limitation may be contributing to the growth phase gene expression changes reported here; however, the actual mechanism contributing to these gene expression changes is not known. Other possible signals contributing to the growth phase gene expression changes reported here include cell density, buildup of culture by-products, or some unknown aspect regarding the age of the culture. Given that bacteria can often integrate more than one signal (18), it is also possible that multiple signals may function cooperatively to induce growth phase gene expression changes in B. bronchiseptica.
When the global growth phase-dependent changes in gene expression of B. bronchiseptica RB50 were analyzed, we found that the expression for most of the characterized virulence genes decreased in a growth phase-dependent manner. Most notably, this included the expression of genes encoding known adhesins. A similar growth-dependent decrease in expression of virulence factors and adhesion-related genes has been reported for B. pertussis Tohama I, along with a growth phase-dependent decrease in adherence (28). In contrast, although the transcription of genes encoding adhesins previously shown to mediate adherence was decreased in late log and stationary phases, we found that the adherence of B. bronchiseptica RB50 did not decrease in these later phases of growth. As biofilm formation is known to be involved with increasing adherence capacity and is growth phase dependent (14), one possible explanation for this difference is differential expression of genes encoding either functional components or regulatory elements involved in biofilm formation. Congruent with this, both FHA and fimbriae have been demonstrated to contribute to biofilm formation in B. bronchiseptica (13), and mutants lacking these adhesins, RB63 (ΔfimBCD) and RBX9 (ΔfhaB), exhibited decreased adherence capacity during stationary phase relative to late-log-phase growth, while the adherence capacity for both RB50 and SP5 (Δprn) did not decrease during these growth phases. However, the role of FHA remains unclear given that we also observed a decreased adherence capacity during mid-log phase relative to the late log phase of growth as well. There were differences between the growth phase-associated transcriptional changes identified in B. pertussis and those reported here for B. bronchiseptica that may be attributed to the large-scale loss and inactivation of genes in B. pertussis compared to B. bronchiseptica. Unfortunately, with the exception of one polysaccharide locus (bps), which has been shown to be necessary for mature biofilm formation (31, 37), genes involved in biofilm formation are not well described for Bordetella, thus limiting the ability to identify and compare expression patterns of such transcripts.
Microarray analysis revealed and qRT-PCR confirmed the increased expression of several TTSS-associated genes in late log and stationary phases. These expression data differ from those in a recent report showing a decrease in expression of several B. pertussis TTSS-related transcripts in these later phases of growth (28). TTSS expression in Bordetella is controlled by a complex, multilayered, transregulatory gene network in which BvgAS lies at the top and activates btrS, the gene encoding the activating sigma factor, which is required for expression of all known TTSS transcripts (25, 43). Due to the hierarchical nature of this regulatory cascade, it is likely that BvgAS activates btrS through an indirect mechanism, involving yet-unidentified regulator elements. Such regulatory elements may be encoded by genes lost or inactivated in B. pertussis and functionally intact in B. bronchiseptica. Alternatively, these regulatory elements could be differentially expressed between B. pertussis and B. bronchiseptica, thus leading to the different patterns of TTSS-related gene expression between these species. Using an in vitro cytotoxicity assay, we demonstrate that the growth-dependent increase in TTSS-related transcripts identified in B. bronchiseptica RB50 leads to a growth-dependent increase in cytotoxicity, a TTSS-dependent function. An analogous virulence phenotype has been demonstrated in Salmonella enterica serovar Typhimurium, where the peak of virulence was determined to be late-log-phase growth and attributed to the peak in expression of an SPI-I-encoded TTSS apparatus (21).
The global transcriptional responses of numerous bacterial pathogens that undergo growth phase-dependent gene regulation have been analyzed by microarray analysis (16, 22, 38, 40). Collectively, these studies have revealed a similar trend among these pathogens in that a major shift in global transcription occurs during the transition from late log phase to stationary phase, resulting in an increase in virulence attributes. Data reported here, and previously reported data (28), demonstrate that both B. bronchiseptica and B. pertussis undergo growth phase-dependent gene regulation, albeit with differences in both virulence gene expression and virulence attributes. For B. pertussis, most known virulence factors, including TTSS and adhesin-related transcripts, decreased in a growth-dependent manner, along with the virulence phenotype adherence (28). In contrast, the data reported here demonstrate that for B. bronchiseptica growth phase-dependent gene regulation results in an increase in the expression of several TTSS genes and leads to a growth phase-dependent increase in cytotoxicity, a TTSS-dependent function. Additionally, while genes encoding adhesins previously shown to mediate adherence were decreased in late log and stationary phases, in contrast to B. pertussis, we found that B. bronchiseptica adherence did not decrease in these later phases of growth.
Given that BvgAS is the major virulence regulator in Bordetella and that many of the classical virulence factor genes exhibited growth phase-dependent transcriptional changes, we tested whether similar global shifts in gene expression would occur in Bvg+ and Bvg− phase-locked mutants. Similar to the growth-dependent prominent shifts in global gene expression in RB50, we identified growth phase-dependent gene regulation in RB53, a Bvg+ phase-locked B. bronchiseptica mutant. This is again in contrast to B. pertussis, where Tohama I-Sc3, a Bvg+ phase-locked B. pertussis mutant, failed to exhibit growth-dependent gene regulation, indicating that a BvgAS system capable of modulating is required for growth-dependent gene regulation (28). The growth phase-dependent transcriptional changes reported here for RB53 indicate that, for B. bronchiseptica, a BvgAS system capable of modulating is not required for growth-dependent gene regulation. Additionally, we tested whether the growth-dependent transcriptional changes identified in both RB50 and RB53 were dependent on a functional BvgAS system by evaluating temporal gene expression changes in RB54, a Bvg− phase-locked derivative of RB50. Our results demonstrated that growth phase-dependent gene regulation occurs in RB54, suggesting that growth phase-dependent shifts in global gene expression are not dependent on a functional BvgAS system. Many of the genes identified as significantly downregulated during both log and stationary phases in RB54 encode transcripts characterized as being negatively regulated by BvgAS and maximally expressed during the Bvg− phase, when BvgAS is inactive (2, 3, 26). Collectively, the gene expression data from all three stains, RB50, RB53, and RB54, suggest that for B. bronchiseptica the regulatory mechanism coordinating these growth-dependent changes in gene expression functions independently of the BvgAS regulatory cascade.
Supplementary Material
Acknowledgments
We thank Crystal Loving, Eric Nicholson, and Thaddeus Stanton for critical review of the manuscript and Sarah Shore for excellent technical support.
We declare no conflicting financial interests.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Editor: A. Camilli
Footnotes
Published ahead of print on 10 August 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., and J. F. Miller. 1993. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J. Bacteriol. 175:3468-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akerley, B. J., D. M. Monack, S. Falkow, and J. F. Miller. 1992. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buboltz, A. M., T. L. Nicholson, M. R. Parette, S. E. Hester, J. Parkhill, and E. T. Harvill. 2008. Replacement of adenylate cyclase toxin in a lineage of Bordetella bronchiseptica. J. Bacteriol. 190:5502-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11:367-373. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. A., and J. F. Miller. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671-685. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durfee, T., A. M. Hansen, H. Zhi, F. R. Blattner, and D. J. Jin. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, J. A., N. A. Groathouse, and S. Boitano. 2005. Bordetella bronchiseptica adherence to cilia is mediated by multiple adhesin factors and blocked by surfactant protein A. Infect. Immun. 73:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fennelly, N. K., F. Sisti, S. C. Higgins, P. J. Ross, H. van der Heide, F. R. Mooi, A. Boyd, and K. H. Mills. 2008. Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect. Immun. 76:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irie, Y., S. Mattoo, and M. H. Yuk. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 186:5692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irie, Y., A. Preston, and M. H. Yuk. 2006. Expression of the primary carbohydrate component of the Bordetella bronchiseptica biofilm matrix is dependent on growth phase but independent of Bvg regulation. J. Bacteriol. 188:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinnear, S. M., P. E. Boucher, S. Stibitz, and N. H. Carbonetti. 1999. Analysis of BvgA activation of the pertactin gene promoter in Bordetella pertussis. J. Bacteriol. 181:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko, K. S., S. Park, W. S. Oh, J. Y. Suh, T. Oh, S. Ahn, J. Chun, and J. H. Song. 2006. Comparative analysis of growth-phase-dependent gene expression in virulent and avirulent Streptococcus pneumoniae using a high-density DNA microarray. Mol. Cells 21:82-88. [PubMed] [Google Scholar]
- 17.Kozak, N. A., S. Mattoo, A. K. Foreman-Wykert, J. P. Whitelegge, and J. F. Miller. 2005. Interactions between partner switcher orthologs BtrW and BtrV regulate type III secretion in Bordetella. J. Bacteriol. 187:5665-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazazzera, B. A. 2000. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 3:177-182. [DOI] [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Livey, I., and A. C. Wardlaw. 1984. Production and properties of Bordetella pertussis heat-labile toxin. J. Med. Microbiol. 17:91-103. [DOI] [PubMed] [Google Scholar]
- 21.Lundberg, U., U. Vinatzer, D. Berdnik, A. von Gabain, and M. Baccarini. 1999. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J. Bacteriol. 181:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangan, M. W., S. Lucchini, V. Danino, T. O. Croinin, J. C. Hinton, and C. J. Dorman. 2006. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:1831-1847. [DOI] [PubMed] [Google Scholar]
- 23.Martinez de Tejada, G., P. A. Cotter, U. Heininger, A. Camilli, B. J. Akerley, J. J. Mekalanos, and J. F. Miller. 1998. Neither the Bvg− phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect. Immun. 66:2762-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattoo, S., J. F. Miller, and P. A. Cotter. 2000. Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect. Immun. 68:2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattoo, S., M. H. Yuk, L. L. Huang, and J. F. Miller. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 52:1201-1214. [DOI] [PubMed] [Google Scholar]
- 26.McMillan, D. J., M. Shojaei, G. S. Chhatwal, C. A. Guzman, and M. J. Walker. 1996. Molecular analysis of the bvg-repressed urease of Bordetella bronchiseptica. Microb. Pathog. 21:379-394. [DOI] [PubMed] [Google Scholar]
- 27.Merkel, T. J., S. Stibitz, J. M. Keith, M. Leef, and R. Shahin. 1998. Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis. Infect. Immun. 66:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura, M. M., S. Y. Liew, C. A. Cummings, M. M. Brinig, C. Dieterich, and D. A. Relman. 2006. Growth phase- and nutrient limitation-associated transcript abundance regulation in Bordetella pertussis. Infect. Immun. 74:5537-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholson, T. L. 2007. Construction and validation of a first-generation Bordetella bronchiseptica long-oligonucleotide microarray by transcriptional profiling the Bvg regulon. BMC Genomics 8:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson, T. L., S. L. Brockmeier, and C. L. Loving. 2009. Contribution of Bordetella bronchiseptica filamentous hemagglutinin and pertactin to respiratory disease in swine. Infect. Immun. 77:2136-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parise, G., M. Mishra, Y. Itoh, T. Romeo, and R. Deora. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189:750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 33.Preston, A., J. Parkhill, and D. J. Maskell. 2004. The bordetellae: lessons from genomics. Nat. Rev. Microbiol. 2:379-390. [DOI] [PubMed] [Google Scholar]
- 34.Roy, C. R., and S. Falkow. 1991. Identification of Bordetella pertussis regulatory sequences required for transcriptional activation of the fhaB gene and autoregulation of the bvgAS operon. J. Bacteriol. 173:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 36.Skulj, M., V. Okrslar, S. Jalen, S. Jevsevar, P. Slanc, B. Strukelj, and V. Menart. 2008. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb. Cell Fact. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sloan, G. P., C. F. Love, N. Sukumar, M. Mishra, and R. Deora. 2007. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 189:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willems, R., A. Paul, H. G. van der Heide, A. R. ter Avest, and F. R. Mooi. 1990. Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 9:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28:945-959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.