Abstract
Human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein modifications over the course of infection have been associated with coreceptor switching and antibody neutralization resistance, but the effect of the changes on replication and host cell receptor usage remains unclear. To examine this question, unique early- and chronic-stage infection envelope V1-toV5 (V1-V5) segments from eight HIV-1 subtype A-infected subjects were incorporated into an isogenic background to construct replication-competent recombinant viruses. In all subjects, viruses with chronic-infection V1-V5 segments showed greater replication capacity than those with early-infection V1-V5 domains in cell lines with high levels of both the CD4 and the CCR5 receptors. Viruses with chronic-infection V1-V5s demonstrated a significantly increased ability to replicate in cells with low CCR5 receptor levels and greater resistance to CCR5 receptor and fusion inhibitors compared to those with early-infection V1-V5 segments. These properties were associated with sequence changes in the envelope V1-V3 segments. Viruses with the envelope segments from the two infection time points showed no significant difference in their ability to infect cells with low CD4 receptor densities, in their sensitivity to soluble CD4, or in their replication capacity in monocyte-derived macrophages. Our results suggest that envelope changes, primarily in the V1-V3 domains, increase both the ability to use the CCR5 receptor and fusion kinetics. Thus, envelope modifications over time within a host potentially enhance replication capacity.
The human immunodeficiency virus type 1 (HIV-1) viral envelope glycoprotein evolves over the course of infection (24, 78), with the portion from constant region 2 to variable loop 5 (C2-V5) diversifying at an approximate rate of 1% per year in the absence of antiretroviral medications (77). The envelope glycoprotein variable loops 1 and 2 (V1-V2) expand and add more glycosylation sites over the course of infection (21, 76). These envelope changes arise primarily due to errors during reverse transcription, the high rate of viral replication, and recombination (25, 42, 58, 86). The rate of mutation fixation in a virus population, however, depends on both the level of viral replication and, more importantly, the selective advantage or disadvantage conferred by the mutation. The host immune response and the replication capacity in the available target cells primarily drive this selection (12, 51). Envelope modifications that confer an advantage in evading the host humoral immune response and/or increase the efficiency of target cell infection and replication are likely favored over the course of an infection within a subject.
Studies with the simian immunodeficiency virus/macaque model, simian human immunodeficiency virus, and HIV-1 have shown that envelope modifications that occur over the course of an infection confer antibody neutralization resistance (8, 9, 72, 85). The host neutralizing antibodies target specific epitopes on the circulating viral envelope glycoproteins, but viruses evolve to escape these responses (70, 85). We have previously shown that in HIV-1 subtype A-infected individuals, changes in the envelope glycoprotein V1-V2 loops account for some of the observed neutralization resistance to autologous plasma (76). Besides influencing the sensitivity to the host neutralizing-antibody response, envelope modifications that occur over the course of infection also potentially affect host cell receptor interactions and replicative capacity in different target cells.
HIV uses the CD4 receptor along with a coreceptor, such as CCR5 and/or CXCR4, for host cell attachment and fusion (11). Early in infection, most HIVs use the CCR5 coreceptor, and over time, HIVs often acquire an ability to use a wider variety of coreceptors, such as CXCR4 (7). In subtype B HIV-1, coreceptor usage has been mapped primarily to the envelope V3 loop (15, 19, 20, 28), while in non-subtype B viruses, other portions of the envelope sequence such as the V1-V2 loops may influence coreceptor usage (26). Because coreceptor switching occurs less frequently in subtype A viruses (26, 29), envelope glycoprotein modifications that occur over the course of infection may affect cell entry efficiency primarily by altering CD4 or the CCR5 receptor utilization. We have previously shown that evolution in the envelope glycoprotein V1-V2 loops can have modest effects on cell entry efficiency in cells with limiting levels of receptors (76). Modifications in the V1-V2 loops and other envelope segments are constrained because antibody neutralization resistance needs to be attained while preserving the ability to bind host receptors and enter cells. The effects of envelope sequence changes that occur over the course of infection on host cell receptor interactions and replicative capacity remain largely undefined, especially for non-subtype B HIV-1.
In the present study, we incorporated previously isolated unique early- and chronic-infection V1-V5 envelope segments into a parental virus. As opposed to the majority of studies which examine HIV-1 envelope glycoprotein phenotypes using viral pseudotypes, we constructed replication-competent recombinant viruses. We found that engineered viruses with V1-V5 segments from the chronic phase of infection had significantly increased replication capacity compared to HIVs with V1-V5 portions from early infection. In addition, variants with chronic-infection V1-V5s also had significantly greater replication capacity in cells with low CCR5 densities and lower sensitivity both to the CCR5 antagonists TAK779 and PSC-RANTES and to the fusion inhibitor T-20, compared to HIVs with V1-V5 portions from early in infection. These properties were associated primarily with sequence changes within the V1-V3 envelope domains. These results in conjunction with our previous studies suggest that envelope sequence changes within the envelope V1-V3 domain may lead to antibody neutralization resistance and a concomitant increase in the ability to use the CCR5 receptor and faster fusion capacity.
MATERIALS AND METHODS
Subjects and envelope sequences.
A first-round PCR product encompassing V1-V5 sequences had been obtained previously from peripheral blood mononuclear cell samples from nine antiretroviral-naïve subtype A-infected women who acquired HIV-1 through heterosexual contact (Table 1) (76). Each sequence was derived from an independent PCR starting with one template copy to minimize resampling bias (74). Previously, we isolated V1-V3 sequences from this first-round product using a second nested PCR. In this study, we used the same first-round amplified product to amplify V1-V5 sequences using primers Env10S (5′-CACTTCTCCAATTGTCCCTCAT-3′; corresponding to nucleotide positions 7647 to 7668 in the HXB2 genome) and Env15 (5′-CCATGTGTAAAGTTAACCCC-3′; HXB2 nucleotide positions 6576 to 6595). Amplified products were cleaned using Qiaquick (Qiagen) and directly sequenced.
TABLE 1.
Subjects, sequences, and clones
| Subject | Early infection
|
Chronic infection
|
||||||
|---|---|---|---|---|---|---|---|---|
| Infection intervala (mo) | Plasma virus (copies/ml) | No. of sequencesb | No. of clonesc | Infection interval (mo) | Plasma virus (copies/ml) | No. of sequences | No. of clones | |
| QA203 | 1 | 39,190 | 3 | 1 | 41 | 45,390 | 4 | 3 |
| QA284 | 1 | 374,160 | 1 | 1 | 47 | 14,080 | 4 | 4 |
| QA779 | 2 | 66,530 | 1 | 1 | 37 | 160,970 | 7 | 6 |
| QB424 | 1 | 330,710 | 3 | 1 | 31 | 352,740 | 5 | 1 |
| QB596 | 6 | 56,050 | 4 | 1 | 24 | 13,800 | 8 | 5 |
| QB670 | 1 | 141,370 | 1 | 1 | 46 | 27,770 | 9 | 0 |
| QC168 | 1 | 993,900 | 1 | 1 | 32 | 1,611,600 | 4 | 3 |
| QC449 | 5 | 782,600 | 1 | 1 | 41 | 185,060 | 2 | 2 |
| QC890 | 2 | 1,044 | 3 | 1 | 42 | 733,080 | 5 | 4 |
Duration from the estimated date of infection to the day of sample collection.
Number of independent sequences analyzed.
Number of replication-competent recombinant viruses with unique V1-V5 segments.
Construction of recombinant viral plasmids.
We modified a previously described yeast gap repair homologous recombination method (43) to incorporate unique V1-V5 PCR-amplified envelope fragments into a full-length subtype A HIV-1 clone (Q23-17) (63) (Fig. 1). To incorporate the Q23 HIV-1 sequences into a Saccharomyces cerevisiae and Escherichia coli shuttle vector, NotI and XhoI digestion was used to isolate the full-length Q23 sequence, and this fragment was cloned into the multiple-cloning site of pRS315 (New England Biolabs). Within Q23, nef sequences were replaced by the HIS3 gene, which allows yeast to synthesize histidine. The selection marker, HIS3, was amplified from pRS313 (New England Biolabs) using primers 5′-AAGTGGTCAAAAAGTAGCATAGTTCTGGTGGCAATGATTGAAATAAATTCCCTTTAAGAGC-3′ and 5′-GTTTATCTAAGTCTTGAGATACTGCTTTAAATAATCGTGTCAC-3′. The underlined portion of each primer denotes the segments homologous to Q23 nef sequences. Yeast (strain S288C) was transformed with the HIS3 gene PCR fragment and BmgBI-linearized pRS315-Q23 plasmid using the lithium acetate (LiAc) technique (13). (BmgBI cuts within the Q23 nef sequence.) Transformed yeast cells were selected and expanded on complete minimal medium (CMM) without leucine or histidine. Yeast plasmids were extracted using glass beads, and a portion of the crude extract was electroporated into E. coli. Transformed E. coli were confirmed to have the plasmid pRS315-Q23Δnef-HIS3 by restriction enzyme digestions. Within pRS315-Q23Δnef-HIS3, the V1-V5 portion of the Q23 envelope gene was replaced by the selection marker URA3 to enable different V1-V5 envelope segments to be shuttled into the plasmid. URA3 encodes the orotidine 5′-phosphate decarboxylase protein involved in uracil biosynthesis. The URA3 gene was amplified from pRS316 (New England Biolabs) using primers 5′-CCATGTGTAAAGTTAACCCCTCTCTGAGATTGTACTGAGAGTGCAC-3′ and 5′-CACTTCTCCAATTGTCCCTCATATCTCCTGGTATTTCACACCGCAGGG-3′. The underlined segment of each primer highlights portions homologous to Q23 envelope sequences (analogous to primers Env15 and Env10S, respectively). Yeast were transformed with the URA3-amplified PCR fragment and BglII-linearized pRS315-Q23Δnef-HIS3 (BglII cuts within the envelope V1-V5 region). Yeast cells were grown on CMM plates lacking leucine, histidine, and uracil. Crude extract from a transformed yeast colony was electroporated into E. coli to isolate and expand the plasmid pRS315-Q23Δnef-HIS3ΔV1-V5-URA3. Different V1-V5 envelope fragments were shuttled into pRS315-Q23Δnef-HIS3ΔV1-V5-URA3 using LiAc transformation. Briefly, pRS315-Q23Δnef-HIS3ΔV1-V5-URA3 was linearized with StuI, which cuts within the URA3 gene. Linearized pRS315-Q23Δnef-HIS3ΔV1-V5-URA3 and amplified V1-V5 envelopes were used to transform yeast, and yeast was grown on CMM plates lacking leucine and histidine and with 5-fluoro-1,2,3,6-tetrahydro-2,6-dioxo-4-pyrimidine carboxylic acid (FOA). FOA inhibits the growth of yeast that expresses the URA3 gene. Yeast colonies on CMM plates lacking leucine and histidine and with FOA were screened for the HIV-1 envelope gene by PCR. Plasmids were expanded in E. coli, and incorporation of the different V1-V5 envelope segments within Q23 was confirmed by sequence analysis.
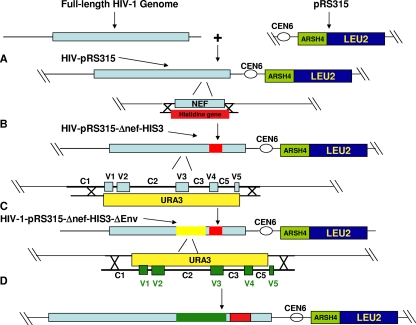
FIG. 1.
Construction of viruses using yeast gap repair homologous recombination. (A) A full-length HIV-1 clone was isolated and ligated into the multiple-cloning site of pRS315 (New England Biolabs). pRS315 sequences permit plasmid replication in both bacteria and yeast, and pRS315 also contains the beta-isopropylmalate dehydrogenase gene for the leucine synthetic pathway (LEU2). Thus, yeast with the recombined plasmid can be selected in leucine dropout media. (B) The yeast selection gene, for histidine (HIS3), was amplified with primers that contained the HIS3 gene sequences at the 3′ end flanked by nef homologous sequences at the 5′ end. This PCR product and the HIV-pRS315 plasmid linearized by endonuclease digestion within the nef gene were used to transform yeast and recover the HIV-pRS315-Δnef-HIS3 plasmid. (C) The URA3 gene was amplified from pRS316 (New England Biolabs) using primers that contained the URA3 gene sequences at the 3′ end flanked by sequences homologous to the HIV-1 envelope. URA3 encodes the orotidine-5′-phosphatase decarboxylase protein involved in the biosynthesis of uracil. Yeast was transformed with HIV-pRS315-Δnef-HIS3 plasmid linearized by endonuclease digestion within the env gene and the URA3 PCR product. Yeast with the recombined plasmid (HIV-pRS315-Δnef-HIS3-ΔEnv) was selected in leucine, histidine, and uracil dropout media. (D) Yeast was transformed with the HIV-pRS315-Δnef-HIS3-ΔEnv plasmid linearized with endonuclease digestion within URA and an envelope PCR product of interest. Recombined plasmid was isolated from yeast selected in leucine and histidine dropout media enriched with FOA. URA3 converts FOA to a toxic product which inhibits yeast with URA3 expression.
Virus stocks and titers.
Viral stocks were prepared by transiently transfecting 293T human embryonic kidney fibroblast cells using the Fugene protocol (Roche Molecular Biochemicals). The 293T cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (DMEM complete). After 48 h in culture, the supernatant was filtered through a 0.45-μm filter, aliquoted, and stored at −80°C until use. The infectious dose of each virus stock was determined by infecting TZM-bl cells with a range of viral dilutions by directly counting β-galactosidase (β-gal)-positive “blue” foci at 48 h postinfection as previously described (32, 76). TZM-bl cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (NIH-ARRRP) (84).
Site-directed mutagenesis.
The E370D mutation was introduced into pHXB-2-env using the QuikChange site-directed mutagenesis kit (Stratagene) with primers 5′-GGAGGGGACCCAGACATTGTAACGCACAG-3′ and 5′-CTGTGCGTTACAATGTCTGGGTCCCCTCC-3′. pHXB-2-env was obtained from the NIH-ARRRP (52). The wild-type HXB-2 envelope plasmid and HXB-2 E370D mutated envelope plasmid were used to make virus pseudotypes with a Q23ΔEnv plasmid as previously described (41, 76). The YU2 K421D mutated virus was constructed by site-directed mutagenesis of pYU2, using primers 5′-CACACTCCCATGTAGAATAGATCAAATTATAAATATGTGGC-3′ and 5′-GCCACATATTTATAATTTGATCTATTCTACATGGGAGTGTG-3′. pYU2 was obtained from the NIH-ARRRP (39). Site-directed mutagenesis was performed on an EcoRI/SalI segment of pYU2 subcloned into the multiple-cloning site of pRS316 (NE Biolabs). The EcoRI/SalI portion with the site-directed mutation was reinserted into the original pYU2 to obtain pYU2 K421D. The HXB-2 E370D and YU2 K421D mutations were confirmed with sequence analysis.
Sensitivity to inhibitors.
Sensitivity to soluble CD4, PRO1008 (Progenics), CCR5 antagonists TAK779 and PSC-RANTES, and fusion inhibitor T-20 was assessed on TZM-bl cells. TAK779 and T-20 were obtained through the NIH-ARRRP (2). Sensitivity was examined in 96-well plates in the presence of twofold serial dilutions of the inhibitor. Soluble CD4 was incubated with 500 infectious particles (IP) of each virus for 1 h at 37°C prior to adding 1 × 104 TZM-bl cells in each well. TAK779, PSC-RANTES, or T-20 was incubated with 1 × 104 TZM-bl cells for 1 h at 37°C prior to adding 500 IP of each virus. After 48 h, all infections were assessed for β-gal expression using Galacton-Light Plus (Applied Biosystems). Relative light units in wells without any infectious virus were used as the background level, and this was subtracted from the relative light units of each well. The level of β-gal expression in the presence of serially diluted inhibitor versus medium alone was used to determine the percentage of inhibition. Data were fitted to estimate the 50% inhibitory concentration (IC50). All IC50s were calculated from a minimum of two independent experiments, and within each experiment all infections were performed in triplicate.
Replication kinetics on cells with various levels of cell surface receptors.
Cell lines with various CD4 and CCR5 surface receptor densities were kindly provided by Emily Platt and David Kabat (62). All cells were plated 1 day prior to infection at a density of 4 × 104 cells per well in a 24-well dish. The next day, the medium was removed and cells were incubated in the presence of 20 μg/ml DEAE-dextran with 500 IP in a total of 100 μl of DMEM complete. After 2 h, cells were washed with phosphate-buffered saline, and 1 ml of fresh DMEM complete was added to each well. Cells were incubated at 37°C, and all of the medium was collected and replaced with fresh DMEM complete on the first and fourth days of infection. Cells were expanded to a T25 flask after the fourth day. Medium was also collected on day 7 after infection. p24 levels were assessed on medium collected from days 1, 4, and 7 after infection using a p24 enzyme-linked immunosorbent assay (Perkin-Elmer). The day 7 p24 levels in the cells with limiting levels of receptors were normalized relative to the day 7 p24 level in the JC53 cells with high levels of CD4 and CCR5.
Determination of coreceptor use.
Coreceptor usage was determined using the GHOST cell assay as previously described (41, 48). Briefly, 4 × 104 GHOST(3)CXCR4 or GHOST(3)X4/R5 cells were plated in a 24-well dish 1 day prior to infection. GHOST(3)X4/R5 and GHOST(3)CXCR4 cells express the CD4 receptor with both the CCR5 and the CXCR4 coreceptors or the CXCR4 coreceptor alone, respectively. In addition, green fluorescent protein (GFP) is under the transcriptional control of the HIV-2 long terminal repeat in these cells. Thus, infection can be assessed by flow cytometric measurement of GFP staining. Cells were exposed to 10,000 infectious virions, and wells were washed 2 h after exposure. Cultures were incubated for 2 days prior to fluorescence-activated cell sorter analysis for GFP staining of 10,000 events.
Replication kinetics on MDMs.
Peripheral blood mononuclear cells (PBMCs) were isolated using the Ficoll-Hypaque density centrifugation method from buffy coats obtained from local blood banks. Monocytes were isolated using the Percoll gradient method (14). Monocytes were incubated in macrophage SFM medium (Gibco BRL) supplemented with 10% human serum, 5% fetal bovine serum, 1 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (complete macrophage medium) for 5 to 7 days prior to infection. Between 0.5 × 106 and 1.0 × 106 monocyte-derived macrophages (MDMs) were plated per well in a 24-well plate. Cells were infected with 5,000 IP of virus in the presence of 20 μg/ml DEAE-dextran. Cells were washed after 2 h of viral exposure. All of the medium was collected and replaced with fresh complete macrophage medium on days 1, 4, 7, 10, and 14. All viruses were evaluated for their ability to replicate in MDMs from a minimum of two different blood donors. Replicative capacities of all recombinant viruses with V1-V5 segments from the same subject were always compared on MDMs obtained from the same donor.
Construction of chimeric V1-V5 envelope segments.
Overlap PCR was used to generate chimeric V1-V5 envelope fragments. The V1-V2 and C2-V5 envelope portions were amplified from the plasmids with the V1-V5 envelope segment of interest using primers Env 15 and 5′-TGAGGTATTACAATTTATTAATCTATA-3′ and Env 10S and 5′-TATAGATTAATAAATTGTAATACCTCA-3′, respectively. The V1-V3 and C3-V5 envelope domains were amplified from the V1-V5 envelope segment of interest using primers Env15 and 5′-GTGTTGTAATTTCTAGATCCCCTCCTG-3′ and Env 10S and 5′-CAGGAGGGGATCTAGAAATTACAACAC-3′, respectively. Amplified segments were gel purified and combined in a PCR with primers Env10S and Env15 to construct the chimeric V1-V5 domains. Construction of all chimeric V1-V5 envelope segments was verified by sequence analysis.
Statistical analysis.
The average from the multiple independent experiments was calculated for each recombinant virus for the replicative-capacity and inhibitor sensitivity assays. Differences among early- and chronic-infection viruses were assessed by two independent statistical tests. First, the value from the early-infection V1-V5 recombinant virus was compared to the median value for the multiple viruses with chronic-infection V1-V5 segments using the Wilcoxon matched-pairs signed-rank test. Second, the Exact Wilcoxon-Mann-Whitney test, stratified by subject, was used for an aggregate comparison between early- and chronic-infection viruses. The two tests showed that the same properties were significantly different (P < 0.05) between the early- and chronic-infection variants except for replication in the cells with low CCR5 levels; in this case the Exact Wilcoxon-Mann-Whitney test demonstrated a significant difference, while with the Wilcoxon matched-pairs signed-rank test a trend was observed (P = 0.1). Only the P values from the Exact Wilcoxon-Mann-Whitney test are presented because it accounts for the measurements from all the multiple chronic-infection variants as opposed to reducing them into a single median value. All P values are based on a two-sided test. All statistical analyses were done with Intercooled Stata version 8.0 (Stata Corporation, College Station, TX) and SAS version 8.2 (SAS Institute, Cary, NC).
RESULTS
V1-V5 sequences.
Sequences were generated from samples taken at 1 to 6 months postinfection and from approximately 24 to 47 months after estimated infection as described previously (Table 1) (76). The median interval of time for the analyzed sequences was about 35 months (range, 18 to 46 months). In previous studies, we have shown that five (QA284, QA779, QB670, QC168, and QC449) of the nine subjects examined in this study had strictly homogenous envelope sequences early in infection (74, 75). Thus, for these subjects, only one early-infection V1-V5 sequence was isolated, and for the remaining four subjects, multiple sequences (median, 3; range, 3 to 4) were isolated from early in infection (Table 1). For all subjects, multiple sequences (median, 4; range, 2 to 9) were isolated and examined from the chronic-infection sample. Similar to our previous results, early-infection sequences showed a significantly lower median number of predicted N-linked glycosylation sites (PNGS) within the V1-V2 domain (median, 5; range, 3 to 8) than did sequences from the chronic phase of infection (median, 6; range, 4 to 9; P = 0.03) (76). There was no significant difference in V1-V2 and V1-V4 length or in the number of V1-V4 PNGS between the early- and chronic-infection sequences. As before, neighbor-joining phylogenetic analysis showed that sequences at both time points within each subject clustered together, suggesting that there was no contamination or reinfection by two different partners (data not shown) (76).
Generation of replication-competent recombinant viruses.
To assess the effects on envelope function from the changes that occur in the V1-V5 segments over time, we constructed replication-competent recombinant viruses incorporating different V1-V5 sequences within a HIV-1 subtype A Q23-17 background. Q23-17 is a replication-competent clone derived from a subject's sample approximately 1 year after estimated infection (63). Because sequences early in infection are relatively homogeneous, only the predominant V1-V5 sequence from early in infection was incorporated into the full-length HIV-1 clone. All unique chronic-infection V1-V5 segments, however, were inserted into the Q23-17 HIV-1 clone. The median titer of the replication-competent recombinant viruses was 5.4 × 105 IP per ml (range, 1.0 × 105 to 2.1 × 106). Some chronic-infection V1-V5 variants in QA203 (one of four), QA779 (one of seven), QB424 (three of four), QB596 (one of eight), QB670 (nine of nine), QC168 (one of five), and QC890 (one of five) did not yield an infectious titer greater than 20 IP/ml, the limit of detection for the titer assay (Table 1). No further studies were pursued with the QB670 envelopes from either time point because no replication-competent recombinant virus with the QB670 V1-V5 chronic-infection sequence could be generated. It should be noted that we introduced V1-V5 segments within a heterologous backbone, and thus these constructs may not accurately reflect the properties of the original full-length parent envelope, which is potentially evident by the nonfunctional envelopes.
Replication capacity.
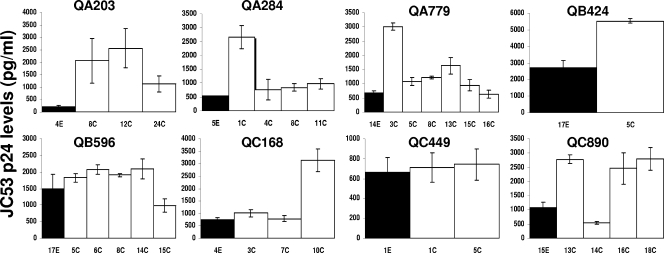
We examined replication efficiency differences among viruses with V1-V5 envelope segments from different times during infection in a cell line with high levels of the CD4 and CCR5 receptors, JC53 (62). Thus, host cell receptors were at a consistently high level in all infections, as opposed to the case for PBMCs, where there is extensive donor variability, especially in CCR5 density (47, 69). All recombinant viruses showed more than a 100-fold increase in p24 antigen levels from day 1 (median, 4.2 pg/ml; range, 0 to 20.8 pg/ml) to day 7 (median, 1,105 pg/ml; range, 222.9 to 5,534.0 pg/ml), suggesting that all viruses replicated in cells with high CD4 and high CCR5 cell surface receptor concentrations. In all subjects, early-infection variants replicated to a lower level than the median for the chronic-infection viruses, although these differences in some individuals (QB596 and QC449) were relatively small (Fig. 2). Early-infection viruses had significantly lower p24 levels (median, 713.0 pg/ml; range, 222.9 to 2,681.4 pg/ml) at day 7 after infection in the JC53 cells than the chronic-infection variants (median, 1,527.8 pg/ml; range, 727.1 to 5,534.0 pg/ml) (P = 0.01).
FIG. 2.
Replication in cells with high levels of CD4 and CCR5 receptors (JC53). Each graph shows the p24 production 7 days after infection for virus with early-infection V1-V5 segments (black bars) and viruses with chronic-infection V1-V5 sequences (white bars). Note that the y axis scale, which depicts p24 levels, is different in each graph. The subject identification is denoted above each graph, and the V1-V5 segment identification is below each column. All infection levels represent mean values from two or more independent experiments. The error bars show the standard deviations.
Receptor utilization.
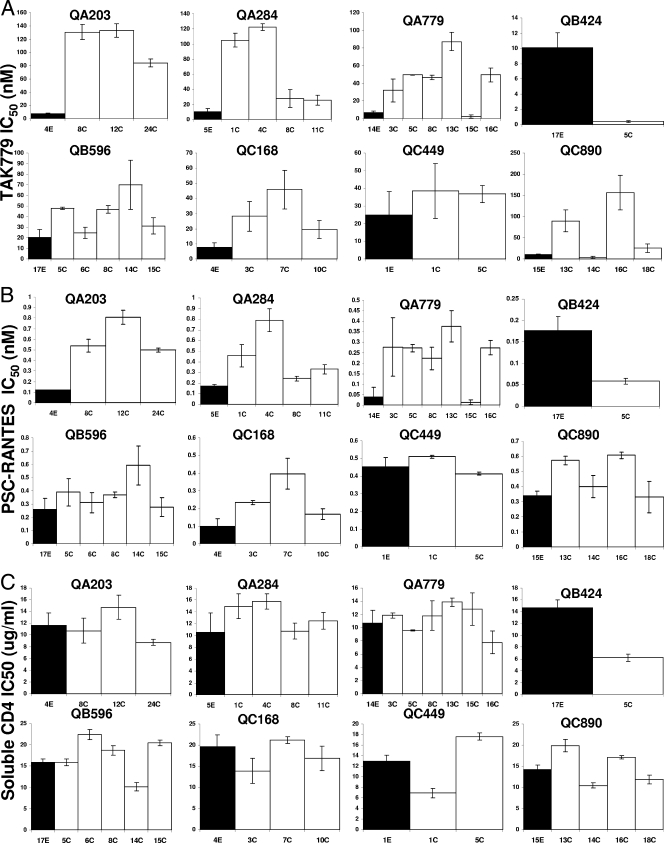
Because host cell entry is the rate-limiting step in HIV-1 replication (3, 44, 65) and entry is potentially limited by receptor binding and fusion (46, 61), we hypothesized that early- and chronic-infection viruses had differences in receptor attachment and fusion kinetics. Previous studies suggest that sensitivity to receptor inhibitors correlates with the affinity of the viral envelope for the host cell receptor (59, 66). Sensitivities to a small-molecule CCR5 inhibitor, TAK779 (2), and to a soluble CD4 molecule, PRO1008-1 (Progenics), were used as surrogate markers to measure CCR5 and CD4 utilization, respectively. To demonstrate that inhibitor sensitivity correlates with binding capacity, we tested viruses with previously documented receptor affinity differences. A YU2 envelope with a lysine (K)-to-aspartic acid (D) mutation at position 421 (K421D) within the envelope bridging sheet region has markedly lower CCR5 binding than the wild-type YU2 (71). The YU2 K421D virus (IC50, 2.1 nM ± 0.6 nM) had an approximately 18-fold greater sensitivity to TAK779 than the wild-type YU2 virus (IC50, 37.7 nM ± 12.6 nM). The HXB2 envelope with a glutamic acid (E)-to-aspartic acid (D) mutation at envelope position 370 (E370D) results in decreased affinity for the CD4 receptor compared to the wild-type HXB2 envelope (49). HXB-2 E370D pseudotypes (IC50, 1.3 μg/ml ± 1.1 μg/ml) had approximately 13-fold greater sensitivity to soluble CD4 than wild-type HXB-2 pseudotypes (IC50, 27.3 μg/ml ± 3.4 μg/ml).
To document differences in receptor utilization between chimeric envelopes with V1-V5 segments from the early and chronic phases of infection, we examined sensitivity to TAK779 and soluble CD4. The various recombinant viruses with different V1-V5 envelope segments demonstrated large variations in TAK779 IC50s, from 0.4 to 156.8 nM (Fig. 3A). In all subjects (except QB424), early-infection HIVs had lower TAK779 IC50s than the median TAK779 IC50s from all the chronic-infection viruses, although in QC449, the difference was less than twofold. In aggregate, early-infection variants (median, 10.0 nM; range, 7.1 to 105.2 nM) showed approximately four- to fivefold greater sensitivity to TAK779 than viruses with chronic-infection V1-V5 domains (median, 46.4 nM; range, 0.4 to 156.8 nM) (P = 0.01).
FIG. 3.
Sensitivity to TAK779 (A), PSC-RANTES (B), and soluble CD4 (C) of viruses with early-infection V1-V5 portions (black bars) or chronic-infection V1-V5 segments (white bars). The y axis shows the IC50s for each inhibitor. Note that the y axis scale is different in each graph. The subject identification is denoted above each graph, and the V1-V5 segment identification is below each column. All IC50s represent mean values from two or more independent experiments. The error bars show the standard deviations.
TAK779 is an allosteric inhibitor (2), and some envelopes with acquired resistance to small-molecule CCR5 inhibitors display continued susceptibility to CCR5 chemokines (64, 81), which suggests that sensitivity to TAK779 may not necessarily correlate with an ability to utilize the native CCR5 receptor. Thus, we examined the sensitivities of the acute- and chronic-infection envelopes to a CCR5 chemokine, PSC-RANTES, a competitive inhibitor. Sensitivity to PSC-RANTES varied from 0.01 to 0.81 nM (Fig. 3B). Similar to TAK779, early-infection variants (median, 0.18 nM; range, 0.04 to 0.45 nM) were significantly more sensitive to PSC-RANTES than the chronic-infection viruses (median, 0.37 nM; range, 0.01 to 0.81 nM) (P = 0.002).
Chronic-infection viruses may potentially have decreased CCR5 inhibitor sensitivity compared to early-infection variants because they may use CXCR4 receptor for cell entry. To exclude this possibility, we investigated coreceptor usage among our recombinant viruses using the GHOST cell assay. GFP staining was observed in more than 5% of the GHOST(3)CXCR4 cells after infection with a known CXCR4-using virus (LAI) (56) and in fewer than 0.2% of the cells with a CCR5-exclusive virus (JRCSF) (6). Recombinant viruses with early- or chronic-infection V1-V5 envelope segments showed a median of 0.2% GFP-positive cells (range, 0.1% to 0.3%). On the other hand, a median of 4% (range, 2% to 16%) of GHOST(3)X4/R5 cells were GFP stained after infection with the same recombinant viruses. This suggests that none of the recombinant viruses utilized the CXCR4 coreceptor for cell entry. Thus, chronic-infection viruses do not have decreased sensitivity to CCR5 antagonists because of the ability to use CXCR4 as a coreceptor.
Among the diverse viruses, soluble CD4 IC50s varied from 6.2 to 22.4 μg/ml (Fig. 3C). Unlike TAK779 and PSC-RANTES sensitivity, in aggregate, early-infection variants showed no significant difference in soluble CD4 sensitivity (median, 13.6 μg/ml; range, 10.5 to 19.7 μg/ml) compared to chronic-infection viruses (median, 13.3 μg/ml; range, 6.2 to 22.4 μg/ml) (P = 0.9).
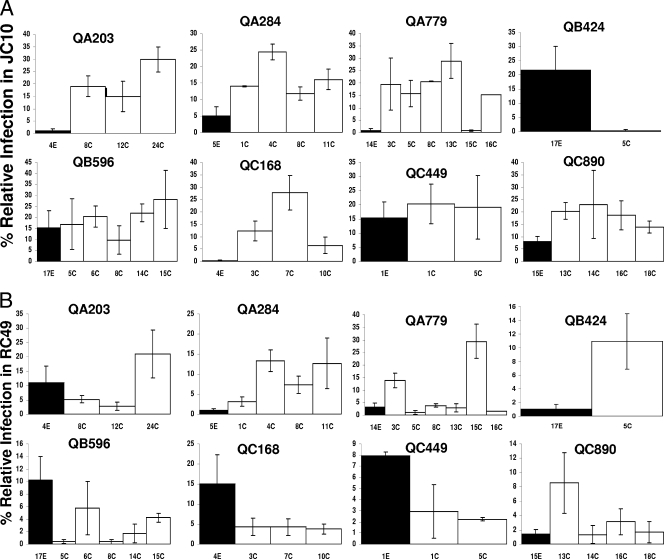
To further confirm that chronic- versus early-infection viruses had an increased ability to use CCR5 and no significant difference in CD4 utilization, we examined replication capacity in cells with limiting levels of receptors. We reasoned that viruses with an envelope glycoprotein that has a greater ability to utilize the CCR5 receptor should be able to replicate efficiently in JC10 cells, which have a low CCR5 density and high levels of CD4. Furthermore, viruses with envelopes that have higher CD4 binding should have better replication in RC49 cells, which express low levels of CD4 and medium density of CCR5. As controls, we observed that the YU2 K421D virus was unable to replicate in the JC10 cells with low CCR5 density. On the other hand, the wild-type YU2 produced 39.4% ± 7.9% p24 in JC10 cells relative to the JC53 cells with high levels of CCR5 at day 7 after infection. Entry in the RC49 cells with low CD4 densities relative to the JC53 cells with high CD4 and high CCR5 concentrations was around fourfold lower for the HXB2 E370D (6.1% ± 1.1%) compared to the wild-type HXB-2 pseudotypes (25.3% ± 3.5%) at day 2 after infection.
In the JC10 cells with low CCR5 receptor concentrations, relative infection levels at day 7 after infection varied from 0.3 to 29.9% for the early- and chronic-infection viruses (Fig. 4A). In all subjects except QB424, early-infection variants had lower relative infection levels in JC10 cells than the median of the relative infection levels of the chronic-infection viruses. These differences were less than twofold in two subjects (QB596 and QC449). Recombinant viruses with early-infection V1-V5 segments (median, 6.6; range, 0.4 to 21.7) showed significantly lower relative infection levels in the JC10 cells than engineered HIVs with V1-V5 portions from the chronic phase of infection (median, 18.3%; range, 0.3 to 29.9%) (P = 0.01). In the RC49 cells with low levels of CD4, relative infection levels varied from 0 to 41.2% (Fig. 4B). There was no significant difference in relative infection levels in the RC49 cells between the early-infection variants (median, 5.5; range, 1.0 to 15.1) and the chronic-infection variants (median, 4.8; range, 1.7 to 10.9) (P = 0.9).
FIG. 4.
Relative replication in cells with high CD4 and low CCR5 levels (JC10) (A) and in cells with low CD4 and medium CCR5 levels (RC49) (B) among viruses with early-infection V1-V5 portions (black bars) or chronic-infection V1-V5 segments (white bars). In each graph, the y axis shows the infection levels in the respective cell lines. Note that the y axis scale is different in each graph. These values are normalized relative to the infection levels in the high-CD4, high-CCR5 (JC-53) cell line. The subject identification is denoted above each graph, and the V1-V5 segment identification is below each column. All infection levels represent mean values from two or more independent experiments. The error bars show the standard deviations.
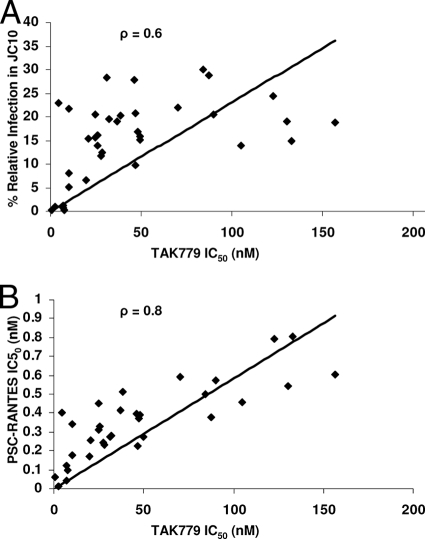
Because early- and chronic-infection envelopes from the eight different subjects showed similar relationships between CCR5 inhibitor IC50s and replicative capacity in the low-CCR5 cells, we examined the correlation between these assays. The TAK779 IC50 was significantly correlated with relative infection levels in the low-CCR5 cells (ρ = 0.6, P = 0.001; Spearman rank correlation) (Fig. 5A) and with sensitivity to PSC-RANTES (ρ = 0.8, P < 0.001; Spearman rank correlation) (Fig. 5B). Although none of these assays directly measures affinity for the CCR5 receptor, collectively our results suggest that they may be highly correlated surrogate markers for an envelope's ability to bind the CCR5 receptor. In contrast, the soluble CD4 IC50 showed no significant correlation with relative infection levels in the low-CD4, medium-CCR5 cell line (ρ = 0.03, P = 0.9; Spearman rank correlation), suggesting that these surrogate markers for CD4 use may be influenced by other factors, such as gp120 shedding or the ability to bind the coreceptor after CD4 engagement.
FIG. 5.
Association between sensitivity to TAK779 and both relative replication in cells with high CD4 and low CCR5 levels (JC10) (A) and sensitivity to PSC-RANTES (B). The y axis shows the relative infection in JC10 cells (A) and sensitivity to PSC-RANTES (B). The x axis shows the TAK779 sensitivity for each chimeric envelope, represented by individual dots. The line shows the best-fit linear regression curve, with the correlation coefficient listed on the top of the graph.
Fusion capacity.
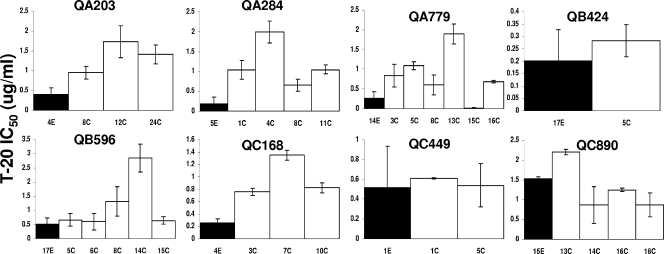
Differences in cell entry and replication between early- and chronic-infection viruses may also be influenced by fusion kinetics. Previous studies have suggested that sensitivity to the fusion inhibitor T-20 correlates with fusion kinetics (66). Although all envelopes harbored an isogenic transmembrane domain, gp41, the diverse recombinant viruses showed great variation in T-20 IC50s from 0.02 to 2.8 μg/ml (Fig. 6). In all subjects except QC890, early-infection HIVs had T-20 IC50s lower than the median T-20 IC50s from all the chronic-infection viruses, although in three subjects (QB424, QB596, and QC449) the difference was less than twofold. Early-infection variants (median, 0.3 μg/ml; range, 0.2 to 1.5 μg/ml) were approximately two- to threefold more sensitive to T-20 than viruses with chronic-infection V1-V5 domains (median, 0.8 μg/ml; range, 0.3 to 1.4 μg/ml) (P = 0.01; Wilcoxon matched-pairs signed-rank test).
FIG. 6.
Sensitivity to fusion inhibitor T-20 among viruses with chronic-infection V1-V5 segments (white bars) versus those with early-infection V1-V5 portions (black bars). The y axis shows the IC50s against the fusion inhibitor. Note that the y axis scale is different in each graph. The subject identification is denoted above each graph, and the V1-V5 segment identification is below each column. All IC50s represent mean values from two or more independent experiments. The error bars show the standard deviations.
Genotypic mapping of the differences to CCR5 and fusion inhibitors.
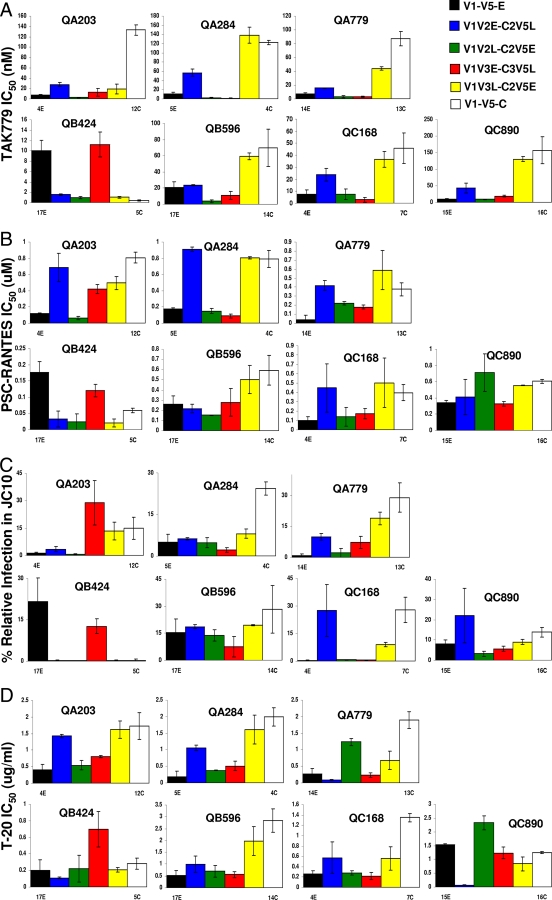
To identify the envelope determinants for the difference in CCR5 utilization and fusion capacity among early- and chronic-infection V1-V5 segments, we constructed chimeric V1-V5 envelope domains. Chimeric V1-V5s were created with the subject's early-infection V1-V5 envelope portion and the chronic-infection V1-V5 envelope segment that demonstrated the highest IC50 against CCR5 inhibitors in seven subjects. No chimeric envelopes were generated from QC449 V1-V5 segments because viruses with early- and chronic-infection envelope regions from this subject showed minimal differences in CCR5 utilization (Fig. 3A and B and 4A). Overlap PCR was used to create four sets of chimeric V1-V5 segments for each subject. We constructed V1-V5s with (i) early-infection V1-V2 and chronic-infection C2-V5 (V1V2E-C2V5L), (ii) chronic-infection V1-V2 and early-infection C2-V5 (V1V2L-C2V5E), (iii) early-infection V1-V3 and chronic-infection C3-V5 (V1V3E-C3V5L), and (iv) chronic-infection V1-V3 and early-infection C3-V5 (V1V3L-C3V5E) segments. These chimeric V1-V5 envelope segments were incorporated in the full-length Q23 clone using the yeast gap repair homologous recombination system. In all cases except QA203, the TAK779 IC50s of the V1-V3/C3-V5 chimeras were within twofold of that of the parent sequence from which the V1-V3 portion was obtained (Fig. 7A) . Thus, the V1V3L-C3V5E chimeras composed of chronic-infection V1-V3 and early-infection C3-V5 segments showed TAK779 IC50s similar to that of the parent virus with the chronic-infection V1-V5 segment, and the V1-V3E-C3V5L showed TAK779 sensitivity similar to that of the virus with early-infection V1-V5 domains. The V1-V3/C3-V5 chimeras’ sensitivity to PSC-RANTES was also similar to that of the parent sequence from which the V1-V3 portion was obtained (Fig. 7B). In the cases where the chronic-infection variant replicated more efficiently than the early-infection variant in the cells with low CCR5 densities (JC10), relative replication in the JC10 cells was higher among the V1V3L-C3V5E than among the V1V3E-C3V5L chimeras, except in QA203 (Fig. 7C). In QB424, where QB424-17E replicated more efficiently than QB424-5C in JC10 cells, the V1V3E-C3V5L produced greater relative amounts of p24 than the V1V3L-C3V5E. Similarly, except for QB424, the V1V3L-C3V5E versus the V1V3E-C3V5L chimeras were less sensitive to fusion inhibitor T-20, if the chronic- compared to the early-infection V1-V5 showed a higher T-20 IC50 (Fig. 7D). In QC890, where QC890-15E was mildly less sensitive to T-20 than QC890-16C, the V1V3E-C3V5L chimera also had a minimally higher T-20 IC50 than the V1V3L-C3V5E chimera. Collectively, this suggests that V1-V3 sequences mainly influence the difference in CCR5 utilization and fusion capacity between early- and chronic-infection viruses. Interestingly, in all cases except QC890, viruses with early-infection V1-V2 and chronic-infection C2-V5 portions consistently demonstrated greater resistance to the CCR5 antagonists and higher replication capacity in cells with low CCR5 receptor densities than viruses with chronic-infection V1-V2 and early-infection C2-V5 segments (Fig. 7A, B, and C). In most cases, these differences among the V1-V2/C2-V5 chimeras, however, were relatively small. A similar pattern was not observed among the V1-V2/C2-V5 chimeras for T-20 sensitivity (Fig. 7D). Thus, the specific envelope segment (V1-V2 or V3) that influence CCR5 usage and fusion capacity differences among early- and chronic-infection variants cannot be identified among these different subjects.
FIG. 7.
Early and chronic chimeric envelope TAK779 IC50s (A), PSC-RANTES IC50s (B), relative replication in high-CD4, low-CCR5 cells (JC10) (C), and T-20 IC50s (D). V1V2E-C2V5L (blue), V1V2L-C2V5E (green), V1V3E-C3V5L (red), and V1V3L-C3V5E (yellow) chimeras are shown. For reference, viruses with early-infection V1-V5 (black bars) and chronic-infection V1-V5 (white bars) segments from which the envelope portions for the chimeras were derived are also shown. The y axis shows the IC50s for each inhibitor (A and C) and relative replication levels (B). The y axis scale is different among the graphs. Subject identifications are denoted above each graph. All values represent mean values from two or more independent experiments with viral stocks from two separate preparations. The error bars show the standard deviations.
Replication capacity in MDMs.
To examine the biological relevance of CCR5 utilization differences among early- and chronic-infection viruses, we examined replication capacity in primary cells with known limiting levels of CD4 and CCR5 receptors. Macrophages express lower surface levels of both CD4 and CCR5 compared to CD4+ T cells (17, 37, 50, 55, 88). In addition, viruses that use the CCR5 coreceptor often have large differences in macrophage tropism (60), and this could potentially relate to enhanced CCR5 usage (40). MDMs were isolated from PBMCs using standard Percoll gradient methods and infected with different viruses. Infections were monitored by p24 antigen levels at days 1, 4, 7, 10, and 14.
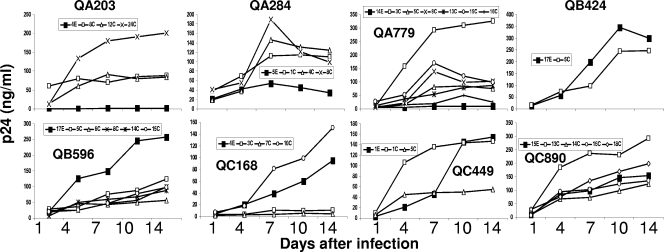
Recombinant viruses with a subject's V1-V5 segments from early and chronic infection displayed various replication capacities in MDMs (Fig. 8). There was no consistent pattern in the replication difference between recombinant viruses with early- versus chronic-infection V1-V5 segments. In some subjects (QA203, QA284, and QA779), all viruses with chronic- versus early-infection V1-V5s showed higher replication in MDMs. In one subject (QB596), the virus with early-infection V1-V5 demonstrated greater replication in MDMs than the recombinant viruses with chronic-infection V1-V5s. In the remaining subjects, there was no significant difference in replication capacity between viruses with early- and chronic-infection V1-V5 segments. In addition, there was no significant correlation between the highest p24 level in MDMs over the course of infection and CCR5 inhibitor IC50s, soluble CD4 IC50s, and replicative capacity in cells with low CD4 or low CCR5 densities. These data suggest that CCR5 utilization differences among early- and chronic-infection viruses do not confer replication capacity differences in MDMs.
FIG. 8.
Replication in MDMs. Each graph shows the p24 production of virus with early-infection V1-V5 segment (filled symbols) and viruses with chronic-infection V1-V5 sequences (open symbols) over time in MDMs. Note that the y axis scale, which depicts p24 levels, is different in each graph. The subject identification is denoted above each graph, and the V1-V5 segment identification is documented in the insets.
DISCUSSION
In this study of sequences from eight individuals with HIV-1 subtype A infection, we showed that modifications within the V1-V5 envelope segments confer increased replication capacity over the course of infection (Fig. 2). Previous studies with subtype B HIV-1 have also demonstrated that ex vivo replication capacity increases over the course of infection (4, 34, 80). Similarly, in the simian immunodeficiency virus/macaque model, longitudinally collected viruses demonstrate greater in vivo replication than variants isolated from early in infection (31). We also showed that longitudinally isolated V1-V5 envelope segments conferred significantly increased resistance to CCR5 inhibitors, TAK779 and PSC-RANTES (Fig. 3A and B). In addition, we demonstrated that in the majority of subjects, V1-V5 domains from the chronic phase of infection, compared to those from early after HIV-1 acquisition, led to increased replication capacity in cells with low CCR5 densities (Fig. 4A). From these significantly correlated measures of CCR5 use (Fig. 5), we concluded that viruses from the chronic phase of infection are more efficient at utilizing CCR5 than variants isolated early after infection. Among the longitudinally collected variants from the different subjects, we also found that resistance to fusion inhibitors increases over time, suggesting that chronic-infection viruses are more fusogenic than isolates from early in infection (Fig. 6). Previous studies and our controls suggest that these measurements could be surrogate markers for coreceptor affinity and fusion kinetics (59, 66). Because cell entry is potentially the rate-limiting step in HIV-1 replication (3, 44, 65), in the aggregate, our data suggest that increases in replication capacity over the course of infection are potentially related to greater affinity for the CCR5 receptor and/or faster fusion kinetics.
A previous study has suggested that acute- and chronic-infection variants are not significantly different in their sensitivities to CCR5 and fusion inhibitors (73). In that study, however, the virus isolates from the acute and chronic phases of infection were obtained from different subjects. In contrast to that publication and similar to our results, other studies which have examined longitudinally collected variants have suggested that viruses become more resistant to CCR5 and fusion inhibitors over time (27, 33, 68). In these studies, PBMC cocultures were used to demonstrate that viruses from the chronic phase of infection have differential susceptibility to receptor and fusion inhibitors compared to late-stage variants. In contrast to these studies, we examined subtype A viruses as opposed to the presumably subtype B HIVs examined in the previous publications. In addition, we avoided in vitro adaptation that may occur among viruses in PBMC cocultures by amplifying V1-V5 envelope segments and incorporating them into a full-length HIV-1 clone using yeast gap repair methodology. One of the major differences between our study and previous publications, however, is that we examined differences among variants from the early and chronic phases of infection as opposed to comparing viruses from the chronic and late stages of disease. Collectively, these studies suggest that both subtype A and B HIVs found early in infection are extremely sensitive to CCR5 antagonists and fusion inhibitors, and sensitivity to these compounds progressively decreases over later times in infection. Studies from our group and others demonstrate that there is a strong correlation between sensitivity to TAK779 and clinically relevant CCR5 antagonists, such as maraviroc and vicriviroc (81, 87; T. Henrich, M. Sagar, and D. Kuritzkes, unpublished data). Potential implications from these studies are that chemokine antagonists and fusion inhibitors may be ideal therapeutic drugs early in infection. In addition, although CCR5 utilization increases over time, HIV-1 infection starts with variants highly sensitive to CCR5 inhibitors, which further justifies the exploration of CCR5 inhibition as a potential means to interrupt transmission (36, 83).
Another major difference between our work and the majority of previously published studies is that we examined envelope phenotypic differences using replication competent viruses as opposed to viral pseudotypes. We employed a modified yeast gap repair homologous recombination system to generate a large number of recombinant replication-competent viruses (Fig. 1). Within the HIV field, the majority of studies examining envelope glycoprotein phenotypic differences employ virus pseudotypes. Although, pseudoviruses are highly conducive to high-throughput analysis of a large number of envelopes, there are a number of inherent limitations. First, the virus pseudotypes are restricted to a single round of replication, and thus, phenotypic differences conferred by the viral envelope glycoproteins, such as replication capacity, cannot be examined over multiple replication cycles. Second, pseudoviruses often display different phenotypes compared to replication-competent viruses. For instance, single-cycle pseudotypes compared to replication-competent chimeric viruses have demonstrated different sensitivities to CCR5 inhibitors on the same target cells (64). Third, the number of envelope glycoproteins expressed on a viral particle may be different among pseudoviruses versus replication-competent viruses (60). This may relate to differences in the number of defective envelopes and glycoprotein processing among the two types of recombinant viruses. Envelope glycoprotein properties need to be examined in more detail among replication-competent recombinant viruses and viral pseudotypes to definitely document the differences among these two virus constructs.
The exact biological mechanism for increased CCR5 utilization among chronic-stage viruses compared to variants early after infection remains unclear. Although, TAK779 is an allosteric (2) and not a competitive inhibitor, TAK779 sensitivity correlates with affinity for the CCR5 receptor (66). Furthermore, TAK779 IC50s correlate with sensitivity to the CCR5 competitive inhibitor RANTES (Fig. 5B) (40, 68). Thus, we suggest that increased CCR5 usage is because of a greater affinity for the CCR5 receptor. Another potential mechanism for higher CCR5 utilization is that chronic-stage envelopes could bind a broader array of CCR5 conformations. Indeed, natural CCR5 ligands, such as RANTES, can trigger internalization, as well occupy and presumably distort the receptor (54). Thus, in vivo, the CCR5 receptor may have different conformations in the presence of chemokines. It has been shown that envelopes from the late phase of infection compared to the variants from the chronic stage of disease display an increased ability to bind a broad range of chimeric CCR5 receptors (30). Therefore, an ability to bind different structural forms of the CCR5 receptor may explain the increased CCR5 usage among chronic-stage variants compared to the early-infection isolates. Finally, it also possible that chronic- and early-stage viruses may have similar binding to the CCR5 receptor, but after CCR5 attachment, viruses with chronic- versus early-stage V1-V5s may have a higher propensity for proceeding to fusion; this may account for the differences in CCR5 utilization. Previous studies have suggested that CCR5 affinity is directly correlated with fusion capacity (66), and thus viruses with chronic- versus early-infection V1-V5s may possess both greater affinity for the CCR5 receptor and increased fusion capacity. Thus, our studies cannot distinguish whether increased replication capacity among the chronic-stage variants is due to increased CCR5 utilization and/or faster fusion kinetics.
Interestingly, both sensitivity to CCR5 inhibitors and differential fusion capacity have been previously mapped to sequence changes within the envelope V3 loop and the bridging sheet, which are important for coreceptor binding (16, 66, 67). Among our eight subjects, differences among the early- and chronic-infection envelope sequences were evident primarily in the V3 loop and not in the bridging sheet, which consists of β strands 2, 3, 20, and 21 (35, 53). V3 loop modifications, however, are not solely responsible for the phenotypic differences observed among early- and chronic-infection envelopes. Our chimeric envelope data suggest that sequence changes within the V3 loop in conjunction with differences in the V1-V2 loops influence CCR5 usage (Fig. 7A, B, and C). Furthermore, although each envelope harbored an isogenic transmembrane domain, notable differences were observed in the sensitivity to fusion inhibitor T-20 among early- and chronic-infection V1-V5 segments, and our envelope chimera data suggest that changes within the V1-V3 domains also affected this phenotype (Fig. 7D). Because of numerous and diverse changes in the V1-V3 domains between the early- and chronic-infection envelope isolates, we were unable to identify canonical envelope modifications as being responsible for the enhanced CCR5 utilization and fusion capacity. Interestingly, previously identified polymorphisms, such as changes at positions 318 and 319 of the V3 loop (HXB2 numbering), which have been associated with differential sensitivity to CCR5 inhibitors (40), were highly conserved among early- and chronic-infection envelope sequences. The 318/319 consensus tyrosine (Y)/alanine (A) motif was modified to serine (S)/A in QA284-8C, Y/threonine (T) in QA779-5C and QA77913C, and Y/glycine (G) among all QC449 variants. Thus, in our study, these exclusive 318/319 modifications within the V3 loop did not influence the majority of observed changes in CCR5 usage. Our studies contrast with other publications potentially because of the differences in the subtype of the virus and because we examined longitudinally isolated viral variants from natural infection as opposed to laboratory-derived viral strains.
Progressively decreasing sensitivity to CCR5 entry inhibitors provides insight into the selection mechanisms acting on the virus during the course of infection within a host. The host antibody neutralizing response is a well-described selection force that drives evolution in the HIV-1 envelope gene (10, 21, 85). Our results imply that receptor use may be another potential selection mechanism driving changes in the viral envelope glycoprotein. After HIV-1 acquisition, a large percentage of memory CD4+ CCR5+ T cells are eliminated from the mucosal tissues (5, 45). In addition, CCR5 receptor levels on the remaining target cells may be downregulated through high-level expression of chemokines such as RANTES, MIP-1α, and MIP-1β (1, 79). Both processes likely decrease the availability of CD4+ T cells with high levels of the CCR5 receptor. The dearth of these cell types potentially forces HIV-1 to evolve envelopes with an increased ability to use low levels of the CCR5 receptor and/or switch to CXCR4 use late in infection. The need to evolve a greater ability to use low levels of CCR5 is likely especially true for subtype A HIV-1, where use of other coreceptors, such as CXCR4, has been infrequently documented (26, 29). It should be noted, however, that not all individuals, such as QB424, displayed an enhanced ability to use CCR5 over time, suggesting that the potential selection forces or virus responses may be different in some hosts.
Chronic-stage viruses showed increased CCR5 utilization compared to early-phase viruses even though envelope glycoproteins expand variable loops and increase the number of glycosylated amino acids. Similar to our previous investigations with the same subjects (76), we found that chronic-stage envelopes had a significantly higher number of glycosylated residues than viruses early after HIV-1 acquisition. Envelope variable loop expansion and increased glycosylation likely evolve to conceal conserved antigenic portions on the viral envelope glycoprotein, such as the CD4 and/or the coreceptor binding site (35, 53), from the host neutralizing antibody response. Shielding these domains, however, may hinder access of the viral envelope glycoprotein for the host cell receptors. Our results imply that increased glycosylation did not adversely affect CCR5 binding. Interestingly, we also found no difference either in sensitivity to a CD4 inhibitor, soluble CD4, or in replication capacity in cells with limiting levels of the CD4 receptor among chronic- and early-stage envelope V1-V5 segments (Fig. 3B and 4B). It has been hypothesized that HIV-1 envelopes with an increased ability to use low levels of CD4 evolve in the absence of humoral immune pressure, such as in the central nervous system, an immunologically privileged site (18, 57). Our data suggest that the converse does not necessarily hold, because neutralizing antibodies, which we have previously documented in these subjects (76), do not lead to envelope modifications that decrease the efficiency of CD4 receptor usage.
After observing increased CCR5 utilization among chronic-stage envelopes, we hypothesized that envelopes from late in infection were more likely to replicate in primary cells with limiting levels of CCR5, such as macrophages. Indeed, some previous studies suggest that isolates from late in disease are more macrophage-tropic than those from early in infection (23, 38, 82). Furthermore, it has been suggested that higher CCR5 binding confers macrophage tropism (22). We, however, observed no significant differences among early envelopes versus chronic-stage variants in their ability to replicate in MDMs (Fig. 8). Therefore, our observations support previous conclusions that replication capacity in macrophages does not correlate with an ability to use CCR5 (59). Because we did not observe significant differences in CD4 receptor use, however, we cannot directly corroborate that CD4 affinity predominately influences macrophage tropism as has been previously suggested (59).
In the same subjects analyzed in this study, we have previously shown that changes within the V1-V2 envelope loops confer significantly increased neutralization resistance to autologous plasma (76). Although we did not directly assess neutralization sensitivity of the chimeric viruses in this study to autologous plasma, collectively our results imply that evolution within the envelope glycoprotein over the course of infection that occurs in response to the host antibody response does not necessarily confer a fitness cost in terms of receptor usage and replication capacity. It should be noted, however, that effects on entry and replication of the specific modifications that confer neutralization escape will need to be examined in detail to validate this hypothesis. In summary, our data suggest that modifications within the envelope V1-V3 lead to antibody neutralization escape, an increased ability to utilize the CCR5 receptor, and faster fusion kinetics.
Acknowledgments
We thank Jawad Kiani, Sonam Sheth, and Laura Cohen for assistance with sequence analysis and PCR amplifications; Nikolaos Chatziandreou and Ines Freitas for help with the PSC-RANTES sensitivity assay; Cammie Lesser for yeast materials and protocols; Emily Platt and David Kabat for cell lines with various levels of CD4 and CCR5; Oliver Hartley for PSC-RANTES; and Julie Overbaugh and the Mombasa cohort staff, who originally provided access to the samples from which these sequences were derived and thus made this research possible.
This study was supported by NIH grant AI1077473 (M.S.), the American Foundation for AIDS Research (amfAR) (M.S.), and a Doris Duke Charitable Foundation Early Career Development Award (M.S.).
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. [DOI] [PubMed] [Google Scholar]
- 2.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, S. C., A. Abraha, K. R. Collins, A. J. Marozsan, H. Baird, M. E. Quinones-Mateu, A. Penn-Nicholson, M. Murray, N. Richard, M. Lobritz, P. A. Zimmerman, T. Kawamura, A. Blauvelt, and E. J. Arts. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaak, H., M. Brouwer, L. J. Ran, F. de Wolf, and H. Schuitemaker. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600-610. [DOI] [PubMed] [Google Scholar]
- 5.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cann, A. J., J. A. Zack, A. S. Go, S. J. Arrigo, Y. Koyanagi, P. L. Green, Y. Koyanagi, S. Pang, and I. S. Chen. 1990. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington, M., M. Dean, M. P. Martin, and S. J. O'Brien. 1999. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol. Genet. 8:1939-1945. [DOI] [PubMed] [Google Scholar]
- 8.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choisy, M., C. H. Woelk, J. F. Guegan, and D. L. Robertson. 2004. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J. Virol. 78:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clapham, P. R., and A. McKnight. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809-1829. [DOI] [PubMed] [Google Scholar]
- 12.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 13.Coligan, J. E., A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, W. Strober, and R. Coico. 1999. Saccharomyces cerevisiae. Wiley, New York, NY.
- 14.de Almeida, M. C., A. C. Silva, A. Barral, and M. Barral Netto. 2000. A simple method for human peripheral blood monocyte isolation. Mem. Inst. Oswaldo Cruz 95:221-223. [DOI] [PubMed] [Google Scholar]
- 15.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Marzio, P., J. Tse, and N. R. Landau. 1998. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res. Hum. Retroviruses 14:129-138. [DOI] [PubMed] [Google Scholar]
- 18.Dunfee, R. L., E. R. Thomas, P. R. Gorry, J. Wang, J. Taylor, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. USA 103:15160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier, R. A., M. Brouwer, S. M. Broersen, and H. Schuitemaker. 1995. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorry, P. R., J. Taylor, G. H. Holm, A. Mehle, T. Morgan, M. Cayabyab, M. Farzan, H. Wang, J. E. Bell, K. Kunstman, J. P. Moore, S. M. Wolinsky, and D. Gabuzda. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray, L., J. Sterjovski, M. Churchill, P. Ellery, N. Nasr, S. R. Lewin, S. M. Crowe, S. L. Wesselingh, A. L. Cunningham, and P. R. Gorry. 2005. Uncoupling coreceptor usage of human immunodeficiency virus type 1 (HIV-1) from macrophage tropism reveals biological properties of CCR5-restricted HIV-1 isolates from patients with acquired immunodeficiency syndrome. Virology 337:384-398. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, B. H., G. M. Shaw, M. E. Taylor, R. R. Redfield, P. D. Markham, S. Z. Salahuddin, F. Wong-Staal, R. C. Gallo, E. S. Parks, and W. P. Parks. 1986. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science 232:1548-1553. [DOI] [PubMed] [Google Scholar]
- 25.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 26.Huang, W., S. H. Eshleman, J. Toma, S. Fransen, E. Stawiski, E. E. Paxinos, J. M. Whitcomb, A. M. Young, D. Donnell, F. Mmiro, P. Musoke, L. A. Guay, J. B. Jackson, N. T. Parkin, and C. J. Petropoulos. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 81:7885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansson, M., M. Popovic, A. Karlsson, F. Cocchi, P. Rossi, J. Albert, and H. Wigzell. 1996. Sensitivity to inhibition by beta-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc. Natl. Acad. Sci. USA 93:15382-15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen, M. A., F. S. Li, A. B. van 't Wout, D. C. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaleebu, P., I. L. Nankya, D. L. Yirrell, L. A. Shafer, J. Kyosiimire-Lugemwa, D. B. Lule, D. Morgan, S. Beddows, J. Weber, and J. A. Whitworth. 2007. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D: results from a rural Ugandan cohort. J. Acquir. Immune Defic. Syndr. 45:28-33. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson, I., L. Antonsson, Y. Shi, M. Oberg, A. Karlsson, J. Albert, B. Olde, C. Owman, M. Jansson, and E. M. Fenyo. 2004. Coevolution of RANTES sensitivity and mode of CCR5 receptor use by human immunodeficiency virus type 1 of the R5 phenotype. J. Virol. 78:11807-11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 32.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koning, F. A., D. Kwa, B. Boeser-Nunnink, J. Dekker, J. Vingerhoed, H. Hiemstra, and H. Schuitemaker. 2003. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 188:864-872. [DOI] [PubMed] [Google Scholar]
- 34.Kwa, D., J. Vingerhoed, B. Boeser, and H. Schuitemaker. 2003. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J. Infect. Dis. 187:1397-1403. [DOI] [PubMed] [Google Scholar]
- 35.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lederman, M. M., R. S. Veazey, R. Offord, D. E. Mosier, J. Dufour, M. Mefford, M. Piatak, Jr., J. D. Lifson, J. R. Salkowitz, B. Rodriguez, A. Blauvelt, and O. Hartley. 2004. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science 306:485-487. [DOI] [PubMed] [Google Scholar]
- 37.Lewin, S. R., S. Sonza, L. B. Irving, C. F. McDonald, J. Mills, and S. M. Crowe. 1996. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res. Hum. Retroviruses 12:877-883. [DOI] [PubMed] [Google Scholar]
- 38.Li, S., J. Juarez, M. Alali, D. Dwyer, R. Collman, A. Cunningham, and H. M. Naif. 1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobritz, M. A., A. J. Marozsan, R. M. Troyer, and E. J. Arts. 2007. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J. Virol. 81:8258-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long, E. M., S. M. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retroviruses 18:567-576. [DOI] [PubMed] [Google Scholar]
- 42.Malim, M. H., and M. Emerman. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469-472. [DOI] [PubMed] [Google Scholar]
- 43.Marozsan, A. J., and E. J. Arts. 2003. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J. Virol. Methods 111:111-120. [DOI] [PubMed] [Google Scholar]
- 44.Marozsan, A. J., D. M. Moore, M. A. Lobritz, E. Fraundorf, A. Abraha, J. D. Reeves, and E. J. Arts. 2005. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J. Virol. 79:7121-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mkrtchyan, S. R., R. M. Markosyan, M. T. Eadon, J. P. Moore, G. B. Melikyan, and F. S. Cohen. 2005. Ternary complex formation of human immunodeficiency virus type 1 Env, CD4, and chemokine receptor captured as an intermediate of membrane fusion. J. Virol. 79:11161-11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore, J. P. 1997. Coreceptors: implications for HIV pathogenesis and therapy. Science 276:51-52. [DOI] [PubMed] [Google Scholar]
- 48.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ometto, L., M. Zanchetta, A. Cabrelle, G. Esposito, M. Mainardi, L. Chieco-Bianchi, and A. De Rossi. 1999. Restriction of HIV type 1 infection in macrophages heterozygous for a deletion in the CC-chemokine receptor 5 gene. AIDS Res. Hum. Retroviruses 15:1441-1452. [DOI] [PubMed] [Google Scholar]
- 51.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 52.Page, K. A., N. R. Landau, and D. R. Littman. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739-769. [DOI] [PubMed] [Google Scholar]
- 54.Pastore, C., G. R. Picchio, F. Galimi, R. Fish, O. Hartley, R. E. Offord, and D. E. Mosier. 2003. Two mechanisms for human immunodeficiency virus type 1 inhibition by N-terminal modifications of RANTES. Antimicrob. Agents Chemother. 47:509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patterson, B. K., A. Landay, J. Andersson, C. Brown, H. Behbahani, D. Jiyamapa, Z. Burki, D. Stanislawski, M. A. Czerniewski, and P. Garcia. 1998. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am. J. Pathol. 153:481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 57.Peeters, M. (ed.). 2002. Recombinant HIV sequences: their role in the global epidemic. Theoretical Biology and Biophysics Group, Los Alamos, NM.
- 58.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 59.Peters, P. J., M. J. Duenas-Decamp, W. M. Sullivan, R. Brown, C. Ankghuambom, K. Luzuriaga, J. Robinson, D. R. Burton, J. Bell, P. Simmonds, J. Ball, and P. R. Clapham. 2008. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology. 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters, P. J., W. M. Sullivan, M. J. Duenas-Decamp, J. Bhattacharya, C. Ankghuambom, R. Brown, K. Luzuriaga, J. Bell, P. Simmonds, J. Ball, and P. R. Clapham. 2006. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J. Virol. 80:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Platt, E. J., J. P. Durnin, and D. Kabat. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79:4347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73:5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pugach, P., A. J. Marozsan, T. J. Ketas, E. L. Landes, J. P. Moore, and S. E. Kuhmann. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rangel, H. R., J. Weber, B. Chakraborty, A. Gutierrez, M. L. Marotta, M. Mirza, P. Kiser, M. A. Martinez, J. A. Este, and M. E. Quinones-Mateu. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 77:9069-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Repits, J., M. Oberg, J. Esbjornsson, P. Medstrand, A. Karlsson, J. Albert, E. M. Fenyo, and M. Jansson. 2005. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J. Gen. Virol. 86:2859-2869. [DOI] [PubMed] [Google Scholar]
- 69.Reynes, J., P. Portales, M. Segondy, V. Baillat, P. Andre, B. Reant, O. Avinens, G. Couderc, M. Benkirane, J. Clot, J. F. Eliaou, and P. Corbeau. 2000. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J. Infect. Dis. 181:927-932. [DOI] [PubMed] [Google Scholar]
- 70.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 72.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rusert, P., H. Kuster, B. Joos, B. Misselwitz, C. Gujer, C. Leemann, M. Fischer, G. Stiegler, H. Katinger, W. C. Olson, R. Weber, L. Aceto, H. F. Gunthard, and A. Trkola. 2005. Virus isolates during acute and chronic human immunodeficiency virus type 1 infection show distinct patterns of sensitivity to entry inhibitors. J. Virol. 79:8454-8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sagar, M., E. Kirkegaard, L. Lavreys, and J. Overbaugh. 2006. Diversity in HIV-1 envelope V1-V3 sequences early in infection reflects sequence diversity throughout the HIV-1 genome but does not predict the extent of sequence diversity during chronic infection. AIDS Res. Hum. Retroviruses 22:430-437. [DOI] [PubMed] [Google Scholar]
- 75.Sagar, M., L. Lavreys, J. M. Baeten, B. A. Richardson, K. Mandaliya, B. H. Chohan, J. K. Kreiss, and J. Overbaugh. 2003. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J. Virol. 77:12921-12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sagar, M., X. Wu, S. Lee, and J. Overbaugh. 2006. HIV-1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Starcich, B. R., B. H. Hahn, G. M. Shaw, P. D. McNeely, S. Modrow, H. Wolf, E. S. Parks, W. P. Parks, S. F. Josephs, R. C. Gallo, et al. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637-648. [DOI] [PubMed] [Google Scholar]
- 79.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Troyer, R. M., K. R. Collins, A. Abraha, E. Fraundorf, D. M. Moore, R. W. Krizan, Z. Toossi, R. L. Colebunders, M. A. Jensen, J. I. Mullins, G. Vanham, and E. J. Arts. 2005. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J. Virol. 79:9006-9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsibris, A. M., M. Sagar, R. M. Gulick, Z. Su, M. Hughes, W. Greaves, M. Subramanian, C. Flexner, F. Giguel, K. E. Leopold, E. Coakley, and D. R. Kuritzkes. 2008. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 82:8210-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tuttle, D. L., C. B. Anders, M. J. Aquino-De Jesus, P. P. Poole, S. L. Lamers, D. R. Briggs, S. M. Pomeroy, L. Alexander, K. W. Peden, W. A. Andiman, J. W. Sleasman, and M. M. Goodenow. 2002. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res. Hum. Retroviruses 18:353-362. [DOI] [PubMed] [Google Scholar]
- 83.Veazey, R. S., R. J. Shattock, M. Pope, J. C. Kirijan, J. Jones, Q. Hu, T. Ketas, P. A. Marx, P. J. Klasse, D. R. Burton, and J. P. Moore. 2003. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 9:343-346. [DOI] [PubMed] [Google Scholar]
- 84.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 86.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 87.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, L., T. He, A. Talal, G. Wang, S. S. Frankel, and D. D. Ho. 1998. In vivo distribution of the human immunodeficiency virus/simian immunodeficiency virus coreceptors: CXCR4, CCR3, and CCR5. J. Virol. 72:5035-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]