Abstract
Ehrlichiae are obligately intracellular bacteria that reside and replicate in phagocytes by circumventing host cell defenses and modulating cellular processes, including host cell gene transcription. However, the mechanisms by which ehrlichiae influence host gene transcription have largely remained undetermined. Numerous ankyrin and tandem repeat-containing proteins associated with host-pathogen interactions have been identified in Ehrlichia species, but their roles in pathobiology are unknown. In this study, we determined by confocal immunofluorescence microscopy and by immunodetection in purified nuclear extracts that the ankyrin repeat-containing protein p200 is translocated to the nuclei of Ehrlichia-infected monocytes. Chromatin immunoprecipitation (ChIP) with DNA sequencing revealed an Ehrlichia chaffeensis p200 interaction located within host promoter and intronic Alu-Sx elements, the most abundant repetitive elements in the human genome. A specific adenine-rich (mid-A-stretch) motif within Alu-Sx elements was identified using electrophoretic mobility shift and NoShift assays. Whole-genome analysis with ChIP and DNA microarray analysis (ChIP-chip) determined that genes (n = 456) with promoter Alu elements primarily related to transcription, apoptosis, ATPase activity, and structural proteins associated with the nucleus and membrane-bound organelles were the primary targets of p200. Several p200 target genes (encoding tumor necrosis factor alpha, Stat1, and CD48) associated with ehrlichial pathobiology were strongly upregulated during infection, as determined by quantitative PCR. This is the first study to identify a nuclear translocation of bacterially encoded protein by E. chaffeensis and to identify a specific binding motif and genes that are primary targets of a novel molecular strategy to reprogram host cell gene expression to promote survival of the pathogen.
Ehrlichia chaffeensis and Ehrlichia canis are obligately intracellular bacteria that reside and replicate within cytoplasmic vacuoles in mononuclear phagocytes. Ehrlichiae are maintained in nature by persistent infection of vertebrate hosts and are transmitted by arthropods (ticks), and thus, their existence requires adaptation to host-specific environments and evasion of both innate and adaptive immune mechanisms (48, 49, 53). The ability of ehrlichiae to evade the vertebrate innate immune response is multifactorial and includes the absence of genes for synthesis of conserved major microbial patterns, including peptidoglycan and lipopolysaccharide, that can elicit destructive antimicrobial responses prior to cell entry (14, 22, 35, 40). E. chaffeensis-infected monocytes are less responsive in mobilizing antimicrobial defenses, indicating that ehrlichiae have evolved strategies to downregulate these cellular responses. During E. chaffeensis infection, transcription levels of many host cell genes are altered, including those involved in early innate and cell-mediated immune responses, apoptosis regulation, membrane trafficking, signal transduction, and cell differentiation (68). Furthermore, ehrlichiae actively block signaling pathways, inhibit lysosomal fusion and superoxide generation, and downregulate innate immune receptors and transcription factors (29, 30, 36, 37) while activating other host cell signaling pathways (38). Some of the cellular processes affected by the bacterium appear to be mediated through host-pathogen protein-protein interactions or protein kinase activities (3, 30, 36, 38). However, the mechanism(s) by which E. chaffeensis directly modulates host cell gene transcription is relatively undefined.
Recently numerous tandem repeat- and ankyrin repeat-containing immunoreactive proteins associated with host-pathogen interactions have been identified and characterized for Ehrlichia species (15, 39, 41, 67). Ankyrin repeat-containing proteins have recently been documented for obligately intracellular bacteria, including Ehrlichia spp. and related rickettsial organisms from the genera Anaplasma, Wolbachia, and Orientia (10, 11, 22, 40, 64). In eukaryotes, the primary function of ankyrin repeats appears to be mediation of protein-protein interactions, and they are present in a wide variety of proteins, including transcriptional regulators, cytoskeletal organizers, developmental regulators, and toxins (54).
Large (200-kDa) ankyrin repeat protein orthologs (p200) have been characterized for E. chaffeensis and E. canis (41) and are major immunoreactive proteins recognized strongly by the host immune response (41-43, 46). E. canis p200 has five species-specific antibody epitopes located primarily in terminal acidic domains of the protein (46). p200 also has a central region containing 21 eukaryote-like ankyrin repeats. Notably, an ankyrin repeat-containing protein (AnkA) has been found in Anaplasma phagocytophilum that is translocated to the nucleus of infected neutrophils, where it appears to form DNA-protein complexes (51). Recently binding of AnkA to regulatory regions of the CYBB gene and interaction with gene regulatory regions containing high AT content have been reported (19). In this study, we determined that the E. chaffeensis ankyrin repeat protein p200 interacts with an adenine-rich motif within Alu-Sx elements, and we used whole-genome analysis to identify a subset of genes associated with metabolic and molecular processes and cell structure that are primary p200 targets.
MATERIALS AND METHODS
Cultivation of E. chaffeensis and E. canis.
E. chaffeensis (Arkansas strain) was cultivated in a human monocyte (THP-1) or canine macrophage (DH82) cell line, and E. canis (Jake strain) was cultivated in DH82 cells only. Uninfected and Ehrlichia-infected THP-1 cells were cultured in Dulbecco's modified Eagle medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 4 mM l-glutamine, and 1% sodium pyruvate at 37°C in a humidified 5%-CO2 atmosphere. DH82 cells were cultured in minimal essential medium (GIBCO-BRL) supplemented with 5% fetal bovine serum (HyClone), nonessential amino acids (Sigma-Aldrich, St. Louis, MO), HEPES buffer (Sigma-Aldrich), and 1% sodium pyruvate (Sigma-Aldrich) at 37°C in a humidified 5%-CO2 atmosphere. Ehrlichial growth was monitored by assessing the presence of morulae using general cytology staining methods.
Antibodies.
Anti-E. canis antibody specific for the amino terminus of recombinant E. canis p200 (p43) and anti-E. chaffeensis Dsb were produced in rabbits as described previously (42, 44). Anti-E. chaffeensis p200 was produced in rabbits after immunization with the recombinant N-terminal region of p200 as described previously (42).
Nuclear extraction.
Cytoplasmic and nuclear fractions were extracted (3 days postinfection) from E. chaffeensis- or E. canis-infected and uninfected DH82 cells using a nuclear extraction kit (Panomics, Fremont, CA). The extractions were performed according to the manufacturer's protocol, except that cells were lysed by 10 passages through a 26-gauge needle prior to incubation.
Immunofluorescence confocal microscopy.
E. chaffeensis-infected (3 days postinfection) and uninfected THP-1 cells were cytocentrifuged onto glass slides and treated for 5 min at room temperature with digitonin (40 μg/ml in phosphate-buffered saline [PBS]; Sigma). The cells were fixed in 4% paraformaldehyde in PBS for 20 min and permeabilized with 1% Triton X-100 and 5% bovine serum albumin in PBS for 1 h. Cells were incubated with polyclonal rabbit sera, anti-E. chaffeensis p200 (1:1,000) and anti-Dsb (1:100), for 1 h and washed thrice with PBS. Cells were then incubated with goat antirabbit Alexa Fluor 488 (1:100) (Invitrogen, Carlsbad, CA) for 1 h and washed three times in PBS. Slides were mounted with medium containing 4′,6′-diamidino-2-phenylindole (Invitrogen), and fluorescent images were obtained at the Optical Imaging Laboratory at the University of Texas Medical Branch using a Zeiss LSM 510 Meta laser-scanning confocal microscope. The confocal images were further analyzed by use of fluorescence intensity profiles (63). A fluorescence intensity profile represents the pixel values in a digitized section along a user-defined area, displayed in a diagram. The electrical signal from the detector was digitized to a numerical value between 0 and 250 arbitrary units. The fluorescent intensity of nuclei was determined from a random selection of 20 nuclei from infected and uninfected cells stained with p200 or Dsb antibody.
Western immunoblotting.
The recombinant E. chaffeensis p200 N-terminal and Dsb proteins were prepared as previously described (46). Cytoplasmic and nuclear extracts were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane using a semidry transfer apparatus (42, 43). Western immunoblotting was performed using one of three polyclonal rabbit sera: anti-E. chaffeensis p200 (1:500; N-terminal), anti-E. canis p200 (1:500; N-terminal; p43), or anti-E. chaffeensis Dsb (1:100). Bound antibodies were detected by incubation with phosphatase-labeled goat anti-rabbit immunoglobulin G (IgG) (H+L) (1:10,000) (Kirkegaard & Perry Laboratories, Gaithersburg, MD) and visualized after incubation with 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium substrate (Kirkegaard & Perry Laboratories).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed on highly infected (95%) THP-1 cells 7 days after infection using the ChIP-IT Express kit (Active Motif, Carlsbad, CA) with anti-E. chaffeensis p200 antibody and normal human IgG according to the manufacturer's instructions. Amplicons were prepared by adapting the standard protocol for whole-genome amplification using the GenomePlex WGA kit (Sigma-Aldrich) as previously described (47), except the initial random fragmentation step was performed by sonication.
ChIP with DNA sequencing (ChIP-Seq).
The immunoprecipitated DNA was amplified by using a whole-genome amplification kit (Sigma-Aldrich), and the amplified DNA was purified by using a GeneElute PCR cleanup kit (Sigma-Aldrich). The purified DNA was cloned into the pCR4-TOPO vector for sequencing according to the manufacturer's protocol (Invitrogen, Carlsbad, CA) and sequenced at the Protein Chemistry Laboratory at the University of Texas Medical Branch. Each sequence was verified and mapped by BLAST using the University of California, Santa Cruz, genome browser.
ChIP-chip.
A two-array set representing whole-genome promoter (5 kb) tiling, containing 385,000 probes (50- to 75-mer) from ∼60,000 human transcripts, was obtained from Nimblegen Systems. Labeling and hybridization of E. chaffeensis ChIP samples were performed by Nimblegen Systems. For each spot on the array, the log2 ratio of Cy5-labeled p200 immunoprecipitation sample versus the Cy3-labeled input DNA sample (not taken through immunoprecipitation steps) was calculated. The intensity ratio of p200 immunoprecipitation to that of input DNA was plotted versus the genomic position to identify regions where increased signal (i.e., DNA fragment enrichment) was observed relative to the control sample. Positions identified as p200 binding sites on the human 5-kb promoter array were identified by using NimbleScan, and peaks were detected by searching for four or more probes above the specified cutoff values, ranging from 90% to 15% using a 500-bp sliding window. The cutoff value is a percentage of a hypothetical maximum (mean ± standard deviation), and the ratio data were randomized 20 times to determine the probability of false positives. Each peak was then assigned a false discovery rate (FDR) score based on the randomization.
EMSA and NoShift assays.
Oligonucleotides probes (biotinylated and unlabeled) (P1 to P4 [see Fig. 5A] and M1 to M4 in [see Fig. 5C]) were resuspended in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), and 10 μg each of sense and antisense probe were added to 100 μl 0.5× SSC (75 mM NaCl, 7.5 mM sodium citrate, pH 7.0); the probes were denatured at 100°C for 10 min and annealed by slowly cooling to room temperature. Nuclear extracts were prepared from uninfected and E. chaffeensis-infected THP-1 cells using a ChIP-IT Express kit (Active Motif). The extractions were performed according to the manufacturer's protocol except that cells were not fixed and were homogenized with 100 strokes of a Dounce homogenizer. The extracts from E. chaffeensis-infected THP-1 cells were incubated with biotinylated DNA probes (0.2 pmol each) for 30 min at 4°C in 20 μl of binding buffer [10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 2.5% glycerol, 0.1% NP-40, 50 ng/μl of poly(dI-dC) or salmon sperm DNA]. As a control, extracts from uninfected THP-1 cells were incubated with biotinylated DNA probes. The electrophoretic mobility shift assay (EMSA) competition experiment was performed with a 50-fold molar excess of an unlabeled double-stranded oligonucleotide. The supershift assay was performed by incubating anti-p200 antibody for 30 min at 4°C. The probe-extract mixture was separated on a 6% DNA retardation gel at 100 V for 1.5 h at 4°C. The DNA was transferred to a precut modified nylon membrane (Biodyne B; Pierce, Rockford, IL) at 20 V for 1 h and cross-linked with UV light for 10 min. Biotin-labeled DNA was detected with a LightShift chemiluminescence EMSA kit (Pierce). The enzyme-linked immunosorbent assay-based EMSA was carried out using a NoShift transcription factor assay kit (EMD Biosciences, San Diego, CA) according to the manufacturer's protocol. Reactions in the absence of extract or using normal rabbit serum were included as controls.
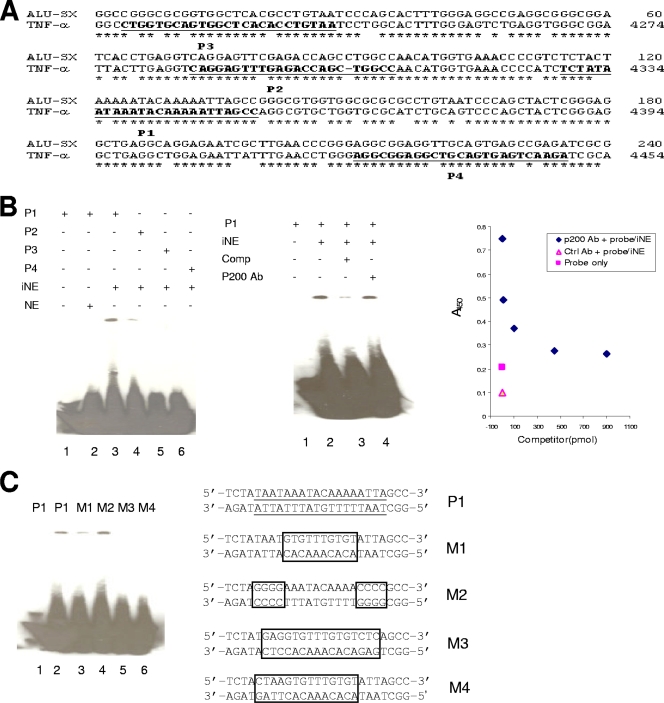
FIG. 5.
Alu mid-A-stretch motif is E. chaffeensis p200 DNA binding motif. (A) Schematic of nucleotides 4214 to 4502 within the 5′ region upstream of the TNF-α gene compared to the human Alu-Sx subfamily consensus sequence (asterisk denotes base identity). Sequences of the double-stranded oligonucleotide probes (P1, P2, P3, and P4) used for EMSAs corresponding to the conserved putative p200 motif of the TNF-α gene are underlined and bold. (B) Left panel: EMSA using biotinylated probes (P1 to P4) and nuclear extracts from infected monocytes (iNE) and uninfected monocytes (NE). Center panel: EMSA using biotinylated P1 and nuclear extracts in the presence of a 50-fold molar excess of unlabeled double-stranded competitor (P1) in the presence of E. chaffeensis p200-specific antibody (Ab). Right panel: NoShift assay performed with iNE and biotinylated P1 probe with detection by anti-E. chaffeensis p200 antibody (1:400). Increasing concentrations of unlabeled P1 probe (x axis) were added, and absorbance (450 nm) was determined. The blank consisted of all assay components without iNE, and the negative control reaction was carried out with normal rabbit serum. (C) EMSA using biotinylated P1 and mutated probes (M1, M2, M3, and M4) and nuclear extract from E. chaffeensis-infected THP-1 monocytes. The putative p200 binding sites are underlined in the P1 probe. The mutated nucleotides in each probe are boxed.
DNA enrichment.
Quantitative real-time PCR (qPCR) was performed on p200 ChIP-enriched DNA and compared to ChIP DNA obtained with normal IgG and input DNA to normalize the enrichment levels of p200 ChIP. Primer pairs were designed to amplify regions (∼150 to 200 bp) within the binding site (FDR peak) identified by ChIP-chip analysis. Amplification of all genes was performed using an initial denaturing step at 94°C for 2 min, followed by 40 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s in triplicate with iQ SYBR green supermix (Bio-Rad Laboratories, Hercules, CA) in a Mastercycler ep realplex2 S instrument (Eppendorf, Hamburg, Germany). A dissociation curve analysis was performed to verify amplicon identity. Relative enrichment for each gene was determined by the comparative CT method.
Host gene expression levels.
Total RNA was extracted from E. chaffeensis-infected and uninfected THP-1 cells (106 cells per flask) using Tri reagent (Applied Biosystems/Ambion, Austin, TX) according to the manufacturer's instructions and treated with DNase I (Applied Biosystems/Ambion) to remove contaminating DNA. cDNA was synthesized from 1 μg of total RNA using an iScript cDNA synthesis kit with oligo(dT) and random primers (Bio-Rad Laboratories), and cDNA for randomly chosen host genes was quantitated by qPCR using iQ SYBR green supermix (Bio-Rad Laboratories) with gene-specific primers (see Table S3 in the supplemental material). The thermal cycling protocol consisted of an initial denaturation step of 95°C for 2 min and 40 cycles of 95° for 10 s, 55°C for 30 s, and 65°C for 30 s. Gene expression values were calculated based on the 2−ΔΔCT method and normalized with glyceraldehyde-3-phosphate dehydrogenase.
Cytokine detection.
The level of tumor necrosis factor alpha (TNF-α) (human) in the supernatant of cultured THP-1 cells infected with E. chaffeensis was determined using an immunoassay according to the manufacturer's protocol (Quantikine; R&D Systems, Minneapolis, MN).
Microarry data accession number.
ChIP-chip microarray data reported here were deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information and assigned the accession number GSE13464 (in accordance with “Minimum Information about a Microarray Experiment” guidelines).
RESULTS
Ehrlichia p200 is translocated to the nuclei of infected monocytes.
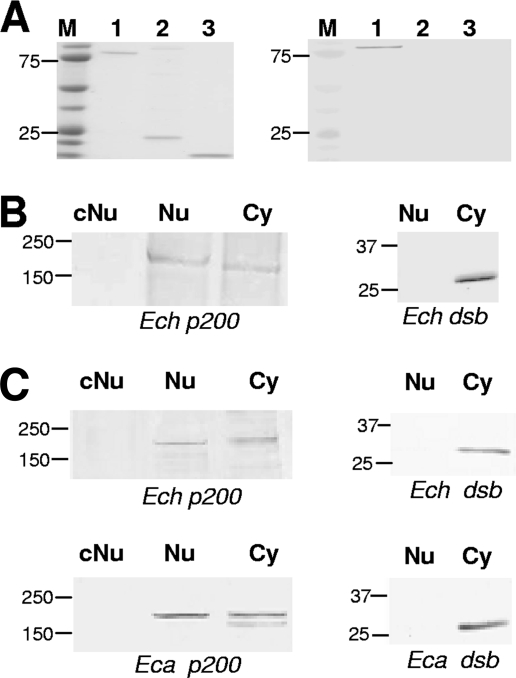
To demonstrate antibody specificity, recombinant E. chaffeensis p200 N-terminal, recombinant Dsb and thioredoxin fusion proteins were separated by gel electrophoresis and probed with anti-E. chaffeensis p200 by Western immunoblotting. Anti-p200 antibody reacted with the recombinant E. chaffeensis p200 N-terminal protein but not with recombinant Dsb or thioredoxin (Fig. 1A). In addition, p200 was detected in both the cytoplasm and nuclear extracts from E. chaffeensis-infected cells but not in the nuclear extracts from uninfected THP-1 cells (Fig. 1B, left panel). As a control to exclude the possibility of contamination of nuclear extracts with ehrlichiae, both nuclear and cytoplasmic extracts were probed with anti-Dsb (ehrlichial periplasmic protein) and Dsb was detected only in cytoplasmic fractions (Fig. 1B and C, right panel), confirming the nuclear extract preparation was not contaminated with ehrlichiae. Since E. canis is not cultivated in THP-1 cells, we also examined cytoplasmic and nuclear fractions obtained from DH82 cells infected with either E. chaffeensis or E. canis and detected p200 but not Dsb in the nuclear extracts (Fig. 1C). We consistently detected p200 in cytoplasmic and nuclear fractions after 3 days of infection; however, p200 was not detected at earlier time points (data not shown).
FIG. 1.
E. chaffeensis p200 antibody specificity and p200 detection in nuclear extracts from E. chaffeensis-infected cells. (A) Coomassie blue staining of recombinant p200 N-terminal protein, recombinant Dsb, and thioredoxin fusion protein, respectively (left panel, lanes 1 to 3). Corresponding Western immunoblot detection using specific anti-E. chaffeensis p200 (right panel, lanes 1 to 3). (B) Western immunoblot detection of p200 and E. chaffeensis periplasmic protein Dsb (not secreted) using nuclear extracts from E. chaffeensis-infected and uninfected THP-1 cells (3 days postinfection). Control nuclear extract from uninfected cells, cNu; nuclear extract from infected monocytes, Nu; cytoplasmic fraction from infected cells, Cy. (C) Western immunoblot detection of p200 and E. chaffeensis and E. canis periplasmic protein Dsb in Nu and Cy fractions of infected and uninfected DH82 cells. Control nuclear extract from uninfected cells, cNu; nuclear extract from infected monocytes, Nu; cytoplasmic fraction from infected cells, Cy. Molecular masses are shown (in kilodaltons).
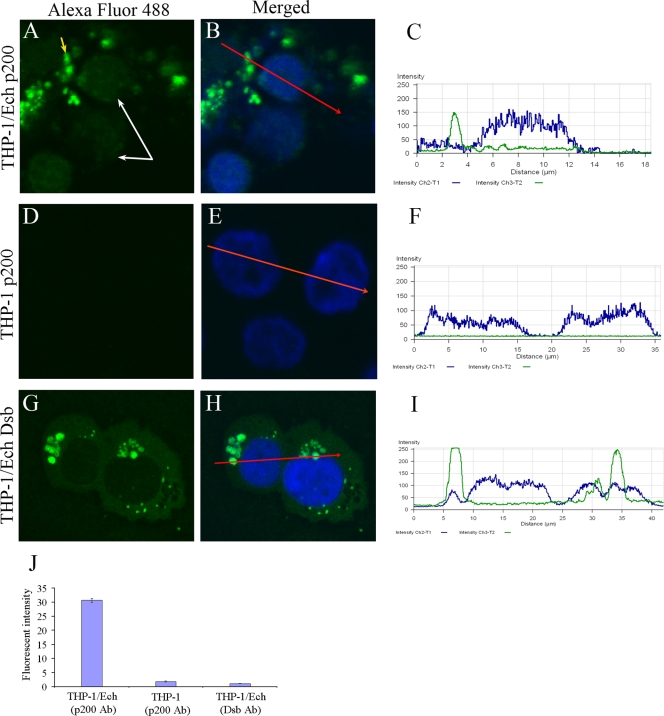
Immunoblot data indicated that E. chaffeensis p200 was present in the nucleus of the E. chaffeensis-infected cells, and this was further investigated by immunofluorescence confocal microscopy with anti-p200 and anti-Dsb antibodies. A low-intensity fluorescent signal representing p200 within the nucleus of E. chaffeensis-infected cells was consistently detected by immunofluorescence microscopy (Fig. 2A and B) and quantified in the corresponding fluorescent-intensity profile (Fig. 2C). Conversely, p200 was not detected in the nuclei of uninfected THP-1 cells (Fig. 2D, E, and F). To further confirm the specificity of the nuclear signal, we examined the nuclear fluorescence associated with Dsb, an ehrlichial periplasmic protein that is not translocated to the nucleus. Dsb was not detected in the nuclei of E. chaffeensis-infected host cells (Fig. 2G, H, and I). The mean level of fluorescent intensity from nuclei represented in these panels is depicted graphically in Fig. 2J.
FIG. 2.
Translocation of Ehrlichia p200 ankryin protein to host cell nucleus. E. chaffeensis-infected (A, B, G, and H) or uninfected (D and E) THP-1 cells were prepared using a cytospin, and cells were fixed with paraformaldehyde, permeabilized with Triton X-100, and incubated with anti-E. chaffeensis p200 and anti-Dsb. Host cell nuclei (blue) were counterstained with 4′,6′-diamidino-2-phenylindole. The nuclei (white arrows) of infected cells (A) and E. chaffeensis morulae (yellow arrow) are identified (A). Serial confocal scan of the nucleus from an E. chaffeensis-infected cell (B, C, H, and I) or uninfected cells (E and F) is shown. A red arrow indicates the direction of the serial scan of the nucleus. The fluorescence intensity of protein nuclear translocation was quantified from infected and uninfected cells reacted with p200 or Dsb antibodies (J).
p200 immunoprecipitation and identification of DNA binding motif.
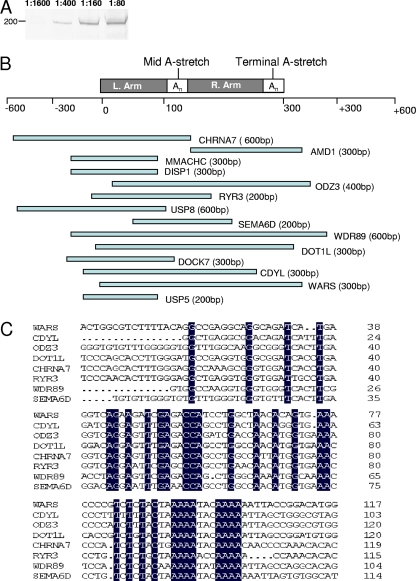
The p200 protein was detected by Western immunoblotting in ChIP protein/DNA complexes from E. chaffeensis-infected host cells (Fig. 3A). To map p200 binding positions, E. chaffeensis p200 target sequences were determined by DNA sequencing (ChIP-Seq) of randomly selected cloned (n = 100) DNA fragments of ChIP host cell DNA. The cloned p200 immunoprecipitated DNA (range, 200 to 800 bp; >75% were ∼300 bp) was amplified by PCR and directly sequenced. First-round sequence data were obtained from 58 clones and analyzed using the University of California, Santa Cruz, genome browser to map p200 binding locations. Fourteen clones (24%) had small inserts (∼200 to 600 bp) homologous to Alu-Sx elements (Fig. 3B). Six clones contained DNA sequences that were homologous to Alu-Sx elements and were mapped to 5′ upstream promoter regions of annotated genes (Table 1). Eight clones had sequences containing Alu-Sx elements but were mapped to introns of the corresponding annotated genes (Table 1). Alignment of the cloned DNA sequences from eight clones identified a 200-bp homologous region containing an adenine-rich motif (5′-AAAATACAAAA-3′) with highest identity to the mid -A-stretch motif (a specific adenine-rich motif) of Alu-Sx elements (Fig. 3C). Common sequence motifs were not identified among the remaining ChIP-Seq clones (n = 44), and they were not investigated further. Clones were not recovered from the negative control (water treated).
FIG. 3.
E. chaffeensis p200 targets repetitive Alu mobile elements. (A) Western immunoblot detection of p200 from ChIP samples prepared from E. chaffeensis-infected cells using specific Antibody. Lanes 1 through 4 contain increasing amounts of antibody (1:1600, 1:400, 1:160, and 1:80). (B) Schematic of the Alu element architecture and the distribution of 14 E. chaffeensis p200 ChIP-Seq clones respective to an Alu element. Alu repeat left and right arms are separated by an A-rich region (Mid A-stretch) and short poly(A) tail (Terminal A-stretch). (C) Nucleic acid alignments of the cloned DNA sequences. Nucleotides identical to the consensus sequence are highlighted.
TABLE 1.
BLAST analysis of p200 ChIP complexes
| Gene | CHRa | Identity (%) | Location | Description | No. of Alu-Sx elements in 5-kb promoter region | Accession no. |
|---|---|---|---|---|---|---|
| SEMA6D | 15 | 99.0 | 5′ upstream | Sema domain, transmembrane domain, and cytoplasmic domain, (semaphorin) 6D | Promoter > 5 kb | NM_153619 |
| ODZ3 | x | 99.3 | 5′ upstream | Odd Oz/ten-m homolog 3 | 2 | NM_001080477 |
| DOT1L | 19 | 99.7 | Intron 23 | DOT1-like, histone methyltransferaseH3 | 7 | NM_032482 |
| USP8 | 15 | 99.3 | 5′ upstream | Ubiquitin-specific peptidase 8 | 6 | NM_005154 |
| CHRNA7 | 15 | 99.0 | 5′ upstream | Cholinergic receptor, nicotinic, alpha 7 | 1 | NM_000746 |
| RYR3 | 15 | 98.8 | 5′ upstream | Ryanodine receptor 3 | 1 | NM_001036 |
| CDYL | 6 | 100 | 5′ upstream | Chromodomain Y-like protein | 2 | NM_004824 |
| AMD1 | 6 | 97.0 | Intron 1 | Adenosylmethionine decarboxylase 1 | 2 | NM_001634 |
| USP5 | 12 | 99.2 | Intron 19 | Ubiquitin-specific peptidase 5 | 1 | NM_003481 |
| WARS | 14 | 98.0 | Intron 5 | Tryptophanyl-tRNA synthetase | 6 | NM_004184 |
| DISP1 | 1 | 100.0 | Intron 3 | Dispatched A | Intron only | NM_032890 |
| WDR89 | 14 | 100.0 | Intron 2 | WD repeat domain 89 | 3 | NM_080666 |
| DOCK7 | 1 | 100.0 | Intron 18 | Dedicator of cytokinesis 7 | 1 | NM_033407 |
| MMACHC | 1 | 97.4 | Intron 1 | Methylmalonic aciduria and homocystinuria-type C protein | 1 | NM_015506 |
CHR, chromosome.
Identification of p200 host gene promoter targets by ChIP-chip.
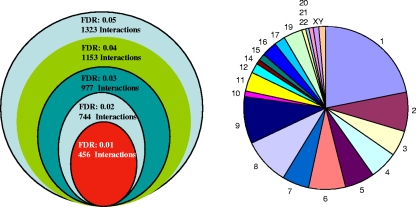
In order to determine p200 target genes using a whole-genome approach, a 5-kb whole-genome promoter microarray (chip) was used to hybridize ChIP p200 DNA (ChIP-chip). A range of FDR (0.05, 0.04, 0.03, 0.02, and 0.01) was applied to identify p200 target genes (Fig. 4, left panel). With the highest FDR (0.05), 1,323 interactions were identified, compared to 456 identified with the lowest FDR (0.01) (Fig. 4, left panel; see also Tables S1 and S2 in the supplemental material), and the target genes were distributed in all chromosomes (except chromosome 18), with the majority located in chromosome 1 (Fig. 4, right panel). For further analysis of specific gene interactions in this study, the lowest FDR (0.01) was applied to minimize the number of false discoveries. Analysis of the 5-kb promoter regions of 200 genes within this subset of identified p200 gene targets revealed that 80% contained at least one promoter Alu-Sx element (Table 2).
FIG. 4.
Summary of p200 URR interactions, displayed as a function of various FDR value thresholds applied to human whole-genome promoter array and total p200 human genome 5-kb promoter gene targets as determined by ChIP-chip analysis and frequency of gene targets in each chromosome (FDR, 0.01).
TABLE 2.
p200 binding sites present within the promoter regions (5 kb) of selected ChIP-chip-identified target genes
| Gene | Location(s) of Alu-Sx element(s) with binding motif in 5-kb promoter region (kb) | Chromosome | GenBank accession no. | Description |
|---|---|---|---|---|
| EFHC1 | 2.0, 2.9 | 6 | NM_018100 | EF-hand domain (C-terminal) containing 1 |
| RHOA | 3.3, 2.3 | 3 | NM_001664 | ras homolog gene family, member A |
| TNF-α | 4.3 | 6 | NM_000594 | Tumor necrosis factor α |
| USF1 | 4.8, 4.5 | 1 | NM_007122 | Upstream transcription factor 1 |
| NDOR1 | 2.8, 2.2, 3.5 | 9 | NM_014434 | NADPH-dependent diflavin oxidoreductase 1 |
| PSMB9 | 4.6 | 6 | NM_002800 | Proteasome (prosome, macropain) subunit, beta type, 9 |
| MAEA | 2.1, 3.1 | 4 | NM_005882 | Macrophage erythroblast attacher |
| PRCC | 2.8, 4.1, 1.9, 4.8, 3.8 | 1 | NM_199416 | Papillary renal cell carcinoma (translocation associated) |
| LY6E | 8 | NM_002346 | Lymphocyte antigen 6 complex, locus E | |
| REXO1L1 | 4.7 | 8 | NM_172239 | REX1, RNA exonuclease 1 homolog (S. cerevisiaea)-like 1 |
| c1orf2 | 4.6, 2.7, 3.8, 3.1 | 1 | NM_006589 | Chromosome 1 open reading frame 2 |
| C4B | 4.1, 4.5 | 6 | NM_001002029 | Complement component 4B |
| MAPKAPK2 | 1 | NM_004759 | Mitogen-activated protein kinase-activated protein kinase 2 | |
| SMAP1L | 2.0 | 1 | NM_022733 | Stromal membrane-associated GTPase-activating protein 2 |
| LTB | 1.0, 2.2, 1.9, 1.5 | 6 | NM_002341 | Lymphotoxin beta (TNF superfamily, member 3) |
| JAK2 | 4.2 | 9 | NM_004972 | Janus kinase 2 |
| ZNF565 | 4.0, 2.7, 2.1, 3.6 | 19 | NM_152477 | Zinc finger protein 565 |
| STAT1 | 3.6 | 2 | NM_139266 | Signal transducer and activator of transcription 1 |
| PTK2B | 5.0 | 8 | NM_004103 | Protein tyrosine kinase 2 beta |
| PPP2R5D | 1.0, 2.2, 1.8, 3.5, 1.4 | 6 | NM_180976 | Protein phosphatase 2, regulatory subunit B', delta isoform |
| CD48 | 0.9 | 1 | NM_001778 | CD48 molecule |
Saccharomyces cerevisiae.
In order to reveal general biological processes that may be affected by p200, a gene functional analysis was performed on the 456 ChIP-chip-identified p200 target genes using Babelomics FatiGO+ software (1, 2). We used this powerful approach to extract gene ontology terms of enriched genes determined by the ChIP-chip whole-genome promoter assay. The cell function for these p200 putative target genes was classified by general function and increasing specificity using hierarchy-based functional annotations. The p200 target genes were identified in three gene ontology databases (biological processes, molecular function, and cell structure) (see Fig. S1 in the supplemental material). At the highest levels of annotation hierarchy within the gene ontology databases, consistent enrichment of genes associated with specific molecular functions, biological processes, and cellular structure identified them as p200 targets (see Fig. S1 in the supplemental material). These included genes associated with transcriptional regulation (DNA and RNA), apoptosis, ATPase activity, and structural associations with the nucleus.
Enrichment of p200 target genes.
Signal enrichment maps are suggestive of binding regions, but in order to further assess the array performance, validation of the targets identified by ChIP-chip experiments was performed. qPCR analysis of randomly selected genes identified as p200 targets by ChIP-chip analysis was performed, and enrichment of these genes by immunoprecipitation relative to the input and the IgG amplicons was determined. Of the 21 genes selected, 19 were enriched (>1-fold) compared to input and negative IgG controls as determined by qPCR (Table 3). At least one Alu-Sx element was identified in the 5-kb promoter region of all enriched genes; however, the two genes (LY6E and MAPKAPK2) that were not enriched lacked Alu-Sx repeats in the promoter region (Table 3).
TABLE 3.
ChIP enrichment of randomly selected ChIP-chip-identified p200 target genes by quantitative PCRa
| Gene | Fold change (vs input) | Fold change (vs IgG) | Gene description |
|---|---|---|---|
| EFHC1 | 4.3 ± 0.1 | 3.1 ± 0.1 | Cell homeostasis |
| RHOA | 3.4 ± 0.1 | 3.2 ± 0.1 | Transport |
| TNF-α | 3.0 ± 0.1 | 3.3 ± 0.3 | Immune system process |
| USF1 | 3.6 ± 0.2 | 2.4 ± 0.1 | Regulation of transcription |
| NDOR1 | 2.4 ± 0.1 | 2.3 ± 0.1 | Cellular metabolic process |
| PSMB9 | 2.2 ± 0.1 | 2.2 ± 0.1 | Immune system process |
| MAEA | 2.2 ± 0.2 | 2.7 ± 0.2 | Cell division |
| PRCC | 2.8 ± 0.1 | 2.1 ± 0.1 | Translocation |
| LY6E | 1.0 ± 0.04 | 1.0 ± 0.1 | Cell communication |
| REXO1L1 | 2.5 ± 0.05 | 1.4 ± 0.1 | Hydrolase activity |
| c1orf2 | 2.5 ± 0.2 | 1.3 ± 0.1 | Intrinsic to membrane |
| C4B | 1.8 ± 0.02 | 2.0 ± 0.1 | Immune system process |
| MAPKAPK2 | 1.2 ± 0.03 | 1.0 ± 0.1 | Regulation of cellular metabolic process |
| SMAP1L | 2.2 ± 0.06 | 1.6 ± 0.1 | GTPase regulator activity |
| LTB | 2.8 ± 0.3 | 1.4 ± 0.1 | Immune system process |
| JAK2 | 2.9 ± 0.3 | 2.0 ± 0.3 | Immune system process |
| ZNF565 | 2.5 ± 0.1 | 2.2 ± 0.1 | Regulation of transcription |
| STAT1 | 2.3 ± 0.1 | 1.5 ± 0.1 | Signal transduction |
| PTK2B | 3.1 ± 0.1 | 2.0 ± 0.1 | Phosphorylation |
| PPP2R5D | 2.9 ± 0.1 | 1.6 ± 0.1 | Signal transduction |
| CD48 | 2.6 ± 0.1 | 1.3 ± 0.1 | Defense response |
Mean n-fold gene enrichment ± standard error was calculated and p200 compared with starting input and IgG control amplicon levels.
Interaction of p200 with Alu-Sx mid-A-stretch motif.
The ChIP-chip assay and analysis of cloned ChIP DNA revealed p200 target genes and identified a possible binding site associated with Alu-Sx elements. In order to identify a specific p200 binding motif, four oligonucleotide probes were designed within a 200-bp region of the Alu-Sx element that was identified by alignment of ChIP DNA sequences (Fig. 3B and C). To investigate p200 binding to the Alu-Sx element, we utilized the promoter region of the TNF-α gene (identified as a target by ChIP-chip analysis), which contained a single Alu-Sx element in the upstream 5-kb promoter region (Fig. 5A). An EMSA was performed using labeled (biotinylated) and unlabeled oligonucleotides representing the adenine-rich binding motif (P1) and two oligonucleotides probes (P2 and P3) corresponding to regions ∼20 bp and 90 bp, respectively, upstream of the adenine-rich motif located in the left arm of the Alu-Sx element and one probe (P4) ∼70 bp downstream of the mid-A-stretch motif in the right arm of the Alu-Sx repeat (Fig. 3B and 5A). Binding and a significant shift in migration of the P1 probe and weaker binding but a similar shift with the P2 probe were observed after incubation with E. chaffeensis-infected nuclear lysates (Fig. 5B, left panel). Probes P3 and P4 did not exhibit binding with infected nuclear lysates, and there was no binding observed for any probes after incubation of nuclear lysates from uninfected cells (Fig. 5B, left panel).
The specificity of the binding motif was further examined using competition experiments with excess unlabeled double-stranded P1 probe, which resulted in a marked reduction in labeled probe-protein complex formation (Fig. 5B, center panel). To examine whether the DNA-protein complex produced in the EMSA assay corresponds to p200, a supershift was performed using p200-specific antibody by EMSA. This experiment demonstrated that the migration of the p200 complex observed with the P1 probe was not retarded by p200-specific antibody (Fig. 5B, center panel). However, due to the limitations in resolution of gel shift assays with proteins of >130 kDa (28), an alternative NoShift assay was used to investigate p200 interaction with the P1 probe (9). Compared with the conventional EMSA, the NoShift assay has the capability of demonstrating the interaction of a specific protein with a specific DNA probe. Sequence-specific binding of the probe P1 with p200 was demonstrated by competition between biotinylated and unlabeled P1 probes (Fig. 5B, right panel). The interaction between p200 and its biotinylated probe in the presence of increased concentrations of unlabeled P1 probe was demonstrated using rabbit anti-E. chaffeensis p200 serum and compared to results with normal rabbit serum. The signal-to-noise (negative control) ratio in the absence of the competing unlabeled capture probe was 3.85:1:0.5, but the signal intensity decreased as the concentration of the unlabeled P1 probe increased (10 pmol to 900 pmol). The addition of a different molar ratio of unlabeled P1 probe to labeled P1 probe resulted in a reduction in labeled-probe binding to the background level, demonstrating specific p200 binding to the P1 probe. To further characterize specific nucleotides within P1 that were important for binding, binding affinity experiments with the mutant probes M1, M2, M3, and M4 were performed (Fig. 5C). DNA-protein complexes were observed with the wild-type probe (P1), the adenine-rich mutant probe (M1), and the paired TAAT mutant probe, M2, but binding was not observed with probes (M3 and M4) containing both paired TAAT and AAATACAAA mutations, demonstrating that both motifs contribute to p200 binding (Fig. 5C).
Expression levels of p200 target genes.
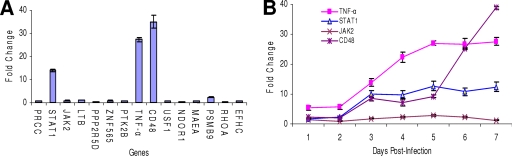
Fifteen genes identified by ChIP-chip analysis as p200 target genes were selected for analysis of gene expression levels in E. chaffeensis-infected cells. Some of these genes (encoding TNF-α, Stat1, and Jak2) are linked to ehrlichial disease pathogenesis, while CD48 is a known receptor for bacterial entry through caveolae (56, 57). The majority of the genes (n = 11) examined exhibited a small decrease (≤1-fold) in expression levels at day 7 postinfection (Fig. 6A). However, three genes (encoding TNF-α, Stat1, and CD48) were strongly upregulated (∼13- to 37-fold) beginning at day 3 and increasing through day 7 postinfection (Fig. 6B). Levels of TNF-α were also determined in cell culture supernatant, and consistent with gene expression, the level of TNF-α was detectable 48 h postinfection (8.6 pg/ml) and increased to a peak of 246 pg/ml (∼30-fold increase) by day 5 postinfection. TNF-α was not detected in supernatants from uninfected THP-1 cell (data not shown).
FIG. 6.
Expression of p200 target genes in E. chaffeensis-infected monocytes. (A) Expression levels of p200 target genes in E. chaffeensis-infected THP-1 cells 7 days postinfection. Data shown are the means (± standard errors) for triplicate experiments. (B) Time course expression analysis of p200 target genes (encoding TNF-α, Stat1, Jak2, and CD48) in E. chaffeensis-infected THP-1 cells. Data shown are the means (± standard errors) for triplicate experiments.
DISCUSSION
Genomic sequencing of multiple Ehrlichia sp. genomes has identified a small subset of proteins containing eukaryote-like ankyrin repeats, suggesting a potential role in host-pathogen interactions. Ankyrin repeats are one of the most common protein sequence motifs in eukaryotes and have recently been identified in human bacterial pathogens from the genera Ehrlichia, Anaplasma, Orientia, Coxiella, Legionella, and Pseudomonas (10, 11, 14, 22, 23, 40, 50, 55). We previously identified and molecularly characterized the p200 orthologs of E. canis and E. chaffeensis and determined that they were primary targets of the humoral immune response (41, 43, 46). The ankyrin repeats of the p200s are found in a centralized region that involves most of the protein, and this region is flanked by two terminal acidic domains that contain major antibody epitopes (46). In this study, we demonstrate that the p200 protein of E. chaffeensis is translocated to the host cell nucleus, ultimately interacting with Alu-Sx element motifs located in promoters and introns of various host cell genes. The translocation of pathogen proteins to the host cell nucleus has been recently linked with other bacterial pathogens, including AnkA of A. phagocytophilum, YopM of Yersinia pestis, and the Osp and IpaH proteins of Shigella flexneri (51, 58, 62, 69, 70).
The E. chaffeensis p200 protein lacks a classical monopartite or bipartite nuclear localization signal (NLS) according to the predictNLS (13) and PSORT software programs (45); thus, its method of entry into the nucleus is not clear. Similarly, the Shigella flexneri nuclear translocated protein, OspB, lacks a classic NLS, suggesting that it has a nonclassical NLS or is transported by a host protein that targets the nucleus (70). There are several known nucleocytoplasmic transport pathways, including the classical nuclear import pathway involving short monopartite and bipartite stretches of basic amino acids (lysine and arginine) (13), which is used by approximately 57% of nuclear transported proteins. The large molecular mass of p200 and the fact that a classical monopartite or bipartite NLS was not present suggest that the mechanism by which it gains access to the monocyte nucleus is nonclassical. Nevertheless, it is possible that p200 has a classical NLS sequence that utilizes importin-α mediated import but is not recognized by standard algorithms. However, other nuclear import pathways exist, including a glycan-dependent nuclear import pathway that has also been described and characterized in vitro (16, 17) and vesicular transport, such as that utilized by Stat3 (7). In addition, recent examples of newly identified nonclassical NLSs suggest that many uncharacterized nonclassical NLSs exist. Two such examples in bacteria are the nuclear translocation of Yersinia Yop M and that of Shigella IpaH, which appear to be mediated by leucine-rich repeats (5, 58, 62).
In this study, a specific motif associated with p200-chromatin interaction was identified using the ChIP-Seq technique, which is now routinely used to investigate transcription factor binding motifs and is a complementary approach to genome-wide assays such as ChIP-chip (18, 25). p200 does not have a known DNA binding domain, but our data suggest that p200 is closely associated with chromatin through a direct interaction with chromatin or indirectly through a protein-protein interaction. ChIP-Seq analysis identified a p200-DNA interaction in a substantial proportion of clones (24%) involving an adenine-rich motif within Alu-Sx elements known as the mid-A-stretch motif (59). Notably, in a related study involving the nuclear translocated protein AnkA of A. phagocytophilum, a consistent DNA binding motif was not identified; such a motif may be related to the choice of cross-linker (cis-DDP) (51). However, in a more recent study, AnkA binding to DNA was examined using formaldehyde as a cross-linker, and of the 10 sequenced clones, ATC-rich sequences were identified in 8. Similarly, we used formaldehyde as a cross-linker to investigate the p200-chromatin interaction because it cross-links amines in proteins and DNA that are in very close proximity (≤2 Å). Using this approach, a specific adenine-rich (mid-A-stretch) motif flanked by AT-rich regions within Alu-Sx elements in the promoter regions of host genes was identified in this study. This finding is somewhat consistent with regard to location (gene regulatory regions) and AT-rich binding sites recently reported for AnkA (19). However, a specific DNA motif, such as Alu, was not identified for AnkA, and it was concluded that the AnkA binding feature was not directly related to primary sequence (19). It is possible that p200 interaction with the chromatin occurs via another protein (via Ank repeats), such as host transcription factors or regulatory proteins. Supporting this conclusion is a previous finding demonstrating the interaction of A. phagocytophilum AnkA with proteins present in host cell nuclei (51). With respect to p200, such proteins may include transcription factors known to interact with the mid-A-stretch motif of Alu elements, such as all MEF2 family members, HNF1.03, OC.2, BARX2, and PAX4 (52).
There are numerous characterized Alu elements classified into 12 major subfamilies according to their relative ages (27), with Alu-Sx being intermediate in age. Interestingly, among Alu elements, the mid-A-stretch motif has many insertions and deletions and is considered to be variable (27). Alu elements are highly represented in the genome, particularly in 5′ upstream regulatory regions (URRs) (52); thus, we employed a now commonly used genome-wide technique, ChIP-chip, to study genome-wide protein-DNA interactions. A relatively small number of 5′ URRs (n = 456) compared to possible genes (∼59,000) were identified as p200 targets (FDR, 0.01). Interestingly, the genes identified by ChIP-Seq were not identified by ChIP-chip. However, recent comparisons of targets identified by ChIP-Seq and ChIP-chip indicate that targets exclusive to each technique are common and are attributed to scoring methods and neighboring repetitive sequences (18). Consequently, p200 targets identified by ChIP-Seq may not overlap with targets identified by ChIP-chip, but the systematic approach of ChIP-chip has been shown to reliably identify gene targets and enrichment levels (26). Although 456 5′ URR targets were identified by ChIP-chip, only 80% of these contained Alu-Sx elements, suggesting that some of these identified targets may constitute false positives or p200 could be involved in other interactions. This is also supported by enrichment data that found 80% of randomly selected genes were enriched and the unenriched genes did not contain Alu-Sx elements in the promoter region. Furthermore, about half of the ChIP-Seq clones identified were mapped to intronic sites, which were not represented (5-kb promoter only) on the microarrays. Therefore, a potentially large number of additional binding sites, primarily those within intronic regions, are not represented in the ChIP-chip analysis performed in this study. This is also supported by the CHIP data that AnkA was found to bind to multiple sites over a 20-kb upstream region of the target gene (19).
The p200 protein has numerous binding sites in both promoter and intronic locations, and whether some or all of these interactions have functional effects is a question that remains to be answered. Recent studies of Drosophila embryos suggest that in general, transcription factors bind thousands of active and inactive regions of the chromosome, but functional interactions in vivo are ultimately governed by higher-order rules (33). Others have also suggested that only a small subset of predicted transcription factor binding sequences are functional in vivo (6). Therefore, further investigation will be needed to understand the effect of p200 on transcription of individual genes and to distinguish functional and nonfunctional p200 binding sites.
The association of the p200 with Alu-Sx elements, which are the most abundant repetitive elements in the human genome, suggests that p200 could affect gene transcription globally through numerous mechanisms associated with Alu element transcriptional and posttranscriptional gene expression regulation. Numerous functions have been linked to Alu elements depending on whether the elements were free or embedded in the genome (20). Although Alu elements are dispersed throughout the genome, a higher proportion are present in the 5-kb regions upstream of gene transcription start sites. The distribution of Alu elements varies between functional gene families, but they are most commonly found in genes related to metabolism, transport, and signaling (52). Two potential functions are associated with Alu elements in 5′ URRs. Alu elements embedded in the 5′ URRs influence gene translation through numerous mechanisms, including mRNA stability, localization, and translation efficiency (21). Translation inhibition has been associated with Alu elements embedded in 5′ URRs. Therefore, p200 binding to these URR Alu elements may play a role in influencing the translational efficiency of these genes. Second, Alu elements may be effectors of transcription by providing binding sites for transcription factors (61). Expression levels of some genes of interest and other randomly selected p200 target genes (n = 15) revealed that most were downregulated while some (encoding TNF-α, Stat1, and CD48) exhibited more than 10-fold upregulation, indicating that p200 may be involved in transcriptional repression and activation. Infection of cells by E. chaffeensis has been shown to alter (up- and downregulate) the transcription levels of host cell genes involved in a variety of functions (68), but it is unlikely that p200 interaction with host cell genes accounts for all the pathogen-orchestrated transcriptional changes that are observed during infection.
The role of p200 in modulation of specific host cell gene transcriptional changes to enhance the survival of E. chaffeensis will require further investigation to demonstrate a direct effect. Gene ontological analysis performed in this study using FatiGO+ software indicates that p200 has a specific role in regulation of numerous biological and molecular processes and structurally related protein genes; particularly prominent were genes related to regulation of transcription ATPase activity and apoptosis. The modulation of apoptosis has previously been reported to occur in Ehrlichia and closely related Anaplasma spp. Apoptosis is delayed in cells infected with Anaplasma phagocytophilum, and recently Ehrlichia ewingii infection has been shown to delay spontaneous neutrophil apoptosis (12, 65, 66). However, the mechanism by which Anaplasma and Ehrlichia spp. modulate cellular apoptosis is not fully understood. Transcriptional modulation of a number of genes associated with apoptosis inhibition and induction is altered to favor cell survival (68). p200 also appears to bind genes associated with transcription, and there is evidence that a large number of genes associated with transcription are modulated during E. chaffeensis infection (68). ATPase activity is present in ehrlichial inclusions (4), and genes associated with ATPase activity also appear to be targets of E. chaffeensis p200 and A. phagocytophilum AnkA (51).
Because of its potentially important role in disease pathogenesis, we used the TNF-α gene to examine binding of p200 to the Alu-Sx motif. In addition, we investigated the temporal expression of this gene and found dramatically increased levels in E. chaffeensis-infected monocytes beginning 3 days postinfection. In agreement with these findings are results in previous studies in which an absence of TNF-α mRNA expression in E. chaffeensis-infected THP-1 cells within 18 h (31, 32) or 24 h (68) postinfection was reported. Hence, the upregulation of TNF-α later in the infection indicates that this response is not mediated by innate immune receptors and suggests that other mechanisms directly related to the pathogen may be responsible. A contribution of cytokine (TNF-α)-associated immunopathology by CD8+ and NK T cells is thought to contribute to fatal ehrlichiosis (8, 24, 60). The new findings in this study suggest that monocytes/macrophages may also contribute substantially to the production of TNF-α and the induction of this gene may be modulated, in part, by p200 interaction with the Alu element.
Numerous genes with known associations with ehrlichial pathobiology and pathogenesis were identified as p200 targets. We found several genes of interest related to IFN-γ regulation and receptor-mediated bacterial uptake that were investigated further. E. chaffeensis blocks phosphorylation of Stat1 and Jak2 during infection (30), and the corresponding genes were also identified as p200 targets, suggesting that E. chaffeensis uses multiple strategies to alter the Jak/Stat pathway. Interestingly, Stat1 was strongly upregulated (>10-fold) 3 days after infection, while expression levels of Jak2 were modestly upregulated (∼4-fold) 5 days postinfection. It is not clear if p200 modulates the expression of these genes, but the fact that these genes are associated with ehrlichial survival and are p200 targets suggests that Ehrlichia modulates these innate immune response effectors by multiple mechanisms. The process of ehrlichia entry into the host cell involves caveolae and glycosylphosphatidylinositol-anchored proteins (34), but the identity of the receptor remains unclear. Thus, the upregulation of CD48, a glycosylphosphatidylinositol-anchored protein associated with caveolae and a receptor for bacterial uptake, was an interesting finding, suggesting that ehrlichial binding and endocytosis may be associated with this protein and modulated by p200 (56, 57).
We have for the first time demonstrated that E. chaffeensis and E. canis p200 proteins are translocated to the nuclei of Ehrlichia-infected monocytes. These results provide strong evidence of specific gene targets of E. chaffeensis p200 and provide new insight into the potential role of p200 in modulation of specific host cell gene expression and related processes. A more comprehensive understanding of the interactions by which p200 affects the host cell and transport in the host cell will provide insight into how obligately intracellular bacteria control their environment and ultimately will provide novel targets for therapeutic intervention.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Allergy and Infectious Diseases grant (AIAI069270), and additional support was provided by the Clayton Foundation for Research.
We thank Gracie Vargas and Jiang Yongquan of the Center for Biomedical Engineering and Rhykka Connelly of the Infectious Disease and Toxicology Optical Imaging Core of UTMB for their assistance with confocal microscope imaging. We also thank David H. Walker and Xue-jie Yu for reviewing the manuscript and providing thoughtful suggestions.
Editor: A. Camilli
Footnotes
Published ahead of print on 3 August 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Al-Shahrour, F., P. Minguez, J. Tarraga, I. Medina, E. Alloza, D. Montaner, and J. Dopazo. 2007. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 35:W91-W96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Shahrour, F., P. Minguez, J. Tarraga, D. Montaner, E. Alloza, J. M. Vaquerizas, L. Conde, C. Blaschke, J. Vera, and J. Dopazo. 2006. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 34:W472-W476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnewall, R. E., N. Ohashi, and Y. Rikihisa. 1999. Ehrlichia chaffeensis and E. sennetsu, but not the human granulocytic ehrlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 67:2258-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnewall, R. E., Y. Rikihisa, and E. H. Lee. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 65:1455-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benabdillah, R., L. J. Mota, S. Lutzelschwab, E. Demoinet, and G. R. Cornelis. 2004. Identification of a nuclear targeting signal in YopM from Yersinia spp. Microb. Pathog. 36:247-261. [DOI] [PubMed] [Google Scholar]
- 6.Biggin, M. D., and R. Tjian. 2001. Transcriptional regulation in Drosophila: the post-genome challenge. Funct. Integr. Genomics 1:223-234. [DOI] [PubMed] [Google Scholar]
- 7.Bild, A. H., J. Turkson, and R. Jove. 2002. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 21:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitsaktsis, C., and G. Winslow. 2006. Fatal recall responses mediated by CD8 T cells during intracellular bacterial challenge infection. J. Immunol. 177:4644-4651. [DOI] [PubMed] [Google Scholar]
- 9.Bruggink, F., and S. Hayes. 2004. Identification of DNA binding proteins using the NoShift transcription factor assay kit. Nat. Methods 1:177-179. [Google Scholar]
- 10.Caturegli, P., K. M. Asanovich, J. J. Walls, J. S. Bakken, J. E. Madigan, V. L. Popov, and J. S. Dumler. 2000. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68:5277-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, N. H., J. M. Kim, E. K. Kwon, S. Y. Kim, S. H. Han, H. Chu, J. H. Lee, M. S. Choi, and I. S. Kim. 2005. Molecular characterization of a group of proteins containing ankyrin repeats in Orientia tsutsugamushi. Ann. N. Y. Acad. Sci. 1063:100-101. [DOI] [PubMed] [Google Scholar]
- 12.Choi, K. S., J. T. Park, and J. S. Dumler. 2005. Anaplasma phagocytophilum delay of neutrophil apoptosis through the p38 mitogen-activated protein kinase signal pathway. Infect. Immun. 73:8209-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, N. E., J. Liebenberg, E. P. de Villiers, K. A. Brayton, E. Louw, A. Pretorius, F. E. Faber, H. H. van, A. Josemans, K. M. van, H. C. Steyn, M. F. van Strijp, E. Zweygarth, F. Jongejan, J. C. Maillard, D. Berthier, M. Botha, F. Joubert, C. H. Corton, N. R. Thomson, M. T. Allsopp, and B. A. Allsopp. 2005. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. USA 102:838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duverger, E., V. Carpentier, A. C. Roche, and M. Monsigny. 1993. Sugar-dependent nuclear import of glycoconjugates from the cytosol. Exp. Cell Res. 207:197-201. [DOI] [PubMed] [Google Scholar]
- 17.Duverger, E., C. Pellerin-Mendes, R. Mayer, A. C. Roche, and M. Monsigny. 1995. Nuclear import of glycoconjugates is distinct from the classical NLS pathway. J. Cell Sci. 108:1325-1332. [DOI] [PubMed] [Google Scholar]
- 18.Euskirchen, G. M., J. S. Rozowsky, C. L. Wei, W. H. Lee, Z. D. Zhang, S. Hartman, O. Emanuelsson, V. Stolc, S. Weissman, M. B. Gerstein, Y. Ruan, and M. Snyder. 2007. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 17:898-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Garcia, J. C., K. E. Rennoll-Bankert, S. Pelly, A. M. Milstone, and J. S. Dumler. 2009. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 77:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasler, J., T. Samuelsson, and K. Strub. 2007. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol. Life Sci. 64:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasler, J., and K. Strub. 2006. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 34:2374-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. A. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howell, M. L., E. Alsabbagh, J. F. Ma, U. A. Ochsner, M. G. Klotz, T. J. Beveridge, K. M. Blumenthal, E. C. Niederhoffer, R. E. Morris, D. Needham, G. E. Dean, M. A. Wani, and D. J. Hassett. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J. Bacteriol. 182:4545-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ismail, N., L. Soong, J. W. McBride, G. Valbuena, J. P. Olano, H. M. Feng, and D. H. Walker. 2004. Overproduction of TNF-alpha by CD8+ type 1 cells and down-regulation of IFN-gamma production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J. Immunol. 172:1786-1800. [DOI] [PubMed] [Google Scholar]
- 25.Ji, H., S. A. Vokes, and W. H. Wong. 2006. A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors. Nucleic Acids Res. 34:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. S., W. Li, D. B. Gordon, A. Bhattacharjee, B. Curry, J. Ghosh, L. Brizuela, J. S. Carroll, M. Brown, P. Flicek, C. M. Koch, I. Dunham, M. Bieda, X. Xu, P. J. Farnham, P. Kapranov, D. A. Nix, T. R. Gingeras, X. Zhang, H. Holster, N. Jiang, R. D. Green, J. S. Song, S. A. McCuine, E. Anton, L. Nguyen, N. D. Trinklein, Z. Ye, K. Ching, D. Hawkins, B. Ren, P. C. Scacheri, J. Rozowsky, A. Karpikov, G. Euskirchen, S. Weissman, M. Gerstein, M. Snyder, A. Yang, Z. Moqtaderi, H. Hirsch, H. P. Shulha, Y. Fu, Z. Weng, K. Struhl, R. M. Myers, J. D. Lieb, and X. S. Liu. 2008. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 18:393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kariya, Y., K. Kato, Y. Hayashizaki, S. Himeno, S. Tarui, and K. Matsubara. 1987. Revision of consensus sequence of human Alu repeats—a review. Gene 53:1-10. [DOI] [PubMed] [Google Scholar]
- 28.Knight, J. S., M. A. Cotter, and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 29.Kumagai, Y., Z. Cheng, M. Lin, and Y. Rikihisa. 2006. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 74:5014-5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E. H., and Y. Rikihisa. 1998. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect. Immun. 66:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, E. H., and Y. Rikihisa. 1996. Absence of tumor necrosis factor alpha, interleukin-6 (IL-6), and granulocyte-macrophage colony-stimulating factor expression but presence of IL-1β, IL-8, and IL-10 expression in human monocytes exposed to viable or killed Ehrlichia chaffeensis. Infect. Immun. 64:4211-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, E. H., and Y. Rikihisa. 1997. Anti-Ehrlichia chaffeensis antibody complexed with E. chaffeensis induces potent proinflammatory cytokine mRNA expression in human monocytes through sustained reduction of IκB-α and activation of NF-κB. Infect. Immun. 65:2890-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, X. Y., S. MacArthur, R. Bourgon, D. Nix, D. A. Pollard, V. N. Iyer, A. Hechmer, L. Simirenko, M. Stapleton, C. L. Luengo Hendriks, H. C. Chu, N. Ogawa, W. Inwood, V. Sementchenko, A. Beaton, R. Weiszmann, S. E. Celniker, D. W. Knowles, T. Gingeras, T. P. Speed, M. B. Eisen, and M. D. Biggin. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, M., and Y. Rikihisa. 2003. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell Microbiol. 5:809-820. [DOI] [PubMed] [Google Scholar]
- 35.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, M., and Y. Rikihisa. 2004. Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-kappa B, ERK 1/2 and p38 MAPK in host monocytes. Cell Microbiol. 6:175-186. [DOI] [PubMed] [Google Scholar]
- 37.Lin, M., and Y. Rikihisa. 2007. Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell Microbiol. 9:861-874. [DOI] [PubMed] [Google Scholar]
- 38.Lin, M., M. X. Zhu, and Y. Rikihisa. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-γ2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, T., X. Zhang, A. Wakeel, V. L. Popov, and J. W. McBride. 2008. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 76:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavromatis, K., C. K. Doyle, A. Lykidis, N. Ivanova, M. P. Francino, P. Chain, M. Shin, S. Malfatti, F. Larimer, A. Copeland, J. C. Detter, M. Land, P. M. Richardson, X. J. Yu, D. H. Walker, J. W. McBride, and N. C. Kyrpides. 2006. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J. Bacteriol. 188:4015-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride, J. W., J. E. Comer, and D. H. Walker. 2003. Novel immunoreactive glycoprotein orthologs of Ehrlichia spp. Ann. N. Y. Acad. Sci. 990:678-684. [DOI] [PubMed] [Google Scholar]
- 42.McBride, J. W., R. E. Corstvet, E. B. Breitschwerdt, and D. H. Walker. 2001. Immunodiagnosis of Ehrlichia canis infection with recombinant proteins. J. Clin. Microbiol. 39:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride, J. W., L. M. Ndip, V. L. Popov, and D. H. Walker. 2002. Identification and functional analysis of an immunoreactive DsbA-like thio-disulfide oxidoreductase of Ehrlichia spp. Infect. Immun. 70:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 46.Nethery, K. A., C. K. Doyle, X. Zhang, and J. W. McBride. 2007. Ehrlichia canis gp200 contains dominant species-specific antibody epitopes in terminal acidic domains. Infect. Immun. 75:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Geen, H., C. M. Nicolet, K. Blahnik, R. Green, and P. J. Farnham. 2006. Comparison of sample preparation methods for ChIP-chip assays. BioTechniques 41:577-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer, G. H. 2002. The highest priority: what microbial genomes are telling us about immunity. Vet. Immunol. Immunopathol. 85:1-8. [DOI] [PubMed] [Google Scholar]
- 50.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 52.Polak, P., and E. Domany. 2006. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC. Genomics 7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rikihisa, Y. 2006. Ehrlichia subversion of host innate responses. Curr. Opin. Microbiol. 9:95-101. [DOI] [PubMed] [Google Scholar]
- 54.Sedgwick, S. G., and S. J. Smerdon. 1999. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24:311-316. [DOI] [PubMed] [Google Scholar]
- 55.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin, J. S., and S. N. Abraham. 2001. Caveolae as portals of entry for microbes. Microbes Infect. 3:755-761. [DOI] [PubMed] [Google Scholar]
- 57.Shin, J. S., and S. N. Abraham. 2001. Glycosylphosphatidylinositol-anchored receptor-mediated bacterial endocytosis. FEMS Microbiol. Lett. 197:131-138. [DOI] [PubMed] [Google Scholar]
- 58.Skrzypek, E., C. Cowan, and S. C. Straley. 1998. Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol. Microbiol. 30:1051-1065. [DOI] [PubMed] [Google Scholar]
- 59.Stevens, T. A., J. S. Iacovoni, D. B. Edelman, and R. Meech. 2004. Identification of novel binding elements and gene targets for the homeodomain protein BARX2. J. Biol. Chem. 279:14520-14530. [DOI] [PubMed] [Google Scholar]
- 60.Stevenson, H. L., E. C. Crossley, N. Thirumalapura, D. H. Walker, and N. Ismail. 2008. Regulatory roles of CD1d-restricted NKT cells in the induction of toxic shock-like syndrome in an animal model of fatal ehrlichiosis. Infect. Immun. 76:1434-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomilin, N. V. 1999. Control of genes by mammalian retroposons. Int. Rev. Cytol. 186:1-48. [DOI] [PubMed] [Google Scholar]
- 62.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH(9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 276:32071-32079. [DOI] [PubMed] [Google Scholar]
- 63.Wakeel, A., J. A. Kuriakose, and J. W. McBride. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77:1734-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walker, T., L. Klasson, M. Sebaihia, M. J. Sanders, N. R. Thomson, J. Parkhill, and S. P. Sinkins. 2007. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC. Biol. 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiong, Q., W. Bao, Y. Ge, and Y. Rikihisa. 2008. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J. Infect. Dis. 197:1110-1118. [DOI] [PubMed] [Google Scholar]
- 66.Yoshiie, K., H. Y. Kim, J. Mott, and Y. Rikihisa. 2000. Intracellular infection by the human granulocytic ehrlichiosis agent inhibits human neutrophil apoptosis. Infect. Immun. 68:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, X. J., J. W. McBride, C. M. Diaz, and D. H. Walker. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, J. Z., M. Sinha, B. A. Luxon, and X. J. Yu. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72:498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zurawski, D. V., C. Mitsuhata, K. L. Mumy, B. A. McCormick, and A. T. Maurelli. 2006. OspF and OspC1 are Shigella flexneri type III secretion system effectors that are required for postinvasion aspects of virulence. Infect. Immun. 74:5964-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zurawski, D. V., K. L. Mumy, C. S. Faherty, B. A. McCormick, and A. T. Maurelli. 2009. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol. Microbiol. 71:350-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.