Abstract
The endoplasmic reticulum resident thyroid hormone-activating type 2 deiodinase (D2) is inactivated by ubiquitination via the hedgehog-inducible WSB-1. Ubiquitinated D2 can then be subsequently taken up by the proteasomal system or be reactivated by USP-33/20-mediated deubiquitination. Given that heterologously expressed D2 accumulates in Saccharomyces cerevisiae lacking the E3 ligase Doa10, we tested whether the human Doa10 ortholog, TEB4, plays a role in D2 ubiquitination and degradation. In a setting of transient coexpression in HEK-293 cells, TEB4 and D2 could be coimmunoprecipitated, and additional TEB4 expression decreased D2 activity by ∼50% (P < 0.05). A highly efficient TEB4 knockdown (>90% reduction in mRNA and protein levels) decreased D2 ubiquitination and increased D2 activity and protein levels by about fourfold. The other activating deiodinase, D1, or a truncated D2 molecule (Δ18-D2) that lacks a critical instability domain was not affected by TEB4 knockdown. Furthermore, TEB4 knockdown prolonged D2 activity half-life at least fourfold, even under conditions known to promote D2 ubiquitination. Neither exposure to 1 μM of the proteasomal inhibitor MG132 for 24 h nor RNA interference WSB-1 knockdown resulted in additive effects on D2 expression when combined with TEB4 knockdown. Similar results were obtained with MSTO-211 cells, which endogenously express D2, after TEB4 knockdown using a lentivirus-based transduction strategy. While TEB4 expression predominates in the hematopoietic lineage, both WSB-1 and TEB4 are coexpressed with D2 in a number of tissues and cell types, except the thyroid and brown adipose tissue, where TEB4 expression is minimal. We conclude that TEB4 interacts with and mediates loss of D2 activity, indicating that D2 ubiquitination and degradation can be tissue specific, depending on WSB-1 and TEB4 expression levels.
3,5,3′-Triiodothyronine (T3) modulates gene expression through the ligand-dependent transcription factor thyroid hormone receptor (15). Despite the presence of T3 (<20%) in the thyroidal secretion, most T3 in the body is generated outside the thyroid parenchyma via deiodination of thyroxine (T4), the main secretory product of the thyroid gland (29). The type 2 iodothyronine deiodinase (D2) is the key thyroid hormone-activating deiodinase, while the type 3 deiodinase (D3) plays the opposite role, inactivating both T3 and T4 also via specific deiodinase reactions, terminating thyroid hormone action. While the D2 pathway is a major source of plasma T3, contributing about 30% in small rodents and at least double that in humans, thyroid hormones are cleared predominantly through the D3 pathway (2). Combined, the roles played by D2 and D3 provided an elegant homeostatic mechanism by which coordinated reciprocal changes in their activities ensure adaptation to iodine availability in the environment (reviewed in reference 24).
Deiodinase expression can also regulate thyroid hormone signaling on a temporal and cell-specific basis. D2 expression results in an additional intracellular source of T3 that ultimately increases the nuclear concentration of T3 and regulates the expression of T3-responsive genes. On the contrary, D3 expression decreases availability of intracellular T3, reducing T3-dependent gene expression (18). The relevance of such mechanisms is illustrated in brown adipose tissue (BAT), where a potent D2 induction increases T3 production during cold exposure (3), and in models of resistance to diet-induced obesity (21, 36). In the chicken developing growth plate, inactivation of D2 is part of the mechanism by which Indian hedgehog induces PTHrP, thereby regulating chondrocyte differentiation (11).
D2 is an endoplasmic reticulum (ER) resident protein (1) that is assembled in a dimeric formation (9) and has a half-life of about 20 min, which can be further reduced by exposure to T4 (25, 31). This substrate-dependent downregulation provides a potent feedback mechanism to efficiently control T3 production (24). Ubiquitination is a key element of D2's posttranslational regulation which inactivates the enzyme and targets it for proteasomal degradation (17). While both UBC6 and UBC7 can function as ubiquitin-conjugating enzymes in D2 ubiquitination (4, 22), the SOCS box-containing protein WSB-1 has been established as an E3 ligase for D2, forming a ubiquitinating catalytic core complex along with elongin B, elongin C, Cul5, and Rbx1 (ECSWSB-1) (11). Furthermore, ubiquitinated D2 (Ub-D2) can be reactivated and rescued from proteasomal degradation by the von Hippel-Lindau protein (pVHL)-interacting deubiquitinating enzyme-1 (VDUI/USP33) (10). In addition to mediating ubiquitination of D2, WSB-1 has also recently been shown to be an E3 ligase for homeodomain protein kinase 2 (HIPK2), a member of the nuclear protein kinase family, which induces both p53- and CtBP-mediated apoptosis (6).
Notably, both WSB-1 and USP33 are physically associated with the D2 protein simultaneously, thus allowing rapid regulatory cycles of ubiquitination and deubiquitination to occur in a dynamic manner (27). WSB-1-mediated D2 ubiquitination takes place at K237 and/or K244 after the substrate (T4 or 3,3′,5′-triiodathyronine [rT3]) binds to the enzyme active center. Given that K244 lies within the D2 dimer interface near the catalytic site, its ubiquitination and/or that of its neighbor K237 provides a putative molecular mechanism for enzyme inactivation to occur (27).
Substantial knowledge about the mechanistic details of D2 ubiquitination was obtained by heterologously expressing human D2 in Saccharomyces cerevisiae (4). In this setting, deletion of Doa10 but not Hrd1, two E3 ligases that target proteins for ER-associated degradation, increases the accumulation of D2 protein (26). The ER-associated degradation system is involved in a number of human diseases, most notably cystic fibrosis (35), and recently has also been reported as a major player in the regulation of metabolism (28). TEB4 (MARCH-VI) is the mammalian homolog of the yeast Doa10 protein (23). This ER resident ubiquitin ligase contains 14 transmembrane helices and a conserved RING-CH finger domain at the N terminus and ubiquitinates ER-associated proteins with a cytoplasmic domain in a UBC7-dependent manner (5, 23, 33), such as Mps2, UBC6, and Ste6 (5, 6, 20, 26, 34). Additionally, as with other RING finger-containing E3 ligases, TEB4 regulates its own UBC7-mediated degradation (19).
In the present work, we investigated the role of TEB4 in the proteasomal degradation of D2 in mammalian cells. We conclude that TEB4 can mediate loss of D2 activity under both basal and substrate-induced D2 ubiquitination, indicating that control of D2 activity and expression via conjugation to ubiquitin are under dual control by both WSB-1 and TEB4. Given that most T3 in the brain is produced via the D2 pathway (16, 24), these findings could have important consequences for our understanding of the Cri-du-chat syndrome, a neurodegenerative disorder associated with a chromosome region containing the TEB4 gene (19).
MATERIALS AND METHODS
DNA constructs.
A total of 0.1 μg of a mammalian expression vector encoding wild-type rat D1 cloned into a CDM8 vector (G21) or human D2 cloned into a D10 vector (hD2SelP) was used for transfection in experiments that included measurement of deiodinase activity. When measuring deiodinase protein levels, cells were transfected with 0.5 or 1 μg of D10/D15 expression vectors encoding human Sec133CysD2 fused to a Flag tag at the carboxyl end (Flag-D2) (10). The green fluorescent protein (GFP)-TEB4 vector was generously provided by Mark Hochstrasser (26). The WSB-1 RNA interference (RNAi) vector and the Δ18-D2 mutant have been previously described (11). The following sequences were cloned using T4 ligase into the HindIII and BamHI sites of a pSilencer 2.1-U6 neo (Ambion, Austin, TX) to generate the TEB4 RNAi vectors: RNAi 1 (top strand), 5′-GTATTGGCACTGCAATACG-3′; RNAi 2 (top strand), 5′-TGATCCAGATTTCAATCCA-3′.
Reagents.
T4 and rT3 were obtained from Sigma (St. Louis, MO) and dissolved in 40 mM NaOH. Outer-ring-labeled [125I]T4 and [125I]rT3 (specific activities of ∼4,400 Ci/mmol and ∼738 Ci/mmol, respectively) were from NEN Life Science Products (Boston, MA), and MG132 was from Calbiochem (San Diego, CA). Anti-Flag M21 was obtained from Sigma.
Cell culture and transfections.
HEK-293 cells were maintained in six-well dishes and transfected using Lipofectamine and Plus reagents (Invitrogen, Carlsbad, CA) as described previously (11). To correct for efficiency of transfection, cells were also cotransfected with 0.5 μg of vectors constitutively expressing either luciferase or β-galactosidase (11). Cells were harvested 48 h after transfection and immediately processed or stored at −80°C. MSTO-211 cells endogenously express D2 activity (8) and were used in TEB4 knockdown experiments (see below). For all TEB4 RNAi studies of the HEK-293 cell line, cells were transfected with 0.3 μg of each RNAi vector or 0.6 μg of nonspecific RNAi vector.
Western blotting and IP.
All cell samples used for Western blotting were sonicated in 0.25 M sucrose-phosphate-EDTA buffer (pH 7.4), and gel loading was corrected based on β-galactosidase activity as described previously (9). Mouse anti-Flag M21 (1:5,000; Sigma, St. Louis, MO) was used as described previously (10) with SuperSignal West Dura extended duration substrate (Thermo Scientific, Rockford, IL) being used for detection. For immunoprecipitations (IPs), cells were transfected with 3 μg GFP-TEB4 and 2 μg Flag-D2, treated with 1 μM MG132 24 h prior to harvest in order to stabilize TEB4 and D2 proteins, and subsequently processed, as described previously, in a buffer containing 0.25 M Tris (pH 7.5), 0.5% Triton X-100, and 300 mM NaCl plus protease inhibitors (Roche, Indianapolis, IN) (9). Cell lysates were incubated with goat anti-GFP (1:300; Abcam, Cambridge, MA), and complexes were pulled down using protein G plus/protein A agarose beads (Oncogene Research Products, San Diego, CA). After precipitation, Flag detection was performed as described above with mouse anti-horseradish peroxidase (1:25,000; Roche, Indianapolis, IN). For ubiquitin-specific bead-based precipitation, HEK-293 cells transiently expressing Flag-D2 were transfected with 2 μg of nonsilencing RNAi vector or 0.4 μg of TEB4 RNAi vectors 1 and 2, as described above, and subsequently processed using the ubiquitinated protein enrichment kit (Calbiochem, CA) by following the manufacturer's recommendations. After precipitation, samples were Western blotted, and detection of Flag-D2 was performed as described above.
Deiodinase assays.
D1 and D2 assays were performed with cell sonicates as described previously (9) using 500 nM rT3 as the substrate for D1 and 0.1 nM T4 as the substrate for D2.
Lentivirus production and transduction of MSTO-211 cells.
Lentivirus generating an RNAi vector targeted against either TEB4 or a nonspecific sequence was produced using the Expression Arrest GIPZ lentiviral system (Open Biosystems, Huntsville, AL), and the V2LHS_12421 and V2LHS_199262 clones, respectively. Lentivirus was produced by following the manufacturer's recommendation after transfecting HEK-293T cells with 32.5 μg of the pGIPZ vector of interest along with 2.5 μg of pHDH-Hgpm2, 2.5 μg pHDM-tat1B, 2.5 μg pRC/CMV-rev1B, and 2.5 μg pHDM-G per 10-cm2 dish. Forty-eight hours after transfection, virus was harvested and stored at −70°C and then titrated in U2OS cells. MST0-211 cells were infected at a multiplicity of infection of 1 with 8 μg/ml of Polybrene (Invitrogen). Forty-eight hours after infection, cells were selected using puromycin at 0.5 μg/ml for 2 weeks prior to use in experiments. Photos of GFP-expressing cells were taken using an Olympus CXK-41 microscope, using QCapture Pro software (Media Cybernetics, Inc., Bethesda, MD).
Real-time qPCR.
Total RNA was extracted from HEK-293 or MSTO-211 cells, using Trizol, following the manufacturer's recommendations (Invitrogen). Real-time quantitative PCR (qPCR) was performed as described previously (7) using a five-point standard curve and the following primers: for TEB4, sense, 5′-TTGTCCTTCCAAGTCCGCCAG-3′, and antisense, 5′-GACTGTGGAGGTGGTGGAGATG-3′; for cyclophilin A, sense, 5′-GGCAAATGCTGGACCCAACAC-3′, and antisense, 5′-TGCCATTCCTGGACCCAAAGC-3′.
Semiquantitative reverse transcriptase PCR.
Total RNA was isolated from the indicated tissues from Wistar rats as described above. cDNA was generated using Superscript II (Invitrogen) with oligo(dT) priming and amplified in 35 cycles, using Taq with the following primers: for WSB-1, sense, 5′-CGAGGGTCAACGAGAAAGAGAT-3′, and antisense, 5′-GACGCAGTAGCTAGTAATGCT-3′; for TEB4, sense, 5′-GTGTCGGTCAGAAGGAACACCTGA-3′, and antisense, 5′-AGTGATCCATCAAGTCCAAGCAT-3′; for cyclophilin A, sense, 5′-TGACTTCACACGCCATAATG-3′, and antisense, 5′-CCACAATGCTCATGCCTTC-3′. Products were then resolved using agarose gel electrophoresis.
In situ probe generation.
An 819-bp-long rat TEB4 fragment was amplified by PCR from rat hypothalamic/cortical cDNA, using the following oligonucleotides: sense, 5′-GTGTCGGTCAGAAGGAACACCTGA-3′; antisense, 5′-AGTGATCCATCAAGTCCAAGCAT-3′. The resulting fragment was cloned into a pGemT vector (Promega, Madison WI) and confirmed by sequencing. The construct was linearized by SacII and transcribed using Sp6 polymerase in the presence of 35S-UTP (Perkin Elmer, Waltham, MA) as described previously to generate the antisense probe, while the sense probe was prepared using NotI and T7 polymerase (14).
Single-label in situ hybridization.
Three adult male Wistar rats were decapitated, and their brains were quickly removed, snap-frozen on dry ice, and stored at −80°C until use. Serial 12-μm-thick coronal sections were cut on a cryostat (Leica Microsystems GmbH, Wetzlar, Germany), mounted on Superfrost Plus slides (Fisher, Hampton, NH), dried at 42°C overnight, and then hybridized as previously described (13). Briefly, after prehybridization, the sections were hybridized under plastic coverslips in a buffer containing 50% formamide, 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10% dextran sulfate, 0.5% sodium dodecyl sulfate, 250 μg/ml denatured salmon sperm DNA, and 105 cpm of the probe of interest for 16 h at 56°C. Slides were then dehydrated in graded dilutions of ethanol and were dipped into Kodak NTB autoradiography emulsion (Eastman Kodak, Rochester, NY). Slides were developed after a 6-week exposure at 4°C. Specificity of hybridization was confirmed using sense probes that resulted in the complete absence of hybridization signal.
Double-label in situ hybridization for TEB4 and immunocytochemistry for GFAP.
Brain sections were prepared and hybridized for TEB4 as described above, and glial fibrillary acidic protein (GFAP) staining was performed as previously described (13). In brief, after posthybridization washes, sections were treated with a mixture of 0.5% Triton X-100 and 0.5% H2O2 for 15 min and then incubated with 1% bovine serum albumin in phosphate-buffered saline (PBS) for 20 min. Sections were then incubated with a mouse GA-5 monoclonal anti-GFAP (1:50; Roche Molecular Biochemicals GmbH, Vienna, Austria) dilution in 1% bovine serum albumin containing PBS overnight at 4°C. Slides were washed with PBS and then incubated in donkey anti-mouse immunoglobulin G (1:500; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for 2 h and ABC Elite (1:1,000; Vector Laboratories, Burlingame, CA) for 1 h. Immunoreactivity was detected with 0.025% 3,3′-diaminobenzidine containing 0.0036% H2O2 in 0.05 M Tris buffer (pH 7.6). After several washes in PBS, the sections were dehydrated, dipped in emulsion, and exposed as described above.
Statistical analysis.
Statistical significance was determined by using an unpaired Student t test when two groups were compared or one-way analysis of variance (ANOVA) with a Newman-Keuls post hoc test when more than two groups were compared, using Prism 3.0 (GraphPad Software, San Diego, CA). In all figures, data shown are means ± the standard error of the mean.
RESULTS
TEB4 interacts with D2 and mediates loss of D2 activity and protein.
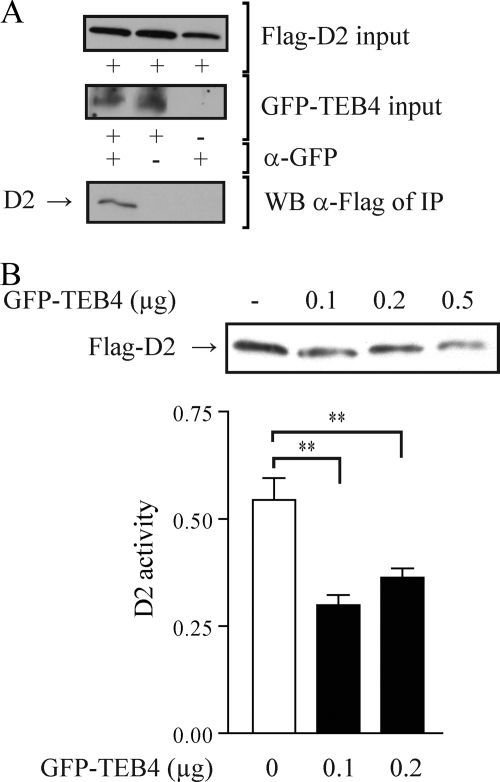
To assess the potential interaction between TEB4 and D2, we used a system in which HEK-293 cells transiently coexpress a GFP-tagged TEB4 (GFP-TEB4) and a Flag-tagged D2 (Flag-D2). In this heterologous system, D2 activity is equivalent to that found in cells that endogenously express this enzyme, and Flag-D2 has been previously shown to display ER distribution and other properties shared by endogenously expressed D2 (27). After harvesting, cells were processed for IP with anti-GFP to pull down TEB4. Subsequent Western blot analysis of the IP pellets with anti-Flag revealed Flag-D2, indicating that both proteins are associated together within the cell (Fig. 1A). Next, their functional relationship was tested by increasing the amount of coexpressed GFP-TEB4. This resulted in a loss of approximately 50% of Flag-D2 activity, and Western blot analysis indicated a progressive loss of Flag-D2 protein that reached 35% of control values (Fig. 1B).
FIG. 1.
Interrelationship between D2 and TEB4 expression. (A) Western blot analysis of Flag-D2 after coimmunoprecipitation with TEB4. HEK-293 cells transiently expressing GFP-TEB4 and Flag-D2, or Flag-D2 alone as a control, were treated with 1 μM MG132 for 24 h prior to harvesting. Cell lysates were then immunoprecipitated with anti-GFP as indicated. IP pellets were Western blotted using anti-Flag. Two percent of the input material Western blotted with either anti-Flag or anti-GFP is also shown. (B) Effects of TEB4 expression on Flag-D2 protein levels and D2 activity. D2 protein levels were assessed by Western blotting with anti-Flag in HEK-293 cells transiently expressing Flag-D2, by increasing amounts of GFP-TEB4 as indicated, and by a vector constitutively expressing β-galactosidase. Loading of cell lysates was normalized to β-galactosidase activity. For D2 enzyme activity, HEK-293 cells transiently expressing D2, increasing amounts of GFP-TEB4 as indicated, and a vector constitutively expressing Renilla luciferase were used, with enzyme activity being normalized by luciferase. Values are mean ± the standard error of the mean (SEM); n = 3; **, P < 0.001 by ANOVA; α, anti. The results shown are those from typical experiments that were repeated at least once.
TEB4 knockdown increases D2 activity and protein levels.
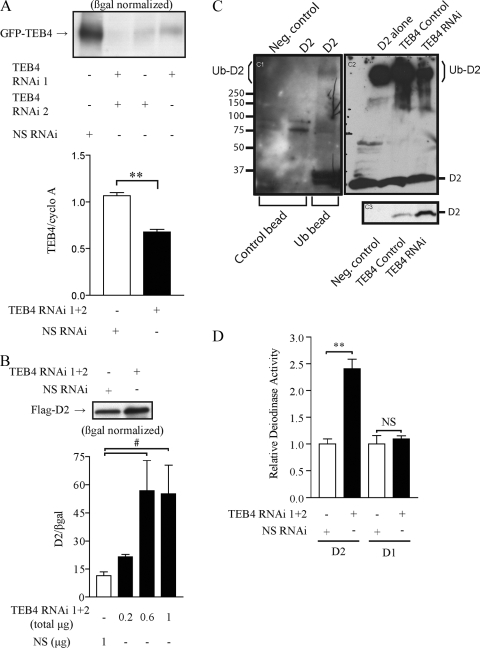
To further investigate the role of TEB4 in D2 expression, we generated two vectors that produce two distinct RNAi vectors targeted against TEB4 (TEB4 RNAi vector 1 and TEB4 RNAi vector 2). Cotransfection of GFP-TEB4 and either TEB4 RNAi vector 1 or 2 in HEK-293 cells markedly decreased GFP-TEB4 protein levels, while cotransfection of both RNAi vectors together decreased GFP-TEB4 protein by >90% (Fig. 2A). In subsequent experiments, real-time qPCR also indicated that endogenous TEB4 mRNA levels were decreased to 63% of control levels when both TEB4 RNAi 1 and 2 vectors where cotransfected (Fig. 2A). The less-efficient knockdown of TEB4 mRNA versus TEB4 protein presumably reflects the transfection efficiency in these cells, which is typically about 50%.
FIG. 2.
TEB4 knockdown and D2 expression. (A) Validation of RNAi constructs targeting TEB4 mRNA. HEK-293 cells transiently expressing GFP-TEB4 and either the TEB4 RNAi vector(s) or a nonspecific RNAi vector as indicated, along with a β-galactosidase expression vector, were treated with 1 μM MG132 for 24 h prior to harvesting to stabilize and increase TEB4 protein levels. GFP-TEB4 levels were detected by Western blotting with anti-GFP, with loading of cell lysates normalized to β-galactosidase levels. For real-time qPCR, endogenous levels of TEB4 in HEK-293 cells were measured in the presence of either both TEB4 RNAi vectors or a nonspecific RNAi vector as indicated. TEB4 mRNA levels are normalized by cyclophilin A (cyclo A) levels. (B) Effects of TEB4 knockdown in D2 protein levels and activity. HEK-293 cells transiently expressing Flag-D2 and the indicated RNAi vectors were Western blotted as described in the legend for Fig. 1B. For D2 enzyme activity, HEK-293 cells transiently expressing D2 and increasing equimolar amounts of TEB4 RNAi vectors or a nonspecific RNAi vector, as indicated, were used. Enzyme activity was normalized to β-galactosidase levels. (C) TEB4 knockdown affects D2 ubiquitination. Sonicates of HEK-293 cells transiently expressing Flag-D2 were processed using the ubiquitin enrichment kit, and the resulting pellet was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with anti-Flag (C1). Negative controls are indicated. Ub-D2 bands (>250 kDa) are indicated and present only in cells expressing D2. In a separate experiment (C2), high-molecular-mass Ub-D2 bands are visualized with decreased intensity during TEB4 knockdown. The effect of TEB4 knockdown on D2 protein levels is shown in panel C3. (D) TEB4 knockdown specifically affects D2. For enzyme activity, HEK-293 cells transiently expressing both D2 and D1 with or without nonspecific RNAi or TEB4 RNAi vectors, as indicated, and a β-galactosidase vector were assayed as described previously (9), and enzyme activity was normalized to β-galactosidase levels. Deiodinase activity is expressed relative to nonspecific RNAi levels. Values are mean ± SEM; n = 3. ** indicates P < 0.001, # indicates P < 0.05, and NS indicates nonsignificant by ANOVA. The results shown are those from typical experiments that were repeated at least once.
Cotransfection of D2 with either RNAi vector alone increased D2 activity by up to 50% in a dose-dependent manner (see Fig. S1A in the supplemental material), while TEB4 knockdown using both RNAi vectors together increased D2 activity by almost fivefold and doubled Flag-D2 protein levels (Fig. 2B). A combined IP-Western blot analysis using ubiquitin-binding beads and anti-Flag identified Ub-D2 as higher-molecular-mass bands, as previously reported (17). Notably, TEB4 knockdown substantially reduced the intensity of these Ub-D2 bands (Fig. 2C) while increasing Flag-D2 protein levels (Fig. 2C, bottom panel). Next, to test the specificity of the D2-TEB4 interaction, we knocked down TEB4 in cells transiently expressing both the type 1 deiodinase (D1), which is not subject to degradation by the ubiquitin-proteasomal system, and D2 (17). While D1 activity remained unaffected in these cells, D2 activity was increased 2.5-fold (Fig. 2C).
TEB4 knockdown interferes with D2 proteasomal degradation.
We next evaluated the role of TEB4 in the proteasomal degradation of D2. Previously, we have shown that D2 interaction with its natural substrate, T4, promotes conformational changes in the D2 molecule that favors D2 ubiquitination and proteasomal degradation, resulting in loss of D2 activity (27). Accordingly, treatment with either cycloheximide or T4 for 2 h decreased D2 activity by 25% (Fig. 3A). However, with TEB4 knockdown, D2 activity almost doubled and was not affected by either of these treatments (Fig. 3A). Additionally, we have shown that treatment with the proteasomal inhibitor MG132 blocks D2 degradation, leading to increased D2 activity (30). Thus, MG132 treatment alone increased D2 activity by about fivefold (Fig. 3B). At the same time, while TEB4 knockdown increased D2 activity by about threefold, a combination of both MG132 and TEB4 knockdown elevated D2 activity only to the same level as that seen with MG132 treatment alone (Fig. 3B).
FIG. 3.
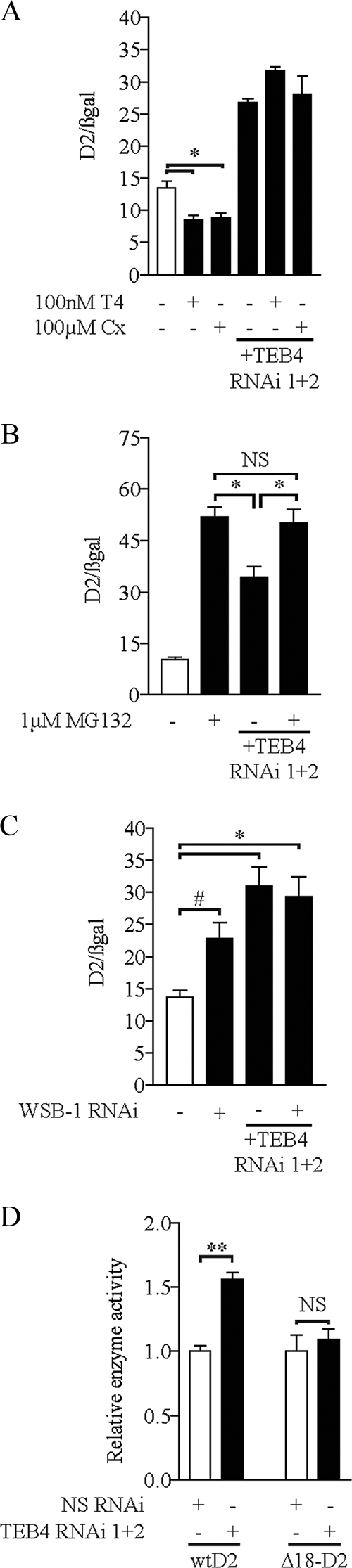
TEB4 knockdown interferes with D2 proteasomal degradation. (A) Loss of substrate-induced D2 degradation with TEB knockdown. HEK-293 cells transiently expressing D2 with or without TEB4 RNAi vectors and a β-galactosidase vector were treated with 100 nM T4 or 100 μM cycloheximide for 2 h prior to harvest, as indicated. Deiodinase activity was measured as described in the legend for Fig. 2B, with activity levels normalized to β-galactosidase levels. (B) HEK-293 cells transiently expressing D2 with or without TEB4 RNAi vectors and a β-galactosidase vector were treated with 1 μM MG132 24 h prior to harvest, as indicated, and deiodinase activity was measured as described in the legend for Fig. 2B, with activity levels normalized to β-galactosidase levels. (C) D2 activity in the absence of TEB4 and/or WSB-1. HEK-293 cells transiently expressing D2 with or without TEB4/WSB-1 RNAi vectors as indicated were assayed for D2 activity as described in the legend for Fig. 2B. Enzyme activity levels were normalized to β-galactosidase levels. (D) TEB4 knockdown does not affect a mutant D2 lacking the instability loop. HEK-293 cells transiently expressing either D2 or Δ18-D2 with or without TEB4 RNAi vectors, as indicated, were assayed for D2 activity as described in the legend for Fig. 2B. Enzyme activity was normalized to nonspecific RNAi levels. Values are mean ± SEM; n = 3. * indicates P < 0.01, # indicates P < 0.05, and NS indicates nonsignificant by ANOVA (A to C) or unpaired Student's t test (D). The results shown are those from typical experiments that were repeated at least once.
It has been previously established that WSB-1 is an E3 ligase adaptor that is involved in both the ubiquitination and subsequent proteasomal degradation of D2 (11). When WSB-1 is knocked down, D2 activity is increased 1.5-fold, while with TEB4 knockdown, D2 activity is increased twofold (Fig. 3C). Interestingly, knockdown of both TEB4 and WSB-1 elevates D2 to levels that are comparable to those seen with TEB4 knockdown alone (Fig. 3C). A destabilization loop of 18 amino acids, from position 92 to 109 in D2, is necessary for WSB-1-mediated ubiquitination of D2 (11, 38). To test if this loop is involved in TEB4-mediated D2 ubiquitination, we transiently expressed the Δ18-D2 mutant, in which the destabilization loop had been deleted, in HEK-293 cells with TEB4 knockdown. While D2 activity increased about 1.5-fold, the D2 mutant (Δ18-D2) remained unaffected (Fig. 3D).
TEB4 knockdown increases endogenous D2 activity.
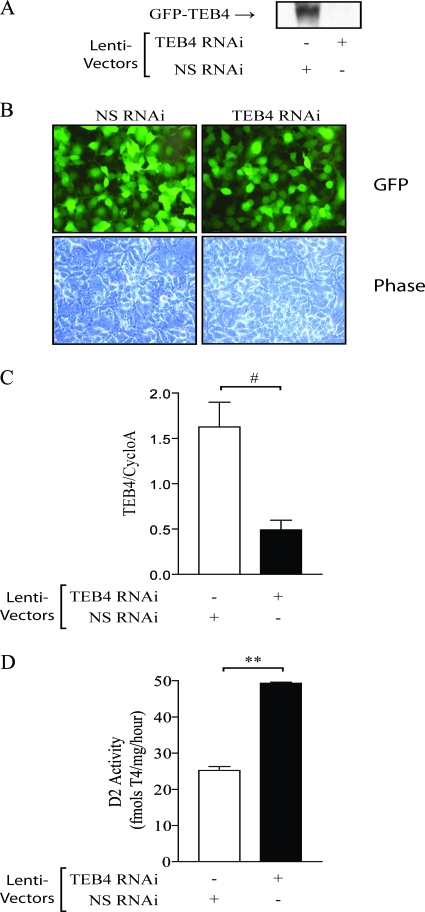
The effects of TEB4 knockdown were also assessed in the mesothelioma cell line MSTO-211 that endogenously expresses D2 (8). Here, we used a lentivirus-mediated transduction strategy to knock down TEB4 expression. Initially, a vector that encodes sequences to generate an RNAi targeting either TEB4 or a nonspecific control sequence, along with genes conferring puromycin resistance and a GFP marker, flanked by lentiviral long-terminal-repeat sequences was obtained from Open Biosystems. To validate the efficacy of the RNAi targeted against TEB4, HEK-293 cells transiently expressing GFP-TEB4 were also transfected with either the vector producing an RNAi against TEB4 or a nonspecific control sequence, as shown in Fig. 2A. Western blotting indicated that GFP-TEB4 expression was reduced to undetectable levels compared to cells transfected with the nonspecific control, confirming the effectiveness of this vector (Fig. 4A). We first transfected MSTO-211 cells with the RNAi-generating vector, and because the transfection efficiency is low, GFP-positive cells were sorted by flow cytometry, and D2 activity was measured. Under these conditions, TEB4 knockdown increased endogenous D2 activity by about threefold (see Fig. S1B in the supplemental material). Second, lentivirus was produced and used to infect MSTO-211 cells, which were subsequently selected with puromycin. After 2 weeks of treatment, it appeared that a majority of the MSTO-211 cells were expressing the GFP marker and consequently also the RNAi of interest (Fig. 4B). Real-time qPCR confirmed that TEB4 mRNA levels were decreased by 75% in the MSTO-211 cells infected with the TEB4 RNAi-producing virus (Fig. 4C). Remarkably, D2 activity was doubled in the same MSTO-211 cells (Fig. 4D).
FIG. 4.
TEB4 knockdown increases endogenous D2 activity. (A) HEK-293 cells transiently expressing GFP-TEB4 and either the lentiviral TEB4 RNAi vector or the lentiviral nonspecific RNAi vector (lenti-vectors), as indicated, were Western blotted for GFP-TEB4 levels, as described in the legend for Fig. 2A. (B) Photos of MSTO-211 cells transduced with either the TEB4 or nonspecific RNAi encoding lentivirus. GFP marker expression in the lentivirus-infected MSTO-211 cells with either the nonspecific lentiviral RNAi vector (left panels) or TEB4 RNAi vector (right panels) after 2 weeks of puromycin selection is shown. Phase contrast images are shown in the bottom row to illustrate the total number of cells. Images were taken with ×400 amplification. (C) Decreased endogenous TEB4 mRNA levels in MSTO-211 cells. TEB4 mRNA levels of the cells shown in panel B were determined by real-time qPCR, as described in the legend for Fig. 2A, and normalized to cyclophilin A levels. (D) D2 activity in MSTO-211 cells ± lentiviral TEB4 knock down was measured as described in reference 9. Values are mean ± SEM; n = 3. ** indicates P < 0.001, and # indicates P < 0.05 by ANOVA. The results shown are those from typical experiments that were repeated at least once.
WSB-1 and TEB4 have distinct patterns of tissue expression.
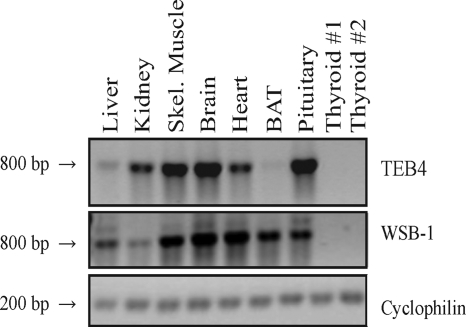
In order to establish the pattern of tissue expression of TEB4 and WSB-1, RNA was extracted from a variety of rat tissues, and the relative amounts of TEB4 and WSB-1 were determined using semiquantitative reverse transcriptase PCR (Fig. 5). Most of the tissues examined expressed TEB4, whereas liver had substantially lower levels of TEB4 mRNA, and the D2-expressing brown adipose tissue (BAT) had almost undetectable levels of TEB4 mRNA. WSB-1 mRNA is also well expressed in most tissues, with somewhat smaller amounts being found in kidney. Unlike TEB4, WSB-1 is well expressed in BAT, and neither WSB-1 nor TEB4 is expressed in the rat thyroid.
FIG. 5.
TEB4 and WSB-1 have different tissue expression profiles. WSB-1 and TEB4 tissue expression in rats. RNA was extracted from the indicated rat tissues and fragments corresponding to either WSB1 or TEB4 were amplified using semiquantitative reverse transcriptase PCR. Cyclophilin A expression was used to control for differences in starting material. Negative controls with no cDNA synthesis had no bands (data not shown).
Expression of both WSB-1 and TEB4 was found in the rat brain, not only in neurons but also in tanycytes and astrocytes, the two cell types that predominantly express D2 in the brain (13). Thus, we used in situ hybridization to study TEB4 expression in the rat brain and found its mRNA to be expressed in most brain regions, including the hypothalamus, hippocampus, cerebellum, and cortex (Fig. 6). Intense TEB4 signal was observed in the floor and lateral wall of the third ventricle, between the rostral pole of the median eminence and the mammillary recess, and in the ependymal cells lining the wall of the third ventricle, which is reminiscent of that of third ventricular tanycytes (Fig. 6A). In control slides prepared in parallel, hybridization with the sense probe resulted in the complete absence of signal (data not shown). Using GFAP staining as a marker for astrocytes, TEB4 and GFAP signal overlapped only rarely in the cortex and hippocampus, suggesting that in these regions, TEB4 is primarily expressed by neurons. In the stratum granulosum of the cerebellum, TEB4 was found in both neurons and GFAP-positive astrocytes (Fig. 7). In contrast to the rather ubiquitous WSB-1 expression in astrocytes (13), TEB4 expression in astrocytes exhibited a region-specific pattern, with only a few scattered GFAP-positive astrocytes containing TEB4 mRNA in the cortex and hippocampus, which is in contrast to the pattern found in the cerebellum (Fig. 7).
FIG. 6.
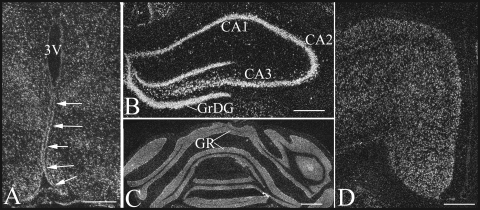
TEB4 mRNA distribution in the brain. Dark-field images of TEB4 mRNA distribution in the ependymal cells lining the wall of the third ventricle (3V) in the mediobasal hypothalamus (A), hippocampus (B), cerebellum (C), and cerebral cortex (D) of rats. Arrows indicate TEB4 hybridization signals detected over ependymal cells lining the floor and wall of the third ventricle (A). In the hippocampus (B), a very strong hybridization signal was present over the pyramidal layer (CA1 to CA3) and the granular layer of the dentate gyrus (GrDG). A very intense hybridization signal was also observed over the granular layer of the cerebellum (GR) (C). TEB4 hybridization signal was detected in the second to sixth layers of the cerebral cortex (D). Scale bars for panels A, B, and D represent 500 μm, and the scale bar for panel C represents 1,000 μm.
FIG. 7.
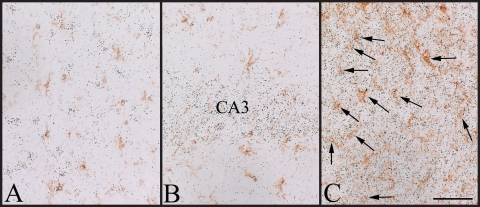
Cell type specificity of TEB4 expression in the rat brain. (A) In the cortex, the TEB4 signal is absent in GFAP-positive astrocytes (brown) but is expressed in many GFAP-negative cells. (B) A strong TEB4 hybridization signal is observed in the GFAP-negative cells of the CA3 region of the hippocampus but is absent in the GFAP-positive astrocytes. (C) A strong hybridization signal is present over both GFAP-positive (indicated by arrows) and -negative cells in the granular layer of the cerebellum. Scale bar, 50 μm.
Given this diverse expression pattern of TEB4 in the rat tissues, we next studied the relative expression of WSB-1 and TEB4 in human tissues in silico, using data from the SymAtlas website (http://symatlas.gnf.org/SymAtlas/) (32). It is clear that TEB4 expression predominates over WSB-1 expression in the immune system (cells and tissues), whereas WSB-1 expression predominates in the central nervous system, with the exception of pons, globus pallidus, amygdala, and cerebellum (see Fig. S2 in the supplemental material). In the other tissues, the relative expression of WSB-1 and TEB4 is variable, without clear tissue-specific predominance.
DISCUSSION
The present investigation provides evidence that D2 and the ER resident E3 ligase TEB4 interact within the cell. First, these two proteins were coimmunoprecipitated (Fig. 1A), suggesting that they either share common interacting surfaces or are part of a larger multiprotein complex. Second, their functional relationship is evident by the fact that increased TEB4 expression reduces D2 activity and protein levels in a dose-dependent manner (Fig. 1B), while TEB4 knockdown increases D2 activity and protein levels by more than 50% (Fig. 2B), while reducing Ub-D2 levels (Fig. 2C). These effects are also observed in cells endogenously expressing D2 (Fig. 4D). Third, the T4-induced D2 ubiquitination and subsequent loss of D2 activity are greatly reduced in cells with TEB4 knockdown, illustrating that this E3 ligase mediates substrate-induced D2 proteolysis (Fig. 3A). These effects seem to be highly D2 specific, given that D1, the other thyroid hormone-activating deiodinase (17), is not affected by TEB4 knockdown, even when coexpressed with D2 in the same cell (Fig. 2D). Taken together, these results indicate that TEB4 mediates the proteasomal degradation of D2 in cells that coexpress both proteins at endogenous levels.
In view of the well-established role played by WSB-1 as an E3 ligase adaptor for D2 (11), the current findings that TEB4 can also mediate D2 degradation illustrates that a critical step in thyroid hormone activation can be regulated by at least two ubiquitin ligases. A similar situation is found with Eps15, a protein that regulates EGF receptor internalization, the ubiquitination of which is mediated by the HECT domain E3 ligase Nedd4 and parkin, a RING finger-containing E3 ligase (12, 37). While we currently do not understand if or how these two pathways interact to regulate D2, it is puzzling that no additional protective effect on D2 activity was noted when both WSB-1 and TEB4 were knocked down at the same time (Fig. 3C). This suggests that both pathways are not necessarily exclusive and in fact could even contribute to D2 ubiquitination in a sequential fashion. While both WSB-1 and TEB4 function in association with UBC7 (11, 19), these ligases differ in their structures and substrate-processing mechanisms. WSB-1 is a small sonic hedgehog-inducible WD40-SOCS box-containing protein that is part of the ECSWSB-1 complex and remains associated with the ER membrane (11). Ubiquitination of D2 via this pathway results in an inactive D2 that remains fully assembled in the ER membrane and can be easily reactivated by USP33/20 deubiquitinases (10, 27). On the other hand, TEB4 is a large CH4C3 RING finger-containing protein that spans the ER membrane multiple times (19, 23) and is thought to associate with Sec61α and -β translocon units and mediate retrotranslocation of multiple ubiquitinated ER proteins to the proteasome complex (19). Thus, given the decrease in Ub-D2 and accumulation of D2 protein during TEB4 knockdown (Fig. 2C), it is likely that ubiquitination of D2 through this pathway is followed by terminal protein disassembly.
Interaction of D2 with its natural substrate, T4, is thought to promote conformational changes that expose critical lysine residues (K237 and K244) in D2 that then can undergo ubiquitination (27). This is lost in Δ18-D2, a mutant D2 lacking an 18-amino-acid loop required for interaction with WSB-1 (11). A similar mechanism is likely to take place with TEB4, given that substrate-induced loss in D2 activity and processing of the Δ18-D2 molecule are both greatly minimized in cells with TEB4 knockdown (Fig. 3A and D). It is remarkable that despite two very different pathways for D2 ubiquitination, the intrinsic mechanisms regulating its conjugation to ubiquitin still seem to be the same in both cases and highly dependent on this instability loop.
The existence of two ubiquitin ligases, WSB-1 and TEB4, to process D2 would gain additional relevance if these proteins had different expression patterns and/or regulation. However, based on data publically available (http://symatlas.gnf.org/SymAtlas/; see also Fig. S2A in the supplemental material) and our own reverse transcriptase PCR studies (Fig. 5), it is clear that both WSB-1 and TEB4 are widely expressed in human and rodent tissues. At the same time, quantitative analyses reveal that TEB4 predominates in cells of the immune system, testes, and some splanchnic tissues, whereas WSB-1 predominates in the brain, ovary, and uterus (see Fig. S2B in the supplemental material). Of particular interest is the isolated expression of WSB-1 in BAT, which normally expresses high levels of D2 (Fig. 5). In the brain, in situ hybridization studies revealed that TEB4, similar to WSB-1, is expressed in tanycytes, while astrocytes in the hippocampus and cortex express WSB1 but not TEB4 (Fig. 6 and 7) (13).
In conclusion, our studies have linked TEB4 to D2 ubiquitination and proteasomal degradation, suggesting that it may be a key regulator of thyroid hormone activation in a number of tissues, including those of the brain. Given the recognized importance of thyroid hormone during brain development, the presence of TEB4 in a chromosomal locus linked to the Cri-du-chat syndrome (19) raises the possibility that patients with this genetic disorder might have altered D2 regulation and thyroid hormone signaling during critical periods of brain development.
Supplementary Material
Acknowledgments
We thank Mark Hochstrasser for providing the GFP-TEB4 vector.
This work was supported by NIDDK grants DK58538 and DK076117, Hungarian Scientific Research Fund Grant OTKA T049081, the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, and the 6th EU Research Framework Program grant LSHM-CT-2003-503041.
Footnotes
Published ahead of print on 3 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baqui, M. M., B. Gereben, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2000. Distinct subcellular localization of transiently expressed types 1 and 2 iodothyronine deiodinases as determined by immunofluorescence confocal microscopy. Endocrinology 141:4309-4312. [DOI] [PubMed] [Google Scholar]
- 2.Bianco, A. C., D. Salvatore, B. Gereben, M. J. Berry, and P. R. Larsen. 2002. Biochemistry, cellular and molecular biology and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev. 23:38-89. [DOI] [PubMed] [Google Scholar]
- 3.Bianco, A. C., and J. E. Silva. 1987. Intracellular conversion of thyroxine to triiodothyronine is required for the optimal thermogenic function of brown adipose tissue. J. Clin. Investig. 79:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botero, D., B. Gereben, C. Goncalves, L. A. de Jesus, J. W. Harney, and A. C. Bianco. 2002. Ubc6p and Ubc7p are required for normal and substrate-induced endoplasmic reticulum-associated degradation of the human selenoprotein type 2 iodothyronine monodeiodinase. Mol. Endocrinol. 16:1999-2007. [DOI] [PubMed] [Google Scholar]
- 5.Carvalho, P., V. Goder, and T. A. Rapoport. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126:361-373. [DOI] [PubMed]
- 6.Choi, D. W., Y. M. Seo, E. A. Kim, K. S. Sung, J. W. Ahn, S. J. Park, S. R. Lee, and C. Y. Choi. 2008. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J. Biol. Chem. 283:4682-4689. [DOI] [PubMed] [Google Scholar]
- 7.Christoffolete, M. A., C. C. Linardi, L. de Jesus, K. N. Ebina, S. D. Carvalho, M. O. Ribeiro, R. Rabelo, C. Curcio, L. Martins, E. T. Kimura, and A. C. Bianco. 2004. Mice with targeted disruption of the Dio2 gene have cold-induced overexpression of the uncoupling protein 1 gene but fail to increase brown adipose tissue lipogenesis and adaptive thermogenesis. Diabetes 53:577-584. [DOI] [PubMed] [Google Scholar]
- 8.Curcio, C., M. M. Baqui, D. Salvatore, B. H. Rihn, S. Mohr, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2001. The human type 2 iodothyronine deiodinase is a selenoprotein highly expressed in a mesothelioma cell line. J. Biol. Chem. 276:30183-30187. [DOI] [PubMed] [Google Scholar]
- 9.Curcio-Morelli, C., B. Gereben, A. M. Zavacki, B. W. Kim, S. Huang, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2003. In vivo dimerization of types 1, 2, and 3 iodothyronine selenodeiodinases. Endocrinology 144:3438-3443. [DOI] [PubMed] [Google Scholar]
- 10.Curcio-Morelli, C., A. M. Zavacki, M. Christofollete, B. Gereben, B. C. de Freitas, J. W. Harney, Z. Li, G. Wu, and A. C. Bianco. 2003. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel-Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J. Clin. Investig. 112:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dentice, M., A. Bandyopadhyay, B. Gereben, I. Callebaut, M. A. Christoffolete, B. W. Kim, S. Nissim, J. P. Mornon, A. M. Zavacki, A. Zeold, L. P. Capelo, C. Curcio-Morelli, R. Ribeiro, J. W. Harney, C. J. Tabin, and A. C. Bianco. 2005. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat. Cell Biol. 7:698-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fallon, L., C. M. Belanger, A. T. Corera, M. Kontogiannea, E. Regan-Klapisz, F. Moreau, J. Voortman, M. Haber, G. Rouleau, T. Thorarinsdottir, A. Brice, P. M. van Bergen En Henegouwen, and E. A. Fon. 2006. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 8:834-842. [DOI] [PubMed] [Google Scholar]
- 13.Fekete, C., B. C. Freitas, A. Zeold, G. Wittmann, A. Kadar, Z. Liposits, M. A. Christoffolete, P. Singru, R. M. Lechan, A. C. Bianco, and B. Gereben. 2007. Expression patterns of WSB-1 and USP-33 underlie cell-specific posttranslational control of type 2 deiodinase in the rat brain. Endocrinology 148:4865-4874. [DOI] [PubMed] [Google Scholar]
- 14.Fekete, C., B. Gereben, M. Doleschall, J. W. Harney, J. M. Dora, A. C. Bianco, S. Sarkar, Z. Liposits, W. Rand, C. Emerson, I. Kacskovics, P. R. Larsen, and R. M. Lechan. 2004. Lipopolysaccharide induces type 2 iodothyronine deiodinase in the mediobasal hypothalamus: implications for the nonthyroidal illness syndrome. Endocrinology 145:1649-1655. [DOI] [PubMed] [Google Scholar]
- 15.Flamant, F., K. Gauthier, and J. Samarut. 2007. Thyroid hormones signaling is getting more complex: STORMs are coming. Mol. Endocrinol. 21:321-333. [DOI] [PubMed] [Google Scholar]
- 16.Galton, V. A., E. T. Wood, E. A. St Germain, C. A. Withrow, G. Aldrich, G. M. St Germain, A. S. Clark, and D. L. St Germain. 2007. Thyroid hormone homeostasis and action in the type 2 deiodinase-deficient rodent brain during development. Endocrinology 148:3080-3088. [DOI] [PubMed] [Google Scholar]
- 17.Gereben, B., C. Goncalves, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2000. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol. Endocrinol. 14:1697-1708. [DOI] [PubMed] [Google Scholar]
- 18.Gereben, B., A. Zavacki, S. Ribich, B. Kim, S. Huang, W. Simonides, A. Zeöld, and A. Bianco. 2008. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr. Rev. 29:898-938. [DOI] [PMC free article] [PubMed]
- 19.Hassink, G., M. Kikkert, S. van Voorden, S. J. Lee, R. Spaapen, T. van Laar, C. S. Coleman, E. Bartee, K. Fruh, V. Chau, and E. Wiertz. 2005. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 388:647-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huyer, G., W. F. Piluek, Z. Fansler, S. G. Kreft, M. Hochstrasser, J. L. Brodsky, and S. Michaelis. 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279:38369-38378. [DOI] [PubMed] [Google Scholar]
- 21.Kalaany, N. Y., K. C. Gauthier, A. M. Zavacki, P. P. Mammen, T. Kitazume, J. A. Peterson, J. D. Horton, D. J. Garry, A. C. Bianco, and D. J. Mangelsdorf. 2005. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 1:231-244. [DOI] [PubMed] [Google Scholar]
- 22.Kim, B. W., A. M. Zavacki, C. Curcio-Morelli, M. Dentice, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2003. Endoplasmic reticulum-associated degradation of the human type 2 iodothyronine deiodinase (D2) is mediated via an association between mammalian UBC7 and the carboxyl region of D2. Mol. Endocrinol. 17:2603-2612. [DOI] [PubMed] [Google Scholar]
- 23.Kreft, S. G., L. Wang, and M. Hochstrasser. 2006. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J. Biol. Chem. 281:4646-4653. [DOI] [PubMed] [Google Scholar]
- 24.Larsen, P. R., J. E. Silva, and M. M. Kaplan. 1981. Relationships between circulating and intracellular thyroid hormones: physiological and clinical implications. Endocr. Rev. 2:87-102. [DOI] [PubMed] [Google Scholar]
- 25.Leonard, J. L., C. A. Siegrist-Kaiser, and C. J. Zuckerman. 1990. Regulation of type II iodothyronine 5′-deiodinase by thyroid hormone. Inhibition of actin polymerization blocks enzyme inactivation in cAMP-stimulated glial cells. J. Biol. Chem. 265:940-946. [PubMed] [Google Scholar]
- 26.Ravid, T., S. G. Kreft, and M. Hochstrasser. 2006. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25:533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sagar, G. D., B. Gereben, I. Callebaut, J. P. Mornon, A. Zeold, W. S. da Silva, C. Luongo, M. Dentice, S. M. Tente, B. C. Freitas, J. W. Harney, A. M. Zavacki, and A. C. Bianco. 2007. Ubiquitination-induced conformational change within the deiodinase dimer is a switch regulating enzyme activity. Mol. Cell. Biol. 27:4774-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheuner, D., and R. J. Kaufman. 2008. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr. Rev. 29:317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schimmel, M., and R. D. Utiger. 1977. Thyroidal and peripheral production of thyroid hormones. Review of recent findings and their clinical implications. Ann. Intern. Med. 87:760-768. [DOI] [PubMed] [Google Scholar]
- 30.Steinsapir, J., A. C. Bianco, C. Buettner, J. Harney, and P. R. Larsen. 2000. Substrate-induced down-regulation of human type 2 deiodinase (hD2) is mediated through proteasomal degradation and requires interaction with the enzyme's active center. Endocrinology 141:1127-1135. [DOI] [PubMed] [Google Scholar]
- 31.St Germain, D. L. 1988. The effects and interactions of substrates, inhibitors, and the cellular thiol-disulfide balance on the regulation of type II iodothyronine 5′-deiodinase. Endocrinology 122:1860-1868. [DOI] [PubMed] [Google Scholar]
- 32.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter, J., J. Urban, C. Volkwein, and T. Sommer. 2001. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20:3124-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121-127. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, M., S. M. Houten, C. Mataki, M. A. Christoffolete, B. W. Kim, H. Sato, N. Messaddeq, J. W. Harney, O. Ezaki, T. Kodama, K. Schoonjans, A. C. Bianco, and J. Auwerx. 2006. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439:484-489. [DOI] [PubMed] [Google Scholar]
- 37.Woelk, T., B. Oldrini, E. Maspero, S. Confalonieri, E. Cavallaro, P. P. Di Fiore, and S. Polo. 2006. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 8:1246-1254. [DOI] [PubMed] [Google Scholar]
- 38.Zeöld, A., L. Pormuller, M. Dentice, J. W. Harney, C. Curcio-Morelli, S. M. Tente, A. C. Bianco, and B. Gereben. 2006. Metabolic instability of type 2 deiodinase is transferable to stable proteins independently of subcellular localization. J. Biol. Chem. 281:31538-31543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.