Abstract
We have identified a spliced transcript that contains sequences from the HCMV UL29 and UL28 open reading frames. It contains amino-terminal UL29 sequences followed by UL28 sequences, and it includes a poly(A) signal derived from the 3′-untranslated region following the UL26 open reading frame. UL29/28 RNA is expressed with early kinetics, and a virus containing a FLAG epitope inserted at the amino terminus of UL29 expressed a tagged ∼79-kDa protein, pUL29/28, that was detected at 6 h postinfection. The virus also expressed a less-abundant tagged 41-kDa protein, which corresponds in size to a protein that could be produced by translation of an unspliced UL29/28 transcript. Consistent with this prediction, both unspliced and spliced UL29/28 transcript was present in RNA isolated from polysomes. FLAG-tagged protein from the UL29/28 locus accumulated within nuclear viral replication centers during the early phase of infection. Late after infection it was present in the cytoplasm as well, and the protein was present and resistant to proteinase treatment in partially purified preparations of viral particles. Disruption of the UL29/28 locus by mutation resulted in a 10-fold decrease in the levels of DNA replication along with a similar reduction in virus yield. Quantitative reverse transcription-PCR analysis revealed an ∼2-fold decrease in immediate-early gene expression at 4 to 10 h postinfection compared to the wild-type virus, and transient expression of pUL29/28 activated the major immediate-early promoter. Our results argue that the UL29/28 locus contributes to activation of immediate-early gene expression.
Human cytomegalovirus (HCMV) is a ubiquitous human pathogen and the prototypical member of the Betaherpesvirus family (26). HCMV infection is generally asymptomatic in healthy adults, but the virus causes disease in immunocompromised adults often leading to pneumonitis, retinitis, or hepatitis. It also is responsible for congenital infections that result in a range of neurological abnormalities. Like all herpesviruses, HCMV infection leads to life-long latency.
Lytic HCMV replication follows a coordinated series of events. The first viral gene products to function within an infected cell are virion tegument proteins (5, 20, 24, 43), some of which facilitate transcription of the immediate-early class of viral genes upon delivery of the DNA genome to the nucleus. As these immediate-early products accumulate, they help to establish a permissive environment for replication and activate expression of the early and late classes of viral genes (26). Proteins encoded by early genes are responsible for viral DNA replication, as well as regulating cellular responses to infection (46), and late proteins include virion constituents (26).
The 230-kbp HCMV genome potentially encodes about 200 open reading frames (ORFs) (30). These ORFs include several families of genes, including the US22 family (7). This gene family is conserved among betaherpesviruses, and in HCMV it is comprised of 13 members: UL23, UL24, UL26, UL28, UL29, UL36, UL43, IRS1, TRS1, US22, US23, US24, and US26 (7, 13, 15, 39). US22 family proteins contain four conserved sequence motifs of uncertain function, which consist of hydrophobic residues interspersed with charged amino acids. Some US22 family members (UL23, UL24, UL36, UL43, IRS1, and US22) are dispensable for HCMV replication in fibroblasts, while others (UL26, UL28, UL29, TRS1, US23, US24, and US26) are not essential but are required for optimal virus yields in these cells (16, 28, 37, 50). Several US22 family members in mouse CMV are required for efficient replication in mouse macrophages (25), suggesting that some of the HCMV family members that are dispensable for replication in fibroblasts might be required for replication in other cell types.
The HCMV US22 family members UL36 and TRS1 have been extensively studied, and their proteins influence cellular pathways similarly to their murine CMV orthologues (11, 47). pUL36 inhibits Fas-mediated apoptosis by blocking caspase-8 activation (44), and pTRS1 binds double-stranded RNA and inhibits activation of the protein kinase R-mediated antiviral response (9, 17). The US22 family members UL26, TRS1, IRS1, and US24 have been shown to influence immediate-early gene expression (16, 42, 45) and, consistent with this role, several US22 family members have been found in preparations of HCMV virions (2, 16, 37, 41, 48). TRS1 and UL26 have also been shown to facilitate virion assembly and influence the stability of viral particles (1, 23, 28).
We recently discovered by mass spectrometry an ∼79-kDa protein containing amino acid sequences derived from two US22 family members: UL28 and UL29 (27). We have now identified a spliced mRNA spanning the UL28 and UL29 ORFs. Disruption of this locus results in reduced immediate-early gene expression, and pUL29/28 can activate expression of the major immediate-early promoter (MIEP) within transfected cells. In addition, we demonstrate that pUL29/28 is packaged into HCMV virions. The transcriptional activity of pUL29/28 and its inclusion in virions gives it the potential to influence gene expression at the very start of infection.
MATERIALS AND METHODS
Cell culture, viruses and plasmids.
Primary human foreskin fibroblasts (HFF) and human MRC-5 embryonic lung fibroblasts cells were propagated in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Wild-type virus, BADwt, was derived from an infectious bacterial artificial chromosome (BAC) clone of the AD169 strain of HCMV, termed pAD/Cre (51). The UL28- and UL29-deficient mutants, BADsubUL28 (originally termed DH54) and BADsubUL29 (originally termed DH56) were produced from the BAC clone and have been described previously (50). Viruses were propagated on fibroblasts and concentrated 10-fold by centrifugation through a sorbitol cushion (20% d-sorbitol, 50 mM Tris-HCl [pH 7.2], 1 mM MgCl2), and titers (infectious units) were determined by using an immunofluorescence assay to quantify pUL123 (IE1)-expressing cells (46). Briefly, serial dilutions of virus samples were plated on MRC-5 fibroblasts, and cells were fixed and permeabilized in methanol at −20°C for 15 min at 36 h postinfection (hpi). pUL123-positive cells were labeled by using a primary mouse antibody to IE1 (1B12) (16) and a secondary goat anti-mouse antibody conjugated with Alexa Fluor 546 or 488 (Molecular Probes), and positive cells were counted by using a fluorescence microscope. Cell-free virus was collected from supernatants of infected cells, and cell-associated virus was prepared by three rounds of freezing and thawing.
The BADinUL29F virus, which expresses pUL29 with an N-terminal FLAG tag, was constructed in the BAC clone of AD169 (51) by using galK selection and counter selection “recombineering” protocols (49) as modified for application to HCMV BAC clones (29). Briefly, PCR products were generated with primers containing both galK sequences and HCMV flanking sequence, which targeted recombination of the amplified galK expression cassette to the desired location on the viral genome. The targeting HCMV-specific primers were 5′-ACTGCTGCTTCTGCTTTTTTGTCTCCTGTGGATCGTCGCGGACTGCCGGCcctgttgacaattaatcatcggcg a-3′ and 5′-GACGAGGAGGAAGACGCCGTGGCCGCCGAGCAGCCCTTGCGACGGCCGGAtcagcactgtcctgctcctt-3′ (capitalized nucleotides correspond to the HCMV sequence, and lowercase nucleotides are galK specific). Competent AD169-BAC-containing SW102 cells were transformed with the HCMV-galK PCR product and selected for growth on M63 minimal salt agar plates supplemented with chloramphenicol and 0.2% galactose. AD169-BAC clones that underwent homologous recombination were screened for growth on MacConkey medium supplemented with galactose, and positive colonies were picked and further screened by PCR to confirm the proper location of the galK insert. Clones that passed the screens were next transformed with linear DNA comprised of three repeats of the FLAG epitope tag coding sequence flanked by the HCMV sequences bracketing the galK integrated sequence. This DNA was generated by annealing the following oligonucleotides: 5′-ACTGCTGCTTCTGCTTTTTTGTCTCCTGTGGATCGCGCGGACTGCCGGCatggactacaaggatgacgacgataaggcagattataaggatgacga-3′ and 5′-GACGAGGAGGAAGACGCCGTGGCCGCCGAGCAGCCCTTGCGAC GGCCGGActtatcatcatcgtctttatagtcggctttgtcatcgtcatccttataat-3′ (with lowercase nucleotides representing the FLAG sequences) and filling in the ends using a high-fidelity polymerase. Recombinants were selected by growth on M63 minimal salt agar plates containing 0.2% 2-deoxygalactose. DNA was isolated and transferred by electroporation into HFF cells (51) to produce BADinUL29F virus. The BADinUL28F virus expresses UL28 with a C-terminal FLAG epitope tag. It was constructed using the galk primers 5′-TGTTTTCGGGCCGTAGCGTGCTGGATAGCGTGAGCGGCACGGGGGCGTCGcctgttgacaattaatcatcggc a-3′ and 5′-GCGGTAAAGCCAGACACCGGCTATATAGCTAGTCATCACAGTCTCCTCCTtcagcactgtcctgctcctt-3′ and the FLAG primers 5′-TTTTCGGGCCGTAGCGTGCTGGATAGCGTGAGCGGCACGGGGGCGTCGgactacaaggatgacgacgataaggcagattataaggatgacgatgacaaag-3′ and 5′-AAAGCCAGACACCGGCTATATAGCTAGTCATCACAGTCTCCTCCTTtcacttatcatcatcgtctttatagtcggctttgtcatcgtcatccttataatct-3′ as described for BADinUL29F.
The MIEP-luciferase plasmid contains 1.1 kb of the MIEP sequence between nucleotide (nt) 74978 and nt 173724 from HCMV strain AD169 (accession no. X17403) using the primers 5′-CTGGAATACGACAAGATAACCCG-3′ and 5′-GGCAGAGATCTGACGGTTCACTAAACGAGCTCTG-3′. The PCR amplicon was digested with MluI and BglII and introduced into the pGL3-Basic (Promega). The expression vector for pUL29/28 was constructed by PCR using the primers for UL29 (5′-ATATCTAGATCCGGCCGTCGCAAGG-3′ and 5′-cacgacgagcCTACGCTTTTTGAACG-3′) and UL28 (5′-gcgtagGCTCGTCGTGGGCTAC-3′ and 5′-ATAGGATCCTCACGACGCCCCCGTGCCGC-3′) (lowercase sequences represent the overlap between genes). UL29 and UL28 amplicons were combined and the UL29/28 sequence was amplified by using outer primers. The product was digested using XbaI and BamHI and introduced into pCGN in-frame with the amino-terminal hemagglutinin (HA) epitope tag. UL29/28 sequence was verified by sequencing analysis. The UL29 gene was amplified by using 5′-ATATCTAGATCCGGCCGTCGCAAGG-3′ and 5′-ATAGGATCCTAAACGCTTTTTGAACGGCAGTC-3′ and introduced into pCGN.
Analysis of viral RNA and DNA.
For 3′RACE (3′ rapid amplification of cDNA ends) analysis, ∼5 × 106 confluent HFF cells were infected at a multiplicity of 3 infectious units/cell, and 6 or 24 h later the total RNA was prepared with TRIzol reagent (Invitrogen) and treated with DNA-free reagent (Ambion). The purified RNA was subjected to 3′RACE analysis (Clontech) in which the two gene-specific primers were located inside the UL28 ORF: 5′-AATCTGCTGCGCGTATGTCAG-3′ and 5′-CACGCCGGTAGCAAGATCCG-3′. The RACE fragments generated by PCR were cloned into pGEM-T Easy (Promega) and screened for UL28 sequences by using PCR, and the positive clones were then sequenced.
RNA levels in infected cells were determined by quantitative real-time reverse transcription-PCR (qRT-PCR) (46). Total RNA was isolated by using TRIzol reagent (Invitrogen), and contaminating DNA was removed by using DNA-free reagent (Ambion). cDNAs were synthesized with TaqMan reverse transcription reagents and random hexamers according to the manufacturer's instructions (Applied Biosystems). Real-time PCR was completed with SYBR green PCR master mix (Applied Biosystems) and primers specific to UL28 (5′-GGATGGATGGAACCCGTGAACA-3′ and 5′-ACGAAACCAGAAGGAGCCCTGAC-3′), UL29 (5′-CCGATGCTCTCGTAGCGAAAGTC-3′ and 5′-GCTGGTGGGGCAGGATAAGTTG-3′), UL83 (5′-GGGACACAACACCGTAAAGCCG-3′ and 5′-CGTGGAAGAGGACCTGACGATGAC-3′), UL123 (5′-GCCTTCCCTAAGACCACCAAT-3′ and 5′-ATTTTCTGGGCATAAGCCATAATC-3′), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-ACCCACTCCTCCACCTTTGAC-3′ and 5′-CTGTTGCTGTAGCCAAATTCGT-3′). For amplification across the splice junction between UL29 and UL28, RT-PCR was carried out using RNA isolated from infected cells as described above with the primers UL29 (5′-TTTCGCTACGAGAGCATCGGC-3′) and UL28 (5′-TTGTGGGTCCAGGCATCACG-3′). Additional primers used for qRT-PCR included UL26 (5′-TCATCGGCACCATCGGACTC-3′ and 5′-GCGCACTACAAAGTTCTCACGGC-3′), UL30 (5′-AGCGGAGGGGGAGAATAAACAGTT-3′ and 5′-GGGCAACACAAGATAGGGAAATACAA-3′), UL32 (5′-GGTTTCTGGCTCGTGGATGTCG-3′ and 5′-CACACAACACCGTCGTCCGATTAC-3′), UL37x1 (5′-GACGAAGTCCGATGAGGAGGATG-3′ and 5′-TGGGACACTGGGCGTTGTTG-3′), UL44 (5′-TACAACAGCGTGTCGTGCTCCG-3′ and 5′-GGCGTGAAAAACATGCGTATCAAC-3′, UL54 (5′-CCCTCGGCTTCTCACAACAAT-3′ and 5′-CGAGTTAGTCTTGGCCATGCAT-3′), UL83 (5′-GGGACACAACACCGTAAAGCCG-3′ and 5′-CGTGGAAGAGGACCTGACGATGAC-3′, UL99 (5′-GTGTCCCATTCCCGACTCG-3′ and 5′-TTCACAACGTCCACCCACC-3′), US3 (5′-TTCCACTCGAAATAGGCTCCGC-3′ and 5′-CGAGAAACACTTTGTGAACGTGGG-3′), and UL27 (5′-ATCAGGCTGTTTAAAGGCGAGGCT-3′ and 5′-AAAGTCGCAGAAGGTCTCCACGAA-3′).
Polysomes were isolated from HCMV-infected cells at 24 and 72 hpi as previously described (3, 21). Briefly, 5 × 106 HFF cells were infected with wild-type virus at a multiplicity of 3.0 infectious units/ml and, 10 min prior to sample isolation at the indicated time points, the cultures were treated with 100 μg of cycloheximide/ml in media. Cells were washed once with phosphate-buffered saline (PBS) containing cycloheximide, collected by scraping and centrifugation at 2,000 rpm for 5 min in PBS containing cycloheximide, and resuspended in 2 ml of lysis buffer (15 mM Tris [pH 7.4], 150 mM NaCl, 1.5 mM MgCl2, 10 mM dithiothreitol, 1% Triton X-100, protease inhibitors, and 50 U of RNasin [Promega]/ml) containing cycloheximide. A control sample was prepared in the absence of cycloheximide and using 50 mM EDTA to dissociate polysomes. Samples were incubated at 4°C for 10 min, and cells were disrupted by using a Dounce homogenizer. Nuclei were removed by centrifugation at 3,000 × g for 5 min at 4°C. Supernatants were subjected to centrifugation at 14,000 rpm for 10 min at 4°C to remove particulate material. Samples were loaded onto a 10 to 50% sucrose gradient and separated by centrifugation at 35,000 × g for 3 h at 4°C. Fractions of 500 μl were collected, and RNA was isolated from 250 μl by using TRIzol reagent (Invitrogen) as described above. Polysomal fractions were determined by separating RNA fractions by agarose gel electrophoresis and identifying fractions containing 18S and 28S rRNA bands in the cycloheximide-treated sample relative to the equivalent fractions from the EDTA-treated control.
Virus DNA was quantified by using qPCR. DNA was prepared from virus particles starting with 100 μl of cell-free virus stock or virus stock that was partially purified by centrifugation through a 20% sorbitol cushion and resuspended in PBS. To remove free DNA, samples were pretreated with DNA-free reagents (Ambion) and then lysed in lysis buffer (400 mM NaCl, 10 mM Tris [pH 8.0], 10 mM EDTA, 0.1 mg of protease K/ml, and 0.2% sodium dodecyl sulfate [SDS]) at 37°C overnight. To isolate viral DNA from infected cells, cells were harvested, pelleted, resuspended in lysis buffer, and incubated at 37°C overnight. DNA from either source was then extracted with phenol-chloroform, treated with RNase A, extracted again with phenol-chloroform, precipitated with ethanol, resuspended in water, and quantified by qPCR using SYBR green (Applied Biosystems) and primers specific for the UL123 gene or cellular β-actin (46).
Analysis of viral proteins.
For immunofluorescence, cells grown on glass coverslips were washed once with PBS and fixed in 2% paraformaldehyde for 15 min. Fixed cells were washed twice with PBS and permeabilized with 0.1% Triton X-100 for 15 min. Cells were washed twice with PBS, incubated in blocking solution (2% bovine serum albumin in PBS) for 30 min, and then labeled with the primary antibody in blocking solution for 1 h. Cells were washed again and then incubated with the secondary antibody in blocking solution for 1 h. Cells were then washed three times with PBS, rinsed once with H2O, and mounted on slides with Slow-Fade solution containing DAPI (4′,6′-diamidino-2-phenylindole) to counterstain the DNA (Molecular Probes). Images were captured by using a Zeiss LSM510 confocal microscope and Nikon Eclipse Ti. The antibodies used for immunofluorescence were an anti-FLAG M2 (Sigma) and anti-HA (Sigma) and a secondary goat anti-mouse antibody conjugated with Alexa Fluor 546 or 488 (Molecular Probes).
To analyze proteins by Western blotting, samples were mixed with sample buffer (0.05 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.1% bromophenol blue, 1.25% [vol/vol] 2-mercaptoethanol) and separated by electrophoresis in an SDS-containing 10% polyacrylamide gel. Resolved proteins were transferred to membranes and blocked with 5% nonfat milk in Tris-buffered saline containing 0.05% Tween 20. Membranes were incubated with a secondary goat anti-mouse antibody (Amersham) and visualized by ECL-Plus detection (Amersham) and a Molecular Dynamics Storm phosphorimager. Proteins were detected with the primary antibodies to pUL69 (1E12 [24]), ppUL83 (8F5 [35]), pUL44 (Virusys), pUL123 (1B12), FLAG M2 (Sigma), pUL99 (10B429), HA (Sigma), and tubulin (Sigma).
To evaluate the protein constituents of virus particles, virus was centrifuged through a sorbitol cushion and then dissolved in 500 μl of TN buffer (50 mM Tris [pH 7.4], 100 mM NaCl). Undiluted virus and serial dilutions of the pellet were frozen at −80°C and subsequently assayed by Western blotting. Some virion samples were treated with protease prior to analysis of their protein constituents. For protease treatment, aliquots (5 μl) of virus were treated with trypsin (25, 40 or 65 μg/ml), in the presence or absence of Triton X-100 (1%) and incubated at 37°C for 1 h before analysis.
Luciferase assays.
293 cells were seeded onto 12-well plates using 5 × 104 cells per well and transfected using Fugene 6 (Roche) according to the manufacturer's instructions. Cells were transfected with 50 ng of pGL3-MIEP reporter plasmid and 10, 100, or 500 ng of pCGN empty vector, pCGN-pUL29/28, or pCGN-pUL29 effector plasmid. Luciferase activity was assayed 48 h posttransfection using a luciferase reporter assay system (Promega) and a Victor3 luminometer (Perkin-Elmer) according to the manufacturers' instructions. Luciferase activity was measured by using equal protein amounts within each lysate and normalized to the luciferase activity from empty vector. Expression of pUL29/28 and pUL29 was assayed by Western blot analysis using the same lysates.
RESULTS
UL28 and UL29 ORFs are expressed from a spliced mRNA.
The AD169 UL28 and UL29 ORFs are transcribed from the same DNA strand between nt 35893 and nt 34754 and between nt 37005 and nt 35924, respectively, with UL29 residing upstream of UL28 (Fig. 1A, top) (7). The UL29 ORF begins with an AUG and potentially encodes a 360-amino-acid protein, while the UL28 ORF potentially encodes a 379-amino-acid protein but lacks a starting methionine. Since we knew that amino acid sequence from UL29 and UL28 are contained in one polypeptide (27), we searched for potential RNA splicing motifs and identified a consensus splice donor site at nt 35927 within the UL29 gene and a consensus acceptor site within the UL28 gene at nt 35780 (Table 1).
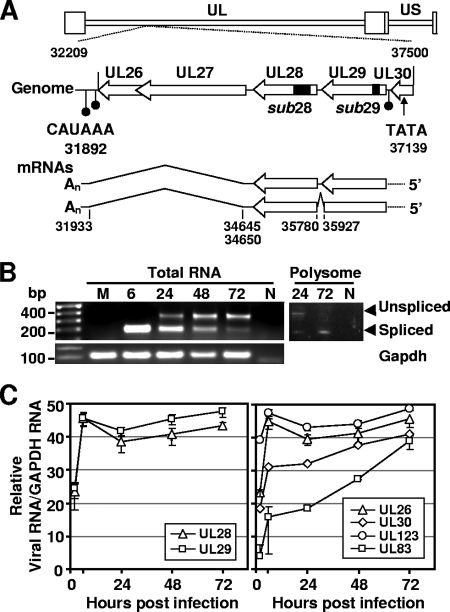
FIG. 1.
HCMV-coded transcripts containing the UL28 and UL29 genes. (A) Location of the UL28 and 29 ORFs on the viral genome and organization of their transcripts. The top of the panel diagrams the coding region showing the location of the UL26-UL30 ORFs within the viral chromosome. The coding sequences are indicated by open arrows with the C terminus of each ORF indicated by an arrowhead. Also indicated are the locations of three polyadenylation signals (filled circles) and a putative TATA box element (arrow). The position of deletions in substitution mutations within the UL28 and UL29 genes of the recombinant viruses, BADsubUL28 and BADsubUL29, are indicated by filled boxes. The bottom of the panel shows UL28 and UL29-related mRNAs with the location of splice donor and acceptor sites noted. Splice sites were identified by 3′ RACE analysis using RNA isolated at 6 and 24 hpi from fibroblasts infected with wild-type HCMV. The nucleotide numbers are from GenBank accession no. X17403. (B) Expression of unspliced and spliced transcripts during infection. Total RNA was isolated at indicated times from fibroblasts infected at a multiplicity of 0.5 infectious unit/cell with wild-type virus. RT-PCR was performed using primers that span a predicted splice donor at nt 35927 and an acceptor at nt 35780 and produced products corresponding to both spliced (188 nt) and unspliced (334 nt) cDNAs. Primers to GAPDH were used for a loading control. PCR products for both spliced and unspliced sequences were also observed using cDNA prepared from purified polysomes but not in the non-RT (N) control. (C) Accumulation of RNAs containing UL28 and UL29 sequences after infection with wild-type virus. Fibroblasts were infected under the conditions described above. Total RNA was isolated at the indicated times after infection, and RNA was quantified by real-time RT-PCR using primers specific to UL28 and UL29 and normalized to GAPDH RNA. RNAs containing the UL26, UL30, UL123, and UL83 ORFs were analyzed as controls for the rate of accumulation of viral RNAs from different kinetic classes.
TABLE 1.
Identified splice donor and acceptor sites in the UL26-to-UL29 regiona
| Splice ORFb | Donor site
|
Acceptor site
|
Junction (coding potential) | Poly(A) addition (nt)c | ||
|---|---|---|---|---|---|---|
| nt | Sequence | nt | Sequence | |||
| UL29^UL28* | 35927 | CGT AG^G TGA GTC | 35780 | GAC AG^G CTC GTC | CGT AG-G CTC GTC (Arg Arg Leu Val) | NA |
| UL27^UL26 3′UTR† | 34645 | CCC GTG G^AT CAG | 31933 | GGGTGGT^GGGGATC | CCC GTG G-GGGGATC (NA) | 31874 (CAUAAA nt 31892) |
| UL27^UL26 3′UTR† | 34650 | ATG AAC CC^C GTG | 31933 | GGGTGGT^GGGGATC | ATG AAC CC-GGGGATC (NA) | 31874 (CAUAAA nt 31892) |
The indicated nucleotide is underlined in each sequence. The nucleotide numbers are from GenBank accession no. X17403: UL26 (nt 32775 to 32209), UL27 (nt 34657 to 32831), UL28 (nt 35893 to 34754), and UL29 (nt 37005 to 35923). The exon is indicated in boldface. NA, not applicable.
*, Identified by PCR and sequence analysis; †, identified by 3′RACE analysis.
The poly(A) signal sequence is indicated in parentheses.
To test for a spliced mRNA, PCR primers were used to amplify sequences from cDNA preparations that were generated at various times after infection of fibroblasts. The primers were designed so that cDNAs derived from unspliced and spliced RNAs would produce 334- and 188-nt products, respectively. Both products were observed by using standard PCR throughout the time course (Fig. 1B, left panel), indicating that both unspliced and spliced RNAs were present. In addition, we observed both the spliced and the unspliced products using cDNA prepared from RNA that was isolated from purified polysomes (Fig. 1B, right panel). This result argues that both RNAs are translated and rules out the possibility that the unspliced product is simply the result of amplification from pre-mRNA sequences. DNA sequence analysis of the spliced PCR-generated product confirmed the existence of a splice junction at the predicted site (Table 1 and Fig. 1A, bottom). This spliced mRNA would produce a 701-amino-acid protein coded by the UL29 and UL28 ORFs (Fig. 2A), and the unspliced mRNA would produce a 360-amino-acid UL29 protein.
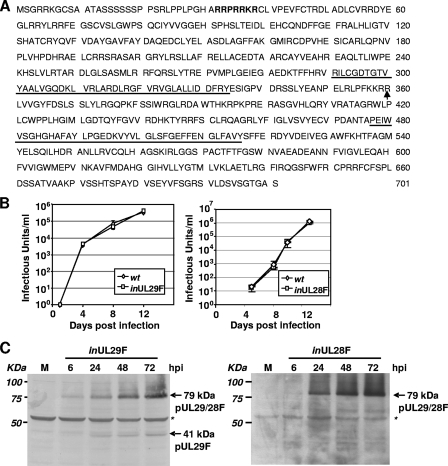
FIG. 2.
Early expression and predominantly nuclear localization of HCMV pUL29/28. (A) Amino acid sequence of HCMV strain AD169 pUL29/28 encoded by the spliced transcript encoding the UL29 and UL28 genes. The location of the junction between UL29 and UL28 is indicated by an arrow. A putative nuclear localization signal is in boldface letters, and the US22 family domains found in both UL29 and UL28 are underlined. (B) Virus expressing epitope-tagged pUL29/28 grows with normal kinetics. Replicate cultures of fibroblasts were infected at a multiplicity of 0.5 infectious unit/cell with wild-type HCMV (wt) or a recombinant virus containing a FLAG epitope at the N terminus of the UL29 ORF (inUL29F) and at the C terminus of the UL28 ORF (inUL28F). Culture supernatants were harvested at the indicated times, and the infectious virus progeny was quantified. (C) Accumulation of tagged proteins in cells infected with inUL29F and inUL28F viruses. Fibroblasts were infected at a multiplicity of 3.0 PFU/cell, harvested at the indicated times, and processed for Western blot assay using a FLAG-specific antibody. A nonspecific band (asterisks) was monitored to confirm equal protein loading.
In order to identify the 3′ ends of RNAs containing the UL29/UL28 ORF, we performed 3′RACE with primers specific to UL28 to amplify cDNA isolated at 6 and 24 hpi. Sequence analysis of multiple clones located the 3′ end of the transcript at nt 31874. The sequences also identified a second splicing event near the 3′ end of the mRNAs: two splice donor sites (one just downstream of the UL28 stop codon at nt 34650 and another just within the 5′ end of the UL27 ORF at nt 34645) and a common splice acceptor site within the UL26 3′-untranslated region (3′UTR) at nt 31933 (Table 1). The UL26 gene is downstream of UL28 and expressed in the same orientation (Fig. 1A, top). Although earlier work identified a UL26-specific AUUAAA polyadenylation signal at nt 32103 (45), it falls within the intron specified by the splice acceptor identified within the UL26 3′UTR. A putative CAUAAA polyadenylation signal is present at nt 31892, 18 nt from the site of poly(A) addition identified in the sequence of the 3′RACE products (Table 1) and within the range expected for the motif to direct processing (8). In sum, our analysis has identified two mRNAs that contain the UL29 and UL28 ORFs (Fig. 1A, bottom). One is spliced to generate a combined UL29/28 coding region, and the other does not contain a splice at this position. Both are spliced near their 3′ end so that they utilize a poly(A) addition signal downstream of UL26.
Previous studies have identified multiple RNAs expressed within the locus spanning from UL29 to UL26 during the early and late phases of infection (6, 45). We used qRT-PCR with primers specific for either UL29 or UL28 to assay DNase-treated total RNA isolated at various times after infection at a multiplicity of 0.1 infectious unit/cell (Fig. 1C). RNA containing UL29 or UL28 was detected at 2 hpi, accumulated to maximal levels by 6 hpi, and remained at that level until 72 hpi, the last time assayed. Probes to the two ORFs detected the accumulation of RNA with identical kinetics, which is consistent with our discovery that the two ORFs are carried on the same RNA. Similar kinetics were evident for accumulation of immediate-early UL123 RNA, and accumulation of early-late UL83 RNA occurred with a delay as expected (Fig. 1C). The RNAs detected at 2 hpi might have been delivered to cells by infecting virions, which contain numerous virus-coded RNAs (7, 51).
pUL29/28 is a 79-kDa protein localized to the nucleus and cytoplasm of infected cells.
The identification of the in-frame splicing event (Table 1) between the UL29 and UL28 ORFs predicts that infected cells contain a 701-amino-acid protein, pUL29/28. Previous in silico structural and functional analysis failed to identify substantial homologies within the UL28 or UL29 protein sequences to mammalian proteins (34). However, we searched for nuclear localization sequences (NLS) in the primary sequence by using two different algorithms, PSORT II (36) and Predict NLS (15) and identified a putative NLS signal, RRPRRKR, at amino acids 32 to 38 (Fig. 2A). This sequence has been previously identified and demonstrated to function as an NLS in the mammalian Sox2 transcription factor (22) and suggests that pUL29/28 accumulates within the nuclei of infected cells. In addition to the putative NLS, the UL29 and UL28 protein sequences contain two US22 domains, and these motifs are also noted in Fig. 2A.
In order to detect expression of the putative pUL29/28 protein, we introduced a FLAG epitope tag at the amino terminus of UL29 within the AD169 strain of HCMV to produce BADinUL29F. The growth of the tagged virus was compared to BADwt after infection of fibroblasts at a multiplicity of 0.1 infectious unit/cell, and the variant grew with the same kinetics as its parent (Fig. 2B). This demonstrates that the introduction of the FLAG epitope at the amino terminus of the UL29 ORF maintains the function of the protein during viral infection, since previous studies have shown that disruption of the ORF results in reduced virus growth (50). We also introduced the FLAG epitope at the carboxyl terminus of the UL28 ORF, and this variant grew with the same kinetics as the parental virus (Fig. 2B).
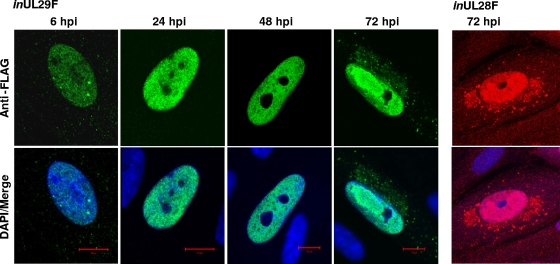
Using Western blot analysis, we detected an ∼79-kDa protein in cells infected with BADinUL29F (Fig. 2C), and this is the predicted size of FLAG-tagged pUL29/28 that would be expressed from the RNA with spliced UL29/28 ORFs (Fig. 1A, bottom). Expression of pUL29/28 was observed as early as 6 hpi and continued throughout infection (Fig. 2C), and this is consistent with the kinetics of RNA expression (Fig. 1C). In addition, we observed a smaller (41-kDa), less-abundant, FLAG-tagged protein specific to infection, which was not detected at 6 h but evident at 24 hpi. The polypeptide migrates at the predicted size for pUL29 expressed from the RNA containing the unspliced UL29 and 28 ORFs (Fig. 1A, bottom), which we found to be associated with polysomes (Fig. 1B, right panel). A nonspecific band was monitored to demonstrate that equal protein was loaded for each sample (Fig. 1C, asterisk). In infections using BADinUL28F, we also detected a 79-kDa protein beginning at 6 hpi (Fig. 2C). However, the smaller 41-kDa band was not observed, as would be expected if the smaller protein terminates at the end of the UL29 ORF. Immunofluorescence using an antibody specific for the FLAG epitope identified protein expression as early as 6 hpi and continuing through the course of infection using BADinUL29F virus (Fig. 3). No differences were observed in protein localization between BADinUL29F and BADinUL28F (Fig. 3). The fluorescent signal must be generated by pUL29/28 in BADinUL28F-infected cells, because pUL29 protein would not be tagged. However, we cannot rule out a contribution from pUL29 to the fluorescent images observed in BADinUL29F-infected cells, even though it was not detected at 6 hpi and is expressed to a significantly lower level than pUL29/28 at later times (Fig. 2C). Expression of the tagged protein was exclusively within the nucleus between 6 and 48 hpi (Fig. 3), a finding consistent with the existence of an NLS within the protein sequence. At 72 hpi, tagged protein was concentrated within replication centers in the nucleus, and it also was present in the cytoplasm (Fig. 3). Taken together, our results demonstrate expression of pUL29/28 as a 79-kDa protein during infection beginning very early after infection and a less abundant pUL29.
FIG. 3.
Localization of pUL29/28F in fibroblasts during infection. Cells were infected at a multiplicity of 0.5 infectious unit/cell using either inUL29F or inUL28F, fixed at the indicated times, and processed for immunofluorescence using a FLAG-specific antibody (inUL29F, green; inUL28F, red) and DAPI (blue).
pUL29/28-deficient viruses produce less viral DNA and progeny.
Previously, our laboratory generated two recombinant viruses, BADsubUL28 and BADsubUL29, which contained transposon insertions between nt 35568 and nt 35780 and between nt 36866 and nt 36896, respectively (50) (Fig. 1A, top). Disruption of either ORF resulted in a small plaque variant that produced a reduced yield in fibroblasts (50). Since the mutations are relatively small and contained entirely within the UL29 and UL28 ORFs, it is unlikely that surrounding ORFs are affected. Nevertheless, as a test for the possibility that the mutations influenced expression of additional ORFs from the complex UL29/28 coding region (Fig. 1A, top), we monitored expression of the downstream UL26 ORF in BADsubUL28-infected cells. Western blot assay demonstrated that pUL26 accumulated at 24 and 72 hpi (Fig. 4A). We also observed by qRT-PCR in BADsubUL28-infected cells expression of RNAs containing both the UL27 and UL30 ORFs (Fig. 4A).
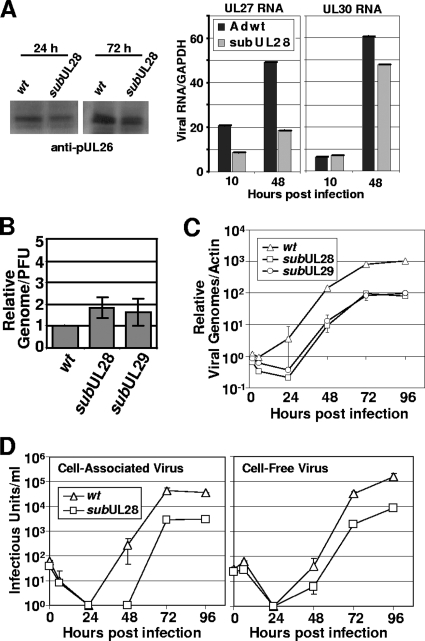
FIG. 4.
Inefficient BADsubUL28 and BADsubUL29 replication. (A) Expression of UL26, UL27, and UL30 in fibroblasts infected with subUL28 compared to wild-type virus. Fibroblasts were infected using equivalent amounts of virus, harvested at the indicated times, and processed for Western blot analysis with a pUL26-specific antibody. Expression of RNAs containing UL27 and UL30 ORFs were quantified by qRT-PCR using total RNA harvested from cells infected by wild-type and subUL28 viruses. (B) Infectivity of wild-type, subUL28, and subUL29 viruses. Genome content was determined by using qPCR and primers against HCMV DNA using partially purified virus, and infectious units were determined by quantifying IE1-positive cells using the same virus stock. The results are from duplicate experiments and are presented relative to wild-type virus. (C) Accumulation of viral DNA. Fibroblasts were infected with wild type, subUL28, or subUL29 at an input genome number equivalent to 0.1 infectious unit of wild-type virus/cell. Total cell-associated DNA was isolated, and viral genomes were quantified by using real-time PCR and normalized to β-actin DNA. (D) Single-step growth analysis of subUL28. Cell-associated (left panel) and cell-free (right panel) virus was assayed. Fibroblasts were infected with wild-type or subUL28 virus at an input genome number equivalent to 0.1 infectious unit of wild-type virus/cell. Infected cell culture medium was collected as cell-free virus samples, and cell-associated virus was isolated by freezing and thawing cell pellets. The amount of virus present in each sample was determined by counting the number of IE1-positive cells and experiment was completed in duplicate. wt, wild type.
To investigate the effect of mutations in UL29 and UL28 during virus growth, we first determined whether BADsubUL29 and BADsubUL28 virions exhibited wild-type infectivity by quantifying their genome to PFU ratios relative to their parent, BADwt. Virions were concentrated and partially purified by centrifugation through a sorbitol cushion. Three samples for each virus prepared in the identical way were assayed for infectivity by plaque assay and genome copy number by qPCR. The infectivity of mutant virions was very similar (within a factor of 2) to that determined for wild-type particles (Fig. 4B). We next determined the kinetics of viral DNA accumulation after infection with aliquots of mutant and wild-type virus stocks containing the same number of genomes and equivalent to a multiplicity of ∼0.1 infectious unit/cell. Viral genome levels during infection were determined through qPCR using primers to HCMV DNA and normalized to cellular actin DNA in each sample (46). Both mutants exhibited a 10-fold decrease in DNA accumulation compared to the wild-type virus (Fig. 4C).
Having demonstrated that BADsubUL28 and BADsubUL29 virions are equally infectious and that both mutants exhibit reduced viral DNA accumulation, we analyzed the growth kinetics of BADsubUL28. During a single round of virus growth after infection with virus aliquots containing the same number of genomes and equivalent to a multiplicity of ∼0.1 infectious unit/cell, BADsubUL28 produced 10-fold less cell-associated and cell-free virus compared to BADwt (Fig. 4D). The reduced yield is likely a direct reflection of reduced viral DNA accumulation since both defects are of the same magnitude.
pUL29/28 is required for efficient immediate-early RNA accumulation.
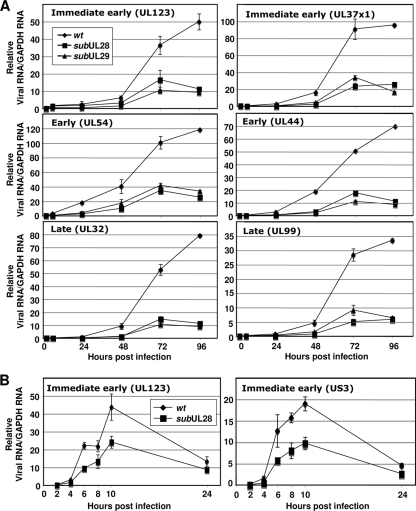
To more precisely identify the step during the viral life cycle compromised by the mutation in the UL29/28 coding region, we determined the expression levels of viral transcripts during the course of infection. HCMV expresses its genes in a regulated cascade of immediate-early, early, and late genes, and we quantified the expression of two members of each kinetic class. qRT-PCR was performed using DNase-treated total RNA isolated from fibroblasts infected with BADwt, BADsubUL28, or BADsubUL29 virus stocks containing the same number of genomes and equivalent to a multiplicity of 0.1 infectious unit/cell. Immediate-early RNAs (UL123 and UL37x1), early RNAs (UL44 and UL54), and the late RNAs (UL32 and UL99) were all reduced by a factor of ∼5 by 96 hpi in mutant compared to wild-type virus-infected cells (Fig. 5A). The levels of the UL123 and US3 immediate-early RNAs produced by BADsubUL28 and BADwt were examined in a second, independent experiment, and RNAs in the mutant virus-infected cells were found to be reduced by a factor of ∼2 at each time assayed between 4 and 10 hpi (Fig. 5B).
FIG. 5.
pUL29/28-deficient viruses express reduced levels of viral transcripts. (A) Accumulation of viral RNAs during a single-step growth analysis. Fibroblasts were infected with wild-type (wt), subUL28, or subUL29 at an input genome number equivalent to 0.1 infectious unit of wild-type virus/cell. Total RNA was isolated at the indicated times after infection, and viral RNA was quantified by real-time RT-PCR using primers specific to the indicated genes and normalized to GAPDH RNA. (B) Accumulation of HCMV RNAs during the immediate-early phase of infection. Total RNA was isolated at the indicated times postinfection, and RNA levels were quantified as described above using primers specific to the immediate-early genes UL123 and US3.
Expression of pUL29/28 activates the MIEP.
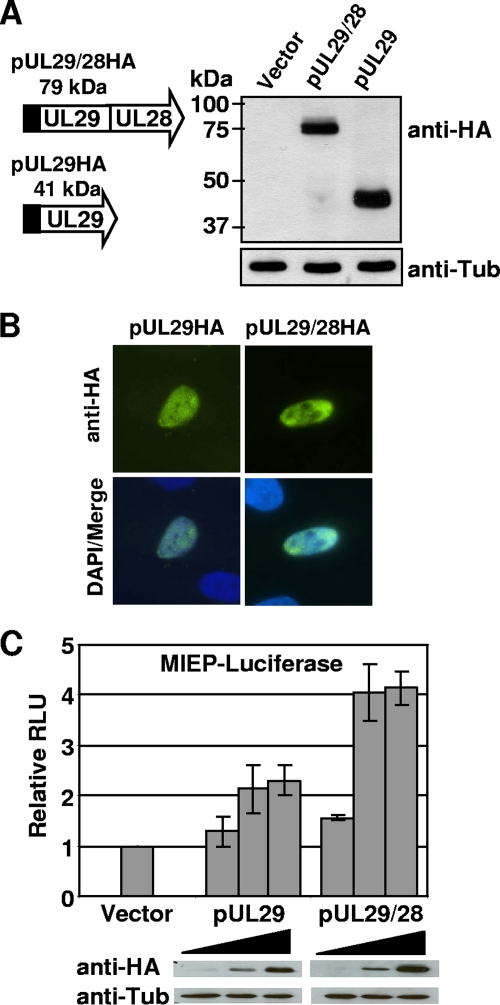
Because we observed a decrease in immediate-early gene expression upon disruption of UL28 during viral replication, we were next interested in determining whether pUL29/28 and possibly pUL29 expression would influence the activity of the viral MIEP outside the context of an HCMV infection. To express these proteins in the absence of infection, we introduced the defined coding region into a mammalian expression vector containing the HA epitope tag (Fig. 6A). Western blot analysis of lysates from transfected 293T cells demonstrated expression of a ∼79-kDa protein upon transfection using the UL29/28 coding sequence and a ∼41-kDa using the UL29 sequence (Fig. 6A), a finding consistent with our observations from cells infected with epitope-tagged viruses (Fig. 2C). Immunofluorescence using the HA antibody of transfected cells showed expression of both proteins within the nucleus (Fig. 6B), which was also observed during viral infection (Fig. 3) and confirms the existence of an NLS within the UL29 coding sequence.
FIG. 6.
pUL29/28 activates the MIEP. (A) Transient expression of pUL29/28 and pUL29. The UL29/28 and UL29 sequences were introduced into mammalian expression vector pCGN in-frame with the HA tag. 293T cells were transfected using empty vector or the pUL29/28HA or pUL29HA expression vectors, and whole-cell lysates were analyzed by Western blotting using an antibody to HA or to cellular tubulin. (B) Localization of pUL29/28HA and pUL29HA was visualized by indirect immunofluorescence on transfected cells using anti-HA (green) and DAPI (blue). (C) 293T cells were transfected using 50 ng of pGL3-MIEP reporter plasmid and 10, 100, or 500 ng of pCGN empty vector, pCGN-pUL29/28, or pCGN-pUL29 effector plasmid. Luciferase activity was assayed 48 h posttransfection using equal protein amounts within each lysate and normalized to luciferase activity from empty vector. The levels of pUL29/28HA and pUL29HA expression were assayed by Western blot analysis using the same lysates and antibody to HA or cellular tubulin.
To determine whether expression of either protein could influence MIEP activity, we performed luciferase assays using a reporter containing the MIEP upstream of the luciferase gene within the pGL3-Basic vector. Increasing amounts of empty vector and pUL29 or pUL29/28 expression vector were transfected into 293T cells. The luciferase activity was determined at 48 h posttransfection using equal protein levels, and these data are presented in Fig. 6C. Increasing amounts of pUL29/28 expression resulted in a fourfold increase in luciferase activity relative to empty vector. Using the same lysate used to measure luciferase activity, we observed by Western blot analysis an increase in the levels of pUL29/28 (Fig. 6C, bottom) relative to cellular tubulin expression. Upon transfection of pUL29, a twofold increase in luciferase activity (Fig. 6C) was observed relative to increasing amounts of empty vector, and Western blot analysis demonstrated pUL29 protein levels were similar to that of pUL29/28 (Fig. 6C). These observations demonstrate that pUL29 and pUL29/28 can both influence MIEP activity, albeit to various degrees.
pUL29/28 is a component of HCMV virions.
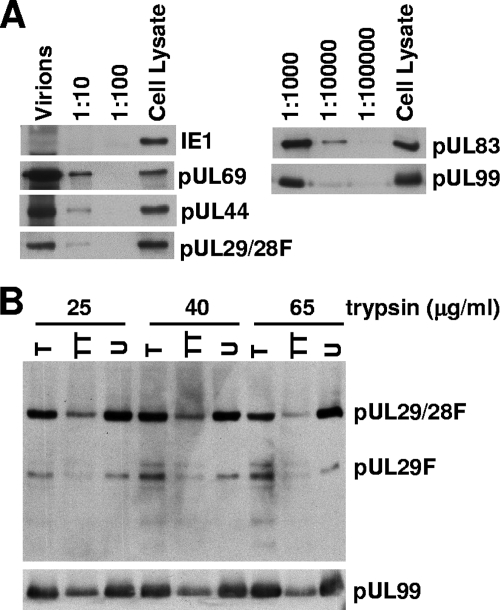
Previous studies have demonstrated that several US22 family members are present in HCMV virion preparations (2, 16, 37, 41, 48). The defect we observed in immediate-early gene expression in the absence of pUL29/28 would be consistent with the loss of a protein delivered to infected cells in virions, so we tested whether pUL29/28 is present in virions. BADinUL29F virions were partially purified by centrifugation through a sorbitol cushion, and proteins associated with the virion preparation were characterized by Western blot analysis. Multiple dilutions of virions were assayed to account for differences in protein abundance and antibody avidity. In Fig. 7A, we demonstrate that several proteins known to be packaged into HCMV virions were present in the BADinUL29F preparation: pUL44, pUL69, pUL83, and pUL99. An anti-FLAG antibody was used to assay for the pUL29/28 protein, and it detected pUL29/28 within the undiluted virion preparation, as well as the 1:10 dilution sample. Infected cell lysates harvested late after infection were used as positive controls. The UL123-coded IE1 protein previously has not been observed in virion preparations (48), and we also failed to demonstrate its presence, even though it was clearly present in the lysate control. The association of pUL29/28 with virions was further analyzed by treatment with trypsin (Fig. 7B). Virions were maintained intact during the treatment or they were incubated with detergent to solubilize their envelopes and expose internal proteins. The known tegument protein, pUL99, was not affected by trypsin treatment of intact virions, but it was partially degraded upon disruption of virions with Triton X-100 (Fig. 7B). Under the same conditions, pUL29/28 was resistant to digestion when virions remained intact, and, like pUL99, it was degraded when virions were disrupted with detergent prior to protease treatment (Fig. 7B). The smaller FLAG-tagged protein (presumably pUL29F) was detected in virion preparations, and it also became susceptible to trypsin after treatment with detergent (Fig. 7B).
FIG. 7.
pUL29 and pUL29/28 are present in HCMV virions. (A) Partially purified BADinUL29F virion preparations contain pUL29/28F. Virions were solubilized, and serial dilutions, as well as an infected cell lysate control, were analyzed by Western blotting with antibodies to the FLAG epitope and the known virion proteins pUL69, pUL44, pUL83, and pUL99. pUL123 was assayed as a negative control. (B) pUL29F and pUL29/28F are located within the virion envelope. inUL29F virions were treated with increasing amounts of trypsin (T), with trypsin and Triton X-100 (TT), or untreated (U). Virions were then solubilized, and proteins were analyzed by Western blotting with a monoclonal antibody specific to the FLAG epitope and, as a control, a monoclonal antibody to pUL99.
Our results demonstrate that pUL29/28 is packaged within virions.
DISCUSSION
HCMV encodes a diverse set of proteins that influence the immediate-early environment of the cell (26). These proteins are either newly expressed upon initiation of viral gene expression or are introduced as components of infectious virions upon virus entry. We have identified here a new HCMV gene product, pUL29/28, that is expressed from the UL29 and UL28 ORFs through the use of multiple splice donor and acceptor sites (Fig. 1A). Previous reports suggested that the mRNAs within the UL29-UL26 region share a common polyadenylation signal, AUUAAA, within the UL26 3′UTR at nt 32103 (10, 45). This motif is not present in the UL29/28 mRNAs that we have mapped, and we have identified a second putative polyadenylation signal at nt 31892, CAUAAA, just upstream of the site of poly(A) addition, as seen in the 3′RACE products (Table 1).
By using a virus with an epitope tag at the N terminus of the UL29 sequence, BADinUL29F, we demonstrated expression of pUL29/28 protein throughout the course of infection at the predicted size of 79 kDa (Fig. 2C). In addition, a less abundant, 41-kDa protein specific to infected cells was observed (Fig. 2C). We also observed these proteins upon transient transfection of expression vectors containing the mapped coding sequences (Fig. 6A). The 41-kDa protein was not observed during infection using a virus with an epitope tag at the C terminus of the UL28 sequence, BADinUL28F (Fig. 2C). This is the predicted size of UL29 protein expressed from an unspliced transcript. We observed both spliced and unspliced RNAs associated with polysomes isolated from HCMV-infected cells (Fig. 1B), which is consistent with the finding that a smaller protein is expressed from this locus. Our results suggest that a 79-kDa protein, pUL29/28, and a 41-kDa protein, pUL29 (Fig. 2C), are expressed through alternative splicing and share a common amino-terminal domain.
Two mutant viruses, BADsubUL29 and BADsubUL28, unable to produce pUL29/28, produced less immediate-early RNA compared to wild-type virus (Fig. 5A). The UL29 and UL28 genes are adjacent to and expressed in the same orientation as the UL30, UL27, and UL26 genes (Fig. 1A), and earlier work has demonstrated that disruption of UL26 results in a delay in immediate-early gene expression and reduced virus yield (28). Given the similarity in phenotypes, we tested whether the disruption of the UL29/28 genes influenced pUL26 expression. Western blot analysis with an antibody to pUL26 demonstrated that the mutations in BADsubUL29 and BADsubUL28 still allow for expression of pUL26 during infection (Fig. 4A). We have monitored the expression of UL27 and UL30 by qRT-PCR and observed RNAs containing both ORFs in BADsubUL28-infected cells (Fig. 4A). Similar levels of expression occurred between viruses for RNAs containing the UL30 ORF. However, we observed slightly reduced levels of UL27 RNA expression. Because pUL29/28-deficient viruses result in reduced gene expression, we cannot distinguish between reduced levels of UL27 RNAs mediated from sequence disruptions in UL29/28 versus an overall reduction in pUL29/28-mediated gene expression. However, other reports have demonstrated that disruption of the UL27 ORF results in only a slight growth attenuation in fibroblasts, and no growth phenotype was observed during virus replication in human tissue implanted in SCID mice (10, 38). In addition, we have recently identified the start site of an RNA containing the UL27 ORF that sits downstream of the UL28 ORF (unpublished observations). Thus, the impaired immediate-early gene expression and growth defect we observed for BADsubUL29 and BADsubUL28 (Fig. 4) almost certainly result from disruption of pUL29/28 and pUL29 function and not from ancillary effects on neighboring ORFs. Further evidence for this conclusion comes from the fact that both of these proteins can induce activity of the MIEP within transfected cells (Fig. 6C).
RNAs from the UL26-29 region have been reported to be expressed with early or late kinetics (6). Given the delay in its expression, how might pUL29/28 and pUL29 be influencing immediate-early gene expression? Although previous characterization of HCMV virion and dense body components by using mass spectrometry did not detect pUL29/28 or pUL29 within particles (48), we found an association by using a virus with an epitope tag at the N terminus of the UL29 sequence. pUL29/28 and pUL29 both copurify with virions and are protected from protease digestion by the virion envelope (Fig. 7A and B). Consistent with their presence in virions, a portion of the tagged proteins is present in the assembly zone late after infection and, as expected for a role in activation of immediate-early gene expression, pUL29/28 is predominantly nuclear at 6 h after infection (Fig. 3). Newly synthesized pUL29/28 might also influence immediate-early gene expression. We have demonstrated the presence of pUL29/28 RNA (Fig. 1C) and protein (Fig. 2C and 3) within infected cells very early after infection and, importantly, the amount of RNA containing UL28 and UL29 sequences increases from 2 to 6 hpi (Fig. 1C). Thus, even though drug sensitivity experiments have categorized this RNA as early-late, it is clearly accumulating during the immediate-early phase of infection. In sum, our data indicate that pUL29/28 and pUL29 are delivered to cells in virions and also expressed very early in infection, and they are predominantly nuclear. Thus, the proteins are present at the start of infection and correctly positioned to assist in the activation of immediate-early genes.
There is ample precedent for tegument proteins such as pUL29/28 and pUL29 to influence immediate-early gene expression. The pUL26 tegument protein was demonstrated to contain a strong transcriptional activation domain (45), and a UL26 deletion virus exhibited a delay in the onset of immediate-early gene expression (28). In this case, it is unclear whether pUL26 is directly activating immediate-early expression, since pUL26 can also influence the stability of viral particles (28, 43). We observed similar levels of infectivity between wild-type virus and the BADsubUL28 and BADsubUL29 mutants (Fig. 4A), so it is not likely that pUL29/28 or pUL29 influences particle stability. The pp71 protein has also been demonstrated to activate immediate-early gene expression (5), and this transactivation activity is enhanced by the tegument protein pUL35 (43). pp71 activation depends upon proteasomal degradation of the cellular Daxx protein, a negative regulator of immediate-early gene expression (18).
HCMV promoters are, of course, regulated by histone modifications (12, 19, 31-33, 36, 40), and we recently discovered that the pUL38 protein interacts with pUL29/28, as well as with the host cell nucleosome remodeling and histone deacetylase (NuRD) complex (27). The NuRD complex contains histone deacetylases and chromatin-remodeling ATPases, and it can repress transcription (4, 14). Although we have not yet shown that pUL29/28 and NuRD reside in the same complex, it is conceivable that pUL29/28 acts to protect the viral chromosome from repressive effects of the NuRD complex.
Acknowledgments
We thank Eain Murphy for help with the galK system, Mariko Aoyagi for providing polysomes fractions, Katherine Faust for the qPCR experiments, and Trish Robinson for help with HCMV antibodies.
This study was supported by a grant from the National Cancer Institute (CA85786).
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Adamo, J. E., J. Schroer, and T. Shenk. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J. Virol. 78:10221-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blais, J. D., V. Filipenko, M. Bi, H. P. Harding, D. Ron, C. Koumenis, B. G. Wouters, and J. C. Bell. 2004. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 24:7469-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowen, N. J., N. Fujita, M. Kajita, and P. A. Wade. 2004. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta 1677:52-57. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate-early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 8.Chen, F., C. C. MacDonald, and J. Wilusz. 1995. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 23:2614-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, S., G. I. Marousek, A. E. Senters, M. G. Davis, and K. K. Biron. 2004. Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir. J. Virol. 78:7124-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cicin-Sain, L., Z. Ruzsics, J. Podlech, I. Bubic, C. Menard, S. Jonjic, M. J. Reddehase, and U. H. Koszinowski. 2008. Dominant-negative FADD rescues the in vivo fitness of a cytomegalovirus lacking an antiapoptotic viral gene. J. Virol. 82:2056-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuevas-Bennett, C., and T. Shenk. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525-9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 14.Denslow, S. A., and P. A. Wade. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26:5433-5438. [DOI] [PubMed] [Google Scholar]
- 15.Efstathiou, S., G. L. Lawrence, C. M. Brown, and B. G. Barrell. 1992. Identification of homologues to the human cytomegalovirus US22 gene family in human herpesvirus 6. J. Gen. Virol. 73(Pt. 7):1661-1671. [DOI] [PubMed] [Google Scholar]
- 16.Feng, X., J. Schroer, D. Yu, and T. Shenk. 2006. Human cytomegalovirus pUS24 is a virion protein that functions very early in the replication cycle. J. Virol. 80:8371-8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakki, M., and A. P. Geballe. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J. Virol. 79:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang, J., and R. F. Kalejta. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334-338. [DOI] [PubMed] [Google Scholar]
- 19.Ioudinkova, E., M. C. Arcangeletti, A. Rynditch, F. De Conto, F. Motta, S. Covan, F. Pinardi, S. V. Razin, and C. Chezzi. 2006. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 384:120-128. [DOI] [PubMed] [Google Scholar]
- 20.Kalejta, R. F., and T. Shenk. 2003. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J. Virol. 77:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai, T., J. Fan, K. Mazan-Mamczarz, and M. Gorospe. 2004. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 24:6773-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, J., G. Pan, K. Cui, Y. Liu, S. Xu, and D. Pei. 2007. A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J. Biol. Chem. 282:19481-19492. [DOI] [PubMed] [Google Scholar]
- 23.Lorz, K., H. Hofmann, A. Berndt, N. Tavalai, R. Mueller, U. Schlotzer-Schrehardt, and T. Stamminger. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80:5423-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mocarski, E., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2702-2772. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 27.Moorman, N. J., I. M. Cristea, S. S. Terhune, M. P. Rout, B. T. Chait, and T. Shenk. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munger, J., D. Yu, and T. Shenk. 2006. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J. Virol. 80:3541-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, E., J. Vanicek, H. Robins, T. Shenk, and A. J. Levine. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. USA 105:5453-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nitzsche, A., C. Paulus, and M. Nevels. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novotny, J., I. Rigoutsos, D. Coleman, and T. Shenk. 2001. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J. Mol. Biol. 310:1151-1166. [DOI] [PubMed] [Google Scholar]
- 35.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325-338. [DOI] [PubMed] [Google Scholar]
- 36.Park, J. J., Y. E. Kim, H. T. Pham, E. T. Kim, Y. H. Chung, and J. H. Ahn. 2007. Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J. Gen. Virol. 88:3214-3223. [DOI] [PubMed] [Google Scholar]
- 37.Patterson, C. E., and T. Shenk. 1999. Human cytomegalovirus UL36 protein is dispensable for viral replication in cultured cells. J. Virol. 73:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prichard, M. N., D. C. Quenelle, D. J. Bidanset, G. Komazin, S. Chou, J. C. Drach, and E. R. Kern. 2006. Human cytomegalovirus UL27 is not required for viral replication in human tissue implanted in SCID mice. Virol. J. 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves, M., J. Murphy, R. Greaves, J. Fairley, A. Brehm, and J. Sinclair. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998-10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schierling, K., T. Stamminger, T. Mertens, and M. Winkler. 2004. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J. Virol. 78:9512-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamminger, T., M. Gstaiger, K. Weinzierl, K. Lorz, M. Winkler, and W. Schaffner. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J. Virol. 76:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terhune, S., E. Torigoi, N. Moorman, M. Silva, Z. Qian, T. Shenk, and D. Yu. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valchanova, R. S., M. Picard-Maureau, M. Budt, and W. Brune. 2006. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J. Virol. 80:10181-10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp, 2nd, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]