Abstract
Human noroviruses (family Caliciviridae) are the leading cause of nonbacterial gastroenteritis worldwide. Despite the prevalence of these viruses within the community, the study of human norovirus has largely been hindered due to the inability to cultivate the viruses ex vivo and the lack of a small-animal model. In 2003, the discovery of a novel murine norovirus (MNV-1) and the identification of the tropism of MNV-1 for cells of a mononuclear origin led to the establishment of the first norovirus tissue culture system. Like other positive-sense RNA viruses, MNV-1 replication is associated with host membranes, which undergo significant rearrangement during infection. We characterize here the subcellular localization of the MNV-1 open reading frame 1 proteins and viral double-stranded RNA (dsRNA). Over the course of infection, dsRNA and the MNV-1 RNA-dependent RNA polymerase (NS7) were observed to proliferate from punctate foci located in the perinuclear region. All of the MNV-1 open reading frame 1 proteins were observed to colocalize with dsRNA during the course of infection. The MNV-1 replication complex was immunolocalized to virus-induced vesicle clusters formed in the cytoplasm of infected cells. Both dsRNA and MNV-1 NS7 were observed to localize to the limiting membrane of the individual clusters by cryo-immunoelectron microscopy. We show that the MNV-1 replication complex initially associates with membranes derived from the endoplasmic reticulum, trans-Golgi apparatus, and endosomes. In addition, we show that MNV-1 replication is insensitive to the fungal metabolite brefeldin A and consistently does not appear to recruit coatomer protein complex I (COPI) or COPII component proteins during replication. These data provide preliminary insights into key aspects of replication of MNV-1, which will potentially further our understanding of the pathogenesis of noroviruses and aid in the identification of potential targets for drug development.
Caliciviruses (family Caliciviridae) are small nonenveloped viruses which are 27 to 35 nm in diameter. They possess a single-stranded, positive-sense RNA genome of 7 to 8 kb. Four genera have been established in the Caliciviridae: Vesivirus, Lagovirus, Sapovirus, and Norovirus (NoV) (21). Sapoviruses and NoVs are enteric pathogens in animals (38) and humans (22, 48). Human NoV (type virus Norwalk) are the major cause of nonbacterial gastroenteritis outbreaks in adults worldwide.
In the United States it is estimated that there are more than 23 million NoV infections per year, constituting 60% of illness caused by enteric pathogens (2). The manifestation of acute gastroenteritis ranges from mild diarrhea to severe disease with vomiting and profuse diarrhea, leading to dehydration and death (39). Significantly, NoVs have been classified by the National Institute of Allergy and Infectious Disease as a class B bioterrorism agent because they are highly contagious, have a low infectious dose, are extremely stable, and are associated with debilitating illness.
Comparatively little is known about human NoV biology, and thus no specific treatments for human NoV infection are available (20). Human NoVs are difficult to study due to the lack of a reliable tissue culture system and a small animal model; thus, the molecular mechanisms of human NoV pathogenesis and immunity are not completely defined. In 2003, MNV-1 was discovered and used to study immunity and pathogenesis of NoV infection in a mouse model (24). Subsequently, we identified a tropism of MNV-1 for mononuclear cells in Stat-1−/− mice (52). This led to the discovery of the first in vivo culture system to study NoV replication—a significant advancement in NoV biology.
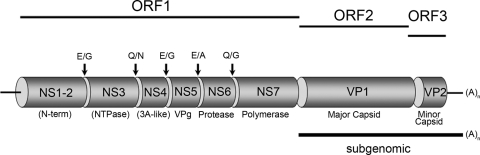
The NoV genome is protein linked at the 5′ end and polyadenylated at the 3′ end and is organized into three (possibly four) open reading frames (ORFs) (46) (Fig. 1). ORF1 encodes a large polyprotein which undergoes proteolytic cleavage by the viral proteinase (Pro) NS6 to produce the mature viral nonstructural proteins in the gene order 5′-NS1-2(N-term)-NS3(NTPase)-NS4(p20)-NS5(VPg)-NS6(Pro)-NS7(Pol)-3′ (14). The two structural proteins have been named VP1 for the major capsid protein and VP2 for the minor capsid protein (or small basic protein) and are encoded by ORF2 and ORF3, respectively (14). Recently, an additional ORF termed ORF4 has been identified within the subgenomic RNA; however, studies have not revealed a functional requirement for this ORF product, nor has an ORF4 product been identified in infected cells. The 5′ end of ORF2 is the most highly conserved region across all known NoV strains (27) and has been shown to be the site of recombination in this virus (11, 12). Independent coexpression of the NoV ORFs 2 and 3 leads to the formation of empty viruslike particles (23).
FIG. 1.
Genome organization of MNV-1. The MNV-1 genome is organized into three ORFs (ORF1 to ORF3). ORF1 encodes for six nonstructural proteins indicated that are cleaved by a virus-encoded cysteine protease (NS6) to produce mature viral proteins. Adjacent to ORF1 and partially overlapping is ORF2 and ORF3, which encode the major (VP1) and minor (VP2) structural proteins, which are produced from a subgenomic RNA species. Some of the identified roles for these proteins are shown, whereas the descriptions in brackets (i.e., NTPase function for NS3) are postulated.
Little is known regarding NoV replication. NoV genome replication is initiated at the 3′ end of the single-strand positive-sense RNA molecule and internally to generate an additional subgenomic positive RNA species. Positive-strand synthesis is initiated at a conserved GTGA nucleotide sequence preceding the first translation initiation codon and is shared by all three ORFs. Initiation of negative-strand synthesis of both genomic and subgenomic RNA occurs from the 3′ end of the positive sense RNA, a strategy invoked by all positive-sense RNA viruses. A subgenomic promoter within the newly synthesized negative sense RNA is recognized by the viral polymerase to generate the positive sense subgenomic RNA for translation of the structural proteins. The promoter site lies partly on the 20-nt overlap of ORF1 and ORF2 and has been identified as a recombination breakpoint (12).
In the present study we show that, like arguably all positive-sense RNA viruses, MNV-1 RNA replication is closely associated with vesicular clusters induced during virus infection. In addition, we show that most, if not all, of the MNV-1 ORF1 proteins are associated with the replication complex (RC). We show that the RC appears derived from cellular membranes originating from the endoplasmic reticulum (ER), some components of the Golgi apparatus, and endosomes. These findings highlight new observations in the viral manipulation of host cell organelles and processes.
MATERIALS AND METHODS
Viruses and cells.
RAW264.7 cells were grown and maintained in Dulbecco modified Eagle medium (Invitrogen, Australia) supplemented with 5% fetal calf serum (Lonza, Basel, Switzerland), 2 mM Glutamax (Gibco-BRL), and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Gibco-BRL). Cells were infected with MNV-1 strain CW1 at a multiplicity of infection (MOI) of 5, as previously described (24), and infected cells were maintained in Dulbecco modified Eagle medium containing 5% fetal calf serum, 2 mM Glutamax, and penicillin (100 U/ml)-streptomycin (100 μg/ml).
Reagents.
MNV specific guinea pig polyclonal antibodies have been described previously (43). Anti-rabbit, anti-guinea pig and anti-mouse specific immunoglobulin G (IgG) Alexa Fluor 488, 549, and 680 were purchased from Molecular Probes (Invitrogen, Leiden, The Netherlands). Anti-calnexin, anti-Giantin, and anti-GM130 were purchased from Merck-Calbiochem (Germany), anti-GalT was a generous gift from Eric Berger (University of Zurich, Zurich, Switzerland), anti-Sec23 was purchased from Sigma Aldrich (St. Louis, MO), anti-p23 was a generous gift from Robert Parton (University of Queensland, Brisbane, Queensland, Australia), anti-LAMP1 was purchased from BD Pharmingen (San Jose, CA), anti-EEA1 was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and anti-double-stranded RNA (anti-dsRNA; clone J2) was purchased from English & Scientific Consulting Bt. (Hungary). Protein A-gold (PAG; 5, 10, and 15 nm) was purchased from Utrecht University (Utrecht, The Netherlands). Brefeldin A (BFA) was purchased from Sigma Aldrich (St. Louis, MO).
Plaque assay.
Plaque assays were performed in six-well plates by seeding 106 RAW264.7 cells the day before and the next day infecting cell monolayers with dilutions of virus-containing culture fluids for 60 min. The cells were overlaid with 2 ml of medium containing 70% Dulbecco modified Eagle medium, 2.5% fetal bovine serum, 15 mM sodium hydrogen carbonate, 5 U of penicillin-streptomycin, 25 mM HEPES, 2 mM Glutamax, and 0.35% low-melting-point agarose/well, followed by incubation for 2 days at 37°C with 5% CO2. Cells were fixed by adding 1 ml of 4% formaldehyde/well directly onto the overlay, followed by incubation for 30 min at room temperature. Cells were rinsed with water and stained with 0.2% crystal violet for 20 min, and the plaques were counted.
Immunofluorescence analysis.
RAW264.7 cell monolayers on coverslips were infected with MNV-1 at an MOI of 5 and incubated at 37°C for different time periods postinfection. The cells were subsequently washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (ProSciTech, Kirwan, Australia) for 10 min at 20°C and permeabilized with 0.1% Triton X-100 in 4% paraformaldehyde for an additional 10 min at 20°C; the cells were washed with PBS, and aldehyde groups were quenched with 0.2 M glycine in PBS and additionally washed twice with PBS before incubation with antibodies. Alternatively, MNV-1-infected cells were fixed with 1:1 mixture of acetone and methanol, followed by incubation at −20°C for 10 min before being washed with PBS. Primary and secondary antibodies were incubated with blocking buffer (PBS containing 1% bovine serum albumin) and washed with PBS containing 0.1% bovine serum albumin between incubation steps. After a final wash with PBS, the coverslips were drained and mounted onto glass slides with a quick-dry mounting medium (United Biosciences, Brisbane, Australia) before visualization on a Zeiss LSM 510 META confocal microscope. Images were collected and collated for publication by using Adobe Photoshop software.
Western blotting.
MNV-1-infected or uninfected cells were lysed in 1× COP buffer (10 mM Tris-HCl [pH 8.2], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml), and proteins were separated on 4 to 12% precast Tris-Bis polyacrylamide gels (Invitrogen). Proteins were subsequently transferred to an Hybond-ECL nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) by using the Bio-Rad wet-transfer blotting module. The membrane was subsequently blocked with 5% skim milk powder (Diploma) in PBS containing 0.1% Tween 20 at room temperature. Incubation with the appropriate antibodies was performed in blocking buffer on a rotating wheel overnight at 4°C. The bound antibodies were subsequently visualized with species-specific IgG conjugated to Alexa Flour 680 (Invitrogen) or IRDye 800CW (Rockland, Inc.) and a Li-Cor Odyssey scanner. The resulting images were digitally scanned and processed in Adobe Photoshop for publication.
Electron microscopy.
The methods for cryofixation, preparation of cryosections, and immunolabeling have been described elsewhere (32-34). Briefly, MNV-1-infected cells were fixed with 4% paraformaldehyde-0.1% glutaraldehyde (ProSciTech) in PBS on ice for 60 min and subsequently embedded within a 10% gelatin block before postfixing with 1% paraformaldehyde in PBS. The sample blocks were infused with a mixture of sucrose and polyvinylpyrrolidone and mounted onto cryostubs (Leica) for cryosectioning. Ultrathin cryosections (∼55 nm) were cut with a Diatome Cryo-P diamond knife and retrieved from the cryo-chamber with a droplet of 14:1 2.3 M sucrose-2% methylcellulose. The recovered sections were subsequently immunolabeled with the appropriate antibodies and PAG and contrasted with 1.8 M methylcellulose containing 0.4% uranyl acetate. The sections were then viewed on JOEL 1010 transmission electron microscope, and images were captured on a MegaView III side-mounted charge-coupled device camera (Soft Imaging Systems) and processed for publication in Adobe Photoshop.
RESULTS
MNV-1 infection induces the formation of unique membranous structures which are associated with the replication complex.
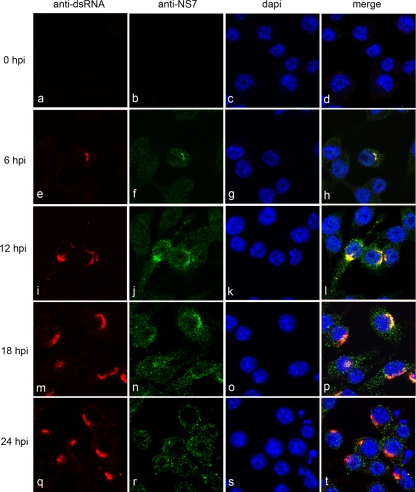
Previously, we showed that MNV-1 induces the formation and proliferation of membrane structures that are closely associated with virions (52). In order to determine the association of the viral RC with these structures, we first compared the subcellular localization of the MNV-1 RNA polymerase (NS7) with that of the replicative intermediate (dsRNA) during the course of infection (Fig. 2). RAW264.7 cells were infected with MNV-1 at an MOI of 5 and fixed at 0, 6, 12, 18, and 24 h postinfection (hpi). Cells were then dual-labeled with antibodies against both NS7 and dsRNA and analyzed by immunofluorescence microscopy. Replication could be detected early during the course of infection (6 hpi, Fig. 2e to h; see also Fig. S1 in the supplemental material), as evidenced by the presence of coincidental punctate cytoplasmic labeling of dsRNA and NS7 adjacent to the nucleus. As the course of infection progressed, this region of perinuclear labeling within infected cells increased in both size and density, with signs of cytopathic effect being detected as early as 18 hpi (see Fig. S2 in the supplemental material). Interestingly, although the localization pattern of dsRNA remained relatively constant throughout infection (Fig. 2e, i, m, and q), localization of NS7 appeared to become more diffuse within the cytoplasm as distinct punctate foci (Fig. 2f, j, n, and r). Although a subset of the labeling remained coincident during the course of infection, we currently cannot explain the change of localization of NS7 during the later stages of infection. It is highly probable that the drastic intracellular rearrangement of cellular ER relates to this anomaly. A major dispersion of the ER at later times postinfection is observed; therefore, it is not surprising to see a more diffuse staining of the protein on the ER. Viral protein translation will still be occurring at these later time points which will be distinct from genome replication. However, the pattern of localization observed during the course of infection correlates with the previously published EM data, which demonstrated the formation and proliferation of membrane structures adjacent to the nucleus.
FIG. 2.
The MNV-1 RC localizes adjacent to the nucleus in infected cells. RAW264.7 were infected with MNV-1 at an MOI of 5; fixed at 0, 6, 12, 18, and 24 hpi; and dual labeled with antibodies to dsRNA and MNV-1 RNA polymerase (NS7). Replication was detected at 6 hpi, as indicated by the presence of coincidental punctuate labeling adjacent to the nucleus. As infection progressed, this area of coincidental labeling increased in both size and density, with signs of cytopathic effect being detected as early as 18 hpi.
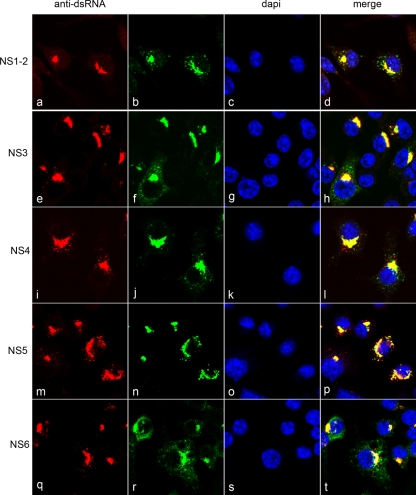
In order to determine whether any of the other nonstructural proteins were constitutively present within the RC, we analyzed the localization of these proteins with dsRNA throughout the course of infection in dual-labeled experiments (Fig. 3). RAW264.7 cells were infected with virus at an MOI of 5 and fixed at 12 hpi. Cells were then labeled with antisera against dsRNA and the MNV-1 nonstructural proteins (NS1-2, NS3, NS4, NS5, NS6, and NS7) and analyzed by immunofluorescence microscopy. All of the nonstructural proteins were found to colocalize with dsRNA, and all exhibited a localization pattern similar to that observed in dsRNA-NS7 dual-labeled experiments. NS3, NS4, and NS5 were consistently observed to show colocalization with anti-dsRNA labeling during the course of infection. This may suggest that these MNV-1 nonstructural proteins play a major role in MNV-1 RNA replication. Interestingly, we detected punctate cytoplasmic labeling of NS1-2, NS6, and NS7 that was not concurrent with dsRNA labeling, indicating a possible function for these proteins outside of the replication complex. However, these results strongly implicate all of the MNV-1 ORF1 proteins in replication of the MNV-1 RNA and as major components of the MNV-1 RC.
FIG. 3.
MNV-1 nonstructural proteins all exhibit a similar subcellular localization pattern in infected cells. RAW264.7 cells were infected with MNV-1 at an MOI of 5; fixed at 12 hpi; and dual labeled with antibodies specific for dsRNA and for MNV-1 nonstructural proteins NS1-2, NS3, NS4, NS5, and NS6. Antibodies were subsequently visualized with species-specific IgG conjugated to Alexa Fluor 488 or 594.
The MNV-1 RC is associated with vesicular clusters.
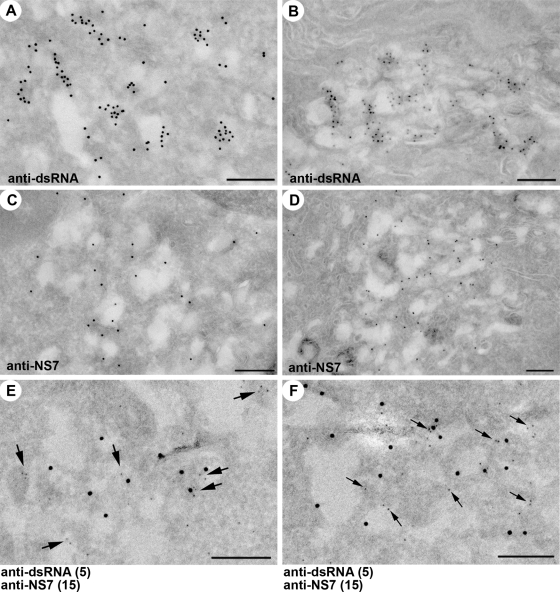
Subsequent to characterizing the RC in terms of viral protein composition, we analyzed immunolabeled cryosections of infected cells in an attempt to visualize the ultrastructure of the virus-induced membranes and gain insight into the topological relationship between the RC and these structures. Cryosections of RAW264.7 cells infected at an MOI of 5 and fixed at 12 hpi were immunolabeled with antibodies to dsRNA and NS7 in single- and dual-labeled experiments and analyzed by transmission electron microscopy (Fig. 4).
FIG. 4.
The MNV-1 RC associates with membrane vesicles in infected cells. Cryosections of RAW264.7 cells infected with MNV-1 at an MOI of 5 and fixed at 12 hpi (A, C, E, and F) or 24 hpi (B and D) were immunolabeled with anti-dsRNA antibodies and 10-nm PAG (A and B) or anti-NS7 antibodies and 10-nm PAG (C and D). In panels E and F, the cryosections from MNV-1-infected RAW264.7 cells were dual labeled with antibodies to dsRNA (5-nm PAG) and NS7 (15-nm PAG). Arrows in panels E and F highlight the smaller 5-nm gold particles. Magnification bars represent 200 nm in all cases.
In both single- and dual-labeled experiments, PAG labeling was shown to be concentrated primarily around the periphery of virus-induced membrane vesicles, with labeling of the outer and inner surface of the membrane being observed. In both single- and dual-labeling experiments the antibodies were observed to bind predominantly to the membrane of the vesicles, with very little labeling observed free within the cytoplasm of infected cells. These vesicles appeared as single-membrane structures ranging in diameter from approximately 100 to 300 nm, a characteristic typical of many virus-induced membrane structures produced during replication of positive-sense RNA viruses. This association of the MNV-1 RC with virus-induced membrane clusters is reminiscent of picornavirus infection (6-9) and consistent with similar membrane rearrangements observed for other positive-sense RNA viruses (18, 28, 29, 33, 49-51).
Replication of MNV-1 is associated with markers for the ER, Golgi apparatus, and endosomes.
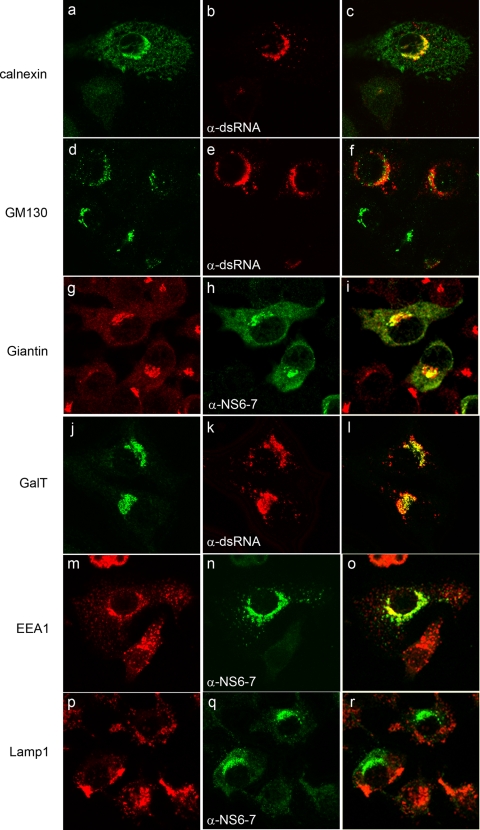
In order to identify the cellular composition of MNV-1-associated membranes we compared the localization of various cellular markers with one of two viral markers (dsRNA or NS6-7) in dual-labeling experiments and confocal microscopy (Fig. 5). Cell markers for the ER (calnexin), cis-Golgi body (GM130), cis/medial-Golgi body (Giantin), trans-Golgi body (GalT), early endosomes (EEA1), and lysosomes (LAMP1) were used in combination with dsRNA or NS6-7 markers based on species specificity (see also Fig. S3 in the supplemental material).
FIG. 5.
MNV-1 associates with markers for the ER, cis/medial-Golgi bodies, and endosomes in infection. RAW264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 12 hpi, and dual labeled with markers for the MNV-1 RC (dsRNA or NS7, shown in green) and various cellular membrane markers (shown in red). The MNV-1 RC was found to partially colocalize with markers for the ER (calnexin), the cis/medial-Golgi apparatus (Giantin) and endosomes (EEA1), visualized as a yellow hue, but showed very little colocalization with the cis Golgi (GM130) and no colocalization with lysosomes (LAMP1).
Markers for several organelles were found to partially, but not exclusively, colocalize with viral markers, including the ER (calnexin; Fig. 5a to c), cis/medial-Golgi (Giantin; Fig. 5g to i), trans-Golgi (GalT; Fig. 5j to l), and early endosomes (EEA1; Fig. 5m to o); however, no colocalization was detected in cells labeled with lysosomes (LAMP1; Fig. 5p to r), with only a very small amount of colocalization observed with the cis-Golgi (GM130; Fig. 5d to f). The distribution of calnexin and EEA1 was observed to become more concentrated in the perinuclear region in infected cells (Fig. 5a and m), whereas the distribution for GM130 and Giantin displayed some dispersion within the cytoplasm; however, virus replication did not appear to have a gross effect on Golgi morphology and localization (Fig. 5d and g). Interestingly, whereas partial colocalization between virus markers and the medial- and trans-Golgi markers Giantin and GalT was observed, we did not detect any strong coincidental labeling with the cis-Golgi marker GM130 and dsRNA (Fig. 5). This association with the medial- and trans-Golgi markers but not the cis-Golgi markers bodies may be dictated by the functions of these proteins or, alternately, may be the result of indirect interactions. The results do suggest that MNV-1 recruits membranes derived from late compartments within the secretory pathway (especially trans-Golgi bodies and endosomes) with some specificity. The recruitment of membranes derived from multiple cellular organelles and/or compartments shares similarity with that of the related picornaviruses (19, 31) and flaviviruses (32).
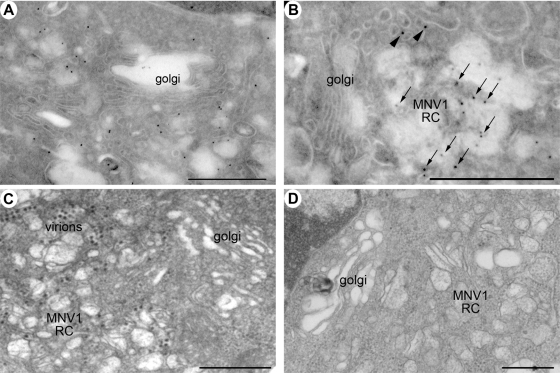
Ultrastructural electron microscopic studies revealed that the MNV-1 RC is positioned adjacent to the Golgi apparatus (Fig. 6). The exact relationship between the Golgi apparatus and MNV-1 replication is not completely understood since MNV-1 does not encode for glycoproteins nor does it dismantle the Golgi to impede protein secretion (cf. poliovirus infection). The images presented in Fig. 6 would suggest that this is the trans-Golgi or trans-Golgi network (TGN). In addition, our initial observations suggested that the Golgi apparatus was disrupted during MNV-1 infection (52). Based on our more detailed immunofluorescence analysis (described above), we undertook a more extensive ultrastructural analysis of potential changes in Golgi morphology during MNV-1 infection. The images presented in Fig. 6 suggest that this is the trans-Golgi apparatus or TGN since the immunofluorescence analysis presented in Fig. 5 indicates an involvement of GalT and endosomes during MNV-1 replication. These results imply that the MNV-1 RC is positioned near or adjacent to the Golgi apparatus, probably near the trans-Golgi body or TGN, during replication and can recruit membranes derived additionally from the ER and endosomes.
FIG. 6.
The MNV-1 RC localizes adjacent to the nucleus near the Golgi apparatus. Ultrastructual analysis of the MNV-1-infected cells at 12 hpi shows the positioning of the MNV-1 RC adjacent to the Golgi apparatus. (A and B) Cryosections immunolabeled with antibodies to MNV-1 NS7 and 10-nm PAG. The identified RC appears to lie in close proximity to an apparently intact Golgi body. (C and D) Resin-embedded sections from MNV-1-infected RAW264.7 cells, again suggesting a close association of the Golgi apparatus with MNV-1 RNA replication.
COPI is not required for MNV-1 replication.
It has been well established that members of the Picornaviridae exhibit differential requirements for coatomer protein complex I (COPI) in the formation of their replication complex (19). Due to the structural motif homology shared by members of the Picornaviridae and Caliciviridae, and our observations that some Golgi markers are recruited to the MNV-1 RC during replication, we sought to determine a possible role for COPI in MNV-1 replication by exploiting the properties of BFA, a fungal metabolite known to inhibit the formation of COPI-coated vesicles in an ARF-dependent manner (16, 30, 47).
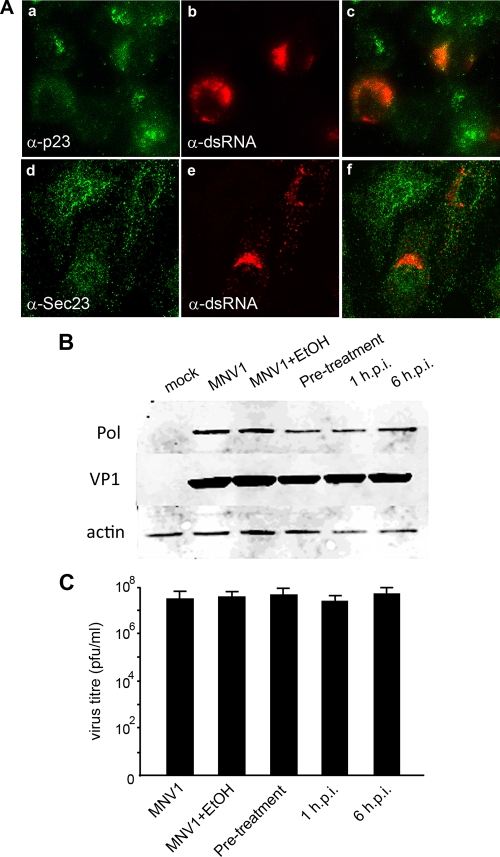
Initially, we investigated the intracellular distribution of both COPI and COPII in MNV-1-infected cells utilizing anti-p23 and anti-Sec23 antibodies, respectively (Fig. 7A). Both of these complexes regulate ER-to-Golgi transport via binding of the coatomer proteins and are directly or indirectly affected by BFA treatment (4, 17, 40, 41). As can be observed, both COPI and COPII did not appear to colocalize with the MNV-1 RC, nor were they redistributed during infection. Thus, it appeared that MNV-1 replication did not involve cellular constituents from the early secretory pathway. Even though we did not observe any involvement of COPI or COPII in MNV-1 replication, we sought to determine the effect of BFA treatment on intracellular replication. An effective BFA concentration, 1 μg/ml, that caused complete dispersion of the Golgi apparatus (see Fig. S3 in the supplemental material) was determined by titration and used to treat cells either prior to infection at 1 hpi or at 6 hpi. Immunoblots of cell lysates harvested subsequently at 12 hpi were labeled with antisera against both the MNV-1 NS7 and the capsid protein and examined for changes in the level of viral protein expression (Fig. 6B). In all samples treated with BFA, no significant difference in the level of either NS7 or capsid expression was detected compared to the untreated control. Subsequently, infected tissue culture fluids were harvested and assessed for production of infectious virions. As can be observed in Fig. 7C no significant loss of virus production was observed in any of the treated samples. These observations suggest that MNV-1 maturation is not dependent on an intact early secretory system.
FIG. 7.
MNV-1 replication is independent of COPI and COPII and insensitive to the action of BFA. (A) RAW264.7 cells were infected with MNV-1 at an MOI of 5, fixed at 12 hpi, and labeled with antibodies specific for dsRNA and for subunits of the COPI (p23; a to c) and COPII (Sec23; d to f) complexes. The antibodies were visualized with species-specific IgG conjugated to Alexa Fluor 488 or 594. No apparent colocalization was observed between dsRNA and either p23 or Sec23. (B) Western blot of lysates prepared from MNV-1-infected RAW264.7 cells that had been treated with BFA (1 μg/ml) prior to infection and at 0 and 6 hpi and cell lysates harvested at 12 hpi. The relative expression levels of MNV-1 NS7 and VP1 were assessed to determine the effect of BFA treatment on replication. None of the BFA-treated samples appear to show any significant difference in the level of either NS7 or VP1 expression. The expression level of the cellular protein actin served as a loading control. (C) Plaque assay of infected tissue culture fluid collected from MNV-1-infected RAW264.7 cells that were treated with BFA as described above at 24 hpi. No significant difference in the level of virus release was detected in any of the treated samples compared to the control.
These results suggest that, in contrast to the related picornaviruses poliovirus and echovirus 11, MNV-1 replication is insensitive to the action of BFA similar to encephalomyocarditis virus and foot-and-mouth-disease virus (26, 36, 37).
DISCUSSION
At present, relatively little is understood regarding the replication of NoVs, a fact largely due to the difficulty in cultivating these viruses and the lack of a small animal model. In the present study we characterized the subcellular localization of the MNV-1 replication complex in infected RAW264.7 cells and identified some host components associated with virus replication, providing preliminary insights into NoV replication.
We have previously demonstrated that MNV-1, like other single-stranded RNA viruses, induces the formation of unique host-derived membrane structures that are closely associated with the virion (52). Using immunofluorescence studies, we first analyzed the localization of the MNV1 proteins encoded by ORF1 and dsRNA, and the ultrastructural morphology of the replication complex in infected cells. Replicating virus was detected early during the course of infection (6 hpi) by both anti-dsRNA and anti-ORF1 antibodies and presented as small localized foci adjacent to the nucleus (Fig. 1). As the time of infection progressed, this region of labeling became larger and more dense within the perinuclear region, correlating with our previous electron microscopic data which shows an accumulation of virus-induced membrane structures over time adjacent to the nucleus (52). All of the six proposed nonstructural proteins encoded by ORF1 were found to associate with dsRNA within the RC and exhibited a very similar localization pattern, indicating a role for all nonstructural proteins in replication. Interestingly, we observed some punctate cytoplasmic labeling of the nonstructural proteins (primarily NS1-2, NS3, and NS6) that did not colocalize with the RC, implying that some of these proteins may not only play a direct role in replication but may also function outside of the RC in processes essential for replication such as membrane induction or cellular manipulation.
Closer observation of the ultrastructure of MNV-1-induced membranes by cryo-electron microscopic demonstrated the association of RC markers (MNV-1 NS7 and dsRNA) with single-membrane vesicles, which were again shown to accumulate adjacent to the nucleus. The immunogold labeling clearly shows the association of both NS7 and dsRNA around the periphery of these vesicles and with the membranes themselves and is reminiscent of the vesicular structures induced during picornavirus infection (8-10, 19, 31). Topologically, the labeling would suggest that replication occurs within the lumen of these vesicles, again analogous to what has been observed for other (+)RNA virus species (25, 28, 31). This topological conundrum would imply that the replicated RNA must pass through a membrane to undergo translation and packaging. In many cases, the presence of small pores facilitates this process, and we are currently undertaking a detailed ultrastructural analysis to identify such pores in the MNV-1-induced vesicles.
When analyzed by immunofluorescence, the viral RC was found to associate with cellular membrane markers for the ER (calnexin), Golgi bodies (Giantin and GalT), and early endosomes (EEA1), but not with late endosomes or lysosomes (LAMP1). Interestingly, whereas MNV-1 was found to associate with Giantin and GalT, no or minimal colocalization was observed between the RC and the GM130. Combined with our observations that COPI and COPII are more than likely not involved in MNV-1 RC formation and the apparent resistance of MNV-1 replication to BFA, it would appear that while MNV-1 utilizes components generated from the ER, cis/medial-Golgi bodies (Giantin), and trans-Golgi bodies (GalT), and endosomes (EEA1), the virus does not appear to utilize components associated with the cis-Golgi apparatus (GM130) or the intermediate compartment, including both COPI and COPII (Fig. 4). This is in stark contrast to what has been observed with certain members of the related Picornaviridae and provides a unique aspect to the intracellular replication of NoVs, which may be equally applicable to human NoVs. The association of MNV-1 with Giantin and GalT but not GM130 may simply indicate a functional requirement for select host proteins in virus replication that have not currently been identified. It is intriguing that both GM130 and Giantin compete for binding on the C terminus of the Golgi tethering protein p115. The current working model is that p115 acts as a “bridge” that connects COPI-containing vesicles to the cis face of the Golgi apparatus via interactions mediated by Rab1 (a GTPase) (5). Interestingly, there is evidence indicating that Giantin is found to interact with p115 on the COPI-containing vesicles (42), whereas p115 binds to GM130 on the Golgi stack. Due to our lack of coincidental staining of MNV-1 and COPI, we suggest that MNV-1 may be preventing the association of Giantin with COPI-positive vesicles. This hypothesis would then suggest that COPI-positive vesicles cannot dock to the Golgi apparatus, presumably inhibiting ER-to-Golgi transport (1) similar to poliovirus (13, 15) but in a BFA-independent manner. We are currently investigating further the involvement of the cis-Golgi body in MNV-1 replication and investigating whether this hypothesis may hold true.
In addition, we observed that the ER and early endosomal markers underwent significant redistribution during the course of infection, whereas the morphology of the Golgi apparatus appeared relatively unchanged. Strikingly, we saw a very close association of MNV-1 replication with the ER and endosomes. A cooperative involvement specifically of the ER and endosomes is not a novel observation during virus replication. Replication of HCV is observed to occur on clusters of membrane vesicles that contain calnexin and Rab5 (44), analogous to our observations with MNV-1. Our results and those with HCV, and many other positive-sense RNA viruses, strongly implicate the manipulation and involvement of multiple cellular organelles and membranes during virus replication. At this time, the nature of this involvement is unclear, but there is increasing evidence to indicate that the lipid composition of such membranes may play a greater role than the protein content (35, 45, 53, 54). However, some recent results have suggested a role for Rab5 in structuring the ER during membrane biogenesis (3). One of the implied roles for Rab5 in ER morphology was the GTPase-mediated control of membrane homotypic fusion to promote the formation of a membrane network rather than large membrane aggregates (3). Interestingly, if one takes an ultrastructural view of virus infection, one of the major hallmarks of infection is the massive proliferation of membrane aggregates that house the viral RC. Thus, it may not be that difficult to envisage a virus-induced manipulation of Rab5 activity to promote such membrane proliferations.
Another consideration is that MNV-1 may be trafficked via an alternate retrograde pathway that bypasses the Golgi apparatus shortly after entry of the incoming virions into infected cells. Many pathogens and/or associated pathogen products utilize retrograde movement to initiate infection in cells. The nonenveloped virus simian virus 40 (SV40) and many bacterial toxins undergo retrograde trafficking through the Golgi apparatus to the ER. Although all of these products eventually localize to the ER, different trafficking strategies are used to reach their destination. The transport of the Pseudomonas exotoxin A occurs from the TGN to the ER in a COPI- and Rab9-dependent manner, whereas Shiga toxin is transported via a COPI-independent but Rab6-dependent manner. In addition, cholera toxin appears independent of COPI, possibly utilizing specific lipid transport mechanisms. In contrast, SV40 reaches the ER bypassing the Golgi stack altogether and traffics directly from the caveosomal early sorting vesicle to the ER. However, SV40 delivery to the ER is sensitive to BFA, so some stages of this transport must be dependent on COPI (13, 14). Further investigation of the pathway utilized by MNV-1 may explain our observation that MNV-1 replicates independently of both COPI and COPII and is insensitive to BFA treatment.
In the present study we have shown that MNV-1 associates with some components of the early and late secretory pathway and replicates independently of COPI. However, the mechanism and pathway by which the virus establishes itself within the host cell still remains unclear. What is clear is that MNV-1 replication displays novel aspects to its replication cycle compared to some members of the Picornaviridae family and yet also shares strategies with other pathogenic positive-sense RNA viruses in the ability to manipulate internal cellular membranes to enrobe its replicating RNA. Further analysis of these virus-host interactions will provide valuable information into the replication of this family of understudied viruses.
Supplementary Material
Acknowledgments
This research was supported by research grants to J.M.M. from the National Health and Medical Research Council of Australia and to J.M.M. and H.W.V. from the National Institutes of Health (grants AI054483 and AI065982). This research was also supported, in part, by the Intramural Research Program of the NIAID, National Institutes of Health (K.Y.G. and S.V.S.).
We acknowledge Alexander Khromykh for advice during early stages of this work.
Footnotes
Published ahead of print on 8 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alvarez, C., R. Garcia-Mata, H.-P. Hauri, and E. Sztul. 2001. The p115-interactive proteins GM130 and Giantin participate in endoplasmic reticulum-Golgi traffic. J. Biol. Chem. 276:2693-2700. [DOI] [PubMed] [Google Scholar]
- 2.Atmar, R. L., and M. K. Estes. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. N. Am. 35:275-290. [DOI] [PubMed] [Google Scholar]
- 3.Audhya, A., A. Desai, and K. Oegema. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlowe, C., L. Orci, T. Yeung, M. Hosobuchi, S. Hamamoto, N. Salama, M. F. Rexach, M. Ravazzola, M. Amherdt, and R. Schekman. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77:895-907. [DOI] [PubMed] [Google Scholar]
- 5.Beard, M., A. Satoh, J. Shorter, and G. Warren. 2005. A cryptic Rab1-binding site in the p115 tethering protein. J. Biol. Chem. 280:25840-25848. [DOI] [PubMed] [Google Scholar]
- 6.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 7.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. 1994(Suppl. 9):147-157. [DOI] [PubMed] [Google Scholar]
- 8.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 10.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bull, R. A., G. S. Hansman, L. E. Clancy, M. M. Tanaka, W. D. Rawlinson, and P. A. White. 2005. Norovirus recombination in ORF1/ORF2 overlap. Emerg. Infect. Dis. 11:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bull, R. A., M. M. Tanaka, and P. A. White. 2007. Norovirus recombination. J. Gen. Virol. 88:3347-3359. [DOI] [PubMed] [Google Scholar]
- 13.Choe, S. S., D. A. Dodd, and K. Kirkegaard. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337:18-29. [DOI] [PubMed] [Google Scholar]
- 14.Clarke, I. N., and P. R. Lambden. 2000. Organization and expression of calicivirus genes. J. Infect. Dis. 181(Suppl. 2):S309-S316. [DOI] [PubMed] [Google Scholar]
- 15.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson, J. G., D. Finazzi, and R. D. Klausner. 1992. Brefeldin A inhibits Golgi membrane-catalyzed exchange of guanine nucleotide onto ARF protein. Nature 360:350-352. [DOI] [PubMed] [Google Scholar]
- 17.Duden, R., G. Griffiths, R. Frank, P. Argos, and T. E. Kreis. 1991. β-COP, a 110-kDa protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell 64:649-665. [DOI] [PubMed] [Google Scholar]
- 18.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gazina, E. V., J. M. Mackenzie, R. J. Gorrell, and D. A. Anderson. 2002. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J. Virol. 76:11113-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green, J., J. Vinje, C. I. Gallimore, M. Koopmans, A. Hale, D. W. Brown, J. C. Clegg, and J. Chamberlain. 2000. Capsid protein diversity among Norwalk-like viruses. Virus Genes 20:227-236. [DOI] [PubMed] [Google Scholar]
- 21.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 22.Hansman, G. S., N. Takeda, K. Katayama, E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Genetic diversity of sapovirus in children, Australia. Emerg. Infect. Dis. 12:141-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin, IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575-1578. [DOI] [PubMed] [Google Scholar]
- 25.Knoops, K., M. Kikkert, S. H. E. van der Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knox, C., K. Moffat, S. Ali, M. Ryan, and T. Wileman. 2005. Foot-and-mouth disease virus replication sites form next to the nucleus and close to the Golgi apparatus, but exclude marker proteins associated with host membrane compartments. J. Gen. Virol. 86:687-696. [DOI] [PubMed] [Google Scholar]
- 27.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 28.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200:307-312. [DOI] [PubMed] [Google Scholar]
- 30.Lippincott-Schwartz, J., L. C. Yuan, J. S. Bonifacino, and R. D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232-240. [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Improved membrane preservation of flavivirus-infected cells with cryosectioning. J. Virol. Methods 56:67-75. [DOI] [PubMed] [Google Scholar]
- 35.Mackenzie, J. M., A. A. Khromykh, and R. G. Parton. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229-239. [DOI] [PubMed] [Google Scholar]
- 36.Martín-Acebes, M. A., M. González-Magaldi, M. F. Rosas, B. Borrego, E. Brocchi, R. Armas-Portela, and F. Sobrino. 2008. Subcellular distribution of swine vesicular disease virus proteins and alterations induced in infected cells: a comparative study with foot-and-mouth disease virus and vesicular stomatitis virus. Virology 374:432-443. [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell, V. K., J. M. Pacheco, T. M. Henry, and P. W. Mason. 2001. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virology 287:151-162. [DOI] [PubMed] [Google Scholar]
- 38.Oliver, S. L., D. W. Brown, J. Green, and J. C. Bridger. 2004. A chimeric bovine enteric calicivirus: evidence for genomic recombination in genogroup III of the Norovirus genus of the Caliciviridae. Virology 326:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, M. M., A. J. Hall, J. Vinje, and U. D. Parashar. 2009. Noroviruses: a comprehensive review. J. Clin. Virol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Peter, F., H. Plutner, H. Zhu, T. E. Kreis, and W. E. Balch. 1993. Beta-COP is essential for transport of protein from the endoplasmic reticulum to the Golgi in vitro. J. Cell Biol. 122:1155-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serafini, T., G. Stenbeck, A. Brecht, F. Lottspeich, L. Orel, J. E. Rothman, and F. T. Wieland. 1991. A coat subunit of Golgi-derived non-clathrin-coated vesicles with homology to the clathrin-coated vesicle coat protein β-adaptin. Nature 349:215-220. [DOI] [PubMed] [Google Scholar]
- 42.Sonnichsen, B., M. Lowe, T. Levine, E. Jamsa, B. Dirac-Svejstrup, and G. Warren. 1998. A role for Giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol. 140:1013-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosnovtsev, S. V., G. Belliot, K.-O. Chang, V. G. Prikhodko, L. B. Thackray, C. E. Wobus, S. M. Karst, H. W. Virgin, and K. Y. Green. 2006. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 80:7816-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone, M., S. Jia, W. D. Heo, T. Meyer, and K. V. Konan. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thackray, L. B., C. E. Wobus, K. A. Chachu, B. Liu, E. R. Alegre, K. S. Henderson, S. T. Kelley, and H. W. t. Virgin. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 81:10460-10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai, S. C., R. Adamik, R. S. Haun, J. Moss, and M. Vaughan. 1993. Effects of brefeldin A and accessory proteins on association of ADP ribosylation factors 1, 3, and 5 with Golgi. J. Biol. Chem. 268:10820-10825. [PubMed] [Google Scholar]
- 48.Tu, E. T., R. A. Bull, G. E. Greening, J. Hewitt, M. J. Lyon, J. A. Marshall, C. J. McIver, W. D. Rawlinson, and P. A. White. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII. 4 variants 2006a and 2006b. Clin. Infect. Dis. 46:413-420. [DOI] [PubMed] [Google Scholar]
- 49.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin. 2004. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 89:11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye, J., C. Wang, R. Sumpter, Jr., M. S. Brown, J. L. Goldstein, and M. Gale, Jr. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.